Background: Glial cell line-derived neurotrophic factor (GDNF) has been effective therapy in laboratory animal models of Parkinson disease.
Results: A human GDNFOS (GDNF opposite strand) gene was discovered and potentially encodes a 105-amino acid peptide.
Conclusion: GDNF sense and GDNFOS antisense genes are differentially regulated in human tissues.
Significance: GDNF and GDNFOS isoforms may have roles in human neurodegenerative diseases.
Keywords: Alternative Splicing, Alzheimer Disease, Evolution, Gene Regulation, Growth Factors, Huntington Disease
Abstract
Primate-specific genes and isoforms could provide insight into human brain diseases. Our bioinformatic analysis revealed that there are possibly five isoforms of human GDNF gene with different pre- and pro-regions by inter- and intra-exon splicing. By using TaqMan primer probe sets, designed between exons, we verified the expression of all isoforms. Furthermore, a novel GDNFOS gene was found to be transcribed from the opposite strand of GDNF gene. GDNFOS gene has four exons that are spliced into different isoforms. GDNFOS1 and GDNFOS2 are long noncoding RNAs, and GDNFOS3 encodes a protein of 105 amino acids. To study human GDNF and GDNFOS regulation in neurodegenerative diseases, the protein and mRNA levels were measured by Western blot and RT-quantitative PCR, respectively, in postmortem middle temporal gyrus (MTG) of Alzheimer disease (AD) and Huntington disease (HD) patients in comparison with those of normal controls. In the MTG of AD patients, the mature GDNF peptide was down-regulated; however, the transcript of GDNF isoform from human exon 2 was up-regulated, whereas that of the conserved isoform from exon 1 remained unchanged in comparison with those of normal controls. In contrast, the mature GDNF peptide and the isoform mRNA levels were not changed in the MTG of HD. The findings of novel GDNF and GDNFOS isoforms and differences in tissue expression patterns dysregulated in AD brains may further reveal the role of endogenous GDNF in human brain diseases.
Introduction
Glial cell line-derived neurotrophic factor (GDNF)4 was initially identified for its ability to promote survival of midbrain dopamine neurons (1), and its neurotrophic actions have been extensively studied in animal models of Parkinson disease (2). However, its actions are not specific for dopamine neurons; GDNF regulates neurite branching, synaptic plasticity, and phenotypes of several neuronal populations (3). Exogenous GDNF supports survival of noradrenergic neurons (4), spinal motor neurons (5, 6), peripheral sensory and autonomic neurons (7), forebrain cholinergic and GABAergic neurons (8), and pancreatic β-cells (9). Furthermore, GDNF protects the brain from ischemic injury (10) and ameliorates neuropathic pain (11). In peripheral tissues, GDNF promotes differentiation of kidney, lung, pancreas, germ cells, myocytes, and thymocytes and influences gastrointestinal inflammation and tumorigenesis (12–19).
GDNF is synthesized in a precursor form, pre-pro-GDNF, that is processed into the mature form, packaged into vesicles, and released upon neuronal activity (1). Previous studies have shown that the human and rodent GDNF genes have three exons encoding two mRNAs that are produced by alternative splicing of exon 2: pre-(α)long pro-GDNF and pre-(β)short-pro-GDNF with the (β)short isoform lacking 26 amino acids in the pro-region (7, 20–22). Recent studies have indicated that both forms are secreted from neurons, but secretion of the (β)short-pro-GDNF and its mature GDNF is activity-dependent, whereas (α)long pro-GDNF and its mature GDNF are secreted constitutively in an adrenal gland pheochromocytoma PC-6.3 cell line (23). Site-directed mutagenesis has shown that the pro-region and C-terminal cysteines are important for GDNF processing and secretion (24). Pre-pro-GDNF processing and secretion are not well studied with respect to the different isoforms, especially in humans where isoforms are more heterogeneous in pre-pro-regions than in those of the rodent. Using an expressed sequence tag (EST) data base search following RT-qPCR in different human tissues, we identified three additional human exons that produce GDNF isoforms and a cis-natural antisense transcript, termed GDNFOS, transcribed from the opposite strand of the GDNF gene (25). One of the GDNFOS isoforms encodes a potentially secreted peptide of 105 amino acids detected in human kidney cell lines and brain by Western blot analysis. Therefore, it may be that human and rodent GDNF isoforms differ in several aspects of the number of exons, alternative splicing, signal peptides, and cis-natural antisense transcript regulation, but also differ at the levels of expression in brain and peripheral tissue.
GDNF is known to be down-regulated in substantia nigra and putamen in human Parkinson disease (26–28); however, GDNF regulation in Alzheimer disease (AD) is less documented (29). A recent report indicated that GDNF concentrations are significantly up-regulated in cerebrospinal fluid and down-regulated in serum in patients with early AD (30). We selected human middle temporal gyrus (MTG) for GDNF study because it is a region consistently affected by Alzheimer disease (31), and neurons of the MTG are also affected in HD (32). We found that the mRNA deriving from human GDNF exon 2 was significantly increased, whereas the mature peptide was decreased in the MTG of AD patients in comparison with MTG of age-matched controls. To the best of our knowledge, this is the first investigation of regulation of human GDNF and GDNFOS isoforms and mature GDNF peptide in the brains of AD and HD patients
EXPERIMENTAL PROCEDURES
Bioinformatic Analysis of GDNF and GDNFOS Isoforms
Based on our study of the role of GDNF in incubation of heroin craving (33), we found that the human GDNF gene contains more spliced ESTs in the University of California, Santa Cruz genome browser (http://genome.ucsc.edu/) than rat. We searched the human EST data base (http://www.ncbi.nlm.nih.gov/dbEST/index.html) to identify additional human GDNF exons. We found human exons 1, 4, and 6 to be homologous to rat exons 1, 2, and 3, respectively. Using 0Sequencher software (Gene Code Corporation, Ann Arbor, MI), we identified additional human exons 2, 3, and 5 that are aligned to the EST clones and human GDNF genomic sequence. Human GDNF isoform nucleotide sequences were translated into amino acid sequences using the ExPASy translation tool (http://us.expasy.org/tools/dna.html), and the peptide sequences were aligned using ClustalW software (http://www.ebi.ac.uk/Tools/msa/clustalw2/). In addition, we also identified a natural antisense gene (GDNFOS for GDNF opposite strand) transcribed from the antisense strand of GDNF gene by EST sequence alignments and Sanger sequencing (Eurofins, Huntsville, AL) of three IMAGE human cDNA clones. The pre-pro-GDNF domains and potential post-translational modification sites were identified by InterProScan (http://www.ebi.ac.uk/interpro/). Based on the possible isoform differences between rat and human, the evolutionary analysis of the splicing junctions was carried out in UCSC syntenic genomic alignment using BLASTZ followed by chaining and netting pipeline (34–36). The protein domains of potential ORF of GDNFOS3 were analyzed by ExPaSy proteomic tools (http://us.expasy.org/tools/#proteome).
