Background: Reports on the role of ceramide to regulate LPS signaling have been inconsistent; thus, we have investigated the role of ceramide metabolites to differentially regulate LPS signaling.
Results: Ceramide-1-phosphate limits LPS-mediated NF-κB activation, MAPK activation, and cytokine secretion.
Conclusion: Ceramide-1-phosphate, but not ceramide, limits LPS signaling.
Significance: Ceramide-1-phosphate may function as an anti-inflammatory lipid.
Keywords: Cytokine, Lipopolysaccharide (LPS), NF-kappaB, Sphingolipid, Toll-like Receptors (TLR), Ceramide, Ceramide-1-phosphate, p65
Abstract
Toll-like receptor 4 (TLR4) is a component of the innate immune system that recognizes a diverse group of molecular structures, such as lipopolysaccharide (LPS) from Gram-negative bacteria. TLR4 signaling ultimately leads to activation of the transcription factor, nuclear factor κB (NF-κB), and the production of cytokines. Ceramide is a bioactive sphingolipid that has been suggested to regulate TLR4-induced NF-κB signaling, although reports on the role of ceramide in TLR4 activation conflict. We investigated the possibility that ceramide metabolites, such as ceramide-1-phosphate (C-1-P), may explain these discrepancies. We now report that exogenous C-1-P, but not ceramide, reduces NF-κB-mediated gene transcription in HEK 293 cells stably transfected with human TLR4, CD14, and MD-2. We demonstrate that inhibition of NF-κB by exogenous C-1-P is dose-dependent and specific to TLR4 in a reporter assay. We further demonstrate a requirement for both the phosphate moiety and the sphingoid backbone to inhibit LPS-activated NF-κB transcription. Specifically, C-1-P prevents the degradation of IκB, the phosphorylation of the p65 subunit of NF-κB, and LPS-stimulated MAPK activation. The functional consequence of C-1-P inhibition of NF-κB is a reduction in LPS-mediated cytokine release from HEK 293 TLR4-expressing cells and human peripheral blood mononuclear cells. Taken together, these data demonstrate that C-1-P may function as an anti-inflammatory lipid mediator of immune response.
Introduction
Toll-like receptors (TLRs)2 are a family of evolutionarily conserved proteins that detect “danger” signals, such as pathogen-associated motifs and endogenous molecules in inappropriate contexts (1, 2). Activation of TLRs induces changes in gene expression and leads to generation of proinflammatory molecules that ultimately induce activation of the adaptive immune system. Aberrant signaling through these receptors has been associated with autoimmune disease development (3). TLRs have also been identified as targets for therapeutic intervention in treating cancer and sepsis (4, 5).
Lipopolysaccharide (LPS) is a component of the Gram-negative bacteria cell wall and is a well known activator of Toll-like receptor 4 (TLR4) (6, 7). LPS responsiveness requires CD14, which is the bona fide LPS receptor (8), and the adaptor protein MD-2 (9). Two distinct pathways are activated by TLR4 signaling: the MyD88-dependent pathway and the TRIF-dependent pathway (10, 11). Both pathways relieve inhibition of the multisubunit transcription factor NF-κB in the cytosol, allowing translocation to the nucleus and production of proinflammatory cytokines (12). LPS also alters lipid signaling, by inducing cytosolic phospholipase A2 (cPLA2) activity, which promotes release of proinflammatory arachidonic acid metabolites in macrophages (13).
Sphingolipid metabolism and localization is important in the biophysical regulation of LPS signaling. In particular, localized enrichment of ceramide in the plasma membrane may preferentially target TLR4 to membrane microdomains, although reports of this phenomenon have been incongruent (14–16). It has been documented that ceramide production via acid sphingomyelinase is required for TLR4 translocation to lipid microdomains and subsequent activation of multiple MAPK signaling cascades, including JNK, p38, and ERK1 and -2 (14). On the other hand, exogenous ceramide was not able to recapitulate recruitment of TLR4 to lipid microdomains containing CD14 (15). It has also been reported that LPS treatment leads to endogenous ceramide production in both LPS-responsive and LPS-hyporesponsive macrophages, arguing that ceramide generation by LPS may be a TLR4-independent event (16). In sum, the role of ceramide and ceramide metabolites in the biophysical regulation of TLR4 signaling requires clarification.
Sphingolipids may also regulate TLR4 activation by alternative, non-biophysical mechanisms. Structural similarity between LPS and ceramide (17) led to the biochemical hypothesis that ceramide may be a TLR4 agonist (18). In one study testing this possibility, ceramide was able to activate TLR4-dependent signaling, suggesting that ceramide was a putative TLR4 agonist (18). However, other reports demonstrated that exogenous ceramide treatment, unlike LPS treatment, was unable to activate ERK and NF-κB in macrophages (16). Conversely, ceramide and ceramide metabolites may reduce LPS-mediated inflammatory processes, suggesting that sphingolipids may be antagonistic to TLR4 signaling. For example, our laboratory has demonstrated that liposomal ceramide limits TLR4-dependent neutrophil recruitment in a murine model of corneal inflammation (19). In addition, both ceramide and ceramide-1-phosphate (C-1-P) negatively regulated TNF-α production by LPS in macrophages (20). These discrepancies may reflect cellular metabolism of ceramide to C-1-P. This is supported by three-dimensional modeling, which suggests that C-1-P is more structurally similar to LPS than ceramide (17). Thus, we have now examined the differential role of ceramide and C-1-P in LPS signaling.
Given the inconsistencies in the literature concerning the putative biophysical and biochemical actions of ceramide and ceramide metabolites in regulating TLR4 signaling, we utilized a cellular model system that expresses the canonical TLR4 signalplex. Using a HEK cell model system that expresses TLR4, CD14, and MD-2 against a null background, we have begun to validate the role of ceramide metabolites and ceramide metabolism in this “defined” system. Using this cellular model, we hoped to define the biophysical and biochemical actions of individual sphingolipid metabolites upon TLR4-dependent NF-κB activation and subsequent cytokine generation. We then confirmed the results from our defined model in a more physiological model, that of human PBMCs.
