Abstract
The cardiomyocyte circadian clock directly regulates multiple myocardial functions in a time-of-day-dependent manner, including gene expression, metabolism, contractility, and ischemic tolerance. These same biological processes are also directly influenced by modification of proteins by monosaccharides of O-linked β-N-acetylglucosamine (O-GlcNAc). Because the circadian clock and protein O-GlcNAcylation have common regulatory roles in the heart, we hypothesized that a relationship exists between the two. We report that total cardiac protein O-GlcNAc levels exhibit a diurnal variation in mouse hearts, peaking during the active/awake phase. Genetic ablation of the circadian clock specifically in cardiomyocytes in vivo abolishes diurnal variations in cardiac O-GlcNAc levels. These time-of-day-dependent variations appear to be mediated by clock-dependent regulation of O-GlcNAc transferase and O-GlcNAcase protein levels, glucose metabolism/uptake, and glutamine synthesis in an NAD-independent manner. We also identify the clock component Bmal1 as an O-GlcNAc-modified protein. Increasing protein O-GlcNAcylation (through pharmacological inhibition of O-GlcNAcase) results in diminished Per2 protein levels, time-of-day-dependent induction of bmal1 gene expression, and phase advances in the suprachiasmatic nucleus clock. Collectively, these data suggest that the cardiomyocyte circadian clock increases protein O-GlcNAcylation in the heart during the active/awake phase through coordinated regulation of the hexosamine biosynthetic pathway and that protein O-GlcNAcylation in turn influences the timing of the circadian clock.
Keywords: Cardiac Metabolism, Circadian Clock, Gene Expression, Glucose Metabolism, O-GlcNAcylation
Introduction
Circadian clocks have emerged as critical regulators of energy metabolism (1, 2). Animal models wherein components of these cell autonomous mechanisms are genetically manipulated invariably exhibit altered energy balance, resulting in overt metabolic phenotypes (e.g. obesity or leanness). This concept is exemplified when either Clock or Bmal1 (two transcription factors at the core of the mammalian clock) are disrupted; ClockΔ19 mutant mice are obesity-prone, whereas Bmal1 null mice are lean (3, 4). Appreciation for links between circadian clocks and metabolism has grown further through demonstration that perturbations in metabolism (e.g. changes in nutrient availability, models of obesity, and diabetes mellitus, etc.) in turn influence the clock mechanism (5–7). Collectively, these observations have fueled identification of a number of post-translational mediators that facilitate the interdependence of circadian clocks with metabolism. These include phosphorylation, ubiquitination, acetylation, and ribosylation of critical clock and/or metabolic components in time-of-day-dependent manners (1, 8–14).
Defining the role of a specific cell autonomous circadian clock in metabolic regulation through the use of animal models wherein clock components are genetically altered in a ubiquitous fashion is often hampered by the fact that time-of-day-dependent rhythms are altered at multiple levels (e.g. behavioral, neurohumoral, cellular). For this reason we have recently utilized a mouse model wherein the circadian clock is genetically disrupted in a specific, metabolically active organ (namely the heart) (13, 15). Cardiomyocyte-specific clock mutant (CCM)4 mice have recently revealed that this cell autonomous clock regulates in a time-of-day-dependent manner 1) expression of a plethora of key genes known to influence both oxidative and non-oxidative metabolism, 2) responsiveness of the heart to fatty acids at transcriptional and metabolic levels, and 3) triglyceride turnover (13, 15, 16). Cardiomyocyte clock control of myocardial metabolism is associated with time-of-day-dependent alterations in both myocardial function (e.g. heart rate, cardiac output) and dysfunction (e.g. ischemia/reperfusion tolerance, cardiac hypertrophy) (13, 17, 18).
Many clock-controlled processes in the heart are similarly influenced by modification of proteins by O-linked β-N-acetylglucosamine (O-GlcNAc), such as gene expression, metabolism, hypertrophic growth, and ischemia/reperfusion tolerance (19–21). Analogous to protein phosphorylation, protein O-GlcNAcylation is an enzyme-catalyzed reversible covalent post-translational modification that affects activity, subcellular location, and/or turnover of the target protein (22, 23). Protein O-GlcNAcylation is often described as a nutrient sensor, signaling to the cell when glucose (and amino acid) availability is elevated (24). The addition of the O-GlcNAc moiety to serine/threonine residues on target proteins is catalyzed by O-GlcNAc transferase (OGT), whereas removal/hydrolysis is catalyzed by O-GlcNAcase (OGA) (24). The substrate for protein O-GlcNAcylation (i.e. UDP-N-acetylglucosamine) is generated via the hexosamine biosynthetic pathway (HBP). The committed step in the HBP is the synthesis of glucosamine 6-phosphate from the glycolytic intermediate fructose 6-phosphate and the amino acid glutamine, a reaction catalyzed by glutamine:fructose 6-phosphate amidotransferase (GFAT). Protein O-GlcNAcylation is, therefore, dependent upon both substrate availability (essentially glucose and glutamine) as well as OGT, OGA, and GFAT intrinsic activity.
The circadian clock and protein O-GlcNAcylation influence overlapping processes in the heart. Both are sensitive to changes in nutrient availability/metabolic fluxes, and both are highly conserved across organisms. Therefore, we hypothesized that a relationship exists between the two processes (i.e. the circadian clock regulates O-GlcNAcylation and/or O-GlcNAcylation regulates the circadian clock). Consistent with this hypothesis, we demonstrate that in the murine heart OGT, OGA, and glutamine synthetase (GLUL) as well as glucose metabolism coordinately vary in a time-of-day-dependent manner to promote protein O-GlcNAcylation in the middle of the active (dark) phase. These diurnal variations appear to be driven by the cardiomyocyte circadian clock as they are absent in CCM hearts. Furthermore, we identify the clock component Bmal1 as an O-GlcNAc-modified protein and identify that pharmacological inhibition of OGA leads to decreased Per2 levels, induction of bmal1 gene expression, and phase shifts in the suprachiasmatic nucleus (SCN) clock. Collectively, these data highlight a novel relationship between protein O-GlcNAcylation and the circadian clock.
MATERIALS AND METHODS
Animals
Male wild-type (WT) and CCM mice were housed at either the Centers for Comparative Medicine at Baylor College of Medicine (PUGNAc administration studies) or at the University of Alabama at Birmingham (all other studies). Male mPER2::LUC knock-in mice (25) were housed at the University of Alabama at Birmingham. Mice were housed in micro-isolator cages under controlled conditions (23 ± 1 °C; 12 h light:12-h dark cycle) and received standard laboratory chow and water ad libitum. To measure diurnal variations in behavior (food intake and activity) and whole body metabolism (energy expenditure), mice were housed within a comprehensive laboratory animal monitoring system (CLAMS, Columbus Instruments, OH). In a subset of studies, elevations in protein O-GlcNAc levels were achieved in vivo by administration of the OGA inhibitor PUGNAc (20 mg/kg intraperitoneally). All studies were reviewed and approved by the respective local IACUC committees before their conduct.
Ex Vivo Heart Perfusions
The ex vivo working mouse heart perfusion was used to assess myocardial metabolism, as described previously (13). Hearts were perfused in the working mode in a non-recirculating manner with a preload of 12.5 mm Hg and an afterload of 50 mm Hg. Standard Krebs-Henseleit buffer was supplemented with 8 mm glucose, 0.4 mm oleate conjugated to 3% BSA (fraction V, fatty-acid free; dialyzed), 10 microunits/ml insulin, 0.05 mm l-carnitine, and 0.13 mm glycerol. The perfusion buffer utilized in studies investigating lactate oxidation was further supplemented with 1.5 mm lactate and 0.15 mm pyruvate. Radiolabeled tracers (0.035 mCi/liter d-[U-14C]glucose, 0.00625 mCi/liter l-[U-14C]lactic acid, 0.02 mCi/liter d-[5-3H]glucose, and 0.04 mCi/liter d-[2-3H]glucose) were used to measure various stages of oxidative and non-oxidative metabolism as described previously. Coronary effluent was collected throughout the perfusion period at 5-min intervals; steady state rates in metabolism are the average of data at perfusion times 30 and 35.
