Background: FSHD is associated with a partial deletion in the D4Z4 repeat array on chromosome 4q. The D4Z4 contains an enhancer.
Results: The D4Z4 enhancer interacts with KLF15 causing overexpression of DUX4c and FRG2 genes.
Conclusion: KLF15 serves as a molecular link between myogenic factors and the D4Z4 enhancer.
Significance: KLF15 contributes to the overexpression of DUX4c and FRG2 genes in FSHD.
Keywords: Chromatin Structure, Muscular Dystrophy, Transcription Enhancers, Transcription Factors, Transcription Regulation, D4Z4, FSHD
Abstract
Facioscapulohumeral muscular dystrophy (FSHD), a dominant hereditary disease with a prevalence of 7 per 100,000 individuals, is associated with a partial deletion in the subtelomeric D4Z4 repeat array on chromosome 4q. The D4Z4 repeat contains a strong transcriptional enhancer that activates promoters of several FSHD-related genes. We report here that the enhancer within the D4Z4 repeat binds the Krüppel-like factor KLF15. KLF15 was found to be up-regulated during myogenic differentiation induced by serum starvation or by overexpression of the myogenic differentiation factor MYOD. When overexpressed, KLF15 activated the D4Z4 enhancer and led to overexpression of DUX4c (Double homeobox 4, centromeric) and FRG2 (FSHD region gene 2) genes, whereas its silencing caused inactivation of the D4Z4 enhancer. In immortalized human myoblasts, the D4Z4 enhancer was activated by the myogenic factor MYOD, an effect that was abolished upon KLF15 silencing or when the KLF15-binding sites within the D4Z4 enhancer were mutated, indicating that the myogenesis-related activation of the D4Z4 enhancer was mediated by KLF15. KLF15 and several myogenesis-related factors were found to be expressed at higher levels in myoblasts, myotubes, and muscle biopsies from FSHD patients than in healthy controls. We propose that KLF15 serves as a molecular link between myogenic factors and the activity of the D4Z4 enhancer, and it thus contributes to the overexpression of the DUX4c and FRG2 genes during normal myogenic differentiation and in FSHD.
Introduction
Facioscapulohumeral muscular dystrophy (FSHD)5 is an autosomal dominant neuromuscular disease with a prevalence of 7 in 100,000 (1). FSHD is characterized by progressive weakness and atrophy of the facial muscles and the shoulder girdle. The disorder is associated with a deletion of an integral number of 3.3-kb tandem repeats (D4Z4) (Fig. 1A) present within the subtelomeric regions of the long arms of chromosomes 4 (4q35) and 10 (10q26) (supplemental Fig. S1A). The D4Z4 repeat copy number varies from 11 to ∼100 in healthy individuals but is consistently less than 11 on at least one chromosome 4 in patients with FSHD (2). Together, a shorter D4Z4 array, a specific simple sequence length polymorphism (SSLP-161) and the presence of the 4qA allele have been specifically associated with FSHD (3).
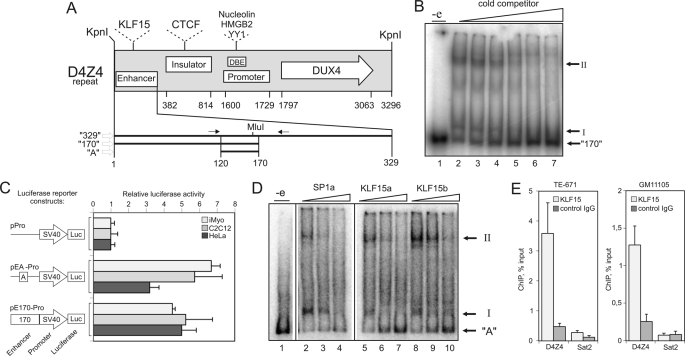
FIGURE 1.
KLF15 interacts with the D4Z4 repeat. A, schematic representation of conserved functional elements within the D4Z4 repeat (nucleotides 1–3296). Enhancer (nucleotides 1–329) (21) containing KLF15 sites (this study); Insulator (nucleotides 382–814) containing CTCF sites (46); Promoter (nucleotides 1600–1729) containing a divergent TATA-box (CATAA) (57); a D4Z4-binding element (DBE) that includes Nucleolin, HMGB2, and YY1 sites (10); DUX4 open reading frame (nucleotides 1797–3063) (57); and fragments 329 (nucleotides 1–329) (21), 170 (nucleotides 1–170), and A (nucleotides 120–170) used in this study are shown. Nucleotide numeration starts from the first nucleotide of the KpnI site that separates individual repeats in the D4Z4 array. Arrows indicate positions of forward and reverse primers used to PCR amplify fragment A. B, fragment 170 forms two complexes (I and II) with proteins in HeLaS3 nuclear extracts. EMSA analysis of a nuclear extract incubated with 32P-labeled fragment 170 in the presence of 3-, 10-, 30-, 100-, 300-, or 1000-fold excess of cold specific competitor; “-e”: no extract control. C, D4Z4 enhancer is active in different cell types. Luciferase (Luc) activity was measured in HeLa cells, C2C12 myoblasts, or human iMyo transfected with reporter constructs that contain the luciferase gene under the control of the SV40 promoter alone (pPro) or downstream of fragments 170 (pE170-Pro) or A (pEA-Pro). Error bars represent S.E. of three independent experiments. D, identification of complexes I and II. EMSA analysis of HeLaS3 nuclear extracts incubated with 32P-labeled fragment A in the presence of 10-, 30-, or 100-fold excess of cold competitors specific for SP1 (SP1a) or KLF15 (KLF15a and -b). E, KLF15 interacts with the D4Z4 repeat in vivo. DNA was immunoprecipitated from TE-671 rhabdomyosarcoma cells or GM10115 hamster cells harboring human chromosome 4 using anti-KLF15 or control antibodies and quantified by qPCR using D4Z4-enhancer specific primers shown in A or Sat2-specific primers.
The transcriptional profiling of FSHD cells grown in vitro and of muscle biopsies has characterized FSHD as a multigenic disorder. Thus, anomalies in the expression of genes involved in the response to oxidative stress (4), vascular smooth muscle-specific and endothelial cell-specific genes (5, 6), as well as a myogenic differentiation program (7–9) have been reported. At the same time, the connection between the myogenic factors and FSHD has never been elucidated.
Gene studies within the 4q35 chromosomal region have shown that FRG1, FRG2, ANT1, DUX4, and DUX4c can be up-regulated in FSHD cells (4, 10–15). The overexpression of FRG1 in skeletal muscles of transgenic mice or that of DUX4 and DUX4c, two proteins encoded by repeated elements at 4q35 in C2C12 myoblasts, recapitulate some of the FSHD features (16–18), but the overall mechanism of their up-regulation in FSHD cells largely remains to be deciphered. The expression of DUX4 in FSHD muscle cells has recently been linked to a unique polymorphism (4qA161) associated with the presence of a previously identified polyadenylation signal in the flanking pLAM region (13) that increases DUX4 transcript stability (19). The mechanism of up-regulation of other genes, including FRG2 and DUX4c, remains unknown.
The D4Z4 repeats and neighboring segments within the 4q35 region are rich in regulatory elements (for review see Ref. 14), whose activity may be perturbed in FSHD. We have recently mapped a potent enhancer within the D4Z4 repeat unit (D4Z4 enhancer) (20, 21). Interestingly, the region homologous to the D4Z4 enhancer that is located proximally to the DUX4c and FRG2 genes (proximal enhancer) (22) is severely mutated (supplemental Fig. S1B). Analysis of the three-dimensional structure of the chromatin in this region has indicated that the D4Z4 enhancer directly contacts the FRG2 and DUX4c promoters (23, 24). D4Z4 enhancer is also able to activate these promoters in vitro (this study and Ref. 11). These observations suggest that the D4Z4 enhancer within the D4Z4 array could control the expression of 42-kb distant DUX4c and FRG2 genes. DUX4c, which is up-regulated in FSHD (25), has been shown to inhibit differentiation of mouse myoblasts (17). FRG2 is overexpressed in myoblasts from FSHD patients after induction of myogenic differentiation, but its function is not known yet (11).
In this study, we identified the Krüppel-like factor KLF15 that directly interacts with the D4Z4 enhancer thereby up-regulating its activity. We also found that KLF15 induces expression of FRG2 and DUX4c. KLF15 is up-regulated during myogenic differentiation, suggesting that the activity of the D4Z4 enhancer may also increase during myogenic differentiation. We also observed that the D4Z4 enhancer activation by MYOD depended on the KLF15 expression suggesting that KLF15 serves as a molecular link between the myogenic factors and the activity of the D4Z4 enhancer during normal myogenic differentiation. Finally, the KLF15 gene was found to be strongly expressed in myoblasts, myotubes, and biopsies from FSHD patients potentially linking aberrant expression of myogenic factors that we observed in these cells to the increase in activity of the D4Z4 enhancer. Taken together, our observations indicate that the KLF15-controlled D4Z4 enhancer could contribute to the up-regulation of FRG2 and DUX4c genes observed during normal myogenic differentiation and in FSHD.
