Abstract
The rapid and accurate diagnosis of active tuberculosis (TB) and its drug susceptibility remain a challenge. Phenotypic assays allow determination of antibiotic susceptibilities even if sequence data are not available or informative. We review 2 emerging diagnostic approaches, reporter phage and breath tests, both of which assay mycobacterial metabolism. The reporter phage signal, Green fluorescent protein (GFP) or β-galactosidase, indicates transcription and translation inside the recipient bacilli and its attenuation by antibiotics. Different breath tests assay, (1) exhaled antigen 85, (2) mycobacterial urease activity, and (3) detection by trained rats of disease-specific odor in sputum, have also been developed. When compared with culture, reporter phage assays shorten the time for initial diagnosis of drug susceptibility by several days. Both reporter phage and breath tests have promise as early markers to determine the efficacy of treatment. While sputum often remains smear and Mycobacterium tuberculosis DNA positive early in the course of efficacious antituberculous treatment, we predict that both breath and phage tests will rapidly become negative. If this hypothesis proves correct, phage assays and breath tests could become important surrogate markers in early bactericidal activity (EBA) studies of new antibiotics.
OVERVIEW
Diagnostic tests for tuberculosis (TB) are most valuable when they rapidly identify both the organism and its antibiotic resistance profile. Although TB remains susceptible to the commonly used drugs in many parts of the world, recently documented increases in multidrug-resistant TB in various countries are of major concern. For example, studies from South Africa have revealed that ∼20% of human immunodeficiency virus (HIV)-associated TB is caused by multidrug-resistant strains [1, 2]. First principles of evolutionary biology strongly suggest that failure to correctly diagnose and treat even rare cases of drug-resistant microbes in regions where they are not common will lead to their contagion and the increased prevalence of resistant strains. Thus, being in a region with low prevalence of drug resistance in the clinic today would be a short-sighted justification for not conducting a complete analysis of drug susceptibility. To ignore the implications of evolutionary biology toward strategies of combating drug resistance would be to compound mistakes that have contributed mightily to the current problems of antibiotic resistance [3].
The currently available methods for detection of M. tuberculosis, the bacillus causing the disease TB, are inadequate [4]. Sputum smears [5, 6] for acid-fast bacteria are limited in sensitivity, missing ∼20% of culture positive samples and 30%–50% in the case of concomitant HIV [7] and are not specific for M. tuberculosis. Culture, which does not require sophisticated resources but is laborious and extremely time-consuming (typically requires 6–8 weeks to deliver results), remains the gold standard. Other approaches assay the host response, such as, radiography, immune response, or M. tuberculosis antigens [8–11]. These approaches are or will become important for TB diagnosis and as biomarkers of the host response. However, they are not informative, at least at the first instance, as to the drug susceptibility of the infecting strain.
Drug susceptibility of mycobacteria can be determined either by genotypic or phenotypic analysis. Genotypic assays, commonly known as nucleic acid amplification tests (NAAT), are polymerase chain reaction (PCR)-based and rely on the identification of defined gene mutations conferring antibiotic resistance [12]. This class of test includes line probe assays: INNO-LiPA Rif. TB kit (Innogenetics NV) and Genotype MTBDR (Hain Lifescence GmbH); molecular beacon analysis [13]; and biprobe analysis [14]. An important advance for rapid clinical diagnosis has recently occurred via Xpert MTB/RIF system (Cepheid) whose power has been well documented in the literature [12, 15–19] and whose use has been endorsed by the World Health Organization (WHO) [20]. The Xpert MTB/RIF system PCR amplifies sequences unique to M. tuberculosis and detects mutations via molecular beacons. Data are obtained as real-time fluorescence and have important advantages over end-point PCR and gel analysis. The Xpert MTB/RIF test is immediately available for application and makes significant improvements in clinical M. tuberculosis diagnosis.
Xpert MTB/RIF correctly diagnoses rifampicin resistance in ∼95% of current clinical isolates; however, microbes continually evolve [21] and the continued efficacy of any assay dependent on specific DNA sequences is not assured. Concerns about microbial evolution should not intrude on the immediate clinical application of Xpert MTB/RIF as the advantage of rapid diagnosis is enormous. However, the fundamental nature of PCR and molecular beacons is that a sample must be queried for the presence or absence of a specific DNA sequence and that specific sequence must be known in advance. In this sense one must “know the answer,” that is, the specific mutation responsible for drug resistance, in order to “ask the question” of whether or not that specific sequence is present in a particular sample. The limitations of PCR and beacon technology are well matched for detection of rifampicin resistance because only 3 well-characterized amino acid changes confer ∼95% of the current clinical cases of resistance. In contrast, resistance to isoniazid (INH) can be due to any of hundreds of different sequence changes. Other antibiotics are between these extremes in terms of the number of resistance alleles that have so far been identified in clinical isolates [22]. The resistance to INH is acquired at a relatively high frequency and is associated with multiple genes, several of which remain unknown [23], and is heightened because of the mutagenic nature of the drug [24]. Subsets of these strains are also rifampicin resistant (MDR/XDR strains). In today’s scenario, most clinical strains that are rifampicin resistant are also INH resistant, and determination of rifampicin resistance is sufficient to classify a strain as MDR. A appreciable percentage of XDR cases in countries like South Africa [25] also exists, and globally it has been reported that ∼8% of MDR cases are XDR [26]. The limitation of detecting only rifampicin resistance also means that MDR and XDR M. tuberculosis cannot be distinguished. Another technical concern in the clinical interpretation of the Xpert MTB/RIF assay is its readout from mixed infections. Depending on the rpoB allele being tested, the assay requires that 65%, or 100% of the DNA must contain the mutant rpoB allele for the rifampicin resistance diagnosis to be made with 95% certainty [16]. Thus, genotypic assays including the rapid Xpert MTB/RIF provide a critical component to control TB, yet have important limitations. Furthermore, restricting susceptibility testing to rifampicin and/or INH could allow the unobserved emergence of strains with totally different drug resistance profiles.