EXPERIMENTAL PROCEDURES
Reagents
HEK 293 cells stably transfected with CD14, MD-2, and TLR4; the pNIFty2-SEAP vector, hygromycin, blasticidin, zeocin, and Quanti-Blue secreted alkaline phosphatase (SEAP) detection reagent were purchased from Invivogen (San Diego, CA). Cell culture medium was purchased from Mediatech (Manassas, VA), and fetal bovine serum from Gemini Bio-Products (West Sacramento, CA). Antibiotic/antimycotic supplement and Lipofectamine 2000 were purchased from Invitrogen. LPS from Escherichia coli O111:B4 and red blood cell lysing buffer were obtained from Sigma. All lipids, including KdO2-lipid A, were purchased from Avanti Polar Lipids (Alabaster, AL). Antibodies to TLR4, CD14, and p-p65 (Ser-536) as well as secondary and isotype control antibodies for PhosflowTM were purchased from BD Biosciences. All antibodies for Western blotting were obtained from Cell Signaling (Danvers, MA) with the exception of p-ERK and ERK antibodies, obtained from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA). Acetylsalicylic acid and celecoxib were purchased from Sigma. Ficoll-Paque PLUS was purchased from GE Healthcare.
Cell Culture
HEK 293 cells stably transfected with CD14, MD-2, and TLR4 were maintained in DMEM with 10% FBS and 1× antibiotic/antimycotic and incubated at 37 °C in 5% CO2. Selection was maintained by supplementation with 10 μg/ml blasticidin and 50 μg/ml hygromycin B. This stably transfected cell line has minimal expression of other TLRs.
Isolation of PBMCs from Human Blood
Heparinized human blood from healthy donors was diluted 2:1 with PBS and then separated via Ficoll-Paque PLUS by density centrifugation. The buffy coat was harvested, and then red blood cells were lysed with red blood cell lysing buffer. Cells were washed twice in PBS and then resuspended and cultured in RPMI.
Creation of Stably Transfected SEAP Reporter Clonal Cell Lines
HEK 293 cells stably transfected with CD14, MD-2, and TLR4 (Invivogen) were transfected with the pNIFty2-SEAP vector with Lipofectamine 2000. The antibiotic zeocin was applied at 100 μg/ml for selection and clone maintenance. Following selection, individual clones were isolated and tested for LPS responsiveness by the Quanti-Blue SEAP reporter assay. As a negative control, HEK 293 cells stably transfected with CD14, MD-2, and TLR4 were transfected with an empty vector in place of the pNIFty2-SEAP. These cells did not express SEAP upon LPS treatment when measured by the Quanti-Blue SEAP reporter assay. Multiple clonal lines of stably transfected HEK TLR4 SEAP-expressing cells were validated and displayed similar activation of NF-κB by LPS.
Quanti-Blue SEAP Reporter Assay
HEK 293 CD14/TLR4/MD-2 SEAP clones were plated in 96-well plates, serum-starved overnight, and then treated with test compounds for 24 h. After treatment, 50 μl of cell culture medium was removed and added to plates containing 150 μl of prewarmed Quanti-Blue detection reagent per well. Color was allowed to develop for 1–8 h, and absorbance was detected at 650 nm.
Liposome Preparation
Ceramide-1-phosphate liposomes were prepared to a DSPC/DOPE/PEG 2000-DSPE/PEG 750-C8 Cer/C-1-P final molar ratio of 4.5/2/1/0.5/2. Ceramide liposomes were prepared to a DSPC/DOPE/PEG 2000-DSPE/PEG 750-C8 Cer/C8 Cer final molar ratio of 3.75/1.75/0.75/0.75/3. Sphingosine-1-phosphate (S-1-P) liposomes contained a DSPC/DOPE/PEG 2000-DSPE/PEG 750-C8 Cer/S-1-P final molar ratio of 4.75/2.75/0.75/0.75/1. Ghost liposomes contained a DSPC/DOPE/PEG 2000-DSPE/PEG 750-C8 Cer molar ratio of 5.36/2.5/1.07/1.07. Note that all liposomes contained a pegylated non-bioactive C8 ceramide species to protect liposomes from aggregation. Lipids dissolved in chloroform or methanol were combined in the desired molar ratios, dried under nitrogen, and then hydrated in endotoxin-free PBS. The solutions were then sonicated until clarity improved and were extruded 11 times through 100-nm polycarbonate membranes.
BSA Carrier Lipid Preparation
Lipids were dried under a stream of nitrogen and then resuspended in DMSO. The lipid/DMSO solution was then added to a fatty acid-free BSA solution to achieve a final concentration of 1 mm lipid, 1 mm BSA in 20 mm HEPES with 10% DMSO. Complexes were allowed to form by rocking at room temperature for 30 min, followed by sonication to clarity.
Sphingolipidomics
Sphingolipids were analyzed by LC/electrospray ionization-MS/MS based on the method described by Merrill et al. (21) with some modifications. These modifications ensure that the samples are properly neutralized to prevent breakdown of sphingomyelin and lysosphingomyelin into other lipids if dried down under alkaline conditions (22). Lipids from HEK 293 cell lysates (500 μg of total protein/sample) were extracted and separated on an Agilent 1100 HPLC system. Mobile phases were as described (21) but with a 0.2% formic acid concentration and a Luna C8 (3 μ) 2-mm inner diameter × 5-cm column maintained at 60 °C for separation of the sphingoid bases and 1-phosphates. The eluate was analyzed with an inline ABI 4000 Q Trap (Applied Biosystems, Foster City, CA) mass spectrometer equipped with a Turbo ion spray source. The peak areas for the different sphingolipid subspecies were compared with that of the internal standards. All data reported are based on monoisotopic mass and are represented as pmol/mg protein.
Phosflow
HEK TLR4 cells were treated for 45 min with PBS, 100 ng/ml LPS, or 100 ng/ml LPS plus 10 μm C8 C-1-P liposomes. Cells were washed with PBS and scraped for collection. Fixation was performed with 2% paraformaldehyde plus 2% BSA in PBS. Cells were permeabilized with PBS plus 0.01% Triton X, blocked with human Fc blocker, and then labeled with antibodies to p-p65, CD14, and TLR4. Staining intensity of p-p65 was measured by flow cytometry. Fluorescence was measured with a BD LSR II flow cytometer, and quantification was performed by FACS DIVA software.
ELISAs
Cells were serum-starved overnight and then treated for 24 h with 100 ng/ml LPS and/or 10 μm liposomal lipid. Conditioned medium was collected and assayed for cytokine levels according to kit instructions. TNF-α and IL-8 ELISA kits were purchased from BD Biosciences (San Diego, CA), the IL-1β ELISA was purchased from Fujirebio (Malvern, PA), and the IL-6 ELISA was purchased from Thermo Scientific (Rockford, IL).