EXPERIMENTAL PROCEDURES
Cell Lines, Culture Conditions, and Transfections
HeLa and HeLaS3 cells (from American Type Culture Collection) and the rhabdomyosarcoma cell lines RD and TE671 (a kind gift of Dr. S. Leibowitz) were grown as described previously (21). Mouse C2C12 cells, human immortalized myoblasts (iMyo) (kind gift of Dr. V. Mouly), and human primary myoblasts were grown and differentiated as described in Refs. 26–28, respectively. The GM10115 hybrid hamster cell line containing human chromosome 4 from Coriell Institute was grown at 34 °C in 8% CO2 on DMEM supplemented with 10% fetal calf serum and 0.2 mm proline. Transient transfection of human immortalized myoblasts with KLF15-SP1 or scrambled siRNA was performed in a 6-well plate format using Lipofectamine 2000 according to the manufacturer's instructions with a minor modification as follows: 600,000 cells were added to the transfection mixture prepared directly in the cell culture plate. For luciferase reporter gene assays, 2.5 × 103 HeLa and 1.25 × 103 RD cells were transfected with 0.1 μg of luciferase reporter plasmids either alone or together with 0.1 μg of KLF15-, SP1-, EGR1-, or GFP-expressing plasmids using jetPEITM (Polyplus) in a 96-well plate format. 2 × 10 4 iMyo cells were transfected with 0.1 μg of luciferase reporter plasmids either alone or together with 0.1 μg of KLF15, MYOD, or shRNA plasmid or 20 μm siRNA using Lipofectamine 2000 (Invitrogen) in a 96-well plate format.
Biopsies
Muscle biopsies were obtained in accordance with the French national regulations. The origin of biopsies is listed in supplemental Table S1.
Reporter Gene Assays
Luciferase activity was determined 48 h after transfection with luciferase reporter plasmid using the Dual-Luciferase assay system (Promega) and normalized to protein concentration (determined by BCA assay, Sigma) and to the activity of the phRL-TK reporter (Promega). All transfections were performed in triplicates and repeated in 3–4 independent experiments. To calculate the relative luciferase activity, the normalized luciferase activity was divided to normalized luciferase activity of the control reporter pPro (Promega). Figures show the average result of three independent experiments.
Western Blotting
Whole cell lysates were prepared using RIPA buffer as described previously (29), separated using 10% SDS-polyacrylamide gel, transferred to Hybond-C extra nitrocellulose membrane (Amersham Biosciences), incubated with primary antibodies against KLF15 (sc-34827X, 1:1000, Santa Cruz Biotechnology), Sp1 (sc-14027X, 1:1000), DUX4c (1:2000) (13), tubulin (sc-8035, 1:5000), actin (MAB1501,1:10000, Millipore), and HRP-conjugated secondary anti-mouse (sc-2005, 1:2000), anti-goat (sc-2768, 1:2000), or anti-rabbit (sc-2313, 1:2000) antibodies according to the standard protocol from Santa Cruz Biotechnology and developed using the ECL+ kit (Amersham Biosciences). Antibodies against DUX4c were described previously (12). Briefly, a 16-residue peptide specific of the DUX4c carboxyl-terminal domain was chosen by accessibility prediction programs, synthesized, coupled to keyhole limpet hemocyanin, and injected into rabbits. The resulting antisera were purified by affinity chromatography on the immobilized peptide (Eurogentec). To detect DUX4c, we used the following conditions. Whole cell extracts of primary cultures of myoblasts were obtained by lysis on ice in 100 μl of 50 mm Tris, pH 7.5, 500 mm NaCl, 0.1% Nonidet P-40, 1 mm DTT, and protease inhibitors mixture (Sigma). 20–30 μg of whole extracts were separated on 4–12% BisTris gels (NuPAGE, Invitrogen) in MOPS buffer and electrotransferred onto a nitrocellulose membrane (Amersham Biosciences). Membranes were incubated with the rabbit anti-DUX4c serum (1:1000) or the mouse anti-GAPDH (1:4000) monoclonal antibody (Ambion) followed by a secondary antibody (goat serum against rabbit immunoglobulins or sheep serum against mouse immunoglobulins) coupled to horseradish peroxidase (HRP) and revealed with the SuperSignal West Femto (Pierce) or Lumi-Light Western blotting substrate (Roche Applied Science).
Plasmids and siRNAs
To obtain the pEA-Pro plasmid, oligonucleotides (Invitrogen) 5′-aattcaatggatccccgccccctccccaccccccaccccccacccccggaaaacgcgtcgtcccca-3′ and 5′-gatctggggacgacgcgttttccgggggtggggggtggggggtggggagggggcggggatccattg-3′ coding for nucleotides 120–170 of the D4Z4 repeat (fragment A) were annealed in 10 mm Tris-HCl, pH 7.4, digested with BamHI, and cloned into the BglII-digested p-Pro vector containing SV40 promoter. pEAmut Pro luciferase reporters with mutated fragment A were cloned in the same way using mutated oligonucleotides. DUX4c was cloned as described previously (25). A 170-bp MluI-digested fragment containing the D4Z4 enhancer was cloned upstream of the DUX4c promoter in the p-ProDUX4c plasmid resulting in pE170-ProDUX4c. The pE170-Pro plasmid containing the SV40 promoter and the enhancer 170 was described before (21). The CMV-SP1 plasmid (Dr. Robert Tjian) pcDNA3-Egr1 (29) was purchased from Addgene (catalog nos. 12097 and 11729, respectively). The hKLF15-pcDNA plasmid (30) was a kind gift of Dr. Deborah Otteson. The pcDNA3-MYOD plasmid was a kind gift of Anna Polesskaya. siRNAs against KLF15 (sc-45567) and a scrambled control (sc-37007) were purchased from Santa Cruz Biotechnology.
One-hybrid Screen
Plasmids and strains required for the one-hybrid screen were kindly provided by Pieter Ouwerkerk, and the screening was performed as described previously (31). Oligonucleotides OPD196 and -197 (Invitrogen) coding for fragment A of the D4Z4 repeat were annealed and directly cloned into BglII/EcoRI-digested pHIS3HX vector (31). The resulting pHIS3HX-1xA plasmid was digested with BamHI/EcoRI and ligated with the OPD196/197 duplex to obtain the pHIS3HX-2xA plasmid. The last step was repeated another time to obtain the pHIS3HX-3xA plasmid that was then digested by XbaI/NotI and cloned into XbaI/NotI-digested integrative vector pINT (31) to obtain the plasmid pINT-HIS-3xA. This was linearized with NcoI/SacI and transformed into the Y187 yeast strain, which contains the control β-galactosidase reporter gene (31) to obtain the reporter strain Y187-A. As a control, we used the Y187 strain transformed with pINT-HIS (Y187-C). Y187-A was used to screen the ProQuest cDNA library from human skeletal muscle (PL10001-02, Invitrogen). White his+ colonies were selected to isolate plasmids coding for cDNA candidates. Y187-C did not produce his+ colonies after transformation with positive plasmids. cDNA inserts of positive plasmids were sequenced (Milligen) and identified using BLAST (NCBI).
Nuclear Extracts and EMSA
Nuclear extracts were prepared from HeLaS3 cells grown to 2 × 106 cells/ml as described previously (32) with the following modifications: after extraction with high salt buffer, extracts were centrifuged twice at 77,000 × g for 30 min and 1 h and kept frozen at −80 °C in small aliquots. The concentration of the nuclear extracts was 7 μg/μl (Bradford assay, Bio-Rad). Probe “A WT” was prepared by annealing oligonucleotides 5′-ccgccccctccccaccccccaccccccacccccggaaaacgcgtcgtcccc-3′ and 5′-ggggacgacgcgttttccgggggtggggggtggggggtggggagggggcgg-3′ in 10 mm Tris-HCl, pH 7.4. Each oligonucleotide was synthesized with a 5′-ctag overhang end to allow 32P labeling. To prepare probe “170,” the pE170-ProDUX4c plasmid was digested by MluI, and the 170-nucleotide fragment was gel-purified using the Nucleospin Extract II kit (Macherey Nagel). As cold competitors, the following oligonucleotide duplexes were used (only one strand is shown 5′ to 3′, and 5′-ctag overhang is not shown): KLF15b, attatgaacacccccaatctcccagatgc (33); KLF15a, agccggggagggggaggggagggtgttg (34); Sp1a, gacgcggggcgcgggggcggggcgcg (35); Sp1b, caccccctccctctcagggagg (36); Amut-all ccgccgcctccgcaccgccgaccgccgaccgccggaaaacgcgtcgtcccc; and Amut-EGR/ZNF ccgaaacctccccaccccccaccccccacccccggaaaacgcgtcgtcccc. The 2.5 pmol of probes were labeled with 26 pmol (80 mCi) (9)) of dCTP (Amersham Biosciences) and Klenow fragment in a final volume of 20 μl, extracted with phenol/chloroform, precipitated with 2 volumes of ethanol, and resuspended in 20 μl of 10 mm Tris-HCl, pH 7.4, to obtain a 35 fmol/μl solution of labeled probe. For each reaction, 3.5 fmol of labeled probe was incubated for 20 min at room temperature in 1× RBM 0.2 buffer (12 mm HEPES-KOH, pH 7.9, 60 mm KCl, 0.2 mm EDTA, 1 mm DTT, 12% glycerin) with 0.43 pmol (600 ng) of poly(dI-dC) (P4929, Sigma) and excess of cold competitors and 3.5 μg of nuclear extract for 20 min. Then the mixture was loaded on 5% 0.75-mm polyacrylamide minigels, subjected to electrophoresis in 0.6× TBE at 92 V. Gels were dried at 80 °C under vacuum and analyzed with a Phosphorimager (Fuji).