Neonatal Cardiomyocyte Cultures
Neonatal rat ventricular myocytes were isolated from 2- to 3-day-old neonatal Sprague-Dawley rats and cultured as described previously (26). A confluent monolayer of spontaneously beating neonatal rat ventricular myocytes had formed within 1–2 days of isolation and cells were treated with NAD+ (0–1 mm) for 3–24 h. Cells were lysed and O-GlcNAc levels determined by Western blot analysis as described below.
Organotypic SCN Cultures
Between 1–4 h after lights on, PER2::LUC mice (transgenic mice expressing the luciferase gene under the control of the PER2 promoter) were killed by cervical dislocation. Organotypic cultures of the SCN were prepared according to previously published methods and culture media (27). Slice cultures containing the SCN were maintained at 36.0 °C, and after stable rhythmicity was established they were treated by adding either 5 μl of 20 mm PUGNAc (100 μm final concentration) or 5 μl of sterile culture water (vehicle) directly into the media. Cultures were treated at various circadian times where CT 12 refers to peak luminescence (25, 27). Bioluminescence was measured with a LumiCycle (Actimetrics, Wilmette, IL). Data were analyzed by using Lumicycle data analysis software (Actimetrics). For each selection of data (two to three cycles before treatment or two cycles after treatment), base-line drift was removed by fitting a polynomial curve with an order of one less than the number of cycles. The base-line-subtracted data were then used to estimate the circadian period by fitting a sine wave multiplied by an exponential decay. A goodness of fit was computed, and only data with at least 80% of the variance accounted for by the curve was used for analysis. Two predictions of the peak of the first cycle after treatment were made; one based on the curve that best fit the cycles preceding treatment and a second based on the curve that best fit the cycles after treatment. The difference between these two predictions was determined to be the phase shift.
Myocardial NAD Levels
Total NAD levels were measured in frozen hearts using a commercially available kit (Bioassay Systems LLC).
Western Blotting
Qualitative analysis of protein expression was performed using standard Western blotting techniques. Briefly, powdered tissue (∼20 mg) was homogenized on ice in T-PER tissue lysis buffer (Pierce) supplemented with 40 μm PUGNAc (Carbogen), 2 mm PMSF (Sigma), 2 mm EDTA, 0.25% Nonidet P-40, and protease and phosphatase inhibitor cocktails (all from EMD Chemicals). Lysates (10–20 μg) were separated on a 7.5% bisacrylamide gel by electrophoresis, transferred to a PVDF membrane, and probed with antibodies to O-GlcNAc (CTD110.6 antibody, Mary-Ann Accavitti, UAB Epitope Recognition and Immunoreagent Core; anti-O-GlcNAc, clone 9D1.E4(10), Millipore), phospho-AMPKα, AMPKα (Cell Signaling Technology), Bmal1, Per1, Per2, Cry2, Glut4, glutamine synthetase (Millipore), Clock, OGA (Santa Cruz), OGT (Sigma), Glut1, or calsequestrin (Abcam) diluted in 1% casein, PBS for 1.5 h at room temperature or overnight at 4 °C. Membranes were incubated with appropriate horseradish peroxidase-conjugated secondary antibodies (1:10,000–1:40,000; Santa Cruz). Bands were visualized with Immunstar Western C detection kit (Bio-Rad) on x-ray film, scanned, quantified using Scion Image Version 4.0.3.2, and normalized to calsequestrin.
For immunoprecipitations, tissue homogenates (described above) were precleared with protein G-agarose fast flow beads (Millipore) for 1 h at 4 °C then removed by centrifugation at 14,000 × g for 10 min. Precleared lysates (500 μg, heart; 1000 μg, liver) were diluted in PBS to a final total tissue protein concentration of 1–1.5 mg/ml then incubated with 2 μg of antibody at 4 °C overnight. Protein G-agarose beads were added to capture the immunocomplex (4 °C for 4 h). Beads were collected by pulse centrifugation (5 s at 14,000 × g), washed twice with ice-cold tissue lysis buffer (described above), and resuspended in 75 μl of 2× Laemmli sample buffer (Bio-Rad). Samples were denatured at 100 °C for 5 min followed by pulse centrifugation to remove immunocomplex from beads then subjected to Western blotting. For a subset of studies, membranes were probed with CTD antibody in the presence of 10 or 100 mm N-acetyl-d-glucosamine (Sigma) for 1.5 h at room temperature to test O-GlcNAc binding specificity.
RNA Isolation and Real-time RT-PCR
RNA was extracted from hearts using standard procedures. Candidate gene expression analysis was performed by quantitative RT-PCR using previously described methods. Specific assays were designed for each gene from mouse sequences available in GenBankTM. Primer and probe sequences have been reported previously (13, 28, 29). RT-PCR data are represented as mRNA molecules/ng total RNA.
Statistical Analysis
Two-way analysis of variance was performed to investigate the main effects of group, e.g. genotype, time, and intervention using Stata Version 10.0 (Stata Corp., San Antonio, TX). The null hypothesis of no model effects was rejected at p < 0.05. For analyses in which the full model was statistically significant (i.e. p < 0.05), Bonferroni post hoc analyses were performed to examine all possible pairwise comparisons. A full model including second-order interactions was conducted for each experiment. Statistically significant differences were determined using Type III sums of squares.
RESULTS
Diurnal Variations in Protein O-GlcNAcylation in Mouse Hearts
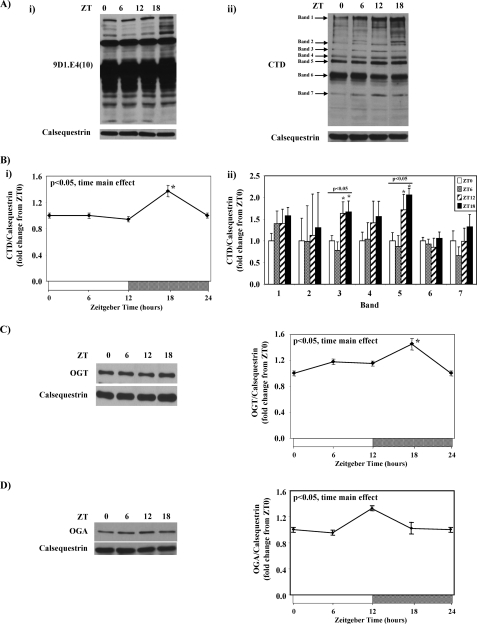
Mouse hearts were collected at 6-h intervals over the course of the day, after which time-of-day-dependent variations in myocardial protein O-GlcNAcylation were determined through immunoblotting. Fig. 1A shows that total protein O-GlcNAcylation levels exhibit diurnal rhythms in mouse hearts, when either the 9D1.E4(10) or CTD antibody is utilized; densitometric analysis of the CTD blot revealed a significant 1.5-fold trough-to-peak difference in total protein O-GlcNAcylation (Fig. 1B). Protein O-GlcNAcylation of seven distinct bands on the CTD immunoblot were next semiquantified; significant time-of-day-dependent variations in O-GlcNAcylation were observed for bands 3 and 5 (Fig. 1B). In all cases, peak protein O-GlcNAcylation was observed in the middle of the dark/active phase (i.e. ZT18). Diurnal variations in the enzymes responsible for the addition (i.e. OGT) and removal (i.e. OGA) of this post-translational modification were next investigated in mouse hearts. Significant time-of-day-dependent variations were observed for both OGT and OGA (Fig. 1, C and D, respectively). Consistent with changes in total protein O-GlcNAcylation (i.e. peak at ZT18; Fig. 1B), OGT protein levels peaked in the middle of the dark phase (i.e. ZT18, Fig. 1C). In contrast, OGA protein levels exhibit a lower amplitude diurnal variation (compared with protein O-GlcNAcylation and OGT) that peaks at the light-to-dark phase transition (i.e. ZT12, Fig. 1D).