Human Postmortem Brain Samples and Rat Tissues
Post mortem MTG samples of controls, AD, and HD (supplemental Table S1, A and B) subjects were obtained from the Department of Pathology of the Division of Neuropathology at Johns Hopkins University School of Medicine, where diagnoses were all confirmed by autopsy (37), and the samples were qualified for an exemption under 45 CFR 46.101 of the DHHS criteria. The RNA integrity of each sample was analyzed by the Agilent RNA 6000 Nano kit with Bioanalyzer (Agilent Technologies). Human brain and peripheral tissue cDNAs were synthesized using pooled total RNA purchased from Clontech. Human pancreas islets were obtained from the Integrated Islet Distribution Program (NIDDK, National Institutes of Health). Male Sprague-Dawley rats (300–400 g; Charles River) were sacrificed by decapitation, and the brains were rapidly frozen in −50 °C isopentane solution and stored in a freezer at −80 °C. Tissue punches of the brain regions were taken from 1-mm coronal sections cut in a cryostat at −20 °C. The peripheral tissues were dissected on ice and immediately frozen by dry ice. The experimental procedures followed the guidelines of the Principles of Laboratory Care (National Institutes of Health publication 86-23, 1996) and were approved by the National Institute on Drug Abuse Animal Care and Use Committee (animal protocol number 07-BNRB-54).
Quantitative RT-PCR Analysis of GDNF Isoforms
Total RNAs were isolated using the TRIzol reagent, and single-stranded cDNAs were synthesized using the Superscript III first strand cDNA synthesis kit according to the manufacturer's protocols (Invitrogen). TaqMan probes (see Fig. 1 and supplemental Table S2) were designed using Primer Express 3.0 (Applied Biosystems, Invitrogen) at the splicing junctions of the different human and rat GDNF and GDNFOS isoforms for RT-qPCR analysis. The rat endogenous control was Fam-labeled Ube2i (supplemental Table S2; synthesized by Integrated DNA Technologies, Inc., Coralville, IA). The human endogenous control was Fam-labeled β-actin (Applied Biosystems). The RT-qPCR assays were carried out with Advanced TaqMan® Fast Universal PCR Master Mix in a 7500 Fast TaqMan instrument (Applied Biosystems, Invitrogen) using a default thermo-cycling program. Technical duplicates and triplicates were carried out in the qPCR assays. Twenty human AD, 8 HD, and 19 and 8 control middle temporal gyrus samples for AD and HD (supplemental Table S1, A and B), respectively, were assayed to compare expression of human GDNF and GDNFOS isoforms. Cycle threshold (Ct) values of more than 36 represented very low mRNA levels and were therefore deleted from the analysis. Mann-Whitney U tests from Prizm software (GraphPad Software, La Jolla, CA) were used to analyze the RT-qPCR data.
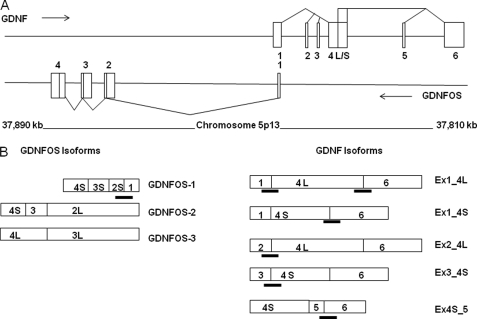
FIGURE 1.
A, human GDNF/GDNFOS genomic locus, gene structures, and splicing patterns. The boxes represent exons, the lines represent introns, and the triangles represent splicing patterns. B, transcripts of GDNF and GDNFOS isoforms and TaqMan probe designs. The boxes represent spliced exons, and the TaqMan probes are marked between exon junctions.
Western Blot Analysis of GDNF and GDNFOS
A polyclonal goat antibody for GDNF (AF-212-NA; R&D Systems, Minneapolis, MN) was used for Western blot to detect GDNF mature peptide (33). We also tried commercially available GDNF antibodies of Santa Cruz Biotechnology B-8 (sc-13147), D-20 (sc-328; Santa Cruz, CA), MAB212 (R & D Systems, Minneapolis, MN), and ANT-014 (Alomone Labs, Jerusalem, Israel). Although those antibodies recognized purified recombinant GDNF dimer and monomer, none of them detected specific bands of the dimer (32 kDa) and monomer (16 kDa) in rat tissue extracts (supplemental Fig. S1A), whereas the goat GDNF antibody (AF-212-NA; R & D Systems, Minneapolis, MN) recognized the specific GDNF dimer and monomer of rat tissue extracts (supplemental Fig. S1B). An affinity-purified anti-GDNFOS3 antibody was developed by injecting rabbit with epitope peptide (CKGMSHGQHFTHT) located at the C terminus (Genemed Synthesis, Inc., San Antonio, TX) and was used for Western blot of HEK293 and SH-SY5Y, CHO cell lines, and tissues of human and rat. The Western blocking solutions for GDNF and GDNFOS were 5 and 7.5% nonfat milk, respectively, to show the specific immunostainings. Student's t test was used to analyze the Western blot data.