Bioinformatics
As a description of the conserved patterns in SP1- and KLF15-binding sites, we used the frequency matrix M00032 from Information Matrix Database (37) and the frequency matrix from Ref. 33, respectively. To search for SP1- and KLF15-binding sites in EMSA probes and competitors, we used the Matrix Search program (37) that assigns a score to each putative transcription factor site and then calculates a match ratio that represents the similarity of each putative Sp1 or KLF15 site to the conserved site (5′-GCCCCGCCC-3′ and 5′-CGCCCCTCC-3′, respectively). Binding sites for Sp1 and KLF15 were visualized using the EnoLOGOS program (38) and are available on line.
Statistical Analysis
The Student's t test and Mann-Whitney test were performed as described previously (39).
Reverse Transcription and qPCR
For KLF15, KLF13, PPARG, FRG1, FRG2, ANT1, TNNT1, MYH1, MYOG, and MYOD expression, analysis of total RNA was isolated from 2 × 106 proliferating myoblasts, differentiated myotubes, or 100 mg of biopsies using TRIzol (Invitrogen) and reverse-transcribed using the High Capacity cDNA archive kit (Applied Biosystems) according to the manufacturer's protocol. cDNA was mixed with 2× Taqman PCR mix (Applied Biosystems) and amplified using Taqman low density array, an Abiprism 7900HT apparatus (Applied Biosystems). Expression was analyzed using the ΔΔCt method (40). The following TaqMan inventoried gene expression assays (Applied Biosystems) were used: KLF15, Hs00362736_m1 (does not discriminate between endogenous and ectopic expression of KLF15); KLF13, Hs00740949_s1; PPARG, Hs00234592_m1; FRG2, Hs03025250_gH; GAPDH, Hs99999905_m1; TNNT1, Hs00162848_m1; FRG1, Hs02387002_g1; ANT1, Hs00154037_m1; MYOG, Hs01072232_m1; and MYOD, MYOD1-Hs00159528_m1. To detect the expression of the endogenous KLF15 and MYH1 using FastStart Universal SYBR Green master mix (Rox) (catalog no. 04913850001, Roche Applied Science) the following primers were used: KLF15-F4 5′-GCTTGAGTTAAATGTGCAGGG-3′ and KLF15-R4 5′-TTCTAAATCAGGGTTGGGAGG-3′, and MYH1-F2 5′-GCACACCCAGAACACCAG-3′ and MYH1-R2 5′-GCTTCTTCCCACCCTTCAG-3′. Primers and conditions for DUX4c and DUX4 RT-PCR were described previously (12).
Chromatin immunoprecipitation was performed using the ChIP-IT Express kit (Active Motif, Carlsbad, CA). 3 × 106 cells were cross-linked using 1% formaldehyde, and chromatin was isolated and enzymatically fragmented according to manufacturer's protocol. Then 5 μg of chromatin (corresponding to ∼300,000 cells) was used in the immunoprecipitation reaction performed using either mouse anti-KLF15 monoclonal antibody 2G8 (catalog no. ab81604, Abcam) or negative control mouse IgG from ChIP-It control human kit (catalog no. 53010, Active Motif). Immunoprecipitated DNA was PCR-amplified using the following primers: D4Z4_F1 (5′-AACTGCCATTCTTTCCTGGG-3′); D4Z4_R1 (5′-TGGTGGAGAGGCAGGAG-3′); Sat2_F1 5′-AGGAGTCATCATCTA-ATGGAATTG-3′ and Sat2_R1 5′-GATGATTCCATTCCATTCCATTTG-3′, and FastStart Universal SYBR Green master mix (Rox) (catalog no. 04913850001, Roche Applied Science). PCR amplification and real time fluorescence measurements were carried out using StepONE plus apparatus (Applied Biosystems), PCR program: 94 °C, 15 min followed by 40 cycles of 94 °C, 15 s; 60 °C, 1 min.
RESULTS
Characterization of the D4Z4 Minimal Enhancer
We have recently shown that the D4Z4 repeat contained a strong transcriptional enhancer, and this activity was mapped to fragment 170 (nucleotides 1–170) of the D4Z4 repeat (Fig. 1A) (20, 21). To better characterize this fragment, we performed electrophoretic mobility shift assays (EMSA) and found that fragment 170 formed two major specific complexes (I and II) with proteins in HeLaS3 nuclear extracts (Fig. 1B). The shorter fragment “A” (nucleotides 120–170 in Fig. 1A) exhibited a similar capacity to interact with proteins in nuclear extracts from HeLaS3 and C2C12 cells as fragment 170 (supplemental Fig. S2, A and B). We then compared the enhancer activity fragments 170 and A in HeLa, C2C12, and human iMyo using reporter constructs where luciferase expression was under the control of the SV40 promoter either alone (p-Pro, negative control) or downstream of fragment 170 (pE170-Pro) or fragment A (pEA-Pro) (Fig. 1C). Quantification of luciferase expression indicated that the enhancer activity of fragment A was similar to that of fragment 170 in all cell lines tested (Fig. 1C). We thus focused on fragment A for further analysis.
KLF15 Interacts with the Minimal D4Z4 Enhancer in Vitro and in Vivo
Analysis of transcription factor-binding sites within the D4Z4 enhancer identified SP1 as a potential binding factor. To identify additional transcription factors potentially interacting with the minimal D4Z4 enhancer (fragment A) in muscle cells, we set up a yeast one-hybrid assay (31) and screened a human skeletal muscle cDNA library using fragment A fused to the HIS3 reporter gene.
We analyzed a total of 22 independent clones isolated from this initial screen. After additional verification by transformation of the isolated plasmids into the control and screening strains, eight plasmids were further analyzed. Five plasmids were considered as true positives: two coding for KLF15, two for EGR1, and one for ZNF444. The other three plasmids coded for CKM, Actin and Sphingomyelinase and were considered as false positives. SP1 was not present among the identified factors possibly because it is down-regulated during myogenesis (41).
We next tested whether factors identified in the one-hybrid screen could form specific complexes with the D4Z4 enhancer. Fragment A contains two SP1 sites that partially overlap with two KLF15 sites, one EGR1 site and one ZNF444 site (Fig. 2B, left panel). To test whether the identified factors could form specific complexes with the D4Z4 enhancer, we mutated the recognition sites of EGR1, SP1, ZNF444, and KLF15 in fragment A and carried out EMSAs using the mutant oligonucleotides as competitors. Fragment A with mutations in EGR1 and ZNF444 sites (A mut-E/Z) retained its ability to bind proteins from nuclear extracts as it still competed efficiently with wild-type (WT) fragment A for the formation of complexes I and II (supplemental Fig. S2C). In contrast, disruption of both KLF15 and SP1 sites (A mut-all) led to an almost complete loss of the competing ability of fragment A (supplemental Fig. S2C), suggesting that these complexes are formed with KLF15 and SP1. We then performed EMSAs using oligonucleotide duplexes that have been shown by others to specifically bind to either KLF15 or SP1 (Fig. 1D) (33–36). KLF15-specific duplexes competed more efficiently with fragment A for formation of complex I. Conversely, SP1-specific duplexes were more efficient in the competition for formation of complex II, suggesting that complex I includes KLF15 and complex II-SP1 (Fig. 1D).
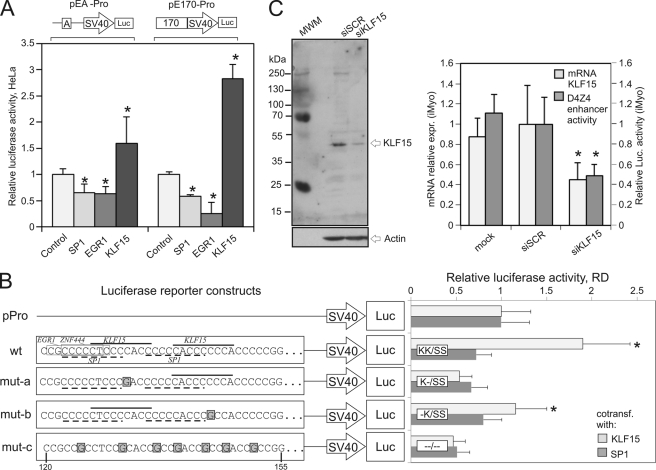
FIGURE 2.