FIGURE 1.
Diurnal variations in mouse heart protein O-GlcNAcylation using the anti-O-GlcNAc antibody 9D1.E4(10) (i) or CTD (ii) (A) is shown. Densitometric analysis of total protein O-GlcNAcylation (i) as well as seven distinct O-GlcNAc protein bands (ii) in the mouse heart (B) is shown. Diurnal variations in OGT (C) and OGA (D) protein levels in the mouse heart are shown. Hearts were isolated at the dark-to-light phase transition (ZT 0), middle of the light phase (ZT 6), light-to-dark phase transition (ZT 12), and middle of the dark phase (ZT 18); ZT 0 and ZT 24 are identical data points. ZT represents zeitgeber time. Data are shown as the mean ± S.E. for between 14 and 20 separate hearts within each group. The main effects for time are indicated in the top left hand corner of the figure panels. *, p < 0.05 for a specific time point versus the trough (i.e. lowest) value.
Diurnal Variations in Protein O-GlcNAcylation and OGT Are Absent in CCM Hearts
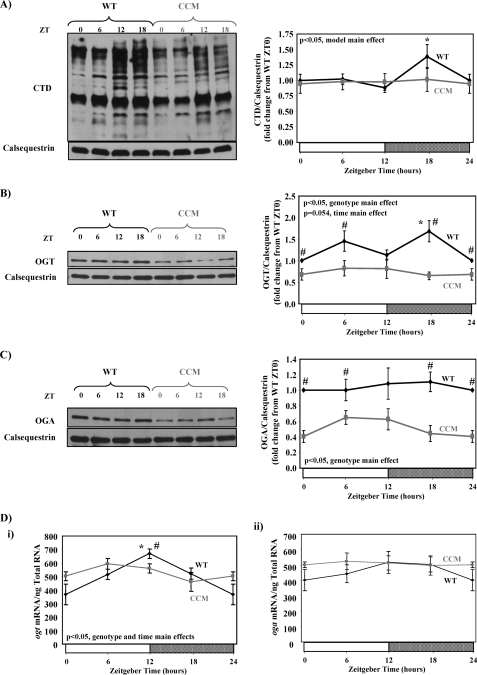
Diurnal variations in myocardial protein O-GlcNAcylation could be mediated by extrinsic (e.g. neurohumoral factors) or intrinsic (i.e. cell autonomous clocks) influences. To investigate the relative contribution of the cardiomyocyte circadian clock, we utilized CCM mice. Similar to the data presented in Fig. 1B, WT mouse hearts exhibit a significant time-of-day dependence in total protein O-GlcNAc levels; there was an approximate 60% increase in overall O-GlcNAcylation between the minimum at ZT12 and the peak at ZT18 (Fig. 2A). In contrast, in CCM hearts no diurnal variation in protein O-GlcNAcylation was observed, and essentially identical levels at ZT12 versus ZT18 were observed (Fig. 2A). Consistent with protein O-GlcNAcylation, in CCM hearts OGT protein levels show no time-of-day-dependent changes and are chronically repressed compared with WT hearts (Fig. 2B). Similarly, OGA protein levels are also chronically repressed in CCM hearts independent of the time-of-day (Fig. 2C). Circadian clocks are primarily transcriptionally based mechanisms such that many target pathways are regulated at a transcriptional level. As such, we next investigated diurnal variations in ogt and oga mRNA levels in WT versus CCM hearts. Fig. 2D shows that ogt, but not oga, mRNA levels vary in a time-of-day-dependent manner in WT hearts; specifically, ogt mRNA levels increase ∼2-fold between ZT0 and ZT12. In contrast, diurnal variations in ogt mRNA levels are completely absent in CCM hearts.
FIGURE 2.
Diurnal variations in protein O-GlcNAcylation (A), OGT (B), and OGA (C) protein levels as well as ogt and oga mRNA levels (D) in WT versus CCM hearts. Hearts were isolated at the dark-to-light phase transition (ZT 0), middle of the light phase (ZT 6), light-to-dark phase transition (ZT 12), and middle of the dark phase (ZT 18); ZT 0 and ZT 24 are identical data points. ZT represents zeitgeber time. Data are shown as the mean ± S.E. for between 11 and 20 separate hearts within each group. Main effects for model, time, or genotype are indicated in the top or bottom left-hand corner of the figure panels. *, p < 0.05 for a specific time point versus the trough (i.e. lowest) value. #, p < 0.05 for WT versus CCM at a distinct zeitgeber time.
Diurnal Variations in Parameters Influencing Carbon Entry into HBP
Protein O-GlcNAcylation is known to be influenced by a number of parameters, including behaviors (e.g. food intake, physical activity) as well as changes in whole body metabolism. To rule out that differences in cardiac protein O-GlcNAcylation were not secondary to these factors, WT and CCM mice were housed within CLAMS cages. Consistent with the cardiomyocyte-specific nature of the CCM model, no differences in diurnal variations in food intake, physical activity, or energy expenditure were observed between WT and CCM littermates (supplemental Fig. 1).
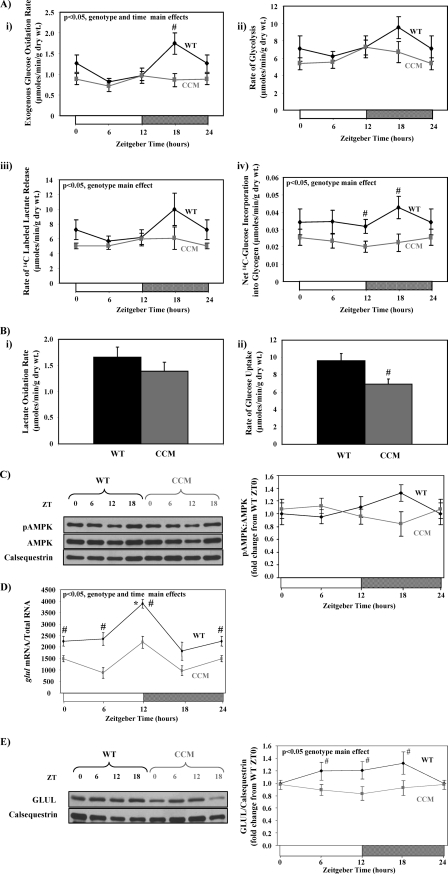
Protein O-GlcNAcylation is regulated in part by the entry of carbon into the HBP, and GFAT catalyzes the committed step for glucose entry into this pathway. GFAT activity is regulated in a number of ways, including expression levels, substrate availability, and phosphorylation. We, therefore, investigated whether GFAT expression, pathways providing GFAT substrates (i.e. glucose uptake and glycolysis), and/or kinases known to phosphorylate GFAT (i.e. AMPK) exhibit a diurnal variation in WT (but not CCM) hearts. Levels of gfat1 and gfat2 mRNA did not exhibit a significant diurnal variation in wild-type hearts, and these levels were unaltered in CCM hearts (supplemental Fig. 2A). Diurnal variations in both oxidative and non-oxidative glucose metabolic fluxes were next investigated in WT versus CCM hearts in a controlled ex vivo setting. WT hearts exhibit a 2.4-fold (trough-to-peak) diurnal variation in the rate of glucose oxidation, peaking during the middle of the active phase (ZT18; Fig. 3Ai). Similar diurnal variations in non-oxidative glucose metabolism were observed in WT hearts (i.e. glycolysis as assessed by [3H]H2O release from [5-3H]glucose or [14C]lactate release from [14C]glucose, as well as net glycogen synthesis; Figs. 3A, ii–iv). Importantly, these diurnal variations are absent in CCM hearts, which exhibit chronically low rates of glucose metabolism (Fig. 3A). Collectively, these observations suggest that the cardiomyocyte circadian clock increases both oxidative and non-oxidative glucose metabolism during the dark phase, which potentially contributes to increased protein O-GlcNAcylation at this time.
FIGURE 3.
