Background: Aquaporin-9 is a glycerol-permeable channel expressed in hepatocytes.
Results: Newly identified chemical inhibitors, as well as aquaporin-9 gene deletion, eliminate glycerol-dependent murine hepatocyte glucose output.
Conclusion: Aquaporin-9 is the primary route of murine hepatocyte glycerol uptake for gluconeogenesis.
Significance: Aquaporin-9 may be a drug target in diabetes treatment
Keywords: Aquaporin, Glycerol, Hepatocyte, Mouse, Small Molecules
Abstract
It has been hypothesized that aquaporin-9 (AQP9) is part of the unknown route of hepatocyte glycerol uptake. In a previous study, leptin receptor-deficient wild-type mice became diabetic and suffered from fasting hyperglycemia whereas isogenic AQP9−/− knock-out mice remained normoglycemic. The reason for this improvement in AQP9−/− mice was not established before. Here, we show increased glucose output (by 123% ± 36% S.E.) in primary hepatocyte culture when 0.5 mm extracellular glycerol was added. This increase depended on AQP9 because it was absent in AQP9−/− cells. Likewise, the increase was abolished by 25 μm HTS13286 (IC50 ∼ 2 μm), a novel AQP9 inhibitor, which we identified in a small molecule library screen. Similarly, AQP9 deletion or chemical inhibition eliminated glycerol-enhanced glucose output in perfused liver preparations. The following control experiments suggested inhibitor specificity to AQP9: (i) HTS13286 affected solute permeability in cell lines expressing AQP9, but not in cell lines expressing AQPs 3, 7, or 8. (ii) HTS13286 did not influence lactate- and pyruvate-dependent hepatocyte glucose output. (iii) HTS13286 did not affect glycerol kinase activity. Our experiments establish AQP9 as the primary route of hepatocyte glycerol uptake for gluconeogenesis and thereby explain the previously observed, alleviated diabetes in leptin receptor-deficient AQP9−/− mice.
Introduction
Gluconeogenesis from glycerol (henceforth glycerol gluconeogenesis) accounts for ∼10% of hepatic glucose production in patients with type 2 diabetes (T2D)3 (1). Increased glycerol gluconeogenesis in T2D patients results not simply from accelerated lipolysis but also from altered hepatic glycerol utilization (1). Hepatic glycerol uptake has been investigated in a number of studies, in primary hepatocyte cell culture and in perfused liver preparations (2–4). Nevertheless, it is still unclear whether facilitated glycerol uptake is required to sustain glycerol gluconeogenesis at physiological rates. Furthermore, the molecular identity of possible glycerol carrier(s) is unknown. It has been hypothesized that the glycerol-permeable channel protein AQP9, a member of the aqua(glycero)porin family, may play a role in hepatic glycerol gluconeogenesis, because it is expressed in the liver and expression is negatively regulated by insulin (5). However, similarly regulated expression of other aquaglyceroporins in the liver has been described (6, 7). Therefore, the relative contributions of each carrier to hepatic glycerol uptake, possible contributions from additional carriers, as well as the relative contribution of lipid bilayer mediated diffusion to glycerol gluconeogenesis remain unclear. AQP9 gene deletion in obese, diabetic mice was found to reduce plasma glucose levels by between 10 and 40% (8). However, oral glycerol caused identical subsequent elevations of blood glucose levels in AQP9+/− and AQP9−/− mice. These experiments suggested an unspecified influence of AQP9 on plasma glucose levels in obese diabetic mice (8). We concluded that these observations required alternative investigation of a possible AQP9 contribution to hepatocyte glucose output. In this study, we have identified novel drugs that specifically inhibit AQP9. We have utilized these substances to investigate whether AQP9 is necessary to sustain hepatic glycerol gluconeogenesis at physiological rates.
EXPERIMENTAL PROCEDURES
DNA Constructs
mAQP8 and mAQP9 were amplified from mouse liver cDNA (NMRI strain); mAQP3 and mAQP7 were amplified from mouse kidney cDNA (C57BL/6 strain). Oligonucleotide sequences are provided in the supplemental Oligonucleotide list. All AQP PCR products were first inserted into Topo-PCRII (Invitrogen) followed by restriction enzyme digest and ligation with pcDNA5/FRT/TO (Invitrogen).
CHO Cell Culture
CHO-FlpIn cells (Invitrogen) were transfected with pcDNA6/TR (Invitrogen), and stable cell lines (CHO-FlpIn-TR) were selected as recommended by the manufacturer. Lipofectamine (Invitrogen) was used to transfect these cells with pcDNA5/FRT/TO-AQP plasmids. Cells were grown at 37 °C, 5% CO2 and selected in hygromycin B (Invitrogen) containing medium (DMEM/F12, containing 5% donor bovine serum; Invitrogen).