Statistical Determinations
Data are presented as means ± S.E. Statistical analyses were performed using one-way analyses of variance with the Tukey post-test or Student's t test where indicated. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
RESULTS
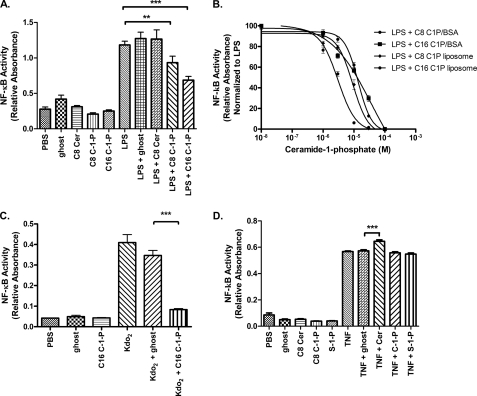
We utilized a fully functional overexpression model of HEK 293 cells stably transfected with TLR4, CD14, and MD-2, hereafter referred to as HEK TLR4 cells. Initially, we assessed the effect of ceramide or ceramide metabolites upon NF-κB-mediated transcription, which is a broad indicator of LPS signaling. We constructed a reporter assay from HEK TLR4 cells transfected with SEAP under the control of an NF-κB-inducible promoter. Cells were treated with 10 μm liposomal lipids alone or concurrently with liposomal lipids and 100 ng/ml LPS. Both short- and long-chain C8 and C16 C-1-P significantly decreased LPS-activated NF-κB transcription (Fig. 1A). In contrast, neither the ghost liposome, which serves as a vehicle control, nor the ceramide liposome treatment altered NF-κB-mediated transcription (Fig. 1A). As a negative control, LPS did not activate SEAP expression in HEK 293 null cells, which are not transfected with TLR4, CD14, and MD-2, but are transfected with the SEAP plasmid, demonstrating TLR4 dependence for SEAP production (data not shown). We next investigated if the inhibitory actions of C-1-P upon LPS-induced NF-κB activity were due to the liposome-based cellular delivery method. We found that for C8 C-1-P, a BSA carrier-C-1-P formulation produced an effect similar to that produced by the liposomal administration: IC50 C8 C-1-P liposome = 9.1 ± 1.1 μm and IC50 C8 C-1-P-BSA = 13.7 ± 1.1 μm (Fig. 1B). For C16 C-1-P, liposomal administration proved a more potent inhibitor of NF-κB activity, which may reflect kinetic or spatial differences in lipid delivery: IC50 C16 C-1-P liposome = 3.1 ± 1.1 μm and IC50 C16 C-1-P-BSA = 35.9 ± 1.9 μm (Fig. 1B). Regardless, C-1-P limited NF-κB-mediated transcription in a dose-dependent manner (Fig. 1B). Thus, based upon the IC50 values, we treated cells with 10 μm liposomal C8 C-1-P for all remaining experiments. We also tested whether C-1-P had the same effect on a structurally homogenous agonist of TLR4. Indeed, C-1-P inhibited KdO2-lipid A-mediated NF-κB activation (Fig. 1C). The effect of C-1-P on NF-κB is specific to TLR4 signaling pathways because TNF-α-mediated NF-κB activation was not inhibited (Fig. 1D). Ceramide induced a modest increase in NF-κB reporter activity with 100 ng/ml TNF-α (Fig. 1D), consistent with a previous report that exogenous C2-ceramide could enhance TNF-α-stimulated NF-κB activity (23). In contrast, S-1-P did not have a measurable effect on TNF-α-stimulated NF-κB activation (Fig. 1D), although S-1-P is required for TRAF2 ubiquitin ligase-dependent NF-κB activation by TNF-α (24). Thus, we conclude that regardless of delivery modality, C-1-P, but not ceramide, dose-dependently reduces TLR4-mediated, but not TNF receptor-mediated, NF-κB activity.
FIGURE 1.
Ceramide-1-phosphate limits TLR4-mediated NF-κB activity, as measured by a SEAP reporter assay. A, ceramide-1-phosphate, but not ceramide, limits LPS-mediated NF-κB activation. HEK TLR4 cells transfected with the SEAP reporter vector were treated for 24 h with a 10 μm concentration of the indicated liposomal lipids with or without 100 ng/ml LPS. Ghost liposome serves as a vehicle control. Relative absorbance is a proxy for NF-κB-mediated transcription. B, inhibition of NF-κB activity is independent of the C-1-P delivery method and chain length. HEK TLR4 cells stably transfected with the SEAP reporter vector were treated for 24 h with 100 ng/ml LPS and varying concentrations of either liposomal C-1-P or C-1-P-BSA carrier. Relative absorbance normalized to LPS treatment is displayed as a function of C-1-P concentration and delivery method. IC50 was determined by GraphPad Prism 5 software using variable fit nonlinear regression. C, C-1-P inhibits Kdo2-lipid A-mediated NF-κB activation. HEK TLR4 cells stably transfected with the SEAP reporter vector were treated for 24 h with 1 μg/ml Kdo2-lipid A and/or liposomal 10 μm C16 C-1-P or lipid mass equivalent ghost liposome control. D, C-1-P does not affect NF-κB activation by TNF-α. HEK TLR4 cells stably transfected with the SEAP reporter vector were treated for 24 h with 100 ng/ml TNF-α and/or 10 μm liposomal lipids. Error bars, S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001. All experiments include a minimum of six replicates and were performed on at least three separate occasions.
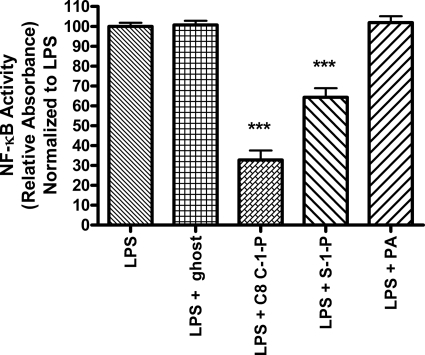
We next explored whether the effect of C-1-P administration upon NF-κB inhibition was dependent upon the phosphate moiety and/or the sphingolipid backbone structure. To this end, we found that phosphatidic acid, a diacylphosphoglycerolipid, had no effect on LPS signaling (Fig. 2), suggesting that the sphingoid backbone is necessary. Further supporting a requirement for the sphingoid backbone, S-1-P, like C-1-P, also inhibited NF-κB-mediated transcription (Fig. 2). Overall, C-1-P was more potent, decreasing LPS-induced NF-κB-mediated transcription by 2-fold over S-1-P at an equimolar concentration. These data support a phosphate group requirement, as ceramide (Fig. 1A) and sphingosine (data not shown) did not affect LPS-induced NF-κB signaling. Overall, these data demonstrate that both the phosphate moiety and the sphingoid backbone are required for inhibition of LPS-mediated NF-κB transcription in HEK TLR4 cells.