Phenotypic tests of microbial growth and metabolism do not depend on foreknowledge of resistance mechanisms that might be involved. Microbial culture, a classical phenotypic test, has up to now been the gold standard for both diagnosis of TB and drug resistance [27]. It is sensitive, and the results are definitive with respect to both identification and characterization. However, conventional culture requires special resources, not the least of which is time. Other phenotypic assays have been developed that shorten the time and volume needed for culture, including: MB/BacT (Organon-Teknika), ESP Culture System II (AccuMed International), MB Redox (Biotest), Nitrate Reductase Assay (NRA), BACTEC 460 and BACTEC MIGIT 960 (Becton Dickinson Microbiology Systems) and Microscopic Observation of Drug Susceptibility (MODS). Direct microscopic observation of growth and its inhibition by antibiotics shortens the time needed for culture assay (9–12 days) while retaining high reliability and low cost [28–35]. A D29 mycobacteriophage based commercial kit (FASTPlaque), which relies on phage amplification on its host M. tuberculosis, offers a phenotypic test with a turnaround time of 4 days. Unfortunately, so far phage amplification to diagnose TB is reported as cumbersome and has high variability [36]. Therefore, there is a pressing need for phenotypic tests that give results with the speed and reliability of the best nucleic acid assays but without the limitation to specific already-known DNA sequence mutations that confer drug resistance.
REPORTER MYCOBACTERIOPHAGES FOR RAPID TB DIAGNOSIS AND PHENOTYPIC ASSAYS
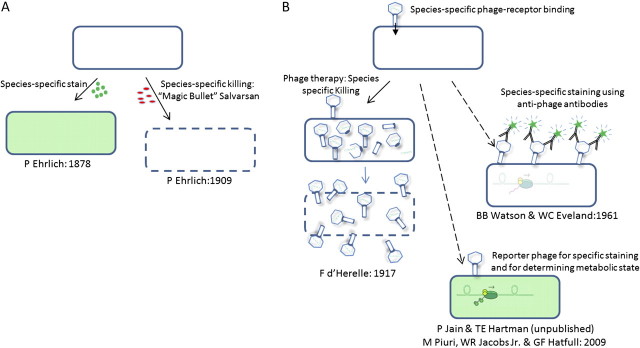
Bacteriophages are viruses that infect bacteria. Bacteriophages have limited host range, that is, each type of phage is specific as to which species of bacteria it will infect. Mycobacteriophages are a subset of bacteriophages that specifically infect mycobacteria. “Mycobacterial reporter phages” inject their DNA into mycobacteria and cause the infected cell to emit a diagnostic signal from a gene cloned into the reporter phage whose expression depends on host machinery. The specificity of injection and reporter expression serves as a specific stain for the infected cell. The use of phages in a diagnostic staining reaction is conceptually similar to other diagnostic stains that depend on differential chemical affinity by small molecules or antibodies. The specificity of chemical staining reactions led to “magic bullet” chemotherapy as first proposed and discovered by Paul Ehrlich in the late nineteenth century [37, 38]. Ehrlich noted that certain dyes stain only specific microorganisms, but not others. Among many other findings, Ehrlich contributed to the acid-fast staining methods for specifically identifying the tubercle bacillus [5, 39]. From his discovery of species-specific “reporters,” Ehrlich made the conceptual leap to the idea of drugs that would be toxic to pathogens but not to the cells of the host (Figure 1). In the case of reporter phages, the conceptual train runs in the reverse direction. The idea that phages could be used to kill specific pathogenic bacteria while sparing host cells was immediately proposed with the discovery of bacteriophages [40]. Phage therapy is still being pursued, although it has yet to objectively justify the enthusiasm it has often engendered [41]. Phage typing of bacterial strains has been used many decades and is still used in epidemiology [42]. However, the first use of phages as a specific cell-staining diagnostic was not published until almost 50 years after the discovery of phage. In the first case phages were essentially employed as a primary antibody against the phage receptor for Bacillus anthracis. Antiphage antibodies labeled with fluorescein were then used as a secondary antibody to develop the signal [43]. A similar antiphage antibody method was developed to identify Listeria monocytogenes [44, 45]. These first “phage-dependent stains” are not “reporter phages” by our definition because they give no information about the metabolic state of the bacterial cells that they identify.
Figure 1.
Comparison of cell staining and chemotherapy by small molecules to reporter phage and phage therapy: Specific staining led to the idea and soon the realization of targeted chemotherapy by small molecules. On the contrary, the idea of phage therapy inspired phage as specific diagnostics, and finally reporter phage for metabolic activity and drug susceptibility. A, Ehrlich first discovered that certain dyes differentially stained only certain microbial species. He made the conceptual leap from this finding to the idea that there must exist “magic bullets” that is, chemotheraputic agents that specifically kill microbes but are harmless to the differing cell types of the host and also to the specificity of the serum reaction, that is, antigen-specific antibiodies. B, Immediately upon their discovery (1917) bacteriophage were proposed as a way to specifically attack vulnerable microbes. Only in 1961 were phage first used in conjunction with antibodies in order to specifically stain target cells to diagnose a species of bacteria (B. anthracis) in analogy to Ehrlich’s use of specific staining reactions. Reporter phage expressing GFP allowed the first single cell visualization of mycobacteria with this diagnostic system in 2009.
Tools for genetic engineering had to be developed to set the stage for mycobacterial reporter phages [46]; the creation and refinement of genetic engineering tools and methods with mycobacteriophages is ongoing. The first recombinant mycobacteriophage reporter phages were based on vectors derived from mycobacteriophage TM4 and used luciferase as the reporter [47]. Luciferase reporter phages (LRPs) allowed assay of the cellular metabolism of M. tuberculosis after its exposure to different antimycobacterial agents because luminescence requires available ATP. Drugs that inhibit host metabolism likewise inhibit expression from the reporter gene. Reporter phages have the advantage of culture but on a shorter timescale. Drug susceptibility or resistance of M. tuberculosis strains was revealed through luminescence: lights on = metabolism and growth potential; lights out = drug susceptibility/inhibited cellular metabolism and inability to grow. The initial luciferase reporter phage serves as a proof of concept, but it is not sensitive enough to be practical clinically. LRP were subsequently generated from D29 and L5 mycobacteriophage as well [48]. Of all the LRPs generated, the TM4 derivative phAE40 has the greatest advantages. Phage TM4 infects all M. tuberculosis strains tested to date. TM4 was least fastidious for host adsorption to all mycobacterial species and with a variety of buffer compositions [49]. The broad host range of TM4 was overcome by the use of p-nitro-alpha-acetylamino-beta-hydroxy propiophenone (NAP) that inhibits other mycobacterial species and renders the phage assay signal specific to M. tuberculosis [50].