AQP9 Inhibitor Screen and Solute Permeability Measurements
Water permeability measurements were as described (9) with modifications: 15,000 CHO-AQP cells were seeded per well of poly-d-lysine (Greiner Bio One)-coated 96-well plates in the presence of tetracycline and grown for 2 days, as described above. Before calcein loading, growth medium was replaced with fresh growth medium (50 μl/well) containing 5 μm calcein AM (BD Biosciences) and 5 mm probenecid (Sigma-Aldrich) for 90 min. Plates were washed in assay buffer (0.8 mm MgSO4, 5 mm KCl, 1.8 mm CaCl2, 25 mm Hepes, pH 7.4, and adjusted to 285 mOsmol with NaCl) and incubated in 75 μl of assay buffer/well, containing 1% DMSO and compounds (Maybridge) at concentrations as indicated in text and figures. Fluorescence intensity was read on a Fluostar Optima plate reader (BMG Labtech) for 30 s/well. Cell shrinkage was induced by adding 75 μl of assay buffer containing 400 mm sucrose 5 s into each read. Half-life time of cell shrinking was determined by fitting cell shrinking curves after sucrose addition to a one-phase exponential function in Prism 5.0 (GraphPad). For qualitative solute permeability measurements, 75 μl of assay buffer containing 400 mm glycerol or 400 mm urea was added to each well.
In Silico Docking and hAQP9 Homology Model
A homology model of hAQP9 was based on the crystal structure of the bacterial glycerol facilitator GlpF (Protein Data Bank code 1FX8). The Swiss-Model server was used to create the model (10). A molecular dynamics simulation was performed for generating an equilibrated receptor structure. The simulation setup was similar to that described by Hub et al. (11). In contrast to Hub's setup, no crystallographic water molecules were included, and no virtual interaction sites were used. Therefore, the integrating time step was set to 2 fs. The system was coupled to an external bath of 300 K with coupling constant of 0.1 ps. Pressure was kept constant by coupling the system to a semi-isotropic Berendsen barostat (12) at 1 bar with a coupling constant of 1 ps. The simulations box contained the protein tetramer embedded in a lipid bilayer of Dipalmitoylphosphatidylcholin lipids and 20531 TIP3P water molecules (13). Eight chloride ions were added to achieve an uncharged simulation system. The system was simulated using amber03 (14). Force field and ion parameters were from Dang (15). The solvent and lipid molecules in the simulation system were equilibrated for 1 ns before production.
A snapshot of a molecular dynamics simulation of the hAQP9 structure after 35-ns equilibration time served as target structure for molecular docking, using FlexX 3.1.4 software (16). Each compound was docked to the intra- and extracellular entrance of each of the four subunits. Four different combination strategies have been applied for ranking ligands: (i) minimum docking score of all poses, (ii) average of docking scores at the extracellular pore entrance, (iii) average of docking scores at the intracellular pore entrance, (iv) minimum of the mean docking scores treating the intracellular and extracellular poses separately. Within the set of inhibiting compounds (t½ cell shrinking >1.0 s), 20 compounds were identified that appear at least one time within the top 20 ranked compounds with regard to the individual ranking strategies.
Primary Hepatocyte Isolation and Culture
Hepatocytes were isolated essentially as described previously (17) with modifications. Perfusion was from a plastic catheter inserted into the portal vein, and outflow was through the abdominal vena cava inferior. Perfusion flow rate was reduced to 6 ml/min. LiberaseTM (Roche Applied Science) was used as collagenase preparation. Hepatocyte viability was assessed after erythrosin B staining and ranged from 72 to 90%, which is comparable with the values obtained previously with this method (17). We seeded between 200,000 and 250,000 viable cells/well in collagenized 24-well plates. Hepatocytes were allowed to attach for 3–4 h in Williams medium E (WME; Invitrogen), supplemented with 5% FBS and 100 nm dexamethasone, before washing twice in prewarmed, serum-free WME and incubating for 16–18 h in WME containing 100 nm dexamethasone and 140 μg/ml Matrigel (BD Biosciences).
Glucose Output Assay
To measure hepatocyte glucose output, cells were washed three times in prewarmed PBS and incubated for 10 min in 250 μl of prewarmed Krebs-Henseleit-Hepes buffer (25 mm NaHepes, 1.2 mm KH2PO4, 1.2 mm MgSO4, 4.7 mm KCl, 25 mm NaHCO3, osmolarity adjusted to 285 mOsmol with NaCl, pH 7.4, at 37 °C) containing 20 μm 1,4-dideoxy-1,4-imino-d-arabinitol (18). HTS13286, dissolved in DMSO, glycerol, and lactic acid/pyruvate mixture, respectively, was included as indicated in the figures. At the end of the incubation period assay buffer was removed from cells, transferred into 1.5-ml tubes, and stored on ice water slurry. Before measurements of glucose content using the glucose oxidase-based AmpliteTM Glucose Quantitation kit (AAT Bioquest), samples were centrifuged briefly and sometimes stored frozen. Glucose-associated fluorescence was measured on a Fluostar Optima plate reader equipped with excitation/emission filters at 540/580 nm.
Glycerol Kinase Assay
Liver extracts were prepared from AQP9+/− heterozygote mice after removal from food for 10 h, as described previously (19). Glycerol in the extraction buffer was omitted. Samples were frozen in liquid nitrogen and stored at −80 °C before use. A radiochemical assay was performed as described previously (20) with the following modifications: 60-μl reactions contained 100 μg of liver extract, 83 mm Tris, pH 7.2, 83 mm KCl, 8.3 mm MgCl2, 4.1 mm ATP, 2.1 mm DTT, 0.83 mm glycerol, 0.83 nCi of 0.8 Ci/mmol [3H]glycerol (PerkinElmer Life Sciences) and were incubated for 1 h at 37 °C.