A, overexpression of the KLF15 gene activates the D4Z4 enhancer. Luciferase activity was measured in HeLa cells cotransfected with pEA-Pro or pE170-Pro reporters along with plasmids expressing SP1, EGR1, KLF15, or GFP (control). Asterisks indicate p value <0.01 (Student's t test). B, mutations in the KLF15 recognition sites abolish the KLF15-dependent activation of the D4Z4 enhancer. Wild-type fragment A containing KLF15, SP1, ZNF44, and EGR1 recognition sites and its mutant versions (mut-a, -b, and -c) were cloned upstream of the SV40 promoter into the luciferase (Luc) reporter vector. Nucleotides 1–35 in the 50-bp-long wild-type and mutant versions of fragment A are shown (corresponding to nucleotides 120–155 within the D4Z4 repeat). K and S refer to the presence of intact KLF15 or SP1 sites, respectively. Luciferase activity was measured in rhabdomyosarcoma (RD) cells cotransfected with the indicated reporters along with the KLF15 or SP1 plasmids. *, p value <0.01 (Student's t test). C, KLF15 silencing inhibits the activity of the D4Z4 enhancer. Left panel, human iMyo were transiently transfected with siRNA against KLF15 or scrambled control. KLF15 expression was revealed by Western blotting. Right panel, KLF15 expression was measured by qRT-PCR in iMyo transiently transfected with siRNA against KLF15 or scrambled siRNA. Luciferase activity was measured in iMyo cells transiently cotransfected with reporter pEA-Pro and siRNA against KLF15 or scrambled siRNA. *, p value <0.02 (Student's t test).
We then used chromatin immunoprecipitation (ChIP) to test whether KLF15 interacted with the D4Z4 enhancer in vivo. Formaldehyde cross-linked chromatin from TE-671 cells was enzymatically fragmented and immunoprecipitated using anti-KLF15 or control antibodies. The precipitated DNA was PCR-amplified with primers specific for the D4Z4 enhancer or centromeric satellite II DNA (Sat2). As shown in Fig. 1E, the D4Z4 enhancer but not Sat2 DNA was specifically immunoprecipitated with the anti-KLF15 antibodies. To confirm that KLF15 interacted with the D4Z4 repeat on chromosome 4, we have repeated KLF15 ChIP in chromatin in the hamster cell line GM10115 that harbors chromosome 4 as a single human chromosome and have shown that the D4Z4 enhancer was enriched ∼5-fold in KLF15 immunoprecipitates as compared with the control (Fig. 1E). We thus conclude that KLF15 interacts specifically with the D4Z4 enhancer.
KLF15 Is Activator of D4Z4 Enhancer
Depending on the context, KLF15 can function as an activator (42, 43) or a repressor (30, 44, 45). To better analyze its role, as well as that of SP1 vis à vis the D4Z4 enhancer, we first tested whether ectopically expressed KLF15 and SP1 could activate the D4Z4 enhancer. We overexpressed KLF15, SP1, or GFP (control) in HeLa cells and analyzed the activity of the D4Z4 enhancer using the luciferase reporter constructs pEA-Pro and pE170-Pro (Fig. 2A). When overexpressed, KLF15 stimulated luciferase expression ∼3-fold although overexpressing SP1 or EGR1 slightly inhibited its expression (Fig. 2A). Similar effects of KLF15 on the D4Z4 enhancer activity were also observed in HeLa cells and in immortalized human myoblasts (supplemental Fig. S3A). To test whether this enhancing effect of KLF15 depends on KLF15 and/or SP1 binding, we introduced mutations that disrupted KLF15- or SP1-binding sites in fragment A (Fig. 2B) and cloned these mutated sequences into the luciferase reporter vector. Disrupting of only one of the two KLF15 sites (mut-a and -b) considerably reduced the enhancer activity, suggesting that the presence of both KLF15 sites is essential for the activity of the D4Z4 enhancer. Conversely, the effect of SP1 on the D4Z4 enhancer was not significantly modified by mutations in the SP1 sites indicating that SP1 does not directly regulate the D4Z4 enhancer activity (Fig. 2B).
To confirm that KLF15 was required for the D4Z4 enhancer activity in human myoblasts, we next silenced KLF15 using an RNAi approach. In iMyo cells transfected with siRNA against KLF15, the endogenous KLF15 expression levels were 2–3-fold lower than in the cells transfected by scrambled siRNA (Fig. 2C). We then tested the D4Z4 enhancer activity in iMyo cells cotransfected with siRNA against KLF15 and pEA-Pro luciferase reporter construct (Fig. 2C, right panel). The activity of the pEA-Pro reporter was significantly lower in the transfected siKLF15 than in control cells suggesting that KLF15 is required to activate the D4Z4 enhancer in proliferating myoblasts (Fig. 2C, right panel). Similar results were obtained in HeLa cells (data not shown). From these results, we conclude that the D4Z4 enhancer activity requires the presence of KLF15 and that KLF15 can stimulate the D4Z4 enhancer in various cells, including human myoblasts.
KLF15 Controls the Expression of DUX4c and FRG2 Genes
The D4Z4 repeat is known to activate the promoters of the FRG2 and FRG1 genes (11, 21). We and others have previously demonstrated that the D4Z4 repeats directly interact with the promoter regions of DUX4c, FRG1, and FRG2 (23, 24). We thus reasoned that the KLF15-regulated D4Z4 enhancer could be a natural activator of these genes. To test this hypothesis, FRG1, FRG2, and ANT1 mRNA expressions were measured in HeLa-overexpressing KLF15. FRG1 and ANT1 expressions remained virtually unchanged, in striking contrast to the overexpression of FRG2 whose expression exhibited a 15-fold increase in KLF15- versus empty vector-transfected HeLa cells (Fig. 3A, left panel, and supplemental Fig. S3C).
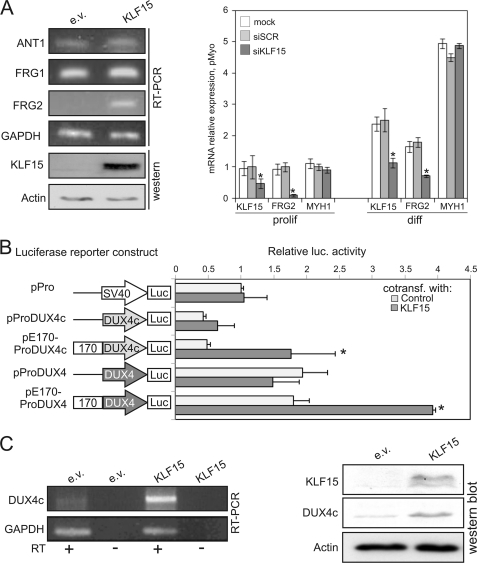
FIGURE 3.
KLF15 activates DUX4c and FRG2. A, left panel, FRG1, FRG2, ANT, and KLF15 expression was measured using RT-PCR in RD cells transiently transfected with the KLF15 plasmid. *, p value <0.01 (Student's t test). Right panel, expression of FRG2 is controlled by KLF15. KLF15 (endogenous and ectopic), FRG2, and MYH1 expression was measured by qRT-PCR in proliferating and differentiated primary human myoblasts from a normal subject transiently transfected with a siRNA against KLF15 or scrambled control siRNA. *, p value <0.01 (Student's t test). B, KLF15 activates DUX4c. The D4Z4 enhancer activates the DUX4c promoter in a KLF15-dependent manner. Luciferase (Luc) activity was measured in iMyo cells cotransfected with KLF15 plasmid or an empty vector control and reporter constructs containing the luciferase gene under the control of the SV40 (p-Pro) or the DUX4c promoter, alone (p-ProDUX4c) or downstream of fragment 170 (pE170-ProDUX4c). *, p value <0.05 (Student's t test). The same experiment was repeated using luciferase reporter constructs, including the DUX4 promoter. C, left panel, DUX4c expression was analyzed using semi-quantitative RT-PCR in proliferating iMyo cells transiently transfected with the KLF15 plasmid or an empty vector control. Full scan of the gel along with necessary controls are shown in supplemental Fig. S3D. Right panel, KLF15, DUX4c, and actin expression was analyzed by Western blot of HeLa cells transfected with the KLF15 plasmid or an empty vector (e.v.).