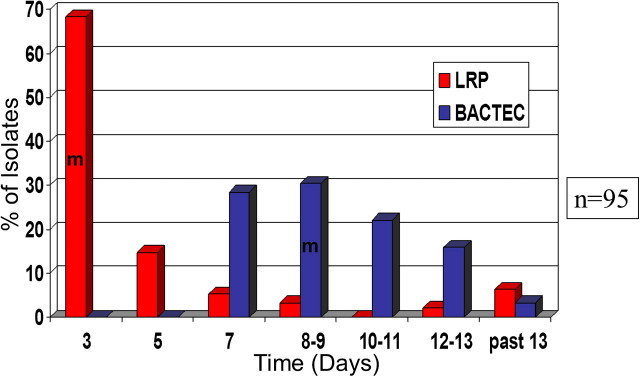
Development and use of temperature sensitive LRP phages, which grow lytically at 30oC but not 37oC, enhances the luciferase activity per cell [51, 52]. In studies performed on clinical samples, the LRP assay required 3 days to deliver the result in comparison to 7–9 days for BACTEC [53–55]. Both systems agreed on the drug susceptibility profile of a set of clinical samples (Figure 2). Agar proportion (AP) was used as gold standard in these studies and required 4–6 weeks to deliver the results. AP means the proportion of colony-forming units (CFUs) on plates with and without antibiotic. If the proportion is <100-fold at a predetermined antibiotic concentration, then the bacteria is defined as “resistant.” Despite improvements, LRP had limitations; it was never possible to visualize single infected cells. Because of this, it was hard to determine the presence of multiple strains of M. tuberculosis with different drug susceptibilities in a sample. The signal obtained per cell was low and required a large number of cells [54].
Figure 2.
Turnaround time to determine the drug susceptibility against first line drugs. Drug susceptibility of 95 clinical samples of M. tuberculosis samples was determined with LRP and BACTECH. Number of cases for which results are available are plotted against time, LRP (red bars) and BACTEC-460 (blue bars). M represents the median time, LRP (3 days), BACTEC (9 days). This figure is reprinted from the published literature [79].
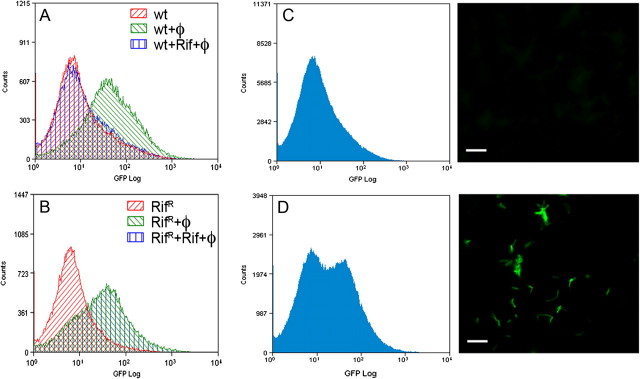
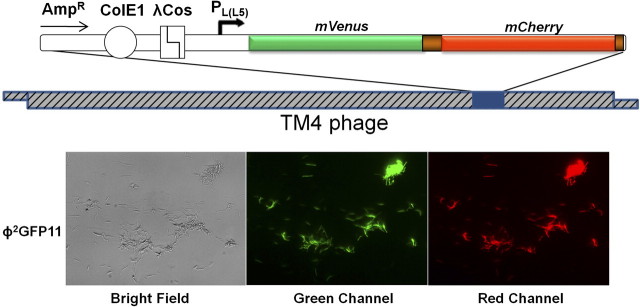
Visualization of the diagnostic signal in individual mycobacterial cells was achieved by means of generating reporter phages expressing GFP and infection at 37oC [56]. Unlike a luciferase reporter, GFP does not require an exogenous substrate; either fluorescence microscopy or flow cytometry can identify and enumerate GFP-expressing bacilli. Bacilli can be concentrated using a membrane filter or centrifugation to increase the sensitivity of detection [56]. The fractional resistant population for a given drug was identifiable by GFP phage (Figure 3). However in our hands phsp60-GFP reporter phage have not been successful in sputum samples (unpublished data), and improvements are required to use reporter phages in the diagnosis of M. tuberculosis. A new phage backbone, phAE159, was derived from PH101 by deleting ∼6.0 kb of nonessential genes to increase the cloning capacity to ∼10 kb. The deletion covers gene gp49, which may be involved in superinfection exclusion [57] and may therefore allow >1 phage to infect the same cell and thereby increase signal strength. Increased cloning capacity allowed us to identify strong promoters and incorporate multiple reporters in the same phage (Figure 4 shows a single phage that contains genes for both GFP and mCherry). Compared to phsp60-GFP reporter phage [56], fluorescence obtained per cell is increased by ∼100-fold due to alterations, including promoter and ribosome binding site (Paras Jain and William R. Jacobs Jr.; unpublished). With the best reporter configuration to date, the minimum time to see a clear fluorescent signal has been reduced to 4 hours in M. tuberculosis. The signal strength accumulates over time and reaches a peak at ∼12–14 hours. The newly derived fluorescent reporter phages are now able to detect M. tuberculosis in sputum samples, and evaluation in clinical samples is in progress. Other reporter phages harboring lacZ or phoA do not require a fluorescent microscope or flow cytometry to detect M. tuberculosis. Assays with these phages offer another way to detect M. tuberculosis in a short time with minimum equipment.
Figure 3.
Determination of drug susceptibility with GFP reporter phage by FACS. A, Drug susceptible cells express GFP after infection with reporter phage. Fluorescence diminishes in the presence of drug. ”ϕ” in the figure designates “phage.” B, Drug resistance cells express GFP both in the presence or absence of drug after phage addition. C, Drug susceptible cells do not show fluorescence in presence of drug (Rif) as seen by FACS or microscopy. D, Sample used in C was spiked with 50% Rif resistant population which results in a 50% GFP positive population, as determined by FACS and seen in microscopy. This figure is reprinted from the published literature [56].