Liver Perfusion
Liver perfusions were essentially performed as described by Kuriyama et al. (5) with minor modifications. Perfusion was with 4 ml/min Krebs-Henseleit-Hepes plus 1% DMSO, kept in a water bath at 42 °C. Samples were collected for 15 consecutive min as indicated in Fig. 4. The dead volume of the system, between buffer reservoir and collection tube, including the blood circulatory system, was 1.5 g of buffer. All animal experiments were approved by the Danish Council for Animal Ethics of the Ministry of Justice.
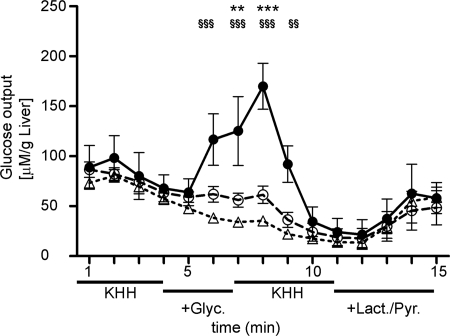
FIGURE 4.
Glucose output in perfused liver. Mouse livers were perfused in situ with Krebs-Henseleit-Hepes (KHH) buffer +1% DMSO containing no gluconeogenic substrate, 0. 5 mm glycerol, or 1.5 mm lactate plus 0.15 mm pyruvate, as indicated. Fractions were collected every minute and analyzed for glucose content. AQP9 deletion (AQP9−/−) or HTS13286 addition to wild-type livers during perfusion with glycerol significantly reduced a glycerol-specific increase in glucose output in wild-type livers. All livers included in the analysis were metabolically comparably active at the end of perfusion, as indicated by enhanced glucose output from lactic acid plus pyruvate as substrates. AQP9+/+ (filled circles), AQP9+/+ plus 25 μm HTS13286 added during perfusion with glycerol (open circles), AQP9−/− (open triangles). Statistical differences: **, p < 0.01; ***, p < 0.001 in AQP9+/+ versus AQP9+/+ plus HTS13286; §§, p < 0.01; §§§, p < 0.001 in AQP9+/+ versus AQP9−/−.
Data Analyses
All statistical analyses and analyses of inhibitor dose responses were conducted in Prism 5. We performed one-way ANOVA and Tukey's post test (lactate/pyruvate substrate), and two-way ANOVA and Bonferroni post tests (glycerol substrate), respectively, to identify differences between groups in glucose output.
RESULTS
AQP9 Small Molecule Inhibitor Identification
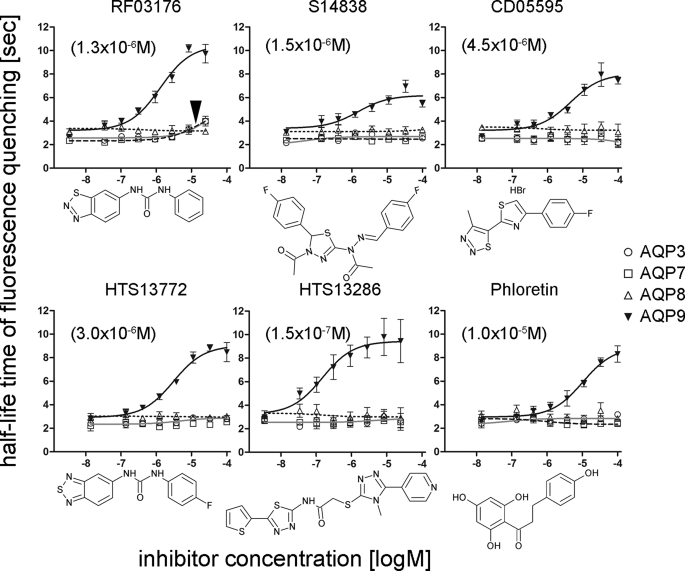
To identify small molecule inhibitors of AQP9, murine (m)AQP9 was introduced into CHO cells. Expression of mAQP9 was inducible by tetracycline addition to the culture medium (21, 22). Utilizing sucrose as an AQP9-impermeable osmolyte, we found that mAQP9 facilitated rapid cell shrinking in a calcein fluorescence intensity-based microplate assay (9, 23, 24). This suggested that mAQP9 expression confers high water permeability to CHO cells. We therefore screened a commercial small molecule library for substances reducing AQP9 water permeability. To increase sensitivity for recovering weak inhibitors we adjusted AQP9 expression to a level where ∼80% of the maximum measureable water permeability was observed in our setup. A set of 1,920 small molecules was screened subsequently at concentrations of 100 μm (results available in supplemental Spreadsheet 1), where each tested microwell plate contained positive (phloretin) and negative (solvent) control wells. We considered the highest ranked compounds with apparently slower cell shrinking for further analyses. Initially, these substances were subjected to in silico molecular docking to a homology model of human AQP9. The 10 best inhibitors of the primary screen and 20 additional substances with good docking properties were purchased individually, and dose-response curves were measured for these substances between 1 and 500 μm. For 23 of these substances inhibition of cell shrinking was only detected at high concentrations, four substances were apparent false positives. Of the remaining three substances, S14838 was suggested by the molecular docking approach, whereas RF03176 and CD05595 were among the most potent compounds identified in the primary screen (Fig. 1). To investigate inhibitor specificity, we constructed three additional CHO cell lines that expressed heterologous murine AQP isoforms: CHO-mAQP3, CHO-mAQP7, and CHO-mAQP8, respectively. Whereas the former two AQPs represent the other known murine aquaglyceroporins (25), AQP8 was chosen as a functionally less related aquaporin that is however expressed in the liver (26–28). We found that RF03176 is a moderate inhibitor of mAQP3 and mAQP7, respectively. No inhibition of cell shrinking was detected when CHO-mAQP8 cells were tested (Fig. 1). In contrast to RF03176, S14838 and CD05595 did not inhibit water permeability in any of the other cell lines tested (Fig. 1). Next we tested 7 structural analogues of RF03176, 12 analogues of S14838, and 22 analogues of CD05595 that were available from the Maybridge compound collection (supplemental Spreadsheet 2). Among several weaker inhibitors of mAQP9, this approach identified HTS13286, which was an improved inhibitor of CHO-mAQP9 cell shrinking, compared with RF03176 (Fig. 1). Furthermore, HTS13286 was an almost 2 orders of magnitude more potent mAQP9 inhibitor than phloretin (Fig. 1).