RESULTS
Human GDNF Gene Has Six Exons and Multiple Isoforms
A search of the human dbEST data base followed by RT-qPCR analysis identified six exons in the human GDNF gene (Fig. 1A). A comparison with the rat GDNF gene structure reveals that three exons, human GDNF exons 1, 4-short, 4-long, and 6, are conserved with rat GDNF exons 1, 2-short, 2-long, and 3, respectively. The human GDNF exon 2, 3, and 5 sequences are aligned (Fig. 1) with human EST clones AJ001897, AJ001899, and DQ235474, respectively. The original translation initiation methionine of the GDNF is located in human exon 4. InterProScan (http://www.ebi.ac.uk/Tools/InterProScan/) indicates that the GDNF isoform with exon 2 (Ex2_4L)5 contains another initiation codon that translates into a 35-amino acid signal peptide in contrast to GDNF isoforms with exons 1 or 3 (Ex1_4S/L and Ex3_4S, respectively), each of which contains 18 amino acid signal peptides (supplemental Fig. S2). The insertion of human exon 5 between exons 4-short and 6 creates a truncated GDNF isoform (Ex4S_5) that potentially utilizes the downstream initiation methionine encoded by exon 6. The truncated GDNF isoform Ex4S_5 contains only 20 amino acids of the pro-region before the furin endoproteinase cleavage site, and InterProScan indicated that the 20-amino acid sequence is not a signal peptide. Therefore, human GDNF isoforms of Ex1_4L, Ex1_4S, Ex3_4S, Ex2_4L, and Ex4S_5 encode 211, 185, 185, 228, and 159 amino acids, respectively, and the peptide sequence differences among the isoforms are located in the pre-pro-regions of GDNF. All of the human GDNF isoforms encode the identical mature GDNF peptide (supplemental Fig. S2).
We investigated human GDNF isoform expression patterns in brain regions and peripheral tissues using isoform-specific TaqMan-DNA minor grove binding protein probes (Fig. 1 and supplemental Table S2). We used the human caudate hGDNF-βS mRNA as a reference. The human hGDNF-αL mRNA levels were highest in intestine, kidney, pancreatic islet, caudate, and putamen, whereas the hGDNF-βS mRNA levels were highest in intestine and kidney (Table 1).
TABLE 1.
Human GDF isoform expression using hGDNF-βS in caudate as a reference
CAU, caudate; PUT, putamen; NAC, nucleus accumbens; SNR, substantia nigra; CTX, cortex; AMG, amygdala; HIP, hippocampus; HTH, hypothalamus; ISL, pancreatic islets; INT, intestine; KID, kidney; MUS, muscle.
| Isoform | CAU | PUT | NAC | SNR | CTX | AMG | HIP | HTH | ISL | INT | KID | MUS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ex1_4S/L | 4.56 | 5.47 | 2.36 | 1.97 | 1.49 | 0.99 | 1.39 | 2.19 | 8.04 | 17.88 | 17.01 | 0.54 |
| Ex2_4La | 0.47 | 0.28 | 0.94 | 0.45 | 0.50 | 0.06 | 0.11 | 0.12 | 12.35 | 0.63 | 0.77 | 0 |
| Ex3_4Sa | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0.24 | 0.56 | 0 |
| Ex4S_5a | 0 | 0.39 | 0.26 | 0 | 0.29 | 0.35 | 0 | 0.40 | 0 | 0.52 | 0 | 0 |
| hGDNF-αL | 50.63 | 35.35 | 19.17 | 21.72 | 10.82 | 3.67 | 7.33 | 4.89 | 86.56 | 40.47 | 30.41 | 0.15 |
| hGDNF-βS | 1 | 1.47 | 0.76 | 0.51 | 0.56 | 0.49 | 0.41 | 0.50 | 1.06 | 7.29 | 4.31 | 0.11 |
| Ratio α:β | 50.6 | 24.1 | 25.2 | 42.6 | 19.3 | 7.5 | 17.9 | 9.8 | 81.7 | 5.6 | 7.1 | 1.4 |
a Primate-specific isoforms.
RT-qPCR showed that the human GDNF exon 1 (Ex1_4L/S) transcript level is highest in brain and peripheral tissues except for pancreatic islets where exon 2 (Ex2_4L) transcript is higher. The exon 3 (Ex3_4S) transcript was not found in most tissues except for low levels in intestine and kidney. The exon 5 (Ex4S_5) transcript was either not detected or was at very low levels in putaman, nucleus accumbens, prefrontal cortex, amygdala, hypothalamus, and intestine (Table 1). The ratios of the human GDNF α-long and β-short isoforms were generally higher in brain regions (except hypothalamus and amygdala) and pancreatic islets than the ratios of α-long and β-short in peripheral tissues (Table 1).
Comparison of Rat GDNF Isoform mRNA and Protein Tissue Expression
Using specific TaqMan probes for rGDNF-αL and rGDNF-βS isoforms (supplemental Table S2) and for reference the rGDNF-βS in rat dorsal striatum, we found that mRNA for rGDNF-αL was expressed severalfold higher than that of the rGDNF-βS across brain regions and peripheral tissues. The highest mRNA expressions were in dorsal striatum, nucleus accumbens, ovary, lung, and stomach, and lower levels were in prefrontal cortex and amygdala (Table 2). Interestingly, the human α-long and β-short ratios are several folds higher than rat α-long and β-short ratios (Tables 1 and 2). Because the long pre-(α)long proGDNF isoform contains the dopamine neuron-stimulating peptide-11 (DNSP-11) (38, 39), this 11-mer peptide-encoded transcript is expressed at a higher level in human than rat brain (supplemental Fig. S2).
TABLE 2.
Rat GDNF isoform expression (n = 3) using rGDNF-βS in dorsal striatum as a reference
DST, dorsal striatum; PFC, prefrontal cortex; STM, stomach; TES, testis; LNG, lung; OVA, ovary; SPL, spleen.