Figure 4.
A dual reporter RFP and GFP mycobacteriophage. Infection of M. tuberculosis with a dual reporter phage. More than 90% of the cells were detected by the use of simple fluorescent microscope. The cells were incubated with phage for 12 hours, and this image is from an exposure time of 15 ms. (Previously unpublished.)
One of the limitations of TM4 derived reporter phages is that TM4 infects multiple mycobacterial species, including nonpathogens. Even though the broad host range of TM4 can be narrowed by the use of NAP, the development of reporter phages that inject specifically into M. tuberculosis would further assure diagnostic specificity. Mycobacteriophage DS6A has been reported to infect only within the TB complex [58]. However, if infection means the simple act of injecting DNA, this claim has been stated more stridently than it has been experimentally proven. What is experimentally clear is that DS6A forms plaques only on lawns within the M. tuberculosis complex [58] (Torin Weisbrod, Oren Mayer, and William R. Jacobs, unpublished). The distinction between injection and full growth of the phage is well documented in other cases. For example, phage lambda can “infect”, that is, inject into receptor-expressing strains of Salmonella typhimurium but not produce a phage burst [59]. The signal from reporter phage depends only on injection and expression of a single gene. The phage “report” does not depend on execution of the coordinated genetic and developmental programs required for a burst of viable phages. New reporter phages are being developed based on mycobacteriophage DS6A; these will allow answering the questions raised concerning specificity of adsorption and injection versus that of a phage burst and plaque formation (Oren Mayer, Torin Weisbrod, and William Jacobs Jr, unpublished).
An important requirement, which is a limitation for rapid diagnosis but sometimes an advantage for reporter phages as well as the MODS system, is the need for active host metabolism in order to produce a signal. The physiological state of bacilli in the sputum is controversial, and reporter phages may be a tool to figuring out what proportion is active or quiescent. However, quiescent cells will not be immediately useful for positively giving a signal diagnostic of M. tuberculosis, and they cannot report on drug susceptibility. We are currently working to combine the MODS method of incubation with reporter phages to allow diagnoses including antibiotic sensitivity assays at earlier times than required by the current MODS approach. (David S. Thaler, Paras Jain, and William R. Jacobs; unpublished). We reason that the lag phase must be required for both assays. However, a signal from a reporter phage requires only host transcription and translation, whereas MODS assays cell growth and division at a microscopic scale. Cells emerging from lag phase would become transcriptionally and translationally active prior to visible growth and division. Once bacilli become metabolically active, the phage signal requires <12 hours, whereas several doublings each requiring ∼24 hours are needed for MODS. The goal of a 1-hour phenotypic report has not yet been reached, but it is closer than it has ever been. The future of phage diagnosis is bright, and its value will be assessed in clinical studies in the near future.
BREATH TESTS
Another approach to rapid diagnosis of active tuberculosis has been to adapt breath tests. Two general categories of breath tests have been proposed for tuberculosis: phenotypic and genotypic. Phenotypic include metabolic signatures as defined by chemical transformations of stable isotopes or the generation of unique species or altered quantities of various compounds (either antigens and metabolites) made by the bacilli, the infected host, or both. Exhaled breath analyzed chemically or by antigen binding has shown promise in the identification of M. tuberculosis [60–66]. Exhaled breath is akin to odor, and rats can diagnosis TB by sniffing autoclaved sputum [67, 68]. Genotypic implies detection of the pathogen’s DNA in the exhaled breath, inspired by examples such as the breath test for altered methylation of DNA in cancer patients [69]. However, DNA detection of M. tuberculosis specific sequences in breath has so far not been successful [70].
Metabolic signatures have been developed as both a rapid diagnostic for Helicobacter pylori infection and as a biomarker of its eradication by drug treatment [71]. This assay uses an oral dose of labeled 13C-urea and then sampling the ratio of 13CO2 to 12CO2 in exhaled air after the urea is hydrolyzed in the stomach by H. pylori urease. This ratio is readily measured using portable infrared spectrometers, and the entire procedure can be completed within 30 minutes (eg, Breathtek test from Otsuka).
M. tuberculosis also expresses urease, a potential virulence factor [72, 73] that is used in classical microbiological assays [74, 75]. It was thus hypothesized that the urease breath test would provide a technology to detect lung burdens of disease and response to therapy. Recent work demonstrated that mycobacterial urease activity can be rapidly detected in exhaled air samples of infected rabbits after direct delivery of 13C-urea to the lung [64]. Furthermore, in both M. tuberculosis and M. bovis infected rabbits, the rate of 13CO2 formation was significantly decreased after INH therapy and mirrored CFUs. Thus, such tests may provide an enabling technology for point-of-care diagnostic or treatment biomarkers for TB, with use of 0.2 μm membrane filters controlling exposure to infectious agents.
Breath tests have several limitations. Breath tests at initial diagnosis are currently unable to distinguish the drug sensitivity profile of infecting strains. The specificity of the urease test to mycobacteria may also be limited by other urease-producing microbial species, such as H. pylori, common in high TB incidence areas. Several approaches have been undertaken to improve specificity: (1) concurrent use of oral nonabsorbable urease inhibitors such as bismuth salts and proton pump inhibitors for selective suppression of GI H. pylori [76]; (2) lung-targeted delivery of the 13C-urea (by inhalation) to enhance specificity in pulmonary cases, especially with rapid measurement after inhalation, before systemic absorption can occur; (3) development of additional markers that are more unique to TB as has been reported [63, 77]. A breath test has been combined with optical detection of antibody binding in the exhaled breath to M. tuberculosis antigen 85 [65]. Rats may detect and correctly interpret chemically complex signals, and perhaps this contributes to their effectiveness in diagnosis [67, 68, 78].
BEYOND THE INITIAL DIAGNOSIS
There is a need for more rapid follow-up to determine treatment success in certain patients. Efficacious treatment is currently followed by sputum conversion from positive to negative that is typically noted at 2 months. Shortening this time might save lives and inhibit the spread of drug-resistant strains. Breath tests might allow assay of changes in M. tuberculosis metabolism that precede sputum conversion. Reporter phages are also likely to become valuable in the same way. A reporter phage signal indicates an active transcription and translation inside the host bacilli. Reporter phages may allow an earlier discrimination of treatment efficacy by the contrast between acid fast bacilli and those that “light up” with the reporter phage. The ability to tell more rapidly if an antibiotic is working could become very important in improving the efficiency of early bactericidal activity (EBA) clinical trials for new candidate antibiotics.