FIGURE 2.
The sphingolipid backbone and phosphate group are required for lipid inhibition of LPS-mediated NF-κB activation. HEK TLR4 cells stably expressing SEAP were treated for 24 h with 100 ng/ml LPS plus a 10 μm concentration of either C8 C-1-P liposome, S-1-P liposome, ghost vehicle control liposome, or phosphatidic acid (PA). NF-κB activation was measured by the SEAP reporter assay. All results shown are normalized to LPS treatment. Error bars, S.E. of at least three replicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
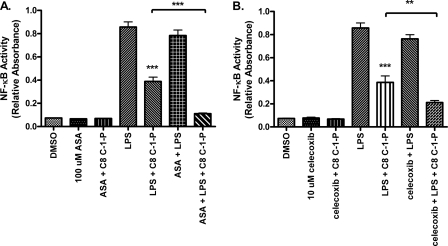
C-1-P has been well characterized as an activator of cPLA2, culminating in the production of proinflammatory arachidonic acid metabolites (25–27). To test whether inhibition of NF-κB via C-1-P was dependent on arachidonic acid metabolite production, we employed cyclooxygenase (COX) inhibitors, blocking a key intermediate in the production of prostanoids. We predicted that if arachidonic acid metabolites were responsible for the inhibition of LPS-activated gene transcription by C-1-P, inhibition of COX isoforms would restore full NF-κB activation by LPS, even in the presence of C-1-P. Instead, we found that the addition of acetylsalicylic acid, an irreversible COX-1 inhibitor, increased inhibition of LPS-mediated NF-κB reporter activity by C-1-P (Fig. 3A). Further, COX-2 inhibition with celecoxib also increased inhibition of LPS-mediated NF-κB reporter activity by C-1-P (Fig. 3B). We therefore ruled out a requirement for cyclooxygenase activation by C-1-P in limiting LPS-mediated NF-κB activity.
FIGURE 3.
Arachidonic acid metabolites are not required for inhibition of NF-κB by C-1-P. HEK TLR4 SEAP cells were pretreated with 100 μm aspirin (ASA) (A) or 10 μm celecoxib (B) for 1 h, followed by 24 h of treatment with 100 ng/ml LPS and/or 10 μm C8 C-1-P liposomes, as indicated. NF-κB activation was measured by the SEAP reporter assay. Error bars, S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001. All experiments include a minimum of six replicates and were performed on three separate occasions.
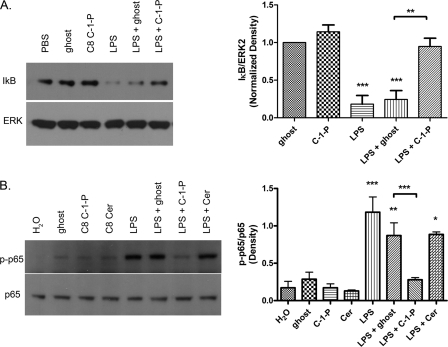
To understand the mechanism(s) by which C-1-P inhibits NF-κB, we probed upstream cytosolic regulatory events that mediate activation and nuclear translocation of NF-κB. NF-κB is a multisubunit transcription factor with many possible subunit combinations and post-translational control mechanisms. Induction of NF-κB transcription can occur when the cytosolic inhibitor IκB is targeted for proteosomal degradation. We monitored the effect of LPS-mediated IκB degradation by C-1-P. Consistent with the literature, we demonstrated that LPS treatment leads to rapid degradation of the IκB protein (Fig. 4A). Importantly, LPS-mediated IκB degradation was inhibited by C8 C-1-P (Fig. 4A). In HEK TLR4 cells, LPS treatment also leads to phosphorylation of the p65 subunit of NF-κB (Fig. 4B). To examine whether p65 activation was affected by C-1-P treatment, we monitored the phosphorylation status of p65. Western blotting demonstrated that concurrent treatment of C8 C-1-P with LPS attenuated p-p65 formation; however, concurrent ceramide or ghost liposome treatments did not (Fig. 4B). As a negative control, HEK 293 null cells, which are not transfected with TLR4, CD14, or MD-2, did not have increased p-p65 levels when treated with LPS, demonstrating TLR4-dependence (data not shown). To confirm the inhibition of phosphorylated p65 by C-1-P, we next employed a PhosflowTM approach to measure intracellular p65 activation. Flow cytometry to measure p-p65 also showed increased p-p65 levels with LPS treatment, which was attenuated by the addition of C8 C-1-P. Specifically, cells positive for p-p65 by treatment regimen were as follows: PBS = 31.0 ± 1.0%, LPS = 51.5 ± 0.5%, and LPS + C8 C-1-P = 40.0 ± 0.1%. Statistical analysis of n = 3 separate experiments revealed that LPS treatment was significantly different from PBS control and that LPS + C-1-P was significantly different from LPS treatment alone at the p < 0.05 confidence level. Thus, taken together, C-1-P, but not ceramide, limits TLR4-induced canonical NF-κB activation via inhibition of phosphorylation and activation of the p65 subunit, which is consistent with diminished IκB degradation.
FIGURE 4.
C8 C-1-P inhibits IκB degradation and phosphorylation of the NF-κB p65 subunit. Cells were treated with 10 μm ghost or C8 C-1-P liposomes with or without 100 ng/ml LPS for 45 min. IκB and ERK protein levels (A) or p-p65 levels (B) were measured by Western blotting. Densitometry results are displayed on the graph on the right. Blots are representative of at least three independently performed experiments. Densitometry was measured using ImageJ software. Error bars, S.E. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
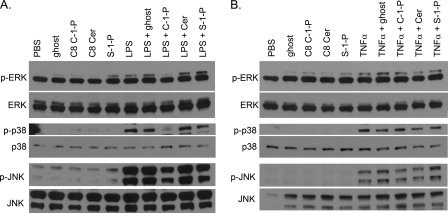
To aid in pinpointing where C-1-P blocked LPS signaling, we monitored the selectivity of sphingolipids to alter MAPK pathways known to be activated by LPS and TNF-α. After 45 min of treatment, LPS activated p-ERK1, p-p38, and p-JNK pathways, but concurrent treatment of 10 μm C8 C-1-P with LPS abrogated phosphorylation of all three MAPKs (Fig. 5A). Notably, none of the other liposomes, such as ghost liposome, ceramide, or S-1-P, were able to limit LPS-stimulated MAPK phosphorylation (Fig. 5A), demonstrating that C-1-P is a unique inhibitor of LPS signaling. In contrast, none of the liposomes were able to limit TNF-α stimulation of MAPKs at 15 min of treatment (Fig. 5B), at which time MAPK activation peaks in this cell line (data not shown). The inability of C-1-P to prevent activation of p-ERK1, p-p38, and p-JNK by TNF-α treatment is evidence that C-1-P discriminately inhibits MAPK signaling via TLR4. The S-1-P liposome, by itself, did increase levels of p-ERK1 and p-JNK at 45 min (Fig. 5A), which is probably caused by ligation of S-1-P with S-1-P receptors. Overall, we have demonstrated that C-1-P selectively inhibited LPS-mediated, but not TNF-α-mediated, MAPK activation and that neither ceramide nor S-1-P could reproduce this unique action of C-1-P.