FIGURE 1.
Inhibition of CHO-AQP cell water permeability. Half-life time of fluorescence quenching (cell shrinking) in the presence of inhibitors was determined after fitting to a one-phase exponential function. CHO-mAQP3, CHO-mAQP7, CHO-mAQP8, and CHO-mAQP9 cells were tested, following addition of 300 mm sucrose. RF03176, S14838, and CD05595 were identified in the primary screen for AQP9 inhibitors and represent the most potent substances (by IC50) in follow-up experiments as exemplified here. HTS13772 and HTS13286 were identified by testing structural analogues of RF03176 and S14838, respectively. Phloretin was tested to compare relative inhibitory potencies. Except for moderate inhibition of CHO-mAQP3 and CHO-mAQP7 cell shrinking by RF03176 (arrowhead), all tested substances were specific inhibitors of CHO-AQP9 cell shrinking. Apparent IC50 values are shown in parentheses. Mean ± S.E. (error bars) are depicted, n = 3. The lowest concentration on all curves is 0 and was edited to fit the log scale of the x axis. Chemical structures of substances are shown below the corresponding dose-response curves.
HTS13286 Affects mAQP9 Water and Solute Permeability
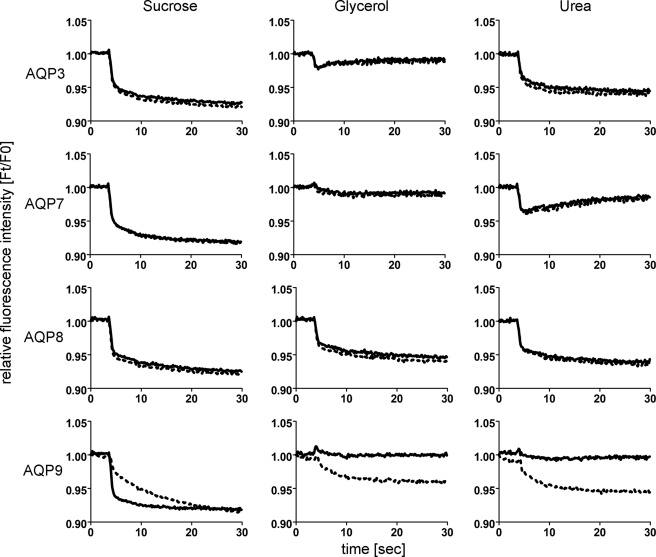
The qualitative effects of HTS13286 were tested on mAQP water, glycerol, and urea permeability (Fig. 2). In these experiments, HTS13286 (25 μm) resulted in markedly slower fluorescence quenching (water permeability) compared with solvent controls, when 200 mm sucrose was added to CHO-mAQP9 cells. In contrast, without inhibitor, these cells showed no fluorescence quenching when 200 mm glycerol or 200 mm urea was added to the extracellular side. In agreement with other studies, this suggested that AQP9 conferred high glycerol and urea permeability, respectively, to CHO cells (29–31). However, in the presence of HTS13286, cell shrinking was observed in CHO-mAQP9 cells in response to glycerol or urea addition (Fig. 2). We interpreted these slow volume changes as a simultaneous HTS13286 inhibition of glycerol influx and water efflux via AQP9, while leaving water and solute diffusion through the lipid bilayer unaffected. No effect of HTS13286 on solute permeability of CHO-AQP3, -7, and -8 cell lines, respectively, was observed.
FIGURE 2.