SUMMARY AND CONCLUSIONS
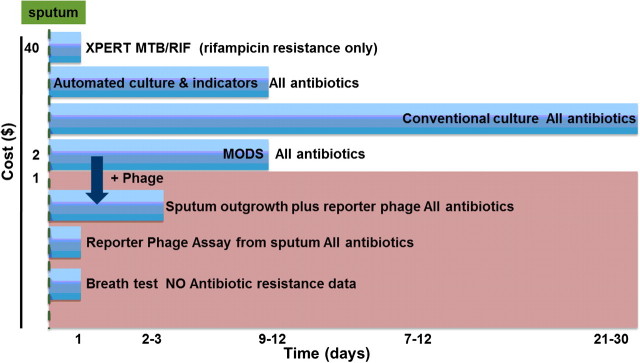
There has been remarkable progress in DNA based assays, and the Xpert MTB/RIF system now allows rapid and sensitive diagnosis of low titer M. tuberculosis in sputum and the sensitive detection of 3 widespread rpoB alleles encoding rifampacin resistance. However, DNA-based tests currently are expensive and inherently limited to diagnosing resistance in cases where only 1 or 2 alleles predominate. It is not predictable if or when the particular epidemiological prevalence of point mutations might change. Therefore, it seems perilous to rely on a diagnostic strategy for drug resistance remaining stable in the face of continuing microbial evolution. Phenotypic tests are not limited to particular alleles because they measure directly what one wants to know: the ability of the bug to metabolize and grow in the presence of the drug. The drawbacks of current phenotypic tests are cost and time. Figure 5 shows a comparison of several current TB diagnosis technologies, with 2 that are moving from the laboratory into clinical settings. The urgent need is for sensitive and quantitative phenotypic assays that approach the short timescale, sensitivity, and accuracy now possible for genotypic analysis but without the limitations to specific alleles inherent in genotypic assays. New phenotypic assays will function either independently or as a complement to the recent impressive progress in DNA sequence analysis. New phenotypic analyses are anticipated to become important in the identification of the full spectrum of antibiotic susceptibility, evaluation during the course of treatment and in EBA trials for new candidate antibiotics.
Figure 5.
Approaches to diagnose active tuberculosis and its drug susceptibility. The 4 methods above the shaded portion are currently available. The methods inside the shaded box, reporter phage and breath tests, are under development. The Y axis is the estimated cost per sample, and the X axis is the amount of time required for assay results. Smear analysis of sputum is not shown in this figure; it is low cost but also lower in reliability than any of the other available methods.
Notes
Acknowledgments.
Dedicated to S. L. Gupta on the first year anniversary of his death. Thanks for helpful comments and suggestions to Fiona K. Doetsch, Nell Eisenberg, Pablo Gonzalez and Travis Hartman.
Financial support.
The work was supported by the National Institutes of Health (AI26170 and AI59877 to W. R. J.); and Albert Einstein College of Medicine Center for AIDS Research (AIO-51519).
Potential conflicts of interest.
All authors: No reported conflicts.
All authors have submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Conflicts that the editors consider relevant to the content of the manuscript have been disclosed.
References
- 1.Hassim S, Shaw PA, Sangweni P, et al. Detection of a substantial rate of multidrug-resistant tuberculosis in an HIV-infected population in South Africa by active monitoring of sputum samples. Clin Infect Dis. 2010;50:1053–9. doi: 10.1086/651119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cohen T, Murray M, Wallengren K, Alvarez GG, Samuel EY, Wilson D. The prevalence and drug sensitivity of tuberculosis among patients dying in hospital in KwaZulu-Natal, South Africa: a postmortem study. PLoS Med. 2010;7:e1000296. doi: 10.1371/journal.pmed.1000296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Relman D, Hamburg M, Choffnes E, Mack A, editors. Microbial evolution and co-adaptation: a tribute to the life and scientific legacies of Joshua Lederberg. Washington, DC: National Academies Press; 2009. 310. [PubMed] [Google Scholar]
- 4.Keeler E, Perkins MD, Small P, et al. Reducing the global burden of tuberculosis: the contribution of improved diagnostics. Nature. 2006;444(Suppl 1):49–57. doi: 10.1038/nature05446. [DOI] [PubMed] [Google Scholar]
- 5.Bishop PJ, Neumann G. The history of the Ziehl-Neelsen stain. Tubercle. 1970;51:196–206. doi: 10.1016/0041-3879(70)90073-5. [DOI] [PubMed] [Google Scholar]
- 6.Allen JL. A modified Ziehl-Neelsen stain for mycobacteria. Med Lab Sci. 1992;49:99–102. [PubMed] [Google Scholar]
- 7.Moore DA, Roper MH. Diagnosis of smear-negative tuberculosis in people with HIV/AIDS. Lancet. 2007;370:1033–4. doi: 10.1016/S0140-6736(07)61474-3. [DOI] [PubMed] [Google Scholar]
- 8.Day J. The roentgen-ray diagnosis of pulmonary tuberculosis. J Nat Med Assoc. 1924;17:21–8. [PMC free article] [PubMed] [Google Scholar]
- 9.Zbinden A, Keller PM, Bloemberg GV. Rapid molecular detection of tuberculosis. N Engl J Med. 2011;364:183. doi: 10.1056/NEJMc1011919. author reply 184-5. [DOI] [PubMed] [Google Scholar]
- 10.Diel R, Goletti D, Ferrara G, et al. Interferon-{gamma} release assays for the diagnosis of latent Mycobacterium tuberculosis infection: a systematic review and meta-analysis. Eur Respir J. 2011;37:88–99. doi: 10.1183/09031936.00115110. [DOI] [PubMed] [Google Scholar]