| Isoform | DST | NAC | PFC | AMG | STM | TES | LNG | OVA | SPL |
|---|---|---|---|---|---|---|---|---|---|
| rGDNF-αL | 4.67 ± 0.46 | 2.63 ± 0.24 | 0.66 ± 0.05 | 0.82 ± 0.16 | 1.31 ± 0.01 | 0.47 ± 0.22 | 2.25 ± 0.12 | 2.97 ± 0.55 | 0.29 ± 0.05 |
| rGDNF-βS | 1.00 ± 0.09 | 0.65 ± 0.05 | 0.11 ± 0.01 | 0.17 ± 0.04 | 0.29 ± 0.04 | 0.19 ± 0.11 | 0.38 ± 0.03 | 0.56 ± 0.06 | 0.05 ± 0.01 |
| Ratio α:β | 4.7 ± 0.39 | 4.0 ± 0.11 | 6.0 ± 0.67 | 4.8 ± 0.52 | 4.5 ± 0.69 | 2.5 ± 0.77 | 5.9 ± 0.23 | 5.3 ± 0.41 | 5.8 ± 0.60 |
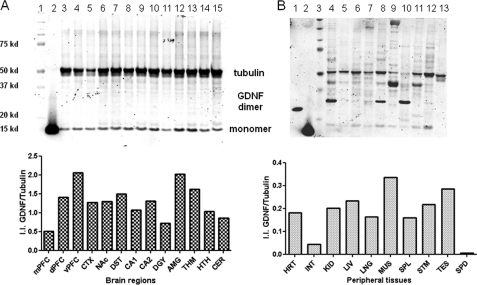
We used Western blot analysis to detect both GDNF monomers and dimers using a goat polyclonal antibody (33). We found that levels of mature GDNF monomer were higher in ventral prefrontal cortex and amygdala; lower in hippocampus CA1, CA2 and dentate gyrus than in other brain regions. We also found the levels of the monomer were higher in muscle and testis than in other peripheral tissues (Fig. 2). The 32-kDa band was observed in rat heart, lung, and spleen, and the 48-kDa band was observed in heart, muscle, and stomach. Because the Western blots were carried out in the same conditions for both brain and peripheral tissues, the formation of GDNF oligomerization could be stronger in peripheral tissues than that of brain (Fig. 2B in reduced and supplemental Fig. S1B in nonreduced conditions). Western blot analysis showed that GDNF peptide levels did not correlate with mRNA levels in brain regions where the striatal levels were several times higher than that of prefrontal cortex. The discordance between GDNF mRNA and the mature peptide in different brain regions could be due to cell type-specific expression of GDNF isoforms and transportation of mature peptide along neuronal processes (40).
FIGURE 2.
A, rat mature GDNF peptide expression in brain regions. First lane, precision molecular mass marker; second lane, recombinant GDNF under reduced conditions; third lane, medial prefrontal cortex; fourth lane, dorsal prefrontal cortex; fifth lane, ventral prefrontal cortex; sixth lane, temporal cortex; seventh lane, nucleus accumbens; eighth lane, dorsal striatum; ninth lane, hippocampus CA1; tenth lane, hippocampus CA2&3; eleventh lane, hippocampus dentate gyrus; twelfth lane, amygdala; thirteenth lane, thalamus; fourteenth lane, hypothalamus; fifteenth lane, cerebellum. The y axis of the bar graph represents normalized (tubulin) integrated intensity (I.I.) of Li-Cor Odyssey scan. B, lane 1, recombinant GDNF under nonreduced condition; lane 2, recombinant GDNF under reduced conditions; lane 3, precision molecular mass marker; lane 4, heart; lane 5, intestine; lane 6, kidney; lane 7, liver; lane 8, lung; lane 9, muscle; lane 10, spleen; lane 11, stomach; lane 12, testis; lane 13, spinal cord. The y axis of the bar graph represents normalized (tubulin) integrated intensity (I.I.) of Li-Cor Odyssey scan.
GDNFOS Is Transcribed from the Opposite Strand of the Human GDNF Gene
By searching the human dbEST data base followed by RT-qPCR analysis and sequencing of the IMAGE cDNA clones, a cis-natural antisense transcript (GDNFOS) gene was identified from the opposite strand of GDNF (Fig. 1A). The GDNFOS gene contains four exons and three initiation sites for transcription, i.e. exon 1, exon 2-long, and exon 3-long for GDNFOS1, GDNFOS2, and GDNFOS3, respectively (Fig. 1B).
The first exon of the GDNFOS1 isoform has 136 nucleotides of reverse complementarity (head-to-head configuration) (25) to the 5′-UTR of the GDNF isoform Ex1_4L/S (Fig. 1A). Exon 1 of the GDNFOS1 isoform is spliced to the exon 2-short, exon 3-short (intra-exonal splicing events), and exon 4-short (alternative poly adenylation event). Exon 2-long of GDNFOS2 isoform is spliced to exon 3-short and exon 4-short. Exon 3-long of GDNFOS3 isoform is spliced to exon 4-long (alternative poly adenylation event; Fig. 1B). The GDNFOS2 and GDNFOS3 transcripts do not overlap with the GDNF sense transcripts. IMAGE clones of 277201, 1762317, and 1637129 were sequenced in two directions to obtain the sequences of GDNFOS1, GDNFOS2, and GDNFOS3 (GenBankTM accession numbers JF824130, JF824131, and JF824129, respectively).
GDNFOS1 and GDNFOS2 IMAGE clones contain the upstream poly(A) site to produce 618- and 2,944-bp transcripts, respectively. GDNFOS1 and GDNFOS2 are predicted long noncoding RNAs (lncRNAs) (41), with no ORF of more than 60 amino acids. The GDNFOS3 IMAGE clone contains the downstream alternative adenylation site to produce a 1,963-bp transcript that potentially encodes an ORF of 105 amino acids (Fig. 3). The calculated molecular mass and isoelectric point of the ORF are 11.86 kDa and pI = 8.47, respectively. The proteome and pathway analysis found that the ORF contains an N-terminal 28-amino acid signal peptide, an internal asparagine-glycosylation site, two cysteines, an endoplastic reticulum targeting sequence (Predotar, http://urgi.versailles.inra.fr/predotar/predotar.html) and in a secreted pathway (Secretome 2.0 server, http://www.cbs.dtu.dk/services/SecretomeP/). There is a single furin-type cleavage site after the signal peptide between residues 28 and 29, and the mature peptide contains interface amino acids of Pro, Ile, Tyr, Trp, and Arg involved in protein-protein interactions (42).
FIGURE 3.
GDNFOS3 nucleotide and peptide sequences. The nucleotide sequence of ORF is represented by bold uppercase letters, and the noncoding sequence is represented by lowercase letters. The nucleotide numbers are marked on the right side, and the amino acid numbers are on the left side. The signal peptide and the consensus glycosylation site sequences are underlined, and the alternative polyadenylation signals are in bold and underlined.