Time-of-day- and cardiomyocyte circadian clock-dependent differences in parameters influencing entry of carbon into the HBP. Diurnal variations in rates of glucose oxidation (i), glycolysis (ii and iii), and glycogen synthesis (iv) in ex vivo perfused in WT versus CCM hearts are shown (A). Rates of lactate oxidation (i) and glucose uptake (ii) for WT versus CCM hearts at ZT18 are shown (B). Diurnal variations in P-AMPK levels in WT versus CCM hearts are shown (C). Diurnal variations in glul mRNA levels in WT versus CCM hearts (D) are shown. Diurnal variations in GLUL protein levels in WT versus CCM hearts (E) are shown. Hearts were isolated at the dark-to-light phase transition (ZT 0), middle of the light phase (ZT6), light-to-dark phase transition (ZT 12), and middle of the dark phase (ZT 18); ZT 0 and ZT 24 are identical data points. ZT represents zeitgeber time. Data are shown as the mean ± S.E. for between 7 and 15 separate hearts within each group. Main effects for model, time, or genotype are indicated in the top left-hand corner of the figure panels. *, p < 0.05 for a specific time point versus the trough (i.e. lowest) value. #, p < 0.05 for WT versus CCM at a distinct ZT.
Myocardial glucose metabolism can be modulated primarily by 1) modulation of pyruvate oxidation by the pyruvate dehydrogenase complex (i.e. the Randle cycle) and/or 2) increasing glucose uptake into the cardiomyocyte. We, therefore, investigated both lactate (which is converted to pyruvate via lactate dehydrogenase) oxidation and glucose uptake in WT versus CCM hearts at ZT18 when oxidative glucose metabolism was at its highest in WT hearts. We report that rates of lactate oxidation were not significantly different between WT and CCM hearts (Fig. 3Bi). In contrast, glucose uptake rates are significantly lower in CCM versus WT hearts (Fig. 3Bii). These data suggest that the cardiomyocyte circadian clock regulates myocardial glucose uptake but not pyruvate oxidation.
Glucose uptake is determined in part by the expression of the glucose transporters (of which GLUT1 and GLUT4 are highly abundant in the heart) as well as signaling pathways promoting GLUT4 translocation to the cell surface such as AMPK. Although glut1 and glut4 mRNAs exhibit diurnal variations in WT, but not CCM hearts, peaking around the middle of the light phase (i.e. ZT 6; supplemental Fig. 2B), neither GLUT1 nor GLUT4 protein levels exhibit time-of-day-dependent variations in WT/CCM hearts (supplemental Fig. 2, C and D). However, consistent with previous reports (16), P-AMPK (which promotes glucose transport) levels tended to peak in WT hearts in the middle of the dark phase (i.e. ZT18; Fig. 3C), when glucose metabolism and O-GlcNAc levels were highest. Furthermore, P-AMPK levels were significantly lower in CCM hearts at ZT18 (p = 0.029; Fig. 3C), consistent with the lower glucose uptake and O-GlcNAc levels in CCM hearts at this time. Collectively these data suggest that the cardiomyocyte circadian clock potentially modulates myocardial glucose uptake through regulation of AMPK activity. In addition, increased AMPK activity during the dark/active phase would also promote HBP flux through phosphorylation and activation of GFAT (30).
Similar to its obligate requirement for the glycolytic intermediate fructose 6-phosphate, GFAT requires glutamine to generate the HBP intermediate glucosamine 6-phosphate. GLUL has been shown to exhibit a diurnal variation in skeletal muscle (31). We found that mRNA levels for glul also oscillate in a time-of-day-dependent manner in WT hearts, peaking at the light-to-dark phase transition (Fig. 3D), and that these diurnal variations are attenuated in CCM hearts. Furthermore, GLUL protein levels are chronically decreased in CCM hearts, suggesting regulation by the cardiomyocyte circadian clock (Fig. 3E). Increased GLUL activity during the dark/active phase would promote HBP flux and, therefore, protein O-GlcNAcylation at this time.
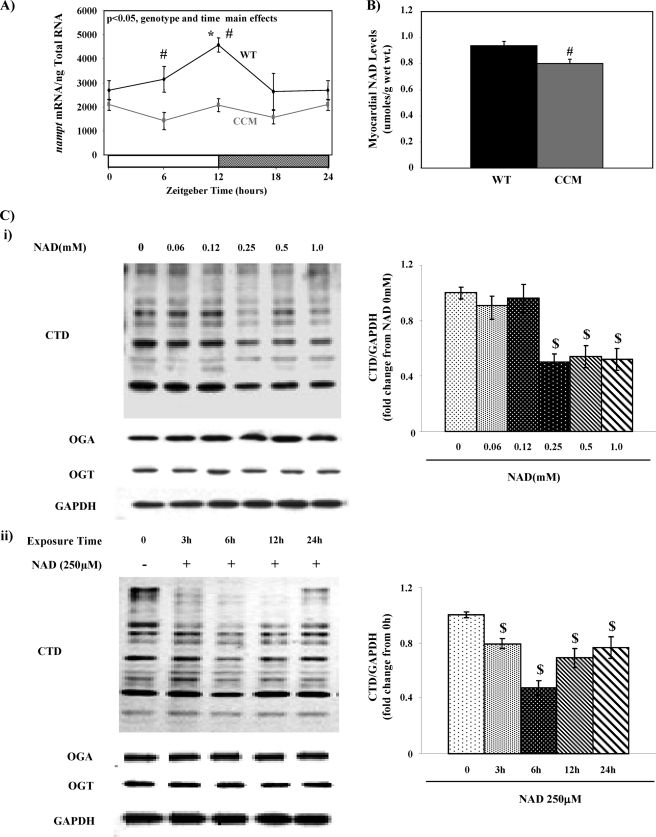
Evidence against NAD as Direct Mediator of Diurnal Variations in Myocardial Protein O-GlcNAcylation
NAD is emerging as an important intermediary link between circadian clocks and metabolism (32). This led us to hypothesize that cardiomyocyte circadian clock-driven oscillations in myocardial NAD levels may mediate diurnal variations in protein O-GlcNAcylation (potentially through modulating the catalytic activity of one or more enzymes in the HBP for example). Consistent with previously published reports, mRNA levels of nampt (nicotinamide phosphoribosyltransferase, a critical enzyme in the NAD salvage pathway) oscillates in WT but not CCM hearts (Fig. 4A) (13). Chronically low expression of nampt in CCM hearts was associated with significantly lower myocardial NAD levels (Fig. 4B). Given that both myocardial NAD and protein O-GlcNAcylation levels are higher in the heart during the dark phase and both are lower in CCM versus WT hearts, we hypothesized that NAD promotes protein O-GlcNAcylation in the heart. To test this hypothesis, neonatal rat ventricular myocytes were challenged with NAD in both a concentration- and time-dependent manner, after which protein O-GlcNAcylation was measured. Fig. 4C shows that NAD (≥250 μm) decreases protein O-GlcNAcylation levels in neonatal rat ventricular myocytes. Furthermore, this effect is relatively rapid (occurs within 3 h) and appears to be independent of changes in either OGT or OGA expression (Fig. 4C). These data demonstrate for the first time that NAD appears to acutely modulate overall cellular O-GlcNAc levels. However, the fact that NAD reduced rather than increased O-GlcNAc levels suggests that NAD does not mediate the increase in cardiac O-GlcNAc levels seen at ZT18.
FIGURE 4.
Influence of NAD on protein O-GlcNAcylation. Diurnal variations in nampt mRNA levels in WT versus CCM hearts (A) are shown. NAD levels in WT versus CCM hearts at ZT18 (B) are shown. Concentration- and time-dependent effects of NAD on protein O-GlcNAcylation as well as OGT and OGA expression in neonatal rat ventricular cardiomyocytes (C) are shown. Hearts were isolated at the dark-to-light phase transition (ZT 0), middle of the light phase (ZT 6), light-to-dark phase transition (ZT 12), and middle of the dark phase (ZT 18); ZT 0 and ZT 24 are identical data points. ZT represents zeitgeber time. Data are shown as the mean ± S.E., for between five and six separate hearts/experiments within each group. Main effects for model, time, or genotype are indicated in the top left-hand corners of figure panels. *, p < 0.05 for a specific time point versus the trough (i.e. lowest) value. #, p < 0.05 for WT versus CCM at a distinct ZT. $, p < 0.05 for a specific NAD challenge compared with control.