HTS13286 effect on AQP9 solute permeability. Average cell fluorescence quenching traces, recorded from CHO-mAQP cells, are shown, n = 3. Water (left), glycerol (middle), and urea (right) permeabilities were tested qualitatively. Addition of membrane-impermeable sucrose (200 mm) induced rapid fluorescence quenching (solid line), consistent with high water permeability. HTS13286 (25 μm; dotted line) decelerated fluorescence quenching in CHO-mAQP9 cells. Addition of 200 mm AQP9-permeable glycerol or urea did not cause volume changes in CHO-mAQP9 cells, indicating rapid glycerol and urea transmembrane equilibration with approximately equal velocity as concomitant water flux in the opposite direction. This equilibration was inhibited by addition of HTS13286 (25 μm), thereby restoring volume changes. Here, slow fluorescence changes indicate equally reduced AQP9-dependent water and solute permeabilities. As a result, the lipid bilayer presumably mediates most water and solute exchange. No effect of HTS13286 was observed in the other tested CHO-mAQP cell lines. Relative to water permeability, the glycerol permeability of CHO-mAQP3 cells appears lower than CHO-mAQP7 and CHO-mAQP9 glycerol permeability. Furthermore, CHO-mAQP3 cells appear impermeable to urea. Finally, relative to water permeability, the urea permeability of CHO-mAQP7 cells appears lower than CHO-mAQP9 urea permeability.
AQP9 Is Crucial for Glycerol-dependent Glucose Production in Primary Hepatocyte Culture
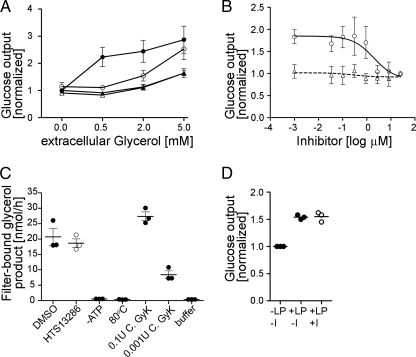
Based on the known function of AQP9 as a glycerol channel, it has been proposed to contribute to hepatic glycerol gluconeogenesis (5). To test this hypothesis, we isolated hepatocytes from wild-type and AQP9−/− mice (8), respectively, and cultured these cells in insulin-free medium. Immunolabeling experiments confirmed robust expression of AQP9 in cells isolated from wild-type mice, whereas no signal was detected in cells isolated from knock-out mice (supplemental Fig. 1). Hepatocytes were incubated in the presence of various concentrations of glycerol subsequently, ranging from 0 to 5 mm (Fig. 3A). AQP9 expression enhanced glucose output by 123% ± 36% when cells were incubated in 0.5 mm glycerol, which is within the physiological range of plasma glycerol levels. When extracellular glycerol was raised to 5 mm, the requirement of AQP9 for efficient glucose production was reduced, suggesting enhanced glycerol availability for glycerol gluconeogenesis from alternative routes, e.g. diffusion through the lipid bilayer. AQP9 inhibition by HTS13286 had effects on hepatocyte glucose production similar to those of AQP9 gene deletion. However, when extracellular glycerol was 5 mm, HTS13286-inhibited glucose production was significantly higher than glucose production in AQP9−/− hepatocytes. These observations are consistent with robust but incomplete inhibition of AQP9 glycerol permeability by HTS13286. No effect of HTS13286 on glucose output in the presence of glycerol was observed in AQP9 knock-out mice. Furthermore, HTS13286 did not inhibit glycerol kinase activity (Fig. 3C) or glucose output from AQP9+/+ hepatocytes provided with lactic acid (1.5 mm) and pyruvate (0.15 mm) as gluconeogenic substrate (Fig. 3D). We also tested the efficacy of HTS13286 on glucose output in primary hepatocyte culture (Fig. 3B). Providing extracellular glycerol at 0.5 mm, we found that half-maximal inhibition of glucose production by HTS13286 was observed at 2.0 μm.
FIGURE 3.
Role of AQP9 in hepatocyte glucose output in primary hepatocyte culture. A, addition of extracellular glycerol at 0. 5 mm resulted in 123% ± 36% increased glucose output in cells isolated from AQP9+/+ wild-type mice (circles) but not in AQP9−/− hepatocytes (triangles; p < 0.001). This increase in glucose output was eliminated by addition of 25 μm HTS13286 (open symbols) to AQP9+/+ hepatocytes at 0.5 mm glycerol (p < 0.001). Similar differences were observed at 2 mm glycerol (p < 0.001 for AQP9+/+ versus AQP9−/− and AQP9+/+ versus AQP9+/+ plus HTS13286). Reduced effects were observed at 5 mm extracellular glycerol. There, HTS13286-treated AQP9+/+ cells produced more glucose (p < 0.05) than inhibitor or solvent-treated AQP9−/− hepatocytes. Filled symbols represent solvent (DMSO) controls; n = 6. All values were normalized to solvent-treated cells without exposure to glycerol. B, dose response of glucose output inhibition by HTS13286 at 0.5 mm glycerol. Half-maximal inhibition in AQP9+/+ hepatocytes was observed at 2 μm HTS13286. Symbols are as in A. Lowest concentration of inhibitor indicated in the logarithmic plot is 0. All values were normalized to glucose output at 25 μm inhibitor. n = 3. C, HTS13286 (25 μm) does not significantly affect glycerol kinase activity in liver extracts (100 μg of protein/sample). p = 0.53, two-tailed, unpaired t test comparing inhibitor-treated sample and solvent control (DMSO). −ATP, no ATP added to extract and sample buffer; 80 °C, extract was heat-inactivated for 10 min at 80 °C; C. GyK, Cellulomonas glycerol kinase, 0.1 and 0.001 units/sample; buffer, sample buffer control. D, HTS13286 (25 μm) does not significantly affect glucose output from lactate (1.5 mm) and pyruvate (0.15 mm) substrate (one-way ANOVA and Tukey's post test). LP, lactate/pyruvate; I, inhibitor (25 μm HTS13286).