FIGURE 5.
C8 C-1-P inhibits LPS-stimulated, but not TNF-α-stimulated, MAPK signaling. HEK TLR4 cells were treated with 100 ng/ml LPS with or without 10 μm liposomal lipids for 45 min (A) or with 100 ng/ml TNF-α ± 10 μm liposomal lipids for 15 min (B). Cell lysates were collected, and p-ERK, p-p38, and p-JNK protein were assessed relative to ERK, p38, and JNK protein by Western blotting. Blots are representative of at least three independently performed experiments.
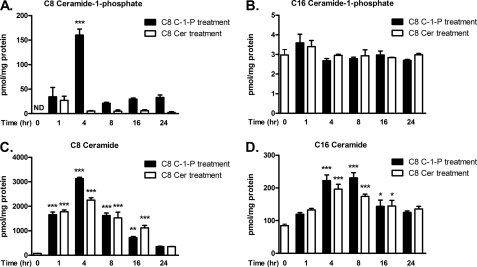
The use of exogenous sphingolipids can be complicated by lipid metabolism. To address this issue, we quantified sphingolipid mass following equimolar liposomal treatments to monitor lipid delivery and to assess global fluctuations in sphingolipid species. For Fig. 6, A–D, we treated HEK TLR4 cells with 10 μm liposomal C8 ceramide or C8 C-1-P lipids over a time course and assessed specific sphingolipid species by mass spectroscopy on separate graphs (as indicated above each graph). First, we confirmed cellular sphingolipid delivery by comparing C8 C-1-P and C8 ceramide mass under basal conditions and following exogenous liposomal treatment. We found that C8 C-1-P and C8 ceramide mass increased substantially following liposomal treatment, with peak concentrations achieved at 4 h (Fig. 6, A and C, respectively). Further, our data indicate that liposomal delivery of C8 C-1-P increases cellular C8 C-1-P mass for up to 24 h (Fig. 6A). Statistical analysis suggested that minimal conversion of C8 ceramide to C8 C-1-P took place following C8 ceramide treatment (Fig. 6A). In contrast, substantial conversion of C8 C-1-P to C8 ceramide was detected for up to 16 h (Fig. 6C), but liposomal C8 ceramide was unable to reproduce the actions of liposomal C8 C-1-P to inhibit LPS-stimulated cell signaling (Figs. 1A, 4B, and 5A). We also assessed physiological C16 C-1-P and C16 ceramide mass as a function of these exogenous lipid treatments. Treatment with liposomal C8 ceramide or C8 C-1-P induced no significant change in C16 C-1-P mass when compared with cells with no lipid treatment (Fig. 6B). Again, in contrast, C16 ceramide mass was increased from 4 to 16 h with both C8 ceramide and C8 C-1-P liposomal treatments; however, no significant difference between the liposomal lipid treatments was detected (Fig. 6D), again arguing that C16 ceramide accumulation may not be responsible for inhibition of NF-κB. Taken together, these mass spectroscopy data further support the unique actions of C-1-P, but not ceramide, to reduce TLR4-dependent NF-κB signaling, despite the significant conversion of C-1-P to ceramide.
FIGURE 6.
An analysis of ceramide and C-1-P mass and metabolism over time after exogenous sphingolipid treatment. HEK TLR4 cells were treated with 10 μm liposomal C8 ceramide or C8 C-1-P for 0, 1, 4, 8, 16, or 24 h. Lipids were extracted and quantified via mass spectrometry and normalized to total cellular protein. For clarity, the mass of each sphingolipid, over time, is presented in a separate graph, as indicated above each graph. Quantifications of C8 C-1-P (A), C16 C-1-P (B), C8 ceramide (C), and C16 ceramide (D) mass by treatment regimen and time are shown. Lipid mass is expressed as pmol of lipid/mg of protein. Error bars, S.E. of at least three replicates when compared with basal levels. *, p < 0.05; **, p < 0.01; ***, p < 0.001; ND, not detectable.
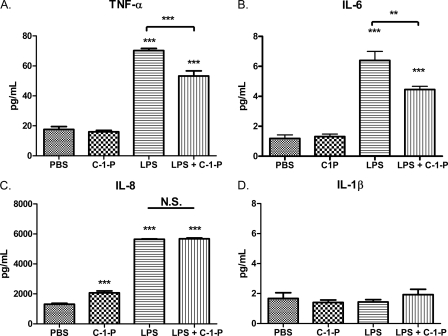
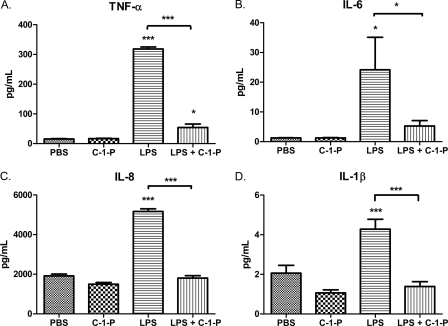
LPS stimulation of TLR4 ultimately leads to the production of cytokines and other inflammatory mediators. To test the potential of C-1-P to limit inflammation, we investigated cytokine secretion following concurrent 100 ng/ml LPS and 10 μm C-1-P treatment. To this end, we found that LPS-mediated TNF-α and IL-6 secretion by HEK TLR4 cells was reduced with C-1-P treatment (Fig. 7, A and B). C8 C-1-P alone induced a modest increase in IL-8 production, which may explain why C-1-P was unable to limit LPS-mediated IL-8 production (Fig. 7C). LPS did not stimulate IL-1β production in this cell line (Fig. 7D). We next assessed the therapeutic potential of liposomal C-1-P to reduce LPS-mediated cytokine secretion in a more physiological model, utilizing healthy donor human PBMCs. Indeed, we determined that concurrent treatment of 10 μm liposomal C-1-P with 100 ng/ml LPS reduced expression of proinflammatory cytokines when compared with LPS alone. C-1-P treatment dampened secretion of TNF-α, IL-6, IL-8, and IL-1β in LPS-stimulated human PBMCs (Fig. 8, A–D). Taken together, C-1-P reduced LPS-stimulated cytokine release in both a TLR4-expressing cell model system and human PBMCs.