Bmal1 Is a Direct Target for Protein O-GlcNAcylation
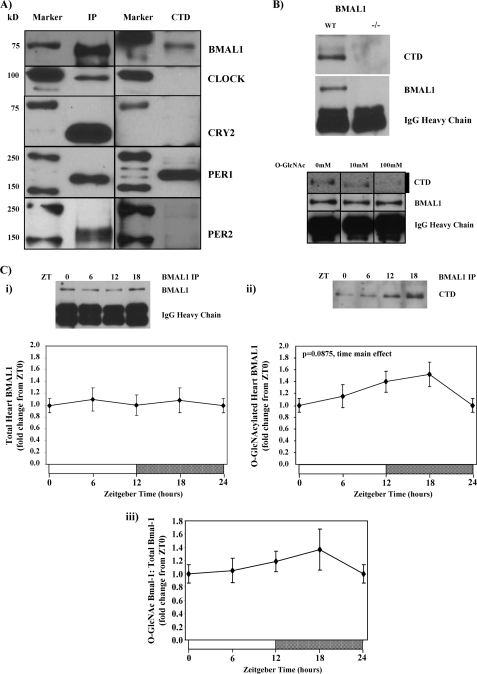
Multiple examples exist wherein those processes influenced by circadian clocks exhibit a feedback relationship with this molecular mechanism. This is particularly true for clock-regulated metabolic processes (1). We, therefore, hypothesized that components of the clock were targets for O-GlcNAc modification. To test this hypothesis we initially examined whether Bmal1, Clock, Cry2, Per1, and/or Per2 were O-GlcNAcylated in the mouse liver. Of these proteins, Bmal1 and Per1 appeared to be positive for O-GlcNAcylation, whereas Clock, Cry2, and Per2 were not (Fig. 5A). Given the critical role of Bmal1 in circadian clock function and that this transcription factor is known to be regulated by a host of post-translational modifications, we focused attention on this protein. We report that the band corresponding to O-GlcNAcylated Bmal1 was absent in Bmal1 null mice and that N-acetyl-d-glucosamine successfully competed for CTD binding (Fig. 5B), thereby supporting the fact that Bmal1 is O-GlcNAc-modified. Fig. 5C shows that Bmal1 protein O-GlcNAcylation may vary in a time-of-day-dependent manner in the heart, with increased levels of this post-translational modification in the middle of the dark phase (i.e. ZT18). These data suggest that circadian clock-mediated diurnal variations in myocardial protein O-GlcNAcylation potentially feed back onto the clock component Bmal1.
FIGURE 5.
Identification of O-GlcNAc-modified clock components. Screening clock components (BMAL1, CLOCK, CRY2, PER1, and PER2) for O-GlcNAc modification in mouse livers is shown (A). The absence of BMAL1 O-GlcNAc modification in BMAL1 null mouse livers or after CTD-binding blockage with O-GlcNAc (10 or 100 mm) (B) is shown. Diurnal variations in Bmal1 O-GlcNAcyation in hearts (D) is shown. Hearts were isolated at the dark-to-light phase transition (ZT 0), middle of the light phase (ZT 6), light-to-dark phase transition (ZT 12), and middle of the dark phase (ZT 18); ZT 0 and ZT 24 are identical data points. ZT represents zeitgeber time. Data are shown as the mean ± S.E. for between 6 and 14 separate hearts/livers within each group. Main effects for time are indicated in the top left-hand corners of figure panels. *, p < 0.05 for a specific time point versus the trough (i.e. lowest) value. IP, immunoprecipitation.
Protein O-GlcNAcylation Influences Circadian Clock
We next investigated whether protein O-GlcNAcylation alters the circadian clock in the heart. Protein O-GlcNAcylation was acutely elevated in WT hearts in vivo through intraperitoneal administration of the OGA inhibitor PUGNAc in the middle of the light phase (i.e. ZT6; Fig. 6A); ZT6 was chosen after our observations that protein O-GlcNAcylation levels are low around this time of day. PUGNAc treatment had no significant effects on CLOCK, BMAL1, CRY2, or PER1 total protein levels (supplemental Fig. 3). In contrast, pharmacological inhibition of OGA resulted in depressed PER2 protein levels during the dark phase (Fig. 6B).
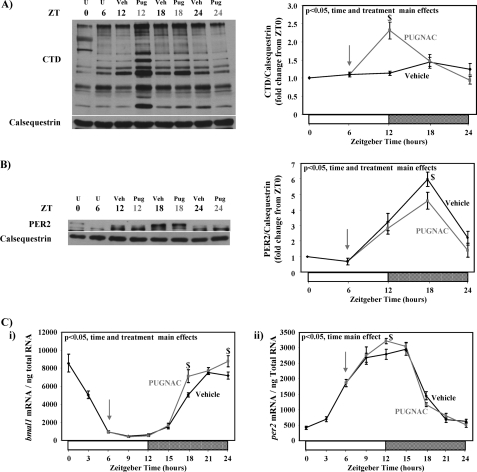
FIGURE 6.
Influence of PUGNAc administration on levels of total protein O-GlcNAcylation (A) and protein expression of PER2 (B) as well as bmal1 and per2 gene expression (C) in mouse hearts. Mice were treated with PUGNAc (20 mg/kg intraperitoneally) at ZT 6 (indicated by an arrow) after which hearts were isolated at ZT 12, ZT 18, and ZT 24. Vehicle-treated mice served as controls. Hearts were isolated at the dark-to-light phase transition (ZT 0), middle of the light phase (ZT 6), light-to-dark phase transition (ZT 12), and middle of the dark phase (ZT 18); ZT 0 and ZT 24 are identical data points. The same samples were utilized for both protein and mRNA measurements. ZT represents zeitgeber time; U represents untreated (i.e. before vehicle or PUGNAc treatment); Veh represents vehicle treatment; Pug represents PUGNAc treatment. Data are shown as the mean ± S.E. for between five and six separate hearts within each group. Main effects for model, time, or treatment are indicated in the top left-hand corners of figure panels. *, p < 0.05 for a specific time point versus the trough (i.e. lowest) value. $, p < 0.05 for a PUGNAc treated versus vehicle at a distinct zeitgeber time.
PER2 is part of a negative feedback loop of the mammalian circadian clock mechanism that represses the transcriptional activity of CLOCK/BMAL1. PUGNAc-mediated depression of PER2 protein levels during the dark phase was associated with a greater induction of bmal1 mRNA (a transcriptional target of CLOCK/BMAL1) at this time (Fig. 6Ci). In contrast, minimal effects were observed on per2 mRNA levels (Fig. 6Cii).
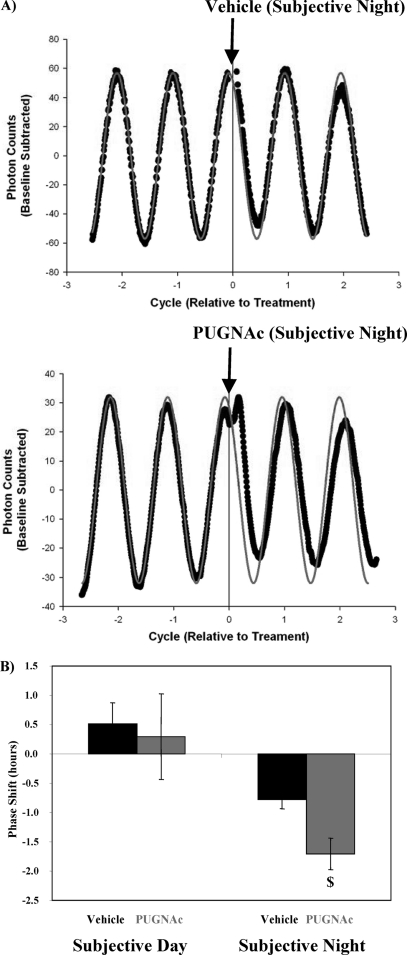
In an attempt to gain better insight into the effects of protein O-GlcNAcylation on circadian clock function, we decided to investigate whether pharmacologically elevating O-GlcNAc levels influenced the SCN (i.e. the central/master) clock in vitro. Fig. 7 shows that PUGNAc (50 μm) administration to SCN slices causes time-of-administration-dependent phase shifts in Per2-luciferase oscillations. Specifically, OGA inhibition during the subjective day had no effect on Per2-luciferase oscillations. In contrast, OGA inhibition during the subjective night results in phase advances (Fig. 7). Taken together, these data suggest that protein O-GlcNAcylation influences the mammalian circadian clock.