AQP9 Is Crucial for Glycerol-dependent Glucose Production in Perfused Murine Liver
Whereas primary hepatocytes represent the most suitable in vitro system for testing hepatocyte function, these cells might nevertheless lose some of their specific identity in culture. Furthermore, expression of AQP3 has been reported in liver of starved mice (6), a condition that cannot directly be reproduced in culture. We therefore investigated the contribution of AQP9 to glycerol-dependent glucose output in in situ perfused livers of starved mice. Similar to the observations in cultured hepatocytes, we found that AQP9 expression greatly enhanced hepatic glucose output when glycerol was provided as a substrate. HTS13286 reduced glucose output in wild-type mice during perfusion with glycerol to levels that were comparable with AQP9−/− mice (Fig. 4).
DISCUSSION
In a previous study, AQP9−/− mice with T2D had lower blood glucose levels than diabetic AQP9+/− littermates (8). Although the cause for lower blood glucose levels in AQP9−/− animals remained unknown, it was hypothesized that small molecule inhibitors of AQP9 could be useful in the treatment of T2D. Previously known AQP9 inhibitors, including phloretin and mercury (30), were not suitable to test this hypothesis. Whereas phloretin is known to inhibit hepatic hexose transporters (32), mercury is, in our opinion, too toxic for conclusive cell-based studies.
We have identified novel inhibitors of murine AQP9 by screening a commercial chemical compound library. With 58 inhibitors of 1,920 screened substances, the frequency of AQP9 inhibitors that were initially considered for further analysis was higher than expected. This may be explained by the chemistry of the identified substances: many compounds contained a diphenyl-urea moiety, which is an easily accessible intermediate in organic synthesis. This moiety seems to contribute at least in part to AQP9 inhibition by several substances. However, at lower concentrations (below 10 μm), only 3 of 30 substances analyzed further inhibited AQP9 water permeability significantly. Subsequent tests of structurally related compounds identified two additional inhibitors with IC50 values in the low micromolar and nanomolar range. Four of the five best identified inhibitors were completely specific for AQP9 and did not affect cells expressing homologous AQPs 3, 7, and 8, respectively, in the tested concentration range. Such specificity has not been reported in previous attempts to identify small molecule AQP inhibitors (24, 33).
It has been hypothesized previously that AQP9 may contribute to hepatic gluconeogenesis by facilitating glycerol uptake (5). This might explain the lowered blood glucose levels in diabetic AQP9−/− mice compared with diabetic AQP9+/− littermates (8). This hypothesis was undermined when AQP9−/− mice were fed glycerol: there, blood glucose levels increased simultaneously in AQP9+/− and AQP9−/− mice, indicating important alternative routes to AQP9 for hepatic glycerol uptake. It was also unclear whether glycerol diffusion through the lipid bilayer could be sufficient to sustain gluconeogenesis from glycerol at physiological rates (34). Furthermore, it was conceivable that AQP9 gene deletion might have affected blood glucose levels in obese diabetic mice in unpredicted ways. In support for the existence of alternative routes, expression of AQP3 was found in liver tissue of starved mice (6) and recently also in human liver (7).
To evaluate whether AQP9 can be a drug target for treating T2D it was necessary to investigate whether AQP9 gene deletion can reduce hepatic glycerol gluconeogenesis. For this reason we have measured hepatocyte glucose production at various glycerol input concentrations. We hypothesized that cellular glycerol utilization would outperform glycerol uptake through the lipid bilayer, if facilitated glycerol uptake should play a role in glycerol gluconeogenesis. Further consideration suggested that both channel-mediated glycerol uptake as well as lipid bilayer-mediated glycerol uptake linearly depend on the inwardly directed glycerol gradient across the plasma membrane. As a consequence, we predicted that glycerol uptake through the lipid bilayer might only be rate-limiting for gluconeogenesis at physiological plasma glycerol concentrations. Elevated plasma glycerol concentrations, however, at high enough levels, would compensate facilitated glycerol uptake deficiency. Interestingly, AQP9 deletion caused elevated plasma glycerol concentrations in normal mice (8), which could indicate an attempt of compensation for AQP9 loss of function. Subsequent experiments in cultured primary hepatocytes showed that these assumptions on the effect of extracellular glycerol concentration were valid. Whereas mAQP9 was necessary for efficient glycerol gluconeogenesis at physiological extracellular glycerol concentrations (0.5 mm), a 10-fold increase in extracellular glycerol concentration alleviated the glycerol gluconeogenesis defect in AQP9−/− hepatocytes or AQP9 inhibitor-treated hepatocytes. With regard to HTS13286 specificity to inhibition of AQP9, these results furthermore indicate an inhibitory mechanism on gluconeogenesis that is restricted to reducing availability of intracellular glycerol. HTS13286 did neither affect utilization of lactic acid and pyruvate as gluconeogenic substrate, nor did it affect the activity of glycerol kinase, further corroborating specificity of this inhibitor to AQP9 in primary hepatocytes and liver extracts. Noteworthy, AQP9 has initially been described as a moderate lactic acid channel, and a physiological role in lactic acid gluconeogenesis as well as lactic acid removal from hepatocytes during hypoxia has been hypothesized (30). Our results suggest that AQP9 is not relevant for lactic acid uptake for gluconeogenesis. We suggest that monocarboxylate transporters constitute the primary route for lactic acid transmembrane movement (35). The measured effects of AQP9 gene deletion on glucose output in primary hepatocytes were further substantiated by the effects of HTS13286 and AQP9 gene deletion on glycerol gluconeogenesis in perfused mouse livers. A glycerol-specific increase in glucose output in wild-type livers was suppressed by HTS13286 and absent in AQP9−/− livers. Together, these results identify AQP9 as the primary glycerol uptake route in murine hepatocytes. The extent of the effect of AQP9 gene deletion on hepatocyte glucose output suggests that no other glycerol channel contributes to glycerol uptake at the hepatocyte basolateral membrane. Previously described AQP3 expression in mouse liver may be specific to other cell types or other cellular compartments (6). Our observations provide a possible explanation for the previously observed parallel rise in blood glucose levels in AQP9−/− mice and heterozygote littermates, after a glycerol meal (8). There, the amounts of provided glycerol might have caused portal plasma glycerol concentrations in large excess of 5 mm, thereby facilitating normal glucose production from glycerol that diffused through the lipid bilayer.