Human Tissue Expression of GDNFOS
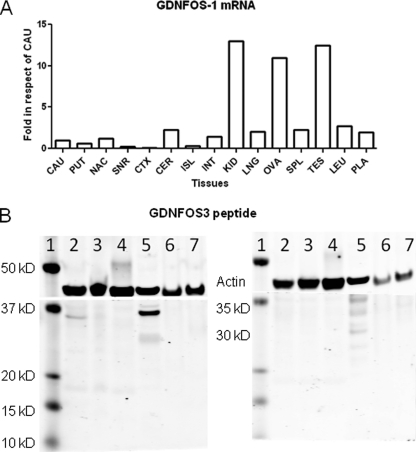
The exon-intron junctions of human GDNFOS are conserved at splicing donor (GT) and acceptor (AG) sites, and the first large intron (31,778 bp) is followed by small introns of 959 and 436 bp for intron 2 and 3, respectively. A TaqMan probe (Fig. 1B and supplemental Table S2) was designed at the splicing junction of GDNFOS gene exons 1 and 2 to investigate the tissue expression patterns of the GDNFOS1 transcript. The highest mRNA levels of GDNFOS1 were found in kidney, ovary, and testis, where GDNFOS1 mRNA is more than 10-fold of that of brain caudate region; higher expression was also observed in cerebellum and nucleus accumbens relative to other brain areas (Fig. 4A). The transcriptional level of GDNFOS1 is lower than that of GDNF isoform Ex1_4L in all brain regions (data not shown).
FIGURE 4.
A, GDNFOS1 mRNA levels in human brain region and peripheral tissue. The abbreviations are the same with the Western blot. B, GDNFOS3 peptide levels in human, hamster, and rat cell lines and tissues. Lane 1, molecular mass marker; lane 2, HEK293 cell; lane 3, SH-SY5Y cell; lane 4, CHO cell; lane 5, human MTG; lane 6, rat kidney; lane 7, rat prefrontal cortex. The left gel is the immunostaining with GDNFOS3 antibody, and the right gel is preabsorption with GDNFOS3 antigenic peptide before incubation with the primary antibody.
The affinity-purified antibody against the C-terminal peptide (CKGMSHGQHFTHT) of GDNFOS3 recognized a 35-kDa band in Western blot (7.5% dry milk blocking solution) of human embryonic kidney (HEK293) and MTG but not in SH-SY5Y and CHO cell lines, rat kidney, and prefrontal cortex (Fig. 4B, left panel). The 35-kDa band could be blocked in HEK293 and MTG by preincubation of the antibody with the antigenic peptide (Fig. 4B, right panel). The 35- and 30-kDa bands might be glycosylated or oligomeric form of GDNFOS3. More background was seen with less concentrated blocking solution (5% dry milk); however, the 30-kDa band was more intense, and 11-kDa bands could also be observed in human HEK293 and SH-SY5Y cells and MTG but not rat kidney and prefrontal cortex and CHO cells (data not shown).
Accelerated Primate Evolution of GDNF/GDNFOS Locus
The finding of human GDNF and GDNFOS isoforms may indicate that the locus is a primate accelerated region (43). We used comparative genomics of the mVISTA and UCSC genome browser of the 46-way vertebrate alignments (44, 45) to examine the conservation and the distribution of the splicing structure of GDNF and GDNFOS in different species and the ORFs on the vertebrate phylogenetic tree. The GDNF/GDNFOS locus predates the vertebrate split, i.e. it is largely shared across fish and humans but absent in invertebrates.
We manually removed insertions, deletions, or stop codons in nonhuman species and fed this codon-based alignment into CODEML (46). As a result, we found that exon 2 of human GDNF is only shared by primates given the narrow distribution of the exon donor sequence GT in these species. Species other than primate either do not contain the translation initiation codon (ATG) or lost their exon donor sequence (Table 3). InterProScan predicted that GDNF exon 2 encodes an extended and primate-specific signal peptide (MQSLPNSNGAAAGRDFK) in frame with exon 4. GDNF exon 3 appears to be shared by multiple mammalian groups given the existence of GT in outgroups, suggesting that it is an ancestral form. The splicing junctions (AG/GT) of GDNF exon 5 appear constrained in primate but divergent in other placental mammals. However, the exon 3 and 5 sequences were not found in EST databases of other species except for that of human.
TABLE 3.
GDNF exon 2 alignment of multiple species in reverse orientation
Amino acids (italic type) are represented as a single-letter code, the nucleotides are regular type, and splicing donor sites (GT) are in bold type. N represents sequences unavailable. Together with the start codon information, the new open reading frame introduced by exon 2 is primate-specific.
| Human | G | A | A | A | T | G | G | N | S | N | P | L | S | Q | M |
| Chimp | G | A | A | A | T | G | G | N | S | N | P | L | S | Q | M |
| Gorilla | G | A | A | G | T | G | G | N | S | N | P | L | S | Q | M |
| Orangutan | G | A | A | A | T | G | G | N | S | N | P | L | S | Q | M |
| Rhesus | G | A | A | A | T | G | G | N | S | N | P | L | S | Q | M |
| Baboon | G | A | A | A | T | G | G | N | S | N | P | L | S | Q | M |
| Marmoset | G | A | A | A | T | G | G | N | S | N | P | L | S | Q | I |
| Mouse lemur | G | A | A | A | T | G | G | N | S | N | P | L | S | Q | T |
| Bushbaby | G | A | A | A | T | G | G | K | S | N | P | L | S | Q | M |
| Tree shrew | G | A | A | A | T | G | G | N | S | N | P | L | S | Q | A |
| Mouse | A | A | A | A | T | G | G | S | N | T | P | P | S | Q | I |
| Rat | A | A | A | A | T | G | G | H | S | T | P | L | S | R | I |
| Kangaroo rat | A | A | A | G | G | G | G | H | G | P | P | S | P | Q | G |
| Guinea pig | A | C | G | G | C | C | K | P | P | L | P | L | L | ||
| Rabbit | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| Alpaca | G | A | A | A | T | C | S | D | S | N | P | L | S | Q | M |
| Dolphin | G | A | A | A | T | C | G | N | S | N | P | L | S | Q | R |
| Cow | G | A | A | A | T | C | G | N | C | N | P | L | S | Q | R |
| Horse | G | A | A | A | T | C | S | N | S | D | P | L | S | Q | L |
| Microbat | N | N | N | N | N | N | N | N | N | N | N | N | N | N | N |
| Megabat | G | A | A | A | T | C | G | K | S | N | P | L | S | Q | M |
| Shrew | G | G | A | A | G | G | G | G | R | S | P | L | S | R | M |
| Elephant | G | A | A | A | T | T | G | N | S | N | P | L | S | R | M |
| Rock hyrax | G | A | A | A | T | T | G | K | S | N | P | L | S | Q | I |
| Tenrec | G | A | C | C | C | A | R | N | S | D | P | L | S | P | L |
| Armadillo | G | A | A | A | T | G | G | N | S | N | P | F | S | Q | M |
GDNFOS exon 1 may be unique in hominoids (human and apes) given the occurrence of the AT-GT substitutions in the splicing donor site in monkey and other mammalian genomes (Table 4). Although GDNFOS exon 2, 3, and 4 splicing acceptor and donor sites are shared across mammals, we observed a TG-AG mutation in the splicing acceptor site of GDNFOS exon 3 in the marmoset genome. Because the ORF of GDNFOS3 was found in human, we should expect that the evolutionary branch toward human demonstrates a different ratio of nonsynonymous substitution rate and synonymous substitution rate (Ka/Ks) compared with other branches. The Ka/Ks value in human branch is 0.9, which demonstrates that GDNFOS is a hominoid young gene with coding potential (46). Based on the species genome alignment of UCSC, GDNFOS3 translation initiation codon (ATG) is disabled in opossum, dog, mouse, and orangutan, and numerous indels occur in rodents (supplemental Fig. S4). Therefore, GDNFOS3 peptide might be human- and chimpanzee-specific.