FIGURE 7.
Influence of OGA inhibition on circadian rhythms in luciferase for SCN isolated from Per2::Luc transgenic mice; representative raw data (gray line indicates predicted oscillation, black circles represent data obtained; A) and calculated phase shifts (B). SCN cultures were treated with vehicle (water) or PUGNAc (50 μm) either during the subjective day or subjective night, after which the effects on the phase of luminescence oscillations were determined (i.e. phase shift). PUGNAc challenge during the subjective night resulted in phase advances, whereas PUGNAc challenge during the subjective day had no effects on phase. Data are shown as the mean ± S.E. for between three and eight separate SCNs within each group. $, p < 0.05 for a PUGNAc treated versus vehicle at a distinct time in the circadian cycle.
DISCUSSION
The aim of this study was to investigate the interrelationship between the cardiomyocyte circadian clock and protein O-GlcNAcylation. We report that protein O-GlcNAc levels exhibit a diurnal variation in the mouse heart, peaking in the middle of the dark/active phase, a rhythm that is absent in CCM hearts. This appears to be driven by cardiomyocyte circadian clock-orchestrated variations in OGT expression (and OGA expression to a lesser extent), concomitant with promotion of carbon flux through the HBP (through coordinated regulation of glucose metabolism, P-AMPK, and GLUL). We also identify for the first time that Bmal1 appears to be a target for O-GlcNAc modification in metabolically active organs (i.e. liver and heart). Furthermore, acutely increasing protein O-GlcNAcylation (through pharmacological inhibition of OGA) attenuates PER2 protein levels, induces bmal1 gene expression, and phase-advances the SCN clock. Therefore, in light of these observations, which are summarized in Fig. 8, we propose that protein O-GlcNAcylation is a novel post-translational link between myocardial metabolism and the cardiomyocyte circadian clock.
FIGURE 8.
Hypothetical model for the interaction between myocardial metabolism, protein O-GlcNAcylation, and the cardiomyocyte circadian clock. Genes/proteins within black boxes are considered cardiomyocyte circadian clock regulated.
For the circadian clock mechanism to operate over the course of the day, post-transcriptional events are essential. Accordingly, various clock components undergo a host of post-translational modifications (including phosphorylation, acetylation, ubiquitination, sumoylation, and ADP-ribosylation), resulting in altered cellular compartmentation, protein-protein interaction, transcriptional activity, and protein stability (9, 10, 32–34). These post-translational events are important not only for normal clock function but also serve as molecular mediators of environmental entrainment factors, enabling synchronization of cell autonomous clocks with their environment. Peripheral circadian clocks (such as those found in the liver and/or heart) are exquisitely sensitive to metabolic cues (2). Conditions of altered metabolic homeostasis, such as restricted feeding, high fat feeding, or diabetes mellitus, result in distinct effects on clock gene phase and/or amplitude (2, 35). At a molecular level, energy/nutrient-sensitive kinases such as AMPK and glycogen synthase kinase-3β directly influence the activity/protein stability of core clock components (36–39). Similarly, the clock mechanism is responsive to direct metabolic signals, including NAD (both total levels and redox status) (40). In the latter case, two NAD-dependent post-translational modifications have recently been shown to be critical for normal clock function, namely (de)acetylation and ADP-ribosylation (41, 42).
Protein O-GlcNAcylation is a post-translational modification known to influence protein function in a number of ways, including activity and protein stability (24, 43). O-GlcNAcylation is highly dependent on nutrient availability, as demonstrated by changes in this post-translational modification after fluctuations in glucose availability. During distinct situations, glutamine may become limiting as well. O-GlcNAcylation of myocardial proteins has been shown to influence many of the same processes as the cardiomyocyte circadian clock. These include myocardial gene expression, metabolism, and contractile function as well as cardiac hypertrophic growth and ischemia/reperfusion tolerance (19–21). Collectively, these observations (i.e. nutrient sensitivity and overlapping functions) led us to hypothesize that an interrelationship may exist between the circadian clock and protein O-GlcNAcylation. Consistent with this hypothesis, total cellular protein O-GlcNAc levels vary in a time-of-day-dependent manner in wild-type hearts, which is absent when the cardiomyocyte circadian clock is genetically ablated (i.e. CCM hearts; Figs. 1B and 2A). Diurnal rhythms in protein O-GlcNAcylation appeared to be orchestrated by the cardiomyocyte circadian clock through coordinated regulation of OGT (and OGA to a lesser extent) as well as carbon availability for the hexosamine biosynthetic pathway (Fig. 2, B and C). The latter includes increased endogenous glutamine synthesis potential (via GLUL) as well as increased oxidative and non-oxidative glucose metabolic flux during the middle of the dark/active period (Fig. 3).
Previous studies have established that circadian clocks influence glucose metabolism (both utilization and production) (44–48). Many of these studies have relied on mouse models of genetic manipulation of circadian clock components in a ubiquitous fashion, leading to questions regarding the relative roles of cell type-specific circadian clocks. Through measurement of diurnal variations in metabolic fluxes in WT and CCM hearts, these studies show that a cell autonomous clock directly regulates both oxidative and non-oxidative utilization of glucose in a time-of-day-dependent manner. Subsequent mechanistic studies (Fig. 3) are consistent with the hypothetical model in Fig. 8 wherein clock-mediated regulation of glucose transport, potentially through AMPK, plays a pivotal role. An underlying role for circadian clocks is to provide the selective advantage of anticipation (49). This leads to speculation that clock-mediated increases in myocardial glucose metabolism during the active phase is in anticipation of increased workload at this time (due to increased physical activity as the animal in the wild forages for food and/or avoids predation). Indeed, the heart specifically generates extra ATP required for contraction in response to increased workload from glucose as opposed to fatty acids (50). To our knowledge, these studies provide the first direct evidence in support of the concept that a peripheral cell autonomous circadian clock directly regulates glucose transport.
Studies originating from a number of laboratories have recently highlighted NAD as an integral component in the mammalian circadian clock (40, 41). NAD levels oscillate in several tissues (including the heart) in a clock-dependent manner via regulation of NAMPT (nicotinamide phosphoribosyltransferase) (13, 51, 52). NAD levels can in turn influence the timing of the circadian clock through post-translational modifications. For example, the transcription factor Clock possesses acetylase activity and has been shown to acetylate Bmal1, subsequently influencing DNA binding (41). This event is reversed by Sirt1, an NAD-dependent deacetylase (41). Similarly, ADP-ribosylation and subsequent inactivation of Bmal1 is catalyzed by an NAD-dependent ribosyltransferase (42). This raised the possibility that NAD mediates clock controlled diurnal variations in protein O-GlcNAcylation. Consistent with this possibility, NAD levels peak in the rodent heart in the middle of the dark phase (when protein O-GlcNAcylation peaks; Figs. 4 and 1, A and B), and NAD levels are low in CCM hearts (as are protein O-GlcNAcylation levels; Figs. 4B and 2A). However, Fig. 4 shows that challenging cardiomyocytes with NAD leads to a concentration- and time-dependent decrease in protein O-GlcNAcylation, which is not consistent with the notion that NAD directly promotes protein O-GlcNAcylation during the dark/active phase. Interestingly, the negative effects of NAD on protein O-GlcNAcylation are rapid and independent of changes in either OGT or OGA protein levels, raising the possibility that this co-enzyme affects one or more of the enzymes in the HBP. A link between NAD and O-GlcNAcylation has not been established previously and clearly requires further investigation.