In summary, we have identified a mechanism that can explain the lowered blood glucose levels caused by AQP9 deletion in mice with T2D (8). Furthermore, we have identified novel small molecule inhibitors of AQP9. One of these substances, HTS13286, caused similarly reduced glycerol-dependent glucose production as did AQP9 deletion. However, the amount of HTS13286 required to inhibit glycerol gluconeogenesis in primary hepatocyte culture was relatively high (IC50 ∼ 2 μm). We expect that HTS13286 is currently not suitable for in vivo experiments because its solubility in 1% DMSO-containing aqueous solution is limited (∼25 μm). It may, however, be possible to identify structural derivatives of this substance for future in vivo studies that are more potent inhibitors of AQP9 and/or are more water-soluble.
Supplementary Material
Acknowledgment
We thank Dr. Nikolaus Bresgen, University of Salzburg, for helpful suggestions on primary hepatocyte isolation.
This work was supported by Lundbeck Foundation Grant R17A1742, a Marie Curie Research Training Network grant, a Nordic Research Grant for Water Imbalance Related Disorders, and European Drug Initiative on Channels and Transporters Grant HEALTH-F4-2007-201924. The Water and Salt Research Center was established and funded by The Danish National Research Foundation.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1, Spreadsheets 1 and 2, and a list of oligonucleotides.
- T2D
- type 2 diabetes
- AQP
- aquaporin
- DMSO
- dimethyl sulfoxide.
REFERENCES
- 1. Puhakainen I., Koivisto V. A., Yki-Järvinen H. (1992) J. Clin. Endocrinol. Metab. 75, 789–794 [DOI] [PubMed] [Google Scholar]
- 2. Li C. C., Lin E. C. (1983) J. Cell. Physiol. 117, 230–234 [DOI] [PubMed] [Google Scholar]
- 3. Westergaard N., Madsen P., Lundgren K. (1998) Biochim. Biophys. Acta 1402, 261–268 [DOI] [PubMed] [Google Scholar]
- 4. Sestoft L., Fleron P. (1975) Biochim. Biophys. Acta 375, 462–471 [DOI] [PubMed] [Google Scholar]
- 5. Kuriyama H., Shimomura I., Kishida K., Kondo H., Furuyama N., Nishizawa H., Maeda N., Matsuda M., Nagaretani H., Kihara S., Nakamura T., Tochino Y., Funahashi T., Matsuzawa Y. (2002) Diabetes 51, 2915–2921 [DOI] [PubMed] [Google Scholar]
- 6. Patsouris D., Mandard S., Voshol P. J., Escher P., Tan N. S., Havekes L. M., Koenig W., März W., Tafuri S., Wahli W., Müller M., Kersten S. (2004) J. Clin. Invest. 114, 94–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rodríguez A., Catalán V., Gómez-Ambrosi J., García-Navarro S., Rotellar F., Valentí V., Silva C., Gil M. J., Salvador J., Burrell M. A., Calamita G., Malagón M. M., Frühbeck G. (2011) J. Clin. Endocrinol. Metab. 96, E586–597 [DOI] [PubMed] [Google Scholar]
- 8. Rojek A. M., Skowronski M. T., Füchtbauer E. M., Füchtbauer A. C., Fenton R. A., Agre P., Frøkiaer J., Nielsen S. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 3609–3614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fenton R. A., Moeller H. B., Nielsen S., de Groot B. L., Rützler M. (2010) Am. J. Physiol. Renal Physiol. 298, F224–230 [DOI] [PubMed] [Google Scholar]
- 10. Schwede T., Kopp J., Guex N., Peitsch M. C. (2003) Nucleic Acids Res. 31, 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hub J. S., Aponte-Santamaría C., Grubmüller H., de Groot B. L. (2010) Biophys. J. 99, L97–99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Berendsen H. J., Postma J. P., Van Gunsteren W. F., DiNola A., Haak J. R. (1984) J. Chem. Physics 81, 3684 [Google Scholar]
- 13. Jorgensen W. L., Chandrasekhar J., Madura J. D., Impey R. W., Klein M. L. (1983) J. Chem. Physics 79, 926 [Google Scholar]