TABLE 4.
GDNFOS exon 1 alignment of multiple species
The splicing donor sites are marked by bold type.
| Human | G | A | A | G | G | A | G | A | G | G | T |
| Chimp | G | A | A | G | G | A | G | A | G | G | T |
| Gorilla | G | A | A | G | G | A | G | A | G | G | T |
| Orangutan | G | A | A | G | G | A | G | A | G | G | T |
| Rhesus | G | A | A | G | G | A | G | A | G | A | T |
| Baboon | G | A | A | G | G | A | G | A | G | A | T |
| Marmoset | G | A | A | G | A | A | G | A | G | G | T |
| Mouse lemur | G | A | A | G | G | A | G | A | G | A | T |
| Bushbaby | G | - | A | G | G | A | G | A | G | A | T |
| Tree shrew | G | G | A | G | G | A | G | C | G | A | T |
| Mouse | G | A | A | G | A | A | G | A | A | G | C |
| Rat | - | A | A | G | A | A | G | A | A | G | C |
| Kangaroo rat | A | A | A | G | G | A | G | A | G | A | T |
| Guinea pig | G | A | A | G | G | A | A | A | G | A | T |
| Squirrel | G | G | A | G | G | A | G | A | G | A | T |
| Rabbit | G | G | G | G | G | A | G | A | G | A | C |
Primate-specific GDNF Transcripts and Mature GDNF Peptide Are Dysregulated in MTG of AD
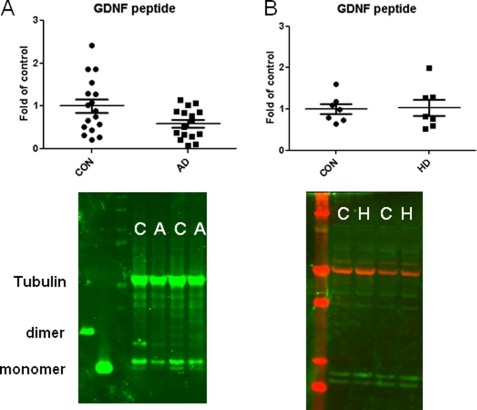
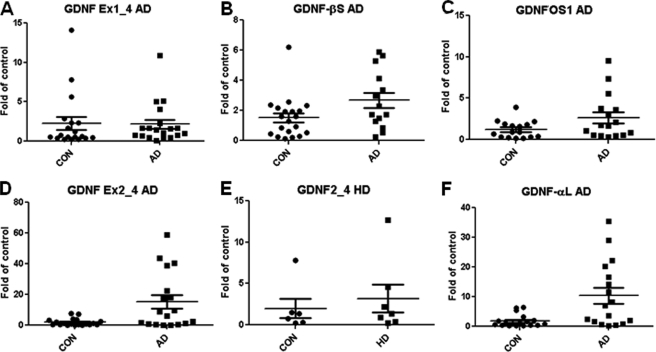
The RNA and protein qualities of the postmortem MTG samples of both controls and AD brains were tested by RNA integrity numbers (average RNA integrity number = 5.04; supplemental Table S1A) and integrity of a single tubulin band on Western blot (Fig. 5 and supplemental Fig. S3, A and B), respectively. A two-tailed Mann-Whitney for Gaussian approximation U test showed that there were no significant changes (Fig. 6, A–C) in the exon 1-driven isoform, pre-(β)short-proGDNF transcript, and GDNFOS1 transcript (Ex1_4S, p = 0.411; hGDNF-βS, p = 0.089 and GDNFOS1, p = 0.101, respectively) at the mRNA level; however, the primate exon 2-driven transcript (Ex2_4L) and pre-(α)long proGDNF transcript (hGDNF-αL) encoding DNSP-11 mRNAs were significantly increased more than 10-fold (Fig. 6, D and F) in MTG of AD in comparison with the matched controls (Ex2_4L, p = 0.013 and hGDNF-αL, p = 0.003). The exon 3-driven isoform mRNA (Ex3_4S) could not be detected in any of the MTG samples using RT-qPCR. Interestingly, an unpaired two-tailed Student t test showed that the mature GDNF peptide of 15 and 16 kDa decreased by 50% (Fig. 5A and supplemental Fig. S3A) in AD MTG in comparison with that of controls (p = 0.0241, t = 2.368, df = 32). We did not find any significant change at both GDNF isoform mRNA or mature peptide levels in the MTG of HD (Figs. 5B and 6E and supplemental Fig. S3B), further indication of the specificity for GDNF dysregulation in MTG of AD.
FIGURE 5.
Western blot analysis of GDNF mature peptide. A, GDNF mature peptide levels in control (C) and AD (A) MTG and scatter plot of AD MTG normalized by control. B, GDNF mature peptide levels in control (C) and HD (H) MTG and scatter plot of HD MTG normalized by control. The y axis is the percentage changes over control.
FIGURE 6.
GDNF and GDNFOS isoform mRNA levels in AD, HD, and control MTG. A, GDNF Ex1_4 in AD. B, GDNF-βS in AD. C, GDNFOS1 in AD. D, GDNF Ex2_4 in AD. E, GDNF Ex2_4 in HD. F, GDNF-αL in AD.