FIGURE 7.
C-1-P selectively reduces cytokine expression in LPS-treated HEK TLR4 cells. HEK TLR4 cells were treated with 10 μm liposomal C-1-P, 100 ng/ml LPS, or C-1-P plus LPS for 24 h. The levels of TNF-α (A), IL-6 (B), IL-8 (C), and IL-1β (D) in conditioned medium were measured by ELISA. Error bars, S.E. of at least six replicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
FIGURE 8.
C-1-P selectively reduces cytokine expression in LPS-treated PBMCs. A, human PBMCs were treated with 10 μm liposomal C-1-P, 100 ng/ml LPS, or C-1-P + LPS for 24 h. The levels of TNF-α (A), IL-6 (B), IL-8 (C), and IL-1β (D) in conditioned medium were measured by ELISA. Error bars, S.E. of at least six replicates. *, p < 0.05; **, p < 0.01; ***, p < 0.001 as determined by analysis of variance except where indicated otherwise.
DISCUSSION
We have demonstrated that exogenous C-1-P limits NF-κB-mediated proinflammatory cytokine secretion in human PBMCs and in a defined HEK cell model of TLR4 activation. Inhibition of TLR4-activated NF-κB by C-1-P is independent of the C-1-P delivery method and acyl chain length because both C8 and C16 C-1-P in liposomal and BSA carrier formulations showed inhibition in the HEK TLR4 cells (Fig. 1B). We also found that both the sphingolipid backbone and the phosphate moiety are required for inhibition of TLR4-mediated NF-κB transcription, because C-1-P and S-1-P diminished NF-κB-dependent SEAP activity, whereas ceramide and phosphatidic acid did not (Fig. 2). Taken together, C-1-P, but not ceramide, reduces TLR4-dependent cytokine secretion by reducing the activation of NF-κB.
Using mass spectrometry, we validated that C-1-P is delivered effectively to cells via liposome, and we ruled out sphingolipid metabolites as the primary agents of NF-κB inhibition (Fig. 6). We did detect significant dephosphorylation of C8 C-1-P to C8 ceramide; however, the inability of ceramide to limit NF-κB-dependent reporter activity (Fig. 1A), p-p65 activation (Fig. 4B), and MAPK activation (Fig. 5A) argues that conversion to ceramide is not the mechanism by which the effects of C-1-P are transduced.
Although we detected inhibition of LPS-mediated NF-κB reporter activity by both C-1-P and S-1-P (Fig. 2), further investigation revealed that these phosphorylated lipid mediators differentially regulate downstream LPS-mediated signaling cascades. C8 C-1-P attenuated LPS-activated formation of p-ERK1, p-p38, and p-JNK, whereas S-1-P did not (Fig. 5A). Conceivably, it is possible that activation of S-1-P receptor signaling acted to mask any inhibitory effect of S-1-P on TLR4 MAPK signaling, but the evidence at hand argues that S-1-P and C-1-P differed mechanistically in their abilities to limit LPS signaling. Given that intracellular S-1-P is required for induction of TNF-α-stimulated NF-κB transcription, through TFAF2 ubiquitin ligase activity (24), we predicted that S-1-P might increase TNF-α-stimulated reporter expression. We did not detect any effect of S-1-P on TNF-α-activated SEAP activity, although such an effect may be discernible with lesser concentrations of TNF-α.
In aggregate, our data indicated that C-1-P reduced LPS-mediated, but not TNF-α-mediated, MAPK and NF-κB activation (Figs. 1D and 5, A and B). This demonstrates that the action of C-1-P is not an indirect effect on the NF-κB pathway, but is selective for TLR4 signaling. Specifically, C-1-P inhibited LPS-mediated degradation of IκB, preventing release of NF-κB from its inhibitor (Fig. 4A). Concomitant with the increased stability of IκB, C-1-P limits the phosphorylation of the p65 subunit induced by LPS by both Western blot (Fig. 4B) and flow cytometry (see “Results”). The functional consequence of the action of C-1-P to limit TLR4-dependent NF-κB activity was decreased secretion of proinflammatory cytokines in the defined HEK model and human PBMCs (Figs. 7 and 8).
Although C-1-P is known to regulate cPLA2 activity, our data indicate that C-1-P inhibition of NF-κB activation is independent of COX metabolites. Although C-1-P enhances the association of cPLA2 with intracellular membranes, allosterically activates cPLA2 (25, 26), and is required for cPLA2-mediated prostaglandin E2 production following inflammatory insult (27), the effect of C-1-P to inhibit NF-κB is independent of cPLA2-dependent prostanoids. However, the effects of C-1-P could still be manifested through lipoxygenase metabolites.
Despite documenting the upstream regulation of NF-κB phosphorylation and activity by C-1-P, the underlying biophysical and biochemical mechanisms or targets for C-1-P are still undefined. It is of interest that we document the selective actions of C-1-P, but not ceramide, in a defined cell model that overexpresses the canonical TLR4 signalosome, suggesting that C-1-P interacts with one or more of these elements. The structural similarity of C-1-P to LPS argues for an antagonistic action of C-1-P at the receptor level. However, this hypothesis is probably too simplistic, because LPS recognition goes far beyond the realm of the MD2, CD14, and TLR4 complex. Additional proteins are known to bind LPS, such as chemokine receptor 4, HSP70, HSP90, and growth differentiation factor 5 (28). The potential biophysical role of C-1-P to alter localization of these signalosome elements can only now be appreciated. In fact, it has been suggested that formation of differing clusters of these LPS receptors could lead to differential signaling via TLR4 (29), possibly by alterations in lipid microenvironments via bioactive lipids.