This study reports that total protein O-GlcNAcylation exhibits a diurnal variation in the wild-type mouse, peaking in the middle of the dark phase with an amplitude (i.e. peak-to-trough ratio) of ∼1.5-fold (Figs. 1B, 2A, and 6A). The potential biological significance of these observations is highlighted from multiple previously published studies. For example, Laczy et al. (53) have recently reported that an approximate 1.5-fold increase in protein O-GlcNAc levels in the heart is associated with a significant decrease in carbohydrate oxidation and an increase in fatty acid oxidation. Animal models of type 2 diabetes mellitus as well as streptozotocin-induced diabetes mellitus exhibit modest increases in protein O-GlcNAcylation in the heart (∼1.2–1.4-fold), and overexpression of O-GlcNAcase normalizes both cardiac protein O-GlcNAcylation levels and contractile abnormalities associated with diabetes (54, 55). Similarly, Zachara et al. (56) have shown that heat shock increases protein O-GlcNAc levels by ∼1.5-fold and that prevention of this post-translational modification response increased heat shock-induced cell death. Collectively, these observations suggest that the diurnal variations in total protein O-GlcNAcylation reported here have the potential to profoundly influence cardiac biology.
Given that the circadian clock influenced protein O-GlcNAcylation, it seemed logical to investigate whether this post-translational modification in turn influences the circadian clock. Indeed, as highlighted above, several nutrient-based mechanisms have been identified previously as modulators of the circadian clock mechanism. Consistent with the concept that O-GlcNAcylation influences the circadian clock, we found that at least two core clock components, namely PER1 and BMAL1, are O-GlcNAc-modified (Fig. 5A); more definitive mass spectroscopic determination of the specific sites of modification will be the subject of future studies. In the case of Bmal1, we observed trends for time-of-day-dependent variations in myocardial Bmal1 O-GlcNAcylation, which were in the same phase as total cellular protein O-GlcNAcylation (Figs. 5B and 1B, respectively). Consistent with this post-translational modification influencing circadian clock function, we found that a pharmacologically induced acute increase in protein O-GlcNAcylation results in lower PER2 protein levels in the heart (Fig. 6) as well as time-of-day-dependent alterations in bmal1 gene expression (Fig. 6). Although our current study does not suggest that PER2 is directly O-GlcNAc-modified, this protein is known to be regulated by additional post-translational modifications, such as phosphorylation and ubiquitination (consistent with the multiple bands observed on the PER2 immunoblot; Fig. 6B) (57). Whether alterations in cellular O-GlcNAcylation in turn influence PER2 phosphorylation/ubiquitination requires future elucidation. However, to investigate the novel relationship between the circadian clock and O-GlcNAcylation further, we next utilized SCN slices isolated from Per2::Luc mice (allowing central clock function monitoring in real time). Challenging SCN slices with the OGA inhibitor PUGNAc during the subjective night resulted in phase advances (an effect that is comparable with light; Fig. 7). Collectively, these observations are consistent with the hypothesis that protein O-GlcNAcylation is a novel post-translational mechanism influencing the timing of the mammalian circadian clock.
In summary, this study has exposed protein O-GlcNAcylation as a novel molecular link between the cardiomyocyte circadian clock and myocardial metabolism. The cardiomyocyte clock influences total protein O-GlcNAcylation in the heart through coordinated changes in OGT and OGA levels as well as substrate supply for the HBP. Furthermore, protein O-GlcNAcylation in turn influences the circadian clock. This relationship likely extends beyond the myocardium, as evidenced by O-GlcNAc modification of BMAL1 in the liver as well as modulation of SCN clock function by the OGA inhibitor PUGNAc. We speculate that protein O-GlcNAcylation may not only mediate several of the effects of circadian clocks on cellular function, but chronic changes in this post-translational modification may account for alterations in clock function during various metabolic disease states.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants HL074259 and HL106199 (NHLBI, to M. E. Y.) and HL101192 and HL079364 (NHLBI, to J. C. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–3.
- CCM
- cardiomyocyte-specific clock mutant
- O-GlcNAc
- O-GlcNAc O-linked β-N-acetylglucosamine
- OGT
- O-GlcNAc transferase
- OGA
- O-GlcNAcase
- HBP
- hexosamine biosynthetic pathway
- GFAT
- glutamine:fructose 6-phosphate amidotransferase
- GLUL
- glutamine synthetase
- SCN
- suprachiasmatic nucleus
- AMPK
- AMP-activated protein kinase
- PUGNAc
- o-(2-acetamido-2-deoxy-d-glucopyranosylidene) amino-N-phenylcarbamate.
REFERENCES
- 1. Durgan D. J., Young M. E. (2010) Circ. Res. 106, 647–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bray M. S., Young M. E. (2009) Obes. Rev. 10, 6–13 [DOI] [PubMed] [Google Scholar]
- 3. Turek F. W., Joshu C., Kohsaka A., Lin E., Ivanova G., McDearmon E., Laposky A., Losee-Olson S., Easton A., Jensen D. R., Eckel R. H., Takahashi J. S., Bass J. (2005) Science 308, 1043–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Shimba S., Ishii N., Ohta Y., Ohno T., Watabe Y., Hayashi M., Wada T., Aoyagi T., Tezuka M. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 12071–12076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Young M. E., Wilson C. R., Razeghi P., Guthrie P. H., Taegtmeyer H. (2002) J. Mol. Cell. Cardiol. 34, 223–231 [DOI] [PubMed] [Google Scholar]
- 6. Oishi K., Kasamatsu M., Ishida N. (2004) Biochem. Biophys. Res. Commun. 317, 330–334 [DOI] [PubMed] [Google Scholar]
- 7. Bray M. S., Young M. E. (2007) Obes. Rev. 8, 169–181 [DOI] [PubMed] [Google Scholar]
- 8. Chatham J., Laczy B., Durgan D., Young M. (2009) Circ. Res. 105, e50 [Google Scholar]
- 9. Hardin P. E., Yu W. (2006) Cell 125, 424–426 [DOI] [PubMed] [Google Scholar]
- 10. Cardone L., Hirayama J., Giordano F., Tamaru T., Palvimo J. J., Sassone-Corsi P. (2005) Science 309, 1390–1394 [DOI] [PubMed] [Google Scholar]
- 11. Katada S., Sassone-Corsi P. (2010) Nat. Struct. Mol. Biol. 17, 1414–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dardente H., Cermakian N. (2007) Chronobiol. Int. 24, 195–213 [DOI] [PubMed] [Google Scholar]
- 13. Bray M. S., Shaw C. A., Moore M. W., Garcia R. A., Zanquetta M. M., Durgan D. J., Jeong W. J., Tsai J. Y., Bugger H., Zhang D., Rohrwasser A., Rennison J. H., Dyck J. R., Litwin S. E., Hardin P. E., Chow C. W., Chandler M. P., Abel E. D., Young M. E. (2008) Am. J. Physiol. Heart Circ. Physiol. 294, H1036–H1047 [DOI] [PubMed] [Google Scholar]