That KLF15 is a specific activator of FRG2 was confirmed by a KLF15 knockdown assay. Human primary myoblasts were transfected with an siRNA against KLF15 or with a scrambled sequence control. As shown in Fig. 3A, right panel, the expression of FRG2 was decreased 5-fold in proliferating myoblasts, although MYH1 expression remained unchanged. These observations suggest that the regulation of FRG2 expression by KLF15 is direct rather than through the action of myogenic factors. We also tested FRG2 and KLF15 expression in serum starvation-induced differentiated myotubes where FRG2, KLF15, and MYH1 expression is higher. In these differentiated myotubes, KLF15 knockdown led to FRG2 repression without affecting MYH1 expression (Fig. 3A, right panel), indicating that KLF15 controls FRG2 expression without affecting myogenic differentiation.
Having demonstrated that KLF15 controls the expression of FRG2, we then asked whether it also controls the expression of two double homeobox genes, DUX4c and DUX4, also located within the 4q35 chromosomal region. Although the DUX4c gene maps within a truncated D4Z4-like element (D4Z4*), its enhancer region is mutated and lacks any KLF15-binding site (supplemental Fig. S1B). The DUX4c and DUX4 promoters and the D4Z4 enhancer were cloned in the luciferase reporter plasmid pPro to produce p-ProDUX4/4c (DUX4 or DUX4c promoter alone) and pE170-ProDUX4/4c (including the D4Z4 enhancer upstream of the DUX4 or DUX4c promoter). Human immortalized myoblasts were cotransfected with these constructs and with a KLF15 plasmid. As seen in Fig. 3B, KLF15 overexpression induced 3–4-fold DUX4 and DUX4c promoters coupled to the D4Z4 enhancer. In the absence of the D4Z4 enhancer, DUX4 and DUX4c promoters were almost insensitive to the KLF15 overexpression indicating that in natural context KLF15 could control the expression of these genes indirectly via the D4Z4 enhancer and not by directly regulating their promoters (Fig. 3B).
It was shown recently that the DUX4 gene is surrounded by CTCF-dependent enhancer blocking elements (46). This observation prompted us to test whether the D4Z4 enhancer is able to activate the expression of DUX4 and DUX4c in vivo. As seen both at the mRNA (Fig. 3C, left panel) and protein (Fig. 3C, right panel) levels, overexpressing KLF15 in human iMyo and HeLa cells resulted in an enhanced expression of DUX4c. Conversely, we did not observe any effect of KLF15 on the DUX4 expression (data not shown) suggesting that the KLF15-controlled D4Z4 enhancer can not activate the DUX4 expression in vivo.
KLF15 Is a Molecular Link between Myogenic Factors and Activation of the D4Z4 Enhancer
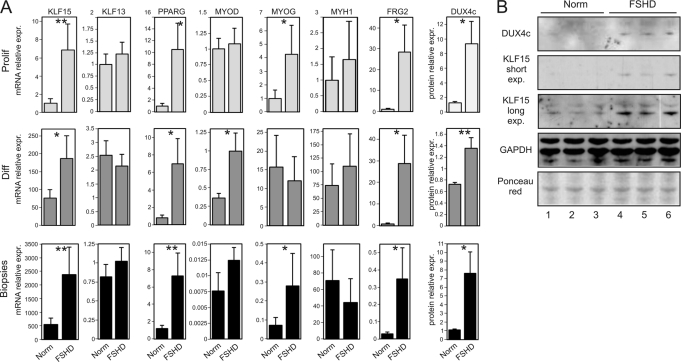
Otteson et al. (33) have shown that KLF15 is up-regulated during differentiation of cardiomyocytes and adipocytes. Indeed, recognition sites for myogenic and adipogenic factors are present in the KLF15 promoter (data not shown). qRT-PCR was used here to quantify KLF15 expression in human myoblasts prior to and following myogenic differentiation induced by serum deprivation. The expression of KLF15 was found to be ∼25-fold higher in differentiated myotubes as compared with proliferating myoblasts (Fig. 4A, left panel). The level of myogenic differentiation was followed by measuring Troponin T and Myogenin mRNA expression. Up-regulation during myogenic differentiation is not a general feature of KLF factors, because the expression of KLF13 was not higher in myotubes compared with myoblasts. The differentiation-dependent induction of KLF15 expression was confirmed by Western blot analysis in both normal and FSHD myoblasts (Fig. 4A, middle panel). KLF15 was further found to be overexpressed in human immortalized myoblasts transfected by MYOD thus confirming that KLF15 expression is indeed controlled by myogenic factors (Fig. 4A, right panel).
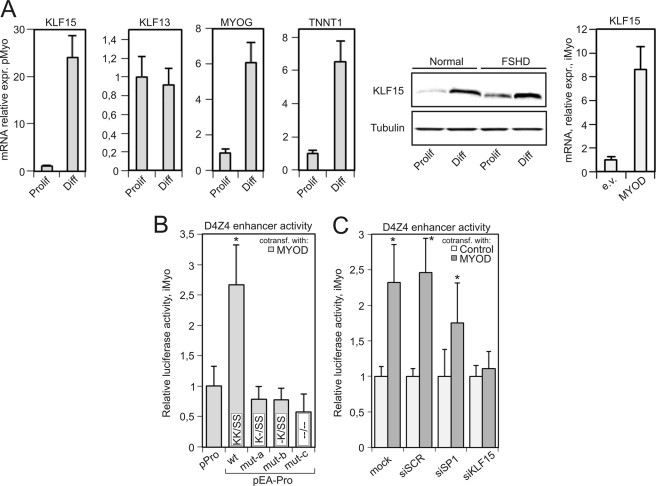
FIGURE 4.
A, KLF15 is up-regulated during myogenic differentiation. Left panel, expression of KLF15, Troponin T1 (TNNT1), and Myogenin (MYOG) was measured using qRT-PCR in primary proliferating human myoblasts (Prolif) and differentiated myotubes (Diff) from a healthy subject (N5 in supplemental Table S1). Middle panel, KLF15 protein was revealed by Western blotting in proliferating myoblasts (Prolif) and differentiated myotubes (Diff) from a healthy control and an FSHD patient (N5 and F1 in supplemental Table S1). Right panel, KLF15 expression was measured using qRT-PCR in immortalized human myoblasts transiently transfected with empty (e.v.) or MYOD-expressing plasmid. B, mutations is KLF15-binding sites abolish the MYOD-dependent activation of the D4Z4 enhancer. Luciferase activity was measured in iMyo cells cotransfected with MYOD plasmid and reporters p-Pro or pEA-Pro containing either wild-type or mutant versions of fragment A. Luciferase activity of the reporter cotransfected with a GFP plasmid was considered as background. *, p value <0.01 (Student's t test). C, KLF15 is essential for MYOD-dependent activation of the D4Z4 enhancer. Luciferase activity was measured in iMyo cells cotransfected with siRNAs against KLF15, SP1, or scrambled control along with MYOD or GFP plasmids and pEA-Pro reporter vector. *, p value <0.01 (Student's t test).
We next tested whether the activity of the D4Z4 enhancer could be induced by an overexpression of MYOD. For this purpose, human immortalized myoblasts were cotransfected with the MYOD plasmid, pPro, pEA-Pro (WT and mutant) luciferase reporters (Fig. 4B). The D4Z4 enhancer activity was found to be ∼2.5–3-fold up-regulated. The enhancing effect of MYOD overexpression was completely abolished by mutations disrupting the KLF15/SP1 sites. Again, disrupting a single KLF15 site was sufficient to make the D4Z4 enhancer completely unresponsive to MYOD overexpression (Fig. 4B). To test whether KLF15 was required for this MYOD-dependent activation of the D4Z4 enhancer, we then cotransfected human immortalized myoblasts with pEA-Pro, MYOD, and siRNAs against either SP1, KLF15, or a scrambled control (Fig. 4C). Only KLF15 silencing led to a complete loss of MYOD-dependent activation of the D4Z4 enhancer (Fig. 4C). Taken together, these data suggest that upon myogenic differentiation, the up-regulation of KLF15 leads to the activation of the D4Z4 enhancer.
KLF15 Is Overexpressed in FSHD
Having demonstrated a possible role of KLF15 in D4Z4 enhancer activation during myogenic differentiation, we then assessed its expression in various samples. As reported in Fig. 5A, KLF15 but not KLF13 expression was found to be considerably higher in myoblasts, myotubes, and muscle biopsies from FSHD patients as compared with healthy controls (Fig. 5A). In line with KLF15 up-regulation, we found that the expression of peroxisome proliferator-activated receptor γ (PPARG), one of the known gene targets of KLF15 (47), was also up-regulated in all FSHD samples. This pattern was similar to that observed for the FRG2 gene (Fig. 5A), a known molecular feature of FSHD (48). The expression of DUX4c has been reported previously to be higher in differentiated myotubes and biopsies from FSHD patients (12) than in controls. Here, DUX4c was further shown to be expressed to a higher level in FSHD myoblasts, thus mimicking KLF15 expression (Fig. 5, A and B). Conversely, neither FRG1 nor ANT1 exhibited any significant changes in their expression patterns when comparing FSHD and control samples confirming a previously published report (data not shown) (48).