- 11.Kunnath-Velayudhan S, Salamon H, Wang HY, et al. Dynamic antibody responses to the Mycobacterium tuberculosis proteome. Proc Natl Acad Sci U S A. 2010;107:14703–8. doi: 10.1073/pnas.1009080107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Helb D, Jones M, Story E, et al. Rapid detection of Mycobacterium tuberculosis and rifampin resistance by use of on-demand, near-patient technology. J Clin Microbiol. 2010;48:229–37. doi: 10.1128/JCM.01463-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Piatek AS, Tyagi S, Pol AC, et al. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol. 1998;16:359–63. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 14.Edwards KJ, Metherell LA, Yates M, Saunders NA. Detection of rpoB mutations in Mycobacterium tuberculosis by biprobe analysis. J Clin Microbiol. 2001;39:3350–2. doi: 10.1128/JCM.39.9.3350-3352.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Banada PP, Sivasubramani SK, Blakemore R, et al. Containment of bioaerosol infection risk by the Xpert MTB/RIF assay and its applicability to point-of-care settings. J Clin Microbiol. 2010;48:3551–7. doi: 10.1128/JCM.01053-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blakemore R, Story E, Helb D, et al. Evaluation of the analytical performance of the Xpert MTB/RIF assay. J Clin Microbiol. 2010;48:2495–501. doi: 10.1128/JCM.00128-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boehme CC, Nabeta P, Hillemann D, et al. Rapid molecular detection of tuberculosis and rifampin resistance. N Engl J Med. 2010;363:1005–15. doi: 10.1056/NEJMoa0907847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melzer M. An automated molecular test for Mycobacterium tuberculosis and resistance to rifampin (Xpert MTB/RIF) is sensitive and can be carried out in less than 2 h. Evid Based Med. 2011;16:19. doi: 10.1136/ebm1163. [DOI] [PubMed] [Google Scholar]
- 19.Van Rie A, Page-Shipp L, Scott L, Sanne I, Stevens W. Xpert((R)) MTB/RIF for point-of-care diagnosis of TB in high-HIV burden, resource-limited countries: hype or hope? Expert Rev Mol Diagn. 2010;10:937–46. doi: 10.1586/erm.10.67. [DOI] [PubMed] [Google Scholar]
- 20.WHO. Roadmap for rolling out Xpert MTB/RIF for rapid diagnosis of TB and MDR TB. [Google Scholar]
- 21.Hamilton-Miller JM. Antibiotic resistance from 2 perspectives: man and microbe. Int J Antimicrob Agents. 2004;23:209–12. doi: 10.1016/j.ijantimicag.2003.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB. Tuberculosis drug resistance mutation database. PLoS Med. 2009;6:e2. doi: 10.1371/journal.pmed.1000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hazbon MH, Brimacombe M, Bobadilla del Valle M, et al. Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2006;50:2640–9. doi: 10.1128/AAC.00112-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siddiqi S, Takhar P, Baldeviano C, Glover W, Zhang Y. Isoniazid induces its own resistance in nonreplicating Mycobacterium tuberculosis. Antimicrob Agents Chemother. 2007;51:2100–4. doi: 10.1128/AAC.00086-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cox HS, McDermid C, Azevedo V, et al. Epidemic levels of drug resistant tuberculosis (MDR and XDR-TB) in a high HIV prevalence setting in Khayelitsha, South Africa. PLoS One. 2010;5:e13901. doi: 10.1371/journal.pone.0013901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nunn P. Institute of Medicine Forum on drug discovery, development and translation. Addressing the threat of drug –resistant TB: a realistic assessment. Geneva: Stop TB Department, WHO; 2008. Global incidence of MDR and XDR-TB. [Google Scholar]
- 27.Sohn H, Minion J, Albert H, Dheda K, Pai M. TB diagnostic tests: how do we figure out their costs? Expert Rev Anti Infect Ther. 2009;7:723–33. doi: 10.1586/eri.09.52. [DOI] [PubMed] [Google Scholar]
- 28.Arias M, Mello FC, Pavon A, et al. Clinical evaluation of the microscopic-observation drug-susceptibility assay for detection of tuberculosis. Clin Infect Dis. 2007;44:674–80. doi: 10.1086/511639. [DOI] [PubMed] [Google Scholar]
- 29.Caviedes L, Lee TS, Gilman RH, et al. Rapid, efficient detection and drug susceptibility testing of Mycobacterium tuberculosis in sputum by microscopic observation of broth cultures. The Tuberculosis Working Group in Peru. J Clin Microbiol. 2000;38:1203–8. doi: 10.1128/jcm.38.3.1203-1208.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Caviedes L, Moore DA. Introducing MODS: a low-cost, low-tech tool for high-performance detection of tuberculosis and multidrug resistant tuberculosis. Indian J Med Microbiol. 2007;25:87–8. doi: 10.4103/0255-0857.32711. [DOI] [PubMed] [Google Scholar]
- 31.Moore DA. Future prospects for the MODS assay in multidrug-resistant tuberculosis diagnosis. Future Microbiol. 2007;2:97–101. doi: 10.2217/17460913.2.2.97. [DOI] [PubMed] [Google Scholar]
- 32.Moore DA, Caviedes L, Gilman RH, et al. Infrequent MODS TB culture cross-contamination in a high-burden resource-poor setting. Diagn Microbiol Infect Dis. 2006;56:35–43. doi: 10.1016/j.diagmicrobio.2006.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moore DA, Evans CA, Gilman RH, et al. Microscopic-observation drug-susceptibility assay for the diagnosis of TB. N Engl J Med. 2006;355:1539–50. doi: 10.1056/NEJMoa055524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Park WG, Bishai WR, Chaisson RE, Dorman SE. Performance of the microscopic observation drug susceptibility assay in drug susceptibility testing for Mycobacterium tuberculosis. J Clin Microbiol. 2002;40:4750–2. doi: 10.1128/JCM.40.12.4750-4752.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shiferaw G, Woldeamanuel Y, Gebeyehu M, Girmachew F, Demessie D, Lemma E. Evaluation of microscopic observation drug susceptibility assay for detection of multidrug-resistant Mycobacterium tuberculosis. J Clin Microbiol. 2007;45:1093–7. doi: 10.1128/JCM.01949-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Minion J, Pai M. Bacteriophage assays for rifampicin resistance detection in Mycobacterium tuberculosis: updated meta-analysis. Int J Tuberc Lung Dis. 2010;14:941–51. [PubMed] [Google Scholar]