Our data are derived from exogenously administered C-1-P, which may or may not mimic the effect of endogenously produced C-1-P. Several lines of evidence argue that endogenous C-1-P may be an important modulator in the regulation of inflammation and restoration of homeostasis. In J774 macrophage cells, C24:1 C-1-P mass increased 30 min after LPS administration (20). LIPID MAPS-Nature Lipidomics Gateway data also demonstrate that Kdo2-lipid A treatment of RAW 264.7 macrophages leads to an increase of multiple C-1-P species over an 8–24-h time course. The increase in C-1-P, however, may be a transient event, because other groups have found that in RAW 264.7 cells, ceramide kinase mRNA and C-1-P specifically generated by ceramide kinase decrease after several h of LPS treatment (30, 31). We propose that fluctuations in C-1-P levels may initially switch cell functions to proinflammatory events, such as cPLA2 activation, and later provide an anti-inflammatory feedback mechanism to reset the system to homeostasis after insult. Exogenous C-1-P treatment may therefore mimic a transient rise in C-1-P that accompanies a return to homeostasis after insult. This could potentially explain the finding from another research group that pretreatment of J774 cells with exogenous C8 C-1-P was able to inhibit TNF-α production at low LPS concentrations (1 ng/ml) but not at higher LPS concentrations (100 ng/ml) (20). Regardless, fine control of cytokine production may lead to useful therapies that dampen chronic inflammatory processes while maintaining immune responsiveness to acute events.
In human PBMCs, C-1-P was able to abrogate the production of TNF-α, IL-6, IL-8, and IL-1β, suggesting that exogenous C-1-P may have the potential to limit positive feedback induced by TLR4-dependent cytokine secretion. Exogenous liposomal C-1-P administration may be a useful therapeutic strategy for disease states with dysregulated TLR signaling, such as sepsis, because it would contribute to the resolution of inflammation. Evidence exists that reduced C-1-P levels in ceramide kinase knock-out mice caused neutropenia and increased their susceptibility to Streptococcus pneumoniae infection (32). Unfortunately, the contribution of C-1-P to the resolution of inflammation still awaits clarification, because C-1-P persists in ceramide kinase-deficient mice, indicating the existence of alternative pathways for C-1-P generation (32, 33).
We believe that ceramide metabolism to C-1-P may account for the differences observed in studies investigating the role of ceramide in TLR4-dependent inflammatory cytokine signaling. For example, one report found that C8 ceramide decreased TNF-α and IL-1β production in macrophages but not in mast cells (34). Further, the authors found that C8 ceramide decreased IL-5, IL-10, and IL-13 in mast cells but not macrophages (34). Differential metabolism of ceramide to C-1-P or other sphingolipids may explain these findings. Alternatively, endogenous C-1-P may act to blunt ceramide accumulation in some cell types or contexts. C-1-P can counter ceramide production through both de novo and recycling pathways as demonstrated by inhibition of serine palmitoyltransferase in alveolar macrophages (35) and inhibition of acid sphingomyelinase in bone marrow-derived macrophages (36).
Our data indicate that, indeed, it may be C-1-P, and not ceramide, that reduces proinflammatory TLR4 signaling in our defined model. However, both ceramide and C-1-P can limit TLR4-dependent cytokine and chemokine release in neutrophils, macrophages, and corneal epithelial cells (19, 20). Discrepancies in the effect of ceramide on TLR4-mediated NF-κB transcription may reflect differential metabolism to C-1-P in various models. Regardless of the mechanisms responsible for tissue-specific selectivity, this accumulated body of work argues for an anti-inflammatory action of ceramide and ceramide metabolites. We have demonstrated that C-1-P modulates LPS signaling by limiting NF-κB activation and proinflammatory cytokine production in a TLR4 expression model and in human PBMCs. Based upon our results, the therapeutic potential of C-1-P to limit acute macrophage and neutrophil inflammatory responses in models of endotoxic shock deserves further study.
Acknowledgment
We acknowledge the Penn State Mass Spectrometry Core Facility for lipid mass measurements.
Note Added in Proof
During the preparation of this manuscript, another report provided evidence of an anti-inflammatory action of C-1-P, through the direct inhibition of TNF-α converting enzyme (37).
This work was supported, in whole or in part, by National Institutes of Health Grant R01HL076789 (to M. K.). The Penn State Research Foundation has previously licensed ceramide nanotechnologies to Keystone Nano Inc. (State College, PA); Dr. Kester is Chief Medical Officer of Keystone Nano Inc.
- TLR
- Toll-like receptor
- NF-κB
- nuclear factor-κB
- C-1-P
- ceramide-1-phosphate
- PBMC
- peripheral blood mononuclear cell
- Cer
- ceramide
- cPLA2
- cytosolic phospholipase A2
- S-1-P
- sphingosine-1-phosphate
- SEAP
- secreted alkaline phosphatase
- COX
- cyclooxygenase
- DSPC
- 1,2-distearoyl-sn-glycero-3-phosphocholine
- DOPE
- 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DSPE
- 1,2-distearoyl-sn-glycero-3-phosphoethanolamine
- Kdo
- 2-keto-3-deoxy-d-manno-octulosonic acid
- p-p65
- p-p38, p-ERK1, and p-JNK, phosphorylated p65, p38, ERK1, and JNK, respectively.