FIGURE 5.
KLF15, DUX4c, and FRG2 are overexpressed in FSHD cells. A, expression of KLF15, FRG2, MYOG, MYH1, KLF13, MYOD, and PPARG was measured by qRT-PCR in proliferating myoblasts (Prolif), differentiated myotubes (Diff) from four healthy subjects (Norm) and four FSHD patients (N1 to N4 and F1 to F4 in supplemental Table S1), and muscle biopsies from two healthy subjects (Norm) and four FSHD patients (Na, Nb, and Fa to Fd in supplemental Table S1); mean results and S.E. are shown for each group. DUX4c expression was measured at the protein level in proliferating myoblasts (quantification of Western blots in B), differentiated myotubes, and muscle biopsies (quantification of Western blots previously published in Ref. 12). *, p value <0.05 (Mann-Whitney test). B, Western blot analysis of DUX4c and KLF15 expression in proliferating myoblasts from healthy subjects (N1, N2, and N5), FSHD patients (F1, F3, and F6). Two exposures are shown for KLF15.
We then asked whether the increased KLF15 expression found in FSHD patients could be explained by an increased level of myogenic differentiation. We analyzed the expression of the myogenic factors MYOD, MYOG, and that of the myosin heavy chain 1 gene (MYH1) known to be up-regulated during myogenic differentiation in proliferating myoblasts, myotubes, and biopsies from FSHD patients and normal individuals. In proliferating myoblasts and differentiated myotubes, the expression of either MYOG or MYOD but not MYH1 was found statistically higher in patients as compared with controls, similar to the enhanced expression of KLF15. In biopsies, the expression of MYOG, but not MYOD or MYH1, was significantly higher in patients (Fig. 5A). From these results, we conclude that the KLF15 gene is overexpressed in FSHD, similar to its target genes PPARG, DUX4c, and FRG2.
DISCUSSION
A reduction in the number of macrosatellite D4Z4 repeats on the long arm of chromosome 4 was one of the first genetic variations found in FSHD patients (49). Functional analyses have demonstrated that D4Z4 repeats can function as silencers (10), insulators (46), or transcriptional enhancers (11, 21). Our group has previously identified and mapped a strong transcriptional enhancer present within each of the D4Z4 repeats (21). In this study, we have identified the transcription factor KLF15 as binding to the D4Z4 enhancer and inducing its activity.
KLF15, a member of the Krüppel-like transcription factors, was first identified as a repressor of kidney-specific chloride channel CLC-K1 (50), but in other works it was demonstrated that KLF15 can also act as an activator of transcription (30, 47). The expression levels of KLF15 are the highest in kidney, liver, pancreas, and cardiac and skeletal muscle (50). It was shown that KLF15 expression is up-regulated during cardiomyogenesis (44) and adipogenesis (47). KLF15 regulates cardiac gene expression by interfering with Myocardin and MEF2 activity (44, 51). Although it was shown that KLF15 regulates glucose metabolism in skeletal muscle (42, 52), KLF15 knock-out mice develop normal skeletal muscles (44) indicating that KLF15 is dispensable for myogenic differentiation of skeletal muscles.
We observed that the expression of KLF15 was up-regulated during differentiation of human skeletal myoblasts and was induced by MYOD ectopic overexpression. Moreover, the activity of the D4Z4 enhancer was also induced by MYOD. This induction was abolished when KLF15 sites were mutated or when KLF15 was inhibited via siRNA. These results suggested that KLF15 links the activity of myogenic factors to the activity of the D4Z4 enhancer.
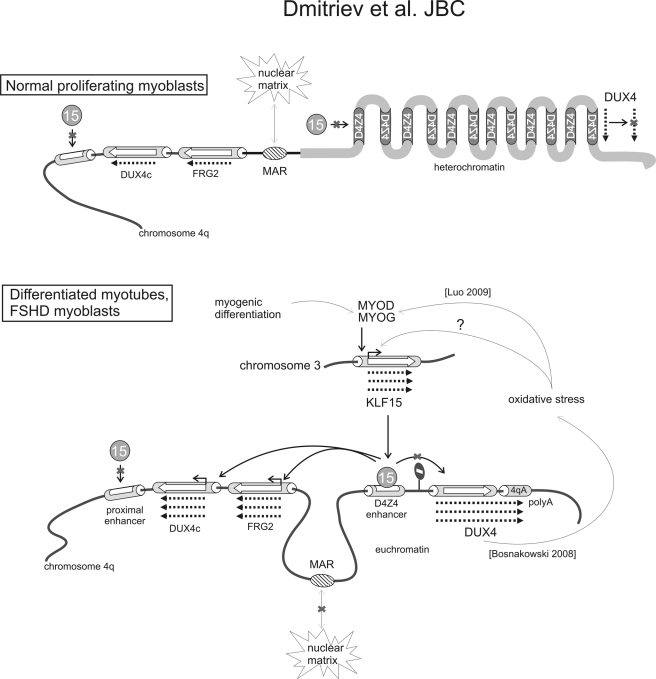
From overexpression and RNAi knockdown experiments, KLF15 was shown here to activate the expression of DUX4c and FRG2, but not FRG1 or ANT1, and all four genes were located within the 4q35 chromosomal region. Previous studies had demonstrated that the D4Z4 enhancer could activate the FRG2 promoter (11, 21) and that it physically interacts with the promoter region of FRG2 as well as that of DUX4c (23, 24). Our present findings indicate that the D4Z4 enhancer is an efficient activator of the DUX4c promoter. We suggest that the D4Z4 enhancer and not the proximal enhancer lacking KLF15 sites within the D4Z4* element contributes to the KLF15-dependent induction of the DUX4c and FRG2 genes providing a possible mechanism for up-regulation of DUX4c and FRG2 during normal myogenic differentiation and in FSHD (schematized in Fig. 6). Our observations that KLF15 expression is higher in proliferating myoblasts, myotubes, and biopsies from FSHD patients than from healthy controls suggest that the activity of the D4Z4 enhancer is higher in these cells. According to this model, the higher activity of the KLF15-dependent D4Z4 enhancer would lead to a higher expression of FRG2 and DUX4c in the patients (Fig. 6). Indeed, FRG2 and DUX4c were found up-regulated not only during the normal myoblast differentiation process but also in FSHD myoblasts and myotubes. DUX4c levels were higher in proliferating FSHD myoblasts (this study), as previously shown for differentiated myocytes and biopsies from FSHD patients (25). FRG2, which was known to be overexpressed in differentiated myotubes and biopsies (11, 48), was found overexpressed as well in proliferating myoblasts from FSHD patients (this study).
FIGURE 6.
Putative model for KLF15-dependent activation of DUX4c and FRG2 expression. In myoblasts from healthy subjects, three factors could interfere with FRG2 and DUX4c expression as follows: (i) low expression of KLF15 keeps the activity of the D4Z4 enhancer low; (ii) MAR is bound to the nuclear matrix separating the DUX4c and FRG2 genes from the D4Z4 repeats; (iii) the heterochromatin structure of the D4Z4 repeats prevents binding of any activating transcription factors. During normal myogenic differentiation and in FSHD, the expression and activity of myogenic factors increase (in case of FSHD this up-regulation may be due to moderate oxidative stress); MYOD activates the expression of KLF15; the structure of D4Z4 repeats is changed to euchromatin facilitating binding of KLF15 to the D4Z4 enhancer; MAR becomes less efficient and allows interaction between the D4Z4 enhancer and the DUX4c and FRG2 promoters. The DUX4 gene, a potential inducer of moderate oxidative stress, is separated from the KLF15-controlled D4Z4 enhancer by enhancer blocking elements thus preventing the D4Z4 enhancer to activate it. The mechanism of DUX4 up-regulation in FSHD is linked to the specific polymorphism (1614qA) stabilizing its mRNA and seems to be KLF15-independent. D4Z4* is truncated D4Z4 repeat.
The role of FRG2 and DUX4c, the two genes regulated by KLF15, in the FSHD dystrophic phenotype is currently under investigation. Originally described as a pseudogene, DUX4c has been shown to inhibit myogenic differentiation suggesting that it might contribute to the FSHD phenotype (14, 17, 25). The function of FRG2 remains previously unknown (11). Both FRG2 and DUX4c are up-regulated during myogenic differentiation (11, 13, 24, 25).