- 37.Schwartz RS. Paul Ehrlich's magic bullets. N Engl J Med. 2004;350:1079–80. doi: 10.1056/NEJMp048021. [DOI] [PubMed] [Google Scholar]
- 38.Daniel TM. Paul Ehrlich and the origins of chemotherapy. Int J Tuberc Lung Dis. 2008;12:113–4. [PubMed] [Google Scholar]
- 39.Ehrlich P. A method for staining the tubercle bacillu Aus dem Verein fur innere Medizin zu Berlin. Sitzung vom 1 Mai. Deutsche Medizinsche Wochenschrift. 1882;8:269–70. [Google Scholar]
- 40.Adams M. Bacteriophages. In: Hershey A, editor. New York: Interscience Publishers Inc; 1959. 592. [Google Scholar]
- 41.Lederberg JS. Smaller Fleas. ad infinitum: Therapeutic bacteriophage redux. Proc Natl Acad Sci U S A. 1996;93:3167–8. doi: 10.1073/pnas.93.8.3167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.van Belkum A, Tassios PT, Dijkshoorn L, et al. Guidelines for the validation and application of typing methods for use in bacterial epidemiology. Clin Microbiol Infect. 2007;13(Suppl 3):1–46. doi: 10.1111/j.1469-0691.2007.01786.x. [DOI] [PubMed] [Google Scholar]
- 43.Dowdle WR, Hansen PA. A phage-fluorescent antiphage staining system for Bacillus anthracis. J Infect Dis. 1961;108:125–35. doi: 10.1093/infdis/108.2.125. [DOI] [PubMed] [Google Scholar]
- 44.Watson BB, Eveland WC. The application of the phage-fluorescent antiphage staining system in the specific identification of Listeria monocytogenes. I. Species specificity and immunofluorescent sensitivity of Listeria monocytogenes phage observed in smear preparations. J Infect Dis. 1965;115:363–9. doi: 10.1093/infdis/115.4.363. [DOI] [PubMed] [Google Scholar]
- 45.Watson BB, Eveland WC. The application of the phage-fluorescent antiphage staining system in the specific identification of Listeria monocytogenes. II. The use of the phage-fluorescent antiphage system in the specific identification of Listeria monocytogenes in tissues from experimentally infected animals. J Infect Dis. 1966;116:155–61. doi: 10.1093/infdis/116.2.155. [DOI] [PubMed] [Google Scholar]
- 46.Jacobs WR, Jr, Tuckman M, Bloom BR. Introduction of foreign DNA into mycobacteria using a shuttle phasmid. Nature. 1987;327:532–5. doi: 10.1038/327532a0. [DOI] [PubMed] [Google Scholar]
- 47.Jacobs WR, Jr, Barletta RG, Udaniv R, et al. Rapid assessment of drug susceptibilities of Mycobacterium tuberculosis by means of luciferase reporter phages. Science. 1993;260:819–22. doi: 10.1126/science.8484123. [DOI] [PubMed] [Google Scholar]
- 48.Pearson RE, Jurgensen S, Sarkis GJ, Hatfull GF, Jacobs WR., Jr Construction of D29 shuttle phasmids and luciferase reporter phages for detection of mycobacteria. Gene. 1996;183:129–36. doi: 10.1016/s0378-1119(96)00530-6. [DOI] [PubMed] [Google Scholar]
- 49.Fullner KJ, Hatfull GF. Mycobacteriophage L5 infection of Mycobacterium bovis BCG: implications for phage genetics in the slow-growing mycobacteria. Mol Microbiol. 1997;26:755–66. doi: 10.1046/j.1365-2958.1997.6111984.x. [DOI] [PubMed] [Google Scholar]
- 50.Riska PF, Jacobs WR, Jr, Bloom BR, McKitrick J, Chan J. Specific identification of Mycobacterium tuberculosis with the luciferase reporter mycobacteriophage: use of p-nitro-alpha-acetylamino-beta-hydroxy propiophenone. J Clin Microbiol. 1997;35:3225–31. doi: 10.1128/jcm.35.12.3225-3231.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Carriere C, Riska PF, Zimhony O, et al. Conditionally replicating luciferase reporter phages: improved sensitivity for rapid detection and assessment of drug susceptibility of Mycobacterium tuberculosis. J Clin Microbiol. 1997;35:3232–9. doi: 10.1128/jcm.35.12.3232-3239.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Riska PF, Su Ya, Bardarov Svetoslav, et al. Rapid film-based determination of antibiotic susceptibilities of Mycobacterium tuberculosis strains by using a luciferase reporter phage and the Bronx Box. J Clin Microbiol. 1999;37:1144–9. doi: 10.1128/jcm.37.4.1144-1149.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bardarov S, Jr, Dou H, Eisenach K, et al. Detection and drug-susceptibility testing of M. tuberculosis from sputum samples using luciferase reporter phage: comparison with the Mycobacteria Growth Indicator Tube (MGIT) system. Diagn Microbiol Infect Dis. 2003;45:53–61. doi: 10.1016/s0732-8893(02)00478-9. [DOI] [PubMed] [Google Scholar]
- 54.Hazbon MH, Guarin N, Ferro BE, et al. Photographic and luminometric detection of luciferase reporter phages for drug susceptibility testing of clinical Mycobacterium tuberculosis isolates. J Clin Microbiol. 2003;41:4865–9. doi: 10.1128/JCM.41.10.4865-4869.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Banaiee N, Bobadilla-Del-Valle M, Bardarov S, Jr, et al. Luciferase reporter mycobacteriophages for detection, identification, and antibiotic susceptibility testing of Mycobacterium tuberculosis in Mexico. J Clin Microbiol. 2001;39:3883–8. doi: 10.1128/JCM.39.11.3883-3888.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Piuri M, Jacobs WR, Jr, Hatfull GF. Fluoromycobacteriophages for rapid, specific, and sensitive antibiotic susceptibility testing of Mycobacterium tuberculosis. PLoS One. 2009;4:e4870. doi: 10.1371/journal.pone.0004870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rybniker J, Nowag A, van Gumpel E, et al. Insights into the function of the WhiB-like protein of mycobacteriophage TM4–a transcriptional inhibitor of WhiB2. Mol Microbiol. 2010;77:642–57. doi: 10.1111/j.1365-2958.2010.07235.x. [DOI] [PubMed] [Google Scholar]