DISCUSSION
Human GDNF gene contains six exons whose alternative splicing creates five human GDNF isoforms (supplemental Fig. S2). We have also identified human GDNFOS isoforms transcribed from opposite strand of GDNF gene. GDNFOS1 is lncRNA and overlaps with the GDNF transcript. The GDNFOS2 transcript is also lncRNA and does not overlap with GDNF transcript. GDNFOS3 has a potential ORF that encodes a protein with no known homologue in GenBank. The GDNF transcript from exon 2 was up-regulated, and the mature GDNF peptide was down-regulated in MTG of AD. The tissue expression patterns of GDNF transcripts differ greatly between human and rat. The alternative intra-exon splicings of both human and rat GDNF produce α-long and β-short proforms differ in 26 amino acids. However, the splicing efficiency could be quite different between the species, given that the ratios of α-long and β-short pro-forms in human brain are much higher than the ratios of α-long and β-short pro-forms in rat brain. Because α-long GDNF isoforms potentially encodes DNSP-11 but not β-short GDNF isoforms (38, 39), the higher expression of the pre-pro(α)long GDNF isoforms in human brain implies also the higher production of DNSP-11. Dysregulation of human GDNF and DNSP-11 and GDNFOS may contribute vulnerability of AD pathogenesis.
The neuroprotective effect of memantine in Alzheimer disease may involve GDNF up-regulation (47), as well as inhibition of the NMDA receptor. In human MTG of AD patients, the reduced mature GDNF peptide secretion might be a feedback signal for the increased human-specific GDNF isoform mRNA expression to compensate for the loss of mature GDNF peptide secretion. On the other hand, DNSP-11 containing α-long isoform up-regulation might be a neuroprotective response of AD brain. We previously observed similar discordant mRNA and protein expression in AD brain for a human-specific de novo gene, C20orf203 (48). Because the RNA integrity numbers of the postmortem MTG samples varied between 2.2 and 7.7 with average RNA integrity number of 5.0, the observed GDNF isoform mRNA changes might not be accurately measured. Because the proteins are more stable in postmortem tissues (49) as indicated by an intact tubulin band in all of the samples (supplemental Fig. S3), the observed down-regulation of mature GDNF peptide should be more reliably measured.
BDNF and GDNF are two essential neurotrophic factors in vertebrates. BDNF plays important roles in neurodegenerative and addictive disorders (50). There are similarities and differences in the gene structures, differential species, tissue expression patterns, pre-pro-forms, and natural antisense transcripts encoded by their opposite DNA strands (51, 52). The BDNF gene alternative splicing generate 26-, 18-, and 33-amino acid signal peptides in BDNF isoforms transcribed from exons 1, 4, and 7, respectively (52, 53). The GDNF gene alternative splicing generates 18- and 36-amino acid signal peptides in GDNF isoforms transcribed from exons 1 and 2, respectively. It can be speculated that the longer signal peptides of BDNF and GDNF may be differential targeting signals in rough endoplasmic reticulum, leading to sequestration in specific secretary compartments of the trans-Golgi network to regulate pro- and mature peptide transport and secretion. Both human BDNF and GDNF loci contain the natural antisense transcripts of BDNFOS and GDNFOS genes (52, 53) that have reverse complementarity to their sense counterparts with 224 nucleotides and 136 nucleotides, respectively. BDNFOS overlaps with the BDNF coding exon within ORF and GDNFOS overlaps with the GDNF 5′-UTR exon (52). Both BDNFOS and GDNFOS genes are transcribed by RNA polymerase II, alternatively spliced, and polyadenylated (25); however, the initiation of BDNFOS transcription is derived from a single promoter (52, 53) composed of hundreds of alternatively spliced isoforms (53), whereas the initiation of GDNFOS gene transcription derives from three exons with fewer alternatively spliced isoforms. GDNFOS transcripts contain various retroposons of Alu, Line, MIRs, and endogenous retroviral sequences (ERVL-MalR, hAT-Charlie and TCMar-Tigger2; RepeatMasker: http://www.repeatmasker.org/), characteristically de novo genes. Unlike human-specific de novo protein coding gene, C20orf203, in which part of the ORF (194 amino acids) is formed by Alu repeats (48), the ORF of GDNFOS does not contain any repeat sequence (RepeatMasker). GDNFOS transcript overlapping with GDNF mRNA might influence the higher ratios of GDNF α-long and β-short proforms in human brain. The cis- and trans-lncRNAs are more species-specific than protein coding genes (25, 54). The GDNFOS3 peptide could indicate a young evolution event integrated into the pre-existing protein network of primates. Recently, Zhang et al. (55) have found that human young genes with novel functions are expressed in early development stages and likely contributed to the evolution of human neocortex.
The existence of primate lncRNAs for BDNFOS and GDNFOS suggests that human plasticity mechanisms controlled by neurotrophic factors evolved rapidly after the split of rodent and primate lineages. Humans are more vulnerable to AD than mice, who are resilient to the development of pathological plaques and tangles after knock-in of one of the dominant AD mutant genes such as PS1M146V, APPswe, and TauP301L (56). A tripartite regulation of GDNF, DNSP-11, and GDNFOS at both mRNA and protein levels might contribute to human intelligence and vulnerability to neurodegenerative diseases. The accelerated primate evolution of the GDNF/GDNFOS locus adds more elements of the gene regulation networks in human brain.
Acknowledgments
We appreciate insightful advice from Dr. Yavin Shaham, Dr. Amina Woods, and Dr. Barry Hoffer at Intramural Research Program/National Institute on Drug Abuse and Dr. Sherry Leonard of the Department of Psychiatry of the University of Colorado at Denver. We thank Dr. Florence Theberge for providing rat brain regional punches; Dr. Brandon Harvey and Dr. Teruo Hayashi for providing HEK293, SH-SY5Y, and CHO cell lines; Anna Stern for editing English; and Xingyu Hou for performing Western blot analysis of rat tissues at National Institute on Drug Abuse.
This work was supported, in whole or in part, by National Institutes of Health NIDA intramural research program. This work was also supported by the Johns Hopkins University Alzheimer's Disease Research Center (NIA P50AG051460), the Huntington's Research Center without Walls (NINDS PO1NS16375), the Integrated Islet Distribution Program, funded by the NIDDK and the JDRF.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables S1 and S2 and Figs. S1–S4.
The nucleotide sequence(s) reported in this paper has been submitted to the DDBJ/GenBankTM/EBI Data Bank with accession number(s) JF824129, JF824130, and JF824131.