- 14. Duan Y., Wu C., Chowdhury S., Lee M. C., Xiong G., Zhang W., Yang R., Cieplak P., Luo R., Lee T., Caldwell J., Wang J., Kollman P. (2003) J. Comput. Chem. 24, 1999–2012 [DOI] [PubMed] [Google Scholar]
- 15. Dang L. X. (1995) J. Am. Chem. Soc. 117, 6954–6960 [Google Scholar]
- 16. Rarey M., Kramer B., Lengauer T., Klebe G. (1996) J. Mol. Biol. 261, 470–489 [DOI] [PubMed] [Google Scholar]
- 17. Klingmuller U., Bauer A., Bohl S., Nickel P. J., Breitkopf K., Dooley S., Zellmer S., Kern C., Merfort I., Sparna T., Donauer J., Walz G., Geyer M., Kreutz C., Hermes M., Gotschel F., Hecht A., Walter D., Egger L., Neubert K., Borner C., Brulport M., Schormann W., Sauer C., Baumann F., Preiss R., MacNelly S., Godoy P., Wiercinska E., Ciuclan L., Edelmann J., Zeilinger K., Heinrich M., Zanger U. M., Gebhardt R., Maiwald T., Heinrich R., Timmer J., von Weizsacker F., Hengstler J. G. (2006) Syst. Biol. 153, 433–447 [DOI] [PubMed] [Google Scholar]
- 18. Andersen B., Rassov A., Westergaard N., Lundgren K. (1999) Biochem. J. 342, 545–550 [PMC free article] [PubMed] [Google Scholar]
- 19. Bublitz C., Kennedy E. P. (1954) J. Biol. Chem. 211, 951–961 [PubMed] [Google Scholar]
- 20. Newsholme E. A., Robinson J., Taylor K. (1967) Biochim. Biophys. Acta 132, 338–346 [DOI] [PubMed] [Google Scholar]
- 21. Hillen W., Berens C. (1994) Annu. Rev. Microbiol. 48, 345–369 [DOI] [PubMed] [Google Scholar]
- 22. Hillen W., Gatz C., Altschmied L., Schollmeier K., Meier I. (1983) J. Mol. Biol. 169, 707–721 [DOI] [PubMed] [Google Scholar]
- 23. Klokkers J., Langehanenberg P., Kemper B., Kosmeier S., von Bally G., Riethmüller C., Wunder F., Sindic A., Pavenstädt H., Schlatter E., Edemir B. (2009) Am. J. Physiol. Renal Physiol. 297, F693–703 [DOI] [PubMed] [Google Scholar]
- 24. Mola M. G., Nicchia G. P., Svelto M., Spray D. C., Frigeri A. (2009) Anal. Chem. 81, 8219–8229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Morinaga T., Nakakoshi M., Hirao A., Imai M., Ishibashi K. (2002) Biochem. Biophys. Res. Commun. 294, 630–634 [DOI] [PubMed] [Google Scholar]
- 26. Koyama Y., Yamamoto T., Kondo D., Funaki H., Yaoita E., Kawasaki K., Sato N., Hatakeyama K., Kihara I. (1997) J. Biol. Chem. 272, 30329–30333 [DOI] [PubMed] [Google Scholar]
- 27. Ma T., Yang B., Verkman A. S. (1997) Biochem. Biophys. Res. Commun. 240, 324–328 [DOI] [PubMed] [Google Scholar]
- 28. Ishibashi K., Kuwahara M., Kageyama Y., Tohsaka A., Marumo F., Sasaki S. (1997) Biochem. Biophys. Res. Commun. 237, 714–718 [DOI] [PubMed] [Google Scholar]
- 29. Carbrey J. M., Gorelick-Feldman D. A., Kozono D., Praetorius J., Nielsen S., Agre P. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 2945–2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tsukaguchi H., Shayakul C., Berger U. V., Mackenzie B., Devidas S., Guggino W. B., van Hoek A. N., Hediger M. A. (1998) J. Biol. Chem. 273, 24737–24743 [DOI] [PubMed] [Google Scholar]
- 31. Tsukaguchi H., Weremowicz S., Morton C. C., Hediger M. A. (1999) Am. J. Physiol. Renal Physiol. 277, F685–696 [DOI] [PubMed] [Google Scholar]
- 32. Ibu J. O., Short A. H. (1986) Scand. J. Gastroenterol. Suppl. 124, 75–81 [DOI] [PubMed] [Google Scholar]
- 33. Detmers F. J., de Groot B. L., Müller E. M., Hinton A., Konings I. B., Sze M., Flitsch S. L., Grubmüller H., Deen P. M. (2006) J. Biol. Chem. 281, 14207–14214 [DOI] [PubMed] [Google Scholar]
- 34. Phillips J. W., Jones M. E., Berry M. N. (2002) Eur. J. Biochem. 269, 792–797 [DOI] [PubMed] [Google Scholar]
- 35. Halestrap A. P., Price N. T. (1999) Biochem. J. 343, 281–299 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.