Intriguingly, all the 4q35 genes that have been postulated to be involved in the pathogenesis of FSHD, including FRG1 (24), FRG2 (11), DUX4c (12), and DUX4 (13), are also up-regulated during normal myogenic differentiation, suggesting that in FSHD myoblasts, the myogenic differentiation program is partly activated. It was conceivable that the up-regulation of KLF15 in FSHD cells be caused by an increased expression of myogenic factors as compared with normal cells. Indeed, we have found that several myogenic factors were abnormally expressed in myoblasts, myotubes, and biopsies from FSHD patients. Proliferating FSHD myoblasts expressed abnormally high levels of MYOG, whereas differentiated FSHD myotubes expressed more MYOD than normal myotubes. Inappropriate up-regulation of several myogenic factors could reflect a defect in the overall myogenic differentiation process. Indeed, a defect in the MYOD pathway has been reported previously in FSHD muscles (9). In agreement with this hypothesis, we speculate that induction of KLF15 expression in FSHD is a consequence of abnormally high expression of myogenic factors in these cells.
The premature expression of some myogenic differentiation markers observed in FSHD myoblasts could be attributed to the oxidative stress, a known molecular feature of FSHD myoblasts (53). Although it is generally considered as a myogenic differentiation blocking factor, moderate oxidative stress was shown to stimulate expression of MYOG and other myogenic factors (54). Interestingly, overexpression of DUX4 was recently shown to inhibit the oxidative stress response thus making cells vulnerable to oxidative stress and suggesting that DUX4 might be the cause of the abnormal expression of myogenic factors in FSHD.
We have also addressed the question whether KLF15-dependent D4Z4 enhancer regulated the expression of DUX4. The D4Z4 enhancer, which is located in the immediate proximity of the DUX4 promoter, is likely to function as a transcriptional activator. Using luciferase reporters, we have shown that the KLF15-stimulated D4Z4 enhancer can activate the DUX4 promoter. However, we did not find any evidence of KLF15-dependent induction of the DUX4 promoter in vitro (luciferase assay) or when tested in its genomic context. This could be due to the presence of enhancer-blocking elements on both sides of the gene (Fig. 6) (46). Thus, in contrast to FRG2 and DUX4c, DUX4 is not controlled by KLF15. Instead, DUX4 expression seems to be controlled by a KLF15-independent mechanism linked to the 4qA polyadenylation signal stabilizing the DUX4 transcript (19). Our model places DUX4 overexpression upstream of DUX4c and FRG2 overexpression in FSHD.
An alternative hypothesis would directly attribute the overexpression of KLF15 to an oxidative stress. It has been previously demonstrated that in FSHD myoblasts, oxidative stress resistance genes are down-regulated suggesting that FSHD myoblasts could wrongly activate an oxidative stress signaling in normal conditions. In oxidative stress conditions, the transcription factor hypoxia-inducible factor 1 (HIF-1) is activated (55). Interestingly, we have noted that HIF-1 recognition sites are present within the promoter region of KLF15 suggesting that the up-regulation of KLF15 in FSHD cells may be mediated, at least in part, by HIF-1. Whichever is the process that leads to KLF15 up-regulation in FSHD cells, we suggest that KLF15 serves as a direct activator of the D4Z4 enhancer, which in turn activates the expression of FRG2 and DUX4c genes.
Other factors besides KLF15 may additionally contribute to the overexpression of FRG2 and DUX4c in FSHD cells. We have reported previously that DUX4c and the D4Z4 enhancer are separated by a matrix attachment region (MAR) that can function as an enhancer blocking element (21, 56). In FSHD cells, the interaction of this MAR with the nuclear matrix is less efficient (21), and the chromatin loop structure is modified as compared with healthy cells. We hypothesize that the D4Z4 enhancer could contact the DUX4c promoter more readily, contributing to the increased DUX4c expression observed in FSHD cells.
In conclusion, we propose a new role for the KLF15 transcription factor that would function as a positive regulator of the expression of FRG2 and DUX4c genes during normal myogenic differentiation by conveying the activity of myogenic factors through D4Z4 enhancer to their promoters. In a similar way, KLF15 links the activity of abnormally expressed myogenic factors to FRG2 and DUX4c overexpression in FSHD.
Supplementary Material
Acknowledgments
We thank Dr. Elisabetta Andermarcher for critical reading of the manuscript; Dr. Anna Polesskaya for human immortalized myoblasts transfection protocol; Dr. D. Otteson for the gift of the KLF15 plasmid, and Dr. V. Mouly for the gift of immortalized human myoblasts.
This work was supported by grants from the Association Française Contre les Myopathies (to Y. S. V., D. L., and A. B.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S3.
- FSHD
- facioscapulohumeral muscular dystrophy
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- iMyo
- immortalized myoblast
- MAR
- matrix attachment region
- qRT
- quantitative RT.
REFERENCES
- 1. Lunt P. W., Harper P. S. (1991) J. Med. Genet. 28, 655–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. van Deutekom J. C., Wijmenga C., van Tienhoven E. A., Gruter A. M., Hewitt J. E., Padberg G. W., van Ommen G. J., Hofker M. H., Frants R. R. (1993) Hum. Mol. Genet. 2, 2037–2042 [DOI] [PubMed] [Google Scholar]
- 3. Lemmers R. J., Wohlgemuth M., van der Gaag K. J., van der Vliet P. J., van Teijlingen C. M., de Knijff P., Padberg G. W., Frants R. R., van der Maarel S. M. (2007) Am. J. Hum. Genet. 81, 884–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Laoudj-Chenivesse D., Carnac G., Bisbal C., Hugon G., Bouillot S., Desnuelle C., Vassetzky Y., Fernandez A. (2005) J. Mol. Med. 83, 216–224 [DOI] [PubMed] [Google Scholar]
- 5. Osborne R. J., Welle S., Venance S. L., Thornton C. A., Tawil R. (2007) Neurology 68, 569–577 [DOI] [PubMed] [Google Scholar]
- 6. Arashiro P., Eisenberg I., Kho A. T., Cerqueira A. M., Canovas M., Silva H. C., Pavanello R. C., Verjovski-Almeida S., Kunkel L. M., Zatz M. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6220–6225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bakay M., Wang Z., Melcon G., Schiltz L., Xuan J., Zhao P., Sartorelli V., Seo J., Pegoraro E., Angelini C., Shneiderman B., Escolar D., Chen Y. W., Winokur S. T., Pachman L. M., Fan C., Mandler R., Nevo Y., Gordon E., Zhu Y., Dong Y., Wang Y., Hoffman E. P. (2006) Brain 129, 996–1013 [DOI] [PubMed] [Google Scholar]
- 8. Celegato B., Capitanio D., Pescatori M., Romualdi C., Pacchioni B., Cagnin S., Viganò A., Colantoni L., Begum S., Ricci E., Wait R., Lanfranchi G., Gelfi C. (2006) Proteomics 6, 5303–5321 [DOI] [PubMed] [Google Scholar]
- 9. Winokur S. T., Chen Y. W., Masny P. S., Martin J. H., Ehmsen J. T., Tapscott S. J., van der Maarel S. M., Hayashi Y., Flanigan K. M. (2003) Hum. Mol. Genet. 12, 2895–2907 [DOI] [PubMed] [Google Scholar]
- 10. Gabellini D., Green M. R., Tupler R. (2002) Cell 110, 339–348 [DOI] [PubMed] [Google Scholar]
- 11. Rijkers T., Deidda G., van Koningsbruggen S., van Geel M., Lemmers R. J., van Deutekom J. C., Figlewicz D., Hewitt J. E., Padberg G. W., Frants R. R., van der Maarel S. M. (2004) J. Med. Genet. 41, 826–836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ansseau E., Laoudj-Chenivesse D., Marcowycz A., Tassin A., Vanderplanck C., Sauvage S., Barro M., Mahieu I., Leroy A., Leclercq I., Mainfroid V., Figlewicz D., Mouly V., Butler-Browne G., Belayew A., Coppée F. (2009) PLoS ONE 4, e7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dixit M., Ansseau E., Tassin A., Winokur S., Shi R., Qian H., Sauvage S., Mattéotti C., van Acker A. M., Leo O., Figlewicz D., Barro M., Laoudj-Chenivesse D., Belayew A., Coppée F., Chen Y. W. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 18157–18162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dmitriev P., Lipinski M., Vassetzky Y. S. (2009) Neuromuscul. Disord. 19, 17–20 [DOI] [PubMed] [Google Scholar]