The GDNF isoforms designations are as follows: Ex1_4S, GDNF isoform with inter-exon splicing of exon 1 to exon 4-short; Ex1_4L, GDNF isoform with inter-exon splicing of exon 1 to exon 4-long; Ex2_4L, GDNF isoform with inter-exon splicing of exon 2 to exon 4-long; Ex3_4S, GDNF isoform with inter-exon splicing of exon 3 to exon 4-short; Ex4S_5, GDNF isoform with inter-exon splicing of exon 4-short to exon 5; GDNF-αL, GDNF isoform with intra-exon splicing of exon 4-long; GDNF-βS, GDNF isoform with intra-exon splicing of exon 4-short.
- GDNF
- glial cell line-derived neurotrophic factor
- GDNFOS
- GDNF opposite strand
- BDNFOS
- BDNF opposite strand
- AD
- Alzheimer disease
- HD
- Huntington disease
- MTG
- middle temporal gyrus
- EST
- expressed sequence tags
- qPCR
- quantitative PCR
- DNSP-11
- dopamine neuron stimulating peptide-11
- lncRNA
- long noncoding RNA
- BDNF
- brain-derived neurotrophic factor.
REFERENCES
- 1. Lin L. F., Doherty D. H., Lile J. D., Bektesh S., Collins F. (1993) Science 260, 1130–1132 [DOI] [PubMed] [Google Scholar]
- 2. Chiocco M. J., Harvey B. K., Wang Y., Hoffer B. J. (2007) Parkinsonism Relat. Disord. 13, (Suppl. 3) S321–S328 [DOI] [PubMed] [Google Scholar]
- 3. Airaksinen M. S., Saarma M. (2002) Nat. Rev. Neurosci. 3, 383–394 [DOI] [PubMed] [Google Scholar]
- 4. Arenas E., Trupp M., Akerud P., Ibáñez C. F. (1995) Neuron 15, 1465–1473 [DOI] [PubMed] [Google Scholar]
- 5. Henderson C. E., Phillips H. S., Pollock R. A., Davies A. M., Lemeulle C., Armanini M., Simmons L., Moffet B., Vandlen R. A., Simpson L. (1994) Science 266, 1062–1064 [DOI] [PubMed] [Google Scholar]
- 6. Trok K., Hoffer B., Olson L. (1996) Neuroscience 71, 231–241 [DOI] [PubMed] [Google Scholar]
- 7. Trupp M., Rydén M., Jörnvall H., Funakoshi H., Timmusk T., Arenas E., Ibáñez C. F. (1995) J. Cell Biol. 130, 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Williams L. R., Inouye G., Cummins V., Pelleymounter M. A. (1996) J. Pharmacol. Exp. Ther. 277, 1140–1151 [PubMed] [Google Scholar]
- 9. Mwangi S., Anitha M., Mallikarjun C., Ding X., Hara M., Parsadanian A., Larsen C. P., Thule P., Sitaraman S. V., Anania F., Srinivasan S. (2008) Gastroenterology 134, 727–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y., Lin S. Z., Chiou A. L., Williams L. R., Hoffer B. J. (1997) J. Neurosci. 17, 4341–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Boucher T. J., Okuse K., Bennett D. L., Munson J. B., Wood J. N., McMahon S. B. (2000) Science 290, 124–127 [DOI] [PubMed] [Google Scholar]
- 12. Farhi J., Ao A., Fisch B., Zhang X. Y., Garor R., Abir R. (2010) Fertil. Steril. 93, 2565–2571 [DOI] [PubMed] [Google Scholar]
- 13. Kondo S., Kishi H., Tokimitsu Y., Muraguchi A. (2003) Eur. J. Immunol. 33, 2233–2240 [DOI] [PubMed] [Google Scholar]
- 14. Little M., Georgas K., Pennisi D., Wilkinson L. (2010) Curr. Top. Dev. Biol. 90, 193–229 [DOI] [PubMed] [Google Scholar]
- 15. Martinelli P. M., Camargos E. R., Morel G., Tavares C. A., Nagib P. R., Machado C. R. (2002) Histochem. Cell Biol. 118, 337–343 [DOI] [PubMed] [Google Scholar]
- 16. von Boyen G. B., Schulte N., Pflüger C., Spaniol U., Hartmann C., Steinkamp M. (2011) BMC Gastroenterol. 11, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Watanabe Y., Kim H. S., Castoro R. J., Chung W., Estecio M. R., Kondo K., Guo Y., Ahmed S. S., Toyota M., Itoh F., Suk K. T., Cho M. Y., Shen L., Jelinek J., Issa J. P. (2009) Gastroenterology 136, 2149–2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fromont-Hankard G., Philippe-Chomette P., Delezoide A. L., Nessmann C., Aigrain Y., Peuchmaur M. (2002) Arch. Pathol. Lab. Med. 126, 432–436 [DOI] [PubMed] [Google Scholar]
- 19. Lucini C., Maruccio L., Antonucci R., Castaldo L. (2008) Eur. J. Histochem. 52, 69–74 [DOI] [PubMed] [Google Scholar]
- 20. Grimm L., Holinski-Feder E., Teodoridis J., Scheffer B., Schindelhauer D., Meitinger T., Ueffing M. (1998) Hum. Mol. Genet. 7, 1873–1886 [DOI] [PubMed] [Google Scholar]
- 21. Matsushita N., Fujita Y., Tanaka M., Nagatsu T., Kiuchi K. (1997) Gene 203, 149–157 [DOI] [PubMed] [Google Scholar]
- 22. Suter-Crazzolara C., Unsicker K. (1994) Neuroreport 5, 2486–2488 [DOI] [PubMed] [Google Scholar]
- 23. Lonka-Nevalaita L., Lume M., Leppänen S., Jokitalo E., Peränen J., Saarma M. (2010) J. Neurosci. 30, 11403–11413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oh-hashi K., Ito M., Tanaka T., Hirata Y., Kiuchi K. (2009) Mol. Cell Biochem. 323, 1–7 [DOI] [PubMed] [Google Scholar]
- 25. Zhang Y., Liu X. S., Liu Q. R., Wei L. (2006) Nucleic Acids Res. 34, 3465–3475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bäckman C. M., Shan L., Zhang Y. J., Hoffer B. J., Leonard S., Troncoso J. C., Vonsatel P., Tomac A. C. (2006) Mol. Cell Endocrinol. 252, 160–166 [DOI] [PubMed] [Google Scholar]
- 