- 14. Tamaru T., Hirayama J., Isojima Y., Nagai K., Norioka S., Takamatsu K., Sassone-Corsi P. (2009) Nat. Struct. Mol. Biol. 16, 446–448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Durgan D. J., Trexler N. A., Egbejimi O., McElfresh T. A., Suk H. Y., Petterson L. E., Shaw C. A., Hardin P. E., Bray M. S., Chandler M. P., Chow C. W., Young M. E. (2006) J. Biol. Chem. 281, 24254–24269 [DOI] [PubMed] [Google Scholar]
- 16. Tsai J. Y., Kienesberger P. C., Pulinilkunnil T., Sailors M. H., Durgan D. J., Villegas-Montoya C., Jahoor A., Gonzalez R., Garvey M. E., Boland B., Blasier Z., McElfresh T. A., Nannegari V., Chow C. W., Heird W. C., Chandler M. P., Dyck J. R., Bray M. S., Young M. E. (2010) J. Biol. Chem. 285, 2918–2929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Durgan D. J., Pulinilkunnil T., Villegas-Montoya C., Garvey M. E., Frangogiannis N. G., Michael L. H., Chow C. W., Dyck J. R., Young M. E. (2010) Circ. Res. 106, 546–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Durgan D. J., Tsai J. Y., Grenett M. H., Pat B. M., Ratcliffe W. F., Villegas-Montoya C., Garvey M. E., Nagendran J., Dyck J. R., Bray M. S., Gamble K. L., Gimble J. M., Young M. E. (2011) Chronobiol. Int. 28, 187–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fülöp N., Marchase R. B., Chatham J. C. (2007) Cardiovasc. Res. 73, 288–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chatham J. C., Marchase R. B. (2010) Biochim. Biophys. Acta 1800, 57–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Marsh S. A., Dell'Italia L. J., Chatham J. C. (2011) Amino Acids 40, 819–828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hanover J. A., Krause M. W., Love D. C. (2010) Biochim. Biophys. Acta 1800, 80–95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hart G. W., Slawson C., Ramirez-Correa G., Lagerlof O. (2011) Annu. Rev. Biochem. 80, 825–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kamemura K., Hart G. W. (2003) Prog. Nucleic Acid Res. Mol. Biol. 73, 107–136 [DOI] [PubMed] [Google Scholar]
- 25. Yoo S. H., Ko C. H., Lowrey P. L., Buhr E. D., Song E. J., Chang S., Yoo O. J., Yamazaki S., Lee C., Takahashi J. S. (2005) Proc. Natl. Acad. Sci. U.S.A. 102, 2608–2613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Zou L., Yang S., Champattanachai V., Hu S., Chaudry I. H., Marchase R. B., Chatham J. C. (2009) Am. J. Physiol. Heart Circ. Physiol. 296, H515–H523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gamble K. L., Allen G. C., Zhou T., McMahon D. G. (2007) J. Neurosci. 27, 12078–12087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Young M. E., Razeghi P., Cedars A. M., Guthrie P. H., Taegtmeyer H. (2001) Circ. Res. 89, 1199–1208 [DOI] [PubMed] [Google Scholar]
- 29. Young M. E., Razeghi P., Taegtmeyer H. (2001) Circ. Res. 88, 1142–1150 [DOI] [PubMed] [Google Scholar]
- 30. Hu Y., Riesland L., Paterson A. J., Kudlow J. E. (2004) J. Biol. Chem. 279, 29988–29993 [DOI] [PubMed] [Google Scholar]
- 31. Yao Z., DuBois D. C., Almon R. R., Jusko W. J. (2006) Pharm. Res. 23, 670–679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Asher G., Schibler U. (2011) Cell Metab. 13, 125–137 [DOI] [PubMed] [Google Scholar]
- 33. Kloss B., Price J. L., Saez L., Blau J., Rothenfluh A., Wesley C. S., Young M. W. (1998) Cell 94, 97–107 [DOI] [PubMed] [Google Scholar]
- 34. Millar A. J. (2000) Curr. Biol. 10, R529–R531 [DOI] [PubMed] [Google Scholar]
- 35. Bray M. S., Young M. E. (2008) Cardiovasc. Res. 79, 228–237 [DOI] [PubMed] [Google Scholar]
- 36. Um J. H., Pendergast J. S., Springer D. A., Foretz M., Viollet B., Brown A., Kim M. K., Yamazaki S., Chung J. H. (2011) PLoS One 6, e18450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Lamia K. A., Sachdeva U. M., DiTacchio L., Williams E. C., Alvarez J. G., Egan D. F., Vasquez D. S., Juguilon H., Panda S., Shaw R. J., Thompson C. B., Evans R. M. (2009) Science 326, 437–440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Iitaka C., Miyazaki K., Akaike T., Ishida N. (2005) J. Biol. Chem. 280, 29397–29402 [DOI] [PubMed] [Google Scholar]
- 39. Yin L., Wang J., Klein P. S., Lazar M. A. (2006) Science 311, 1002–1005 [DOI] [PubMed] [Google Scholar]
- 40. Rutter J., Reick M., Wu L. C., McKnight S. L. (2001) Science 293, 510–514 [DOI] [PubMed] [Google Scholar]
- 41. Doi M., Hirayama J., Sassone-Corsi P. (2006) Cell 125, 497–508 [DOI] [PubMed] [Google Scholar]
- 42. Asher G., Reinke H., Altmeyer M., Gutierrez-Arcelus M., Hottiger M. O., Schibler U. (2010) Cell 142, 943–953 [DOI] [PubMed] [Google Scholar]
- 43. Love D. C., Hanover J. A. (2005) Sci. STKE 2005, re13. [DOI] [PubMed] [Google Scholar]
- 44. Durgan D. J., Moore M. W., Ha N. P., Egbejimi O., Fields A., Mbawuike U., Egbejimi A., Shaw C. A., Bray M. S., Nannegari V., Hickson-Bick D. L., Heird W. C., Dyck J. R., Chandler M. P., Young M. E. (2007) Am. J. Physiol. Heart Circ. Physiol. 293, H2385–H2393 [DOI] [PubMed] [Google Scholar]
- 45. Marcheva B., Ramsey K. M., Buhr E. D., Kobayashi Y., Su H., Ko C. H., Ivanova G., Omura C., Mo S., Vitaterna M. H., Lopez J. P., Philipson L. H., Bradfield C. A., Crosby S. D., JeBailey L., Wang X., Takahashi J. S., Bass J. (2010) Nature 466, 627–631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Doi R., Oishi K., Ishida N. (2010) J. Biol. Chem. 285, 22114–22121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sadacca L. A., Lamia K. A., deLemos A. S., Blum B., Weitz C. J. (2011) Diabetologia 54, 120–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Leighton B., Kowalchuk J. M., Challiss R. A., Newsholme E. A. (1988) Am. J. Physiol. 255, E41–E45 [DOI] [PubMed] [Google Scholar]
- 49. Edery I. (2000) Physiol. Genomics 3, 59–74 [DOI] [PubMed] [Google Scholar]
- 50. Goodwin G. W., Taylor C. S., Taegtmeyer H. (1998) J. Biol. Chem. 273, 29530–29539 [DOI] [PubMed] [Google Scholar]
- 51. Powanda M. C., Wannemacher R. W., Jr. (1970) J. Nutr. 100, 1471–1478 [DOI] [PubMed] [Google Scholar]
- 52. Storch K. F., Lipan O., Leykin I., Viswanathan N., Davis F. C., Wong W. H., Weitz C. J. (2002) Nature 417, 78–83 [DOI] [PubMed] [Google Scholar]
- 53. Laczy B., Fülöp N., Onay-Besikci A., Des Rosiers C., Chatham J. C. (2011) PLoS One 6, e18417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Fülöp N., Mason M. M., Dutta K., Wang P., Davidoff A. J., Marchase R. B., Chatham J. C. (2007) Am. J. Physiol. Cell Physiol. 292, C1370–1378 [DOI] [PubMed] [Google Scholar]
- 55. Hu Y., Belke D., Suarez J., Swanson E., Clark R., Hoshijima M., Dillmann W. H. (2005) Circ. Res. 96, 1006–1013 [DOI] [PubMed] [Google Scholar]
- 56. Zachara N. E., O'Donnell N., Cheung W. D., Mercer J. J., Marth J. D., Hart G. W. (2004) J. Biol. Chem. 279, 30133–30142 [DOI] [PubMed] [Google Scholar]
- 57. Takahashi J. S., Hong H. K., Ko C. H., McDearmon E. L. (2008) Nat. Rev. Genet. 9, 764–775 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.