- 15. Cabianca D. S., Gabellini D. (2010) J. Cell Biol. 191, 1049–1060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bosnakovski D., Xu Z., Gang E. J., Galindo C. L., Liu M., Simsek T., Garner H. R., Agha-Mohammadi S., Tassin A., Coppée F., Belayew A., Perlingeiro R. R., Kyba M. (2008) EMBO J. 27, 2766–2779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bosnakovski D., Lamb S., Simsek T., Xu Z., Belayew A., Perlingeiro R., Kyba M. (2008) Exp. Neurol. 214, 87–96 [DOI] [PubMed] [Google Scholar]
- 18. Gabellini D., D'Antona G., Moggio M., Prelle A., Zecca C., Adami R., Angeletti B., Ciscato P., Pellegrino M. A., Bottinelli R., Green M. R., Tupler R. (2006) Nature 439, 973–977 [DOI] [PubMed] [Google Scholar]
- 19. Lemmers R. J., van der Vliet P. J., Klooster R., Sacconi S., Camaño P., Dauwerse J. G., Snider L., Straasheijm K. R., van Ommen G. J., Padberg G. W., Miller D. G., Tapscott S. J., Tawil R., Frants R. R., van der Maarel S. M. (2010) Science 329, 1650–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Petrov A. P., Laoudj D., Vassetzky Y. S. (2003) Genetics 39, 147–151 [Google Scholar]
- 21. Petrov A., Allinne J., Pirozhkova I., Laoudj D., Lipinski M., Vassetzky Y. S. (2008) Genome Res. 18, 39–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Coppée F., Mattéotti C., Anseau E., Sauvage S., Leclercq I., Leroy A., Marcowycz A., Gerbaux C., Figlewicz D., Ding H., Belayew A. (2004) in Facioscapulohumeral Muscular Dystrophy: Clinical Medicine and Molecular Biology (Upadhyaya M., Cooper D., eds) pp. 1–250, Garland:BIOS Scientific Publishers, Abingdon, UK [Google Scholar]
- 23. Pirozhkova I., Petrov A., Dmitriev P., Laoudj D., Lipinski M, Vassetzky Y. S. (2008) PLoS ONE 3, e3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Bodega B., Ramirez G. D., Grasser F., Cheli S., Brunelli S., Mora M., Meneveri R., Marozzi A., Mueller S., Battaglioli E., Ginelli E. (2009) BMC Biol. 7, 41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ansseau E., Marcowycz A., Laoudj-Chenivesse D., Tassin A., Sauvage S., Vanderplanck C., Barro M., Leroy A., Leclercq I., Mainfroid V., Figlewicz D., Belayew A., Coppée F. (2009) PLoS ONE, e7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hsu D. K., Guo Y., Alberts G. F., Copeland N. G., Gilbert D. J., Jenkins N. A., Peifley K. A., Winkles J. A. (1996) J. Biol. Chem. 271, 13786–13795 [DOI] [PubMed] [Google Scholar]
- 27. Zhu C. H., Mouly V., Cooper R. N., Mamchaoui K., Bigot A., Shay J. W., Di Santo J. P., Butler-Browne G. S., Wright W. E. (2007) Aging Cell 6, 515–523 [DOI] [PubMed] [Google Scholar]
- 28. Barro M., Carnac G., Flavier S., Mercier J., Vassetzky Y., Laoudj-Chenivesse D. (2010) J. Cell. Mol. Med. 14, 275–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yu J., de Belle I., Liang H., Adamson E. D. (2004) Mol. Cell 15, 83–94 [DOI] [PubMed] [Google Scholar]
- 30. Otteson D. C., Liu Y., Lai H., Wang C., Gray S., Jain M. K., Zack D. J. (2004) Invest. Ophthalmol. Vis. Sci. 45, 2522–2530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Meijer A. H., Ouwerkerk P. B., Hoge J. H. (1998) Yeast 14, 1407–1415 [DOI] [PubMed] [Google Scholar]
- 32. Ausubel F. M. (2003) Current Protocols in Molecular Biology, Section 12.1 (Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K., eds) John Wiley & Sons, Hoboken, NJ [Google Scholar]
- 33. Otteson D. C., Lai H., Liu Y., Zack D. J. (2005) BMC Mol. Biol. 6, 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uchida S., Sasaki S., Marumo F. (2001) Kidney Int. 60, 416–421 [DOI] [PubMed] [Google Scholar]
- 35. Lemaire P., Vesque C., Schmitt J., Stunnenberg H., Frank R., Charnay P. (1990) Mol. Cell. Biol. 10, 3456–3467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Adachi H., Tsujimoto M. (2002) J. Biol. Chem. 277, 24014–24021 [DOI] [PubMed] [Google Scholar]
- 37. Chen Q. K., Hertz G. Z., Stormo G. D. (1995) Comput. Appl. Biosci. 11, 563–566 [DOI] [PubMed] [Google Scholar]
- 38. Workman C. T., Yin Y., Corcoran D. L., Ideker T., Stormo G. D., Benos P. V. (2005) Nucleic Acids Res. 33, W389–W392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. McDonald J. H. (2009) Handbook of Biological Statistics, pp. 1–313, Sparky House Publishing, Baltimore, MD [Google Scholar]
- 40. Livak K. J., Schmittgen T. D. (2001) Methods 25, 402–408 [DOI] [PubMed] [Google Scholar]
- 41. Viñals F., Fandos C., Santalucia T., Ferré J., Testar X., Palacín M., Zorzano A. (1997) J. Biol. Chem. 272, 12913–12921 [DOI] [PubMed] [Google Scholar]
- 42. Gray S., Feinberg M. W., Hull S., Kuo C. T., Watanabe M., Sen-Banerjee S., DePina A., Haspel R., Jain M. K. (2002) J. Biol. Chem. 277, 34322–34328 [DOI] [PubMed] [Google Scholar]
- 43. Yamamoto J., Ikeda Y., Iguchi H., Fujino T., Tanaka T., Asaba H., Iwasaki S., Ioka R. X., Kaneko I. W., Magoori K., Takahashi S., Mori T., Sakaue H., Kodama T., Yanagisawa M., Yamamoto T. T., Ito S., Sakai J. (2004) J. Biol. Chem. 279, 16954–16962 [DOI] [PubMed] [Google Scholar]
- 44. Fisch S., Gray S., Heymans S., Haldar S. M., Wang B., Pfister O., Cui L., Kumar A., Lin Z., Sen-Banerjee S., Das H., Petersen C. A., Mende U., Burleigh B. A., Zhu Y., Pinto Y. M., Pinto Y., Liao R., Jain M. K. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 7074–7079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang B., Haldar S. M., Lu Y., Ibrahim O. A., Fisch S., Gray S., Leask A., Jain M. K. (2008) J. Mol. Cell. Cardiol. 45, 193–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Ottaviani A., Rival-Gervier S., Boussouar A., Foerster A. M., Rondier D., Sacconi S., Desnuelle C., Gilson E., Magdinier F. (2009) PLoS Genet. 5, e1000394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Mori T., Sakaue H., Iguchi H., Gomi H., Okada Y., Takashima Y., Nakamura K., Nakamura T., Yamauchi T., Kubota N., Kadowaki T., Matsuki Y., Ogawa W., Hiramatsu R., Kasuga M. (2005) J. Biol. Chem. 280, 12867–12875 [DOI] [PubMed] [Google Scholar]
- 48. Klooster R., Straasheijm K., Shah B., Sowden J., Frants R., Thornton C., Tawil R., van der Maarel S. (2009) Eur. J. Hum. Genet. 17, 1615–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wijmenga C., Hewitt J. E., Sandkuijl L. A., Clark L. N., Wright T. J., Dauwerse H. G., Gruter A. M., Hofker M. H., Moerer P., Williamson R., Van Ommen G. B., Padberg G., Frants R. (1992) Nat. Genet. 2, 26–30 [DOI] [PubMed] [Google Scholar]
- 50. Uchida S., Tanaka Y., Ito H., Saitoh-Ohara F., Inazawa J., Yokoyama K. K., Sasaki S., Marumo F. (2000) Mol. Cell. Biol. 20, 7319–7331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Leenders J. J., Wijnen W. J., Hiller M., van der Made I., Lentink V., van Leeuwen R. E., Herias V., Pokharel S., Heymans S., de Windt L. J., Høydal M. A., Pinto Y. M., Creemers E. E. (2010) J. Biol. Chem. 285, 27449–27456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Gray S., Wang B., Orihuela Y., Hong E. G., Fisch S., Haldar S., Cline G. W., Kim J. K., Peroni O. D., Kahn B. B., Jain M. K. (2007) Cell Metab. 5, 305–312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Winokur S. T., Barrett K., Martin J. H., Forrester J. R., Simon M., Tawil R., Chung S. A., Masny P. S., Figlewicz D. A. (2003) Neuromuscul. Disord. 13, 322–333 [DOI] [PubMed] [Google Scholar]
- 54. Luo S. W., Zhang C., Zhang B., Kim C. H., Qiu Y. Z., Du Q. S., Mei L., Xiong W. C. (2009) EMBO J. 28, 2568–2582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang C., Han Y., Wang Y., Sun X., Yan S., Yeh E. T., Chen Y., Cang H., Li H., Shi G., Cheng J., Tang X., Yi J. (2009) EMBO J. 28, 2748–2762 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Petrov A., Pirozhkova I., Carnac G., Laoudj D., Lipinski M., Vassetzky Y. S. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 6982–6987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Gabriëls J., Beckers M. C., Ding H., De Vriese A., Plaisance S., van der Maarel S. M., Padberg G. W., Frants R. R., Hewitt J. E., Collen D., Belayew A. (1999) Gene 236, 25–32 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.