- 58.Stager CE, Gangadharam PR. Comparison of several culture media used for studies on mycobacteriophages. J Med Microbiol. 1978;11:187–91. doi: 10.1099/00222615-11-2-187. [DOI] [PubMed] [Google Scholar]
- 59.Friedman D, Olson E, Georgopoulos C, Tilly K, Herskowitz I, Banuett F. Interactions of bacteriophage and host macromolecules in the growth of bacteriophage lambda. Microbiol Rev. 1984;48:299–325. doi: 10.1128/mr.48.4.299-325.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwiatkowska S, Szkudlarek U, Luczynska M, Nowak D, Zieba M. Elevated exhalation of hydrogen peroxide and circulating IL-18 in patients with pulmonary tuberculosis. Respir Med. 2007;101:574–80. doi: 10.1016/j.rmed.2006.06.015. [DOI] [PubMed] [Google Scholar]
- 61.Phillips M, Cataneo RN, Condos R, et al. Volatile biomarkers of pulmonary tuberculosis in the breath. Tuberculosis (Edinb) 2007;87:44–52. doi: 10.1016/j.tube.2006.03.004. [DOI] [PubMed] [Google Scholar]
- 62.Adams KL, Steele PT, Bogan MJ, et al. Reagentless detection of Mycobacteria tuberculosis H37Ra in respiratory effluents in minutes. Anal Chem. 2008;80:5350–7. doi: 10.1021/ac8002825. [DOI] [PubMed] [Google Scholar]
- 63.Syhre M, Manning L, Phuanukoonnon S, Harino P, Chambers ST. The scent of Mycobacterium tuberculosis–part II breath. Tuberculosis (Edinb) 2009;89:263–6. doi: 10.1016/j.tube.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 64.Jassal MS, Nedeltchev GG, Lee JH, et al. 13[C]-urea breath test as a novel point-of-care biomarker for tuberculosis treatment and diagnosis. PLoS One. 2010;5:e12451. doi: 10.1371/journal.pone.0012451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McNerney R, Wondafrash BA, Amena K, Tesfaye A, McCash EM, Murray NJ. Field test of a novel detection device for Mycobacterium tuberculosis antigen in cough. BMC Infect Dis. 2010;10:161. doi: 10.1186/1471-2334-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Phillips M, Basa-Dalay V, Bothamley G, et al. Breath biomarkers of active pulmonary tuberculosis. Tuberculosis (Edinb) 2010;90:145–51. doi: 10.1016/j.tube.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 67.Weetjens BJ, Mgode GF, Machang'u RS, et al. African pouched rats for the detection of pulmonary tuberculosis in sputum samples. Int J Tuberc Lung Dis. 2009;13:737–43. [PubMed] [Google Scholar]
- 68.Poling A, Weetjens BJ, Cox C, et al. Using giant African pouched rats to detect tuberculosis in human sputum samples: 2009 findings. Am J Trop Med Hyg. 2010;83:1308–10. doi: 10.4269/ajtmh.2010.10-0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Han W, Wang T, Reilly AA, Keller SM, Spivack SD. Gene promoter methylation assayed in exhaled breath, with differences in smokers and lung cancer patients. Respir Res. 2009;10:86. doi: 10.1186/1465-9921-10-86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jain R, Schriever CA, Danziger LH, Cho SH, Rubinstein I. The IS6110 repetitive DNA element of Mycobacterium tuberculosis is not detected in exhaled breath condensate of patients with active pulmonary tuberculosis. Respiration. 2007;74:329–33. doi: 10.1159/000101786. [DOI] [PubMed] [Google Scholar]
- 71.Bell GD, Weil J, Harrison G, et al. 14C-urea breath analysis, a non-invasive test for Campylobacter pylori in the stomach. Lancet. 1987;1:1367–8. doi: 10.1016/s0140-6736(87)90664-7. [DOI] [PubMed] [Google Scholar]
- 72.Gordon AH, Hart PD, Young MR. Ammonia inhibits phagosome-lysosome fusion in macrophages. Nature. 1980;286:79–80. doi: 10.1038/286079a0. [DOI] [PubMed] [Google Scholar]
- 73.Hart PD, Young MR. Ammonium chloride, an inhibitor of phagosome-lysosome fusion in macrophages, concurrently induces phagosome-endosome fusion, and opens a novel pathway: studies of a pathogenic mycobacterium and a nonpathogenic yeast. J Exp Med. 1991;174:881–9. doi: 10.1084/jem.174.4.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Damato JJ, Collins MT, McClatchy JK. Urease testing of mycobacteria with BACTEC radiometric instrumentation. J Clin Microbiol. 1982;15:478–80. doi: 10.1128/jcm.15.3.478-480.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Toda T, Hagihara Y, Takeya K. A simple urease test for the classification of mycobacteria. Am Rev Respir Dis. 1961;83:757–61. doi: 10.1164/arrd.1961.83.5.757. [DOI] [PubMed] [Google Scholar]
- 76.Zhang L, Mulrooney SB, Leung AF, et al. Inhibition of urease by bismuth(III): implications for the mechanism of action of bismuth drugs. Biometals. 2006;19:503–11. doi: 10.1007/s10534-005-5449-0. [DOI] [PubMed] [Google Scholar]
- 77.Syhre M, Chambers ST. The scent of Mycobacterium tuberculosis. Tuberculosis (Edinb) 2008;88:317–23. doi: 10.1016/j.tube.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 78.Daigeler A. The cotton rat (Sigmodon hispidus hispidus) as an experimental animal in the diagnosis of tuberculosis. Z Hyg Infektionskr. 1952;135:588–91. [PubMed] [Google Scholar]
- 79.Banaiee N, Bobadilla-del-Valle M, Riska PF, et al. Rapid idenstification and susceptibility testing of Mycobacterium tuberculosis from MGIT cultures with luciferase reporter mycobacteriophages. J Med Microbiol. 2003;52:557–61. doi: 10.1099/jmm.0.05149-0. [DOI] [PubMed] [Google Scholar]