REFERENCES
- 1. Matzinger P. (1998) Semin. Immunol. 10, 399–415 [DOI] [PubMed] [Google Scholar]
- 2. Akira S., Takeda K. (2004) Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 3. Rifkin I. R., Leadbetter E. A., Busconi L., Viglianti G., Marshak-Rothstein A. (2005) Immunol. Rev. 204, 27–42 [DOI] [PubMed] [Google Scholar]
- 4. Wolska A., Lech-Marańda E., Robak T. (2009) Cell Mol. Biol. Lett. 14, 248–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Wittebole X., Castanares-Zapatero D., Laterre P. F. (2010) Mediators Inflamm. 2010, 568396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Poltorak A., He X., Smirnova I., Liu M. Y., Van Huffel C., Du X., Birdwell D., Alejos E., Silva M., Galanos C., Freudenberg M., Ricciardi-Castagnoli P., Layton B., Beutler B. (1998) Science 282, 2085–2088 [DOI] [PubMed] [Google Scholar]
- 7. Hoshino K., Takeuchi O., Kawai T., Sanjo H., Ogawa T., Takeda Y., Takeda K., Akira S. (1999) J. Immunol. 162, 3749–3752 [PubMed] [Google Scholar]
- 8. Wright S. D., Ramos R. A., Tobias P. S., Ulevitch R. J., Mathison J. C. (1990) Science 249, 1431–1433 [DOI] [PubMed] [Google Scholar]
- 9. Shimazu R., Akashi S., Ogata H., Nagai Y., Fukudome K., Miyake K., Kimoto M. (1999) J. Exp. Med. 189, 1777–1782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yamamoto M., Sato S., Hemmi H., Hoshino K., Kaisho T., Sanjo H., Takeuchi O., Sugiyama M., Okabe M., Takeda K., Akira S. (2003) Science 301, 640–643 [DOI] [PubMed] [Google Scholar]
- 11. Andreakos E., Sacre S. M., Smith C., Lundberg A., Kiriakidis S., Stonehouse T., Monaco C., Feldmann M., Foxwell B. M. (2004) Blood 103, 2229–2237 [DOI] [PubMed] [Google Scholar]
- 12. Kawai T., Akira S. (2007) Trends Mol. Med. 13, 460–469 [DOI] [PubMed] [Google Scholar]
- 13. Qi H. Y., Shelhamer J. H. (2005) J. Biol. Chem. 280, 38969–38975 [DOI] [PubMed] [Google Scholar]
- 14. Cuschieri J., Bulger E., Billgrin J., Garcia I., Maier R. V. (2007) Surg. Infect. 8, 91–106 [DOI] [PubMed] [Google Scholar]
- 15. Pfeiffer A., Böttcher A., Orsó E., Kapinsky M., Nagy P., Bodnár A., Spreitzer I., Liebisch G., Drobnik W., Gempel K., Horn M., Holmer S., Hartung T., Multhoff G., Schütz G., Schindler H., Ulmer A. J., Heine H., Stelter F., Schütt C., Rothe G., Szöllôsi J., Damjanovich S., Schmitz G. (2001) Eur. J. Immunol. 31, 3153–3164 [DOI] [PubMed] [Google Scholar]
- 16. MacKichan M. L., DeFranco A. L. (1999) J. Biol. Chem. 274, 1767–1775 [DOI] [PubMed] [Google Scholar]
- 17. Joseph C. K., Wright S. D., Bornmann W. G., Randolph J. T., Kumar E. R., Bittman R., Liu J., Kolesnick R. N. (1994) J. Biol. Chem. 269, 17606–17610 [PubMed] [Google Scholar]
- 18. Fischer H., Ellström P., Ekström K., Gustafsson L., Gustafsson M., Svanborg C. (2007) Cell Microbiol. 9, 1239–1251 [DOI] [PubMed] [Google Scholar]
- 19. Sun Y., Fox T., Adhikary G., Kester M., Pearlman E. (2008) J. Leukoc. Biol. 83, 1512–1521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Józefowski S., Czerkies M., Łukasik A., Bielawska A., Bielawski J., Kwiatkowska K., Sobota A. (2010) J. Immunol. 185, 6960–6973 [DOI] [PubMed] [Google Scholar]
- 21. Merrill A. H., Jr., Sullards M. C., Allegood J. C., Kelly S., Wang E. (2005) Methods 36, 207–224 [DOI] [PubMed] [Google Scholar]
- 22. Wijesinghe D. S., Allegood J. C., Gentile L. B., Fox T. E., Kester M., Chalfant C. E. (2010) J. Lipid Res. 51, 641–651 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Dbaibo G. S., Obeid L. M., Hannun Y. A. (1993) J. Biol. Chem. 268, 17762–17766 [PubMed] [Google Scholar]
- 24. Alvarez S. E., Harikumar K. B., Hait N. C., Allegood J., Strub G. M., Kim E. Y., Maceyka M., Jiang H., Luo C., Kordula T., Milstien S., Spiegel S. (2010) Nature 465, 1084–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Subramanian P., Stahelin R. V., Szulc Z., Bielawska A., Cho W., Chalfant C. E. (2005) J. Biol. Chem. 280, 17601–17607 [DOI] [PubMed] [Google Scholar]
- 26. Stahelin R. V., Subramanian P., Vora M., Cho W., Chalfant C. E. (2007) J. Biol. Chem. 282, 20467–20474 [DOI] [PubMed] [Google Scholar]
- 27. Lamour N. F., Subramanian P., Wijesinghe D. S., Stahelin R. V., Bonventre J. V., Chalfant C. E. (2009) J. Biol. Chem. 284, 26897–26907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Triantafilou K., Triantafilou M., Dedrick R. L. (2001) Nat. Immunol. 2, 338–345 [DOI] [PubMed] [Google Scholar]
- 29. Triantafilou M., Triantafilou K. (2002) Trends Immunol. 23, 301–304 [DOI] [PubMed] [Google Scholar]
- 30. Rovina P., Graf C., Bornancin F. (2010) Biochem. Biophys. Res. Commun. 399, 150–154 [DOI] [PubMed] [Google Scholar]
- 31. Rovina P., Jaritz M., Bornancin F. (2010) Biochem. Biophys. Res. Commun. 401, 164–167 [DOI] [PubMed] [Google Scholar]
- 32. Graf C., Zemann B., Rovina P., Urtz N., Schanzer A., Reuschel R., Mechtcheriakova D., Müller M., Fischer E., Reichel C., Huber S., Dawson J., Meingassner J. G., Billich A., Niwa S., Badegruber R., Van Veldhoven P. P., Kinzel B., Baumruker T., Bornancin F. (2008) J. Immunol. 180, 3457–3466 [DOI] [PubMed] [Google Scholar]
- 33. Mitsutake S., Yokose U., Kato M., Matsuoka I., Yoo J. M., Kim T. J., Yoo H. S., Fujimoto K., Ando Y., Sugiura M., Kohama T., Igarashi Y. (2007) Biochem. Biophys. Res. Commun. 363, 519–524 [DOI] [PubMed] [Google Scholar]
- 34. Chiba N., Masuda A., Yoshikai Y., Matsuguchi T. (2007) J. Cell Physiol. 213, 126–136 [DOI] [PubMed] [Google Scholar]
- 35. Granado M. H., Gangoiti P., Ouro A., Arana L., Gómez-Muñoz A. (2009) Biochim. Biophys. Acta 1791, 263–272 [DOI] [PubMed] [Google Scholar]
- 36. Gómez-Muñoz A., Kong J. Y., Salh B., Steinbrecher U. P. (2004) J. Lipid Res. 45, 99–105 [DOI] [PubMed] [Google Scholar]
- 37. Lamour N. F., Wijesinghe D. S., Mietla J. A., Ward K. E., Stahelin R. V., Chalfant C. E. (2011) J. Biol. Chem. 286, 42808–42817 [DOI] [PMC free article] [PubMed] [Google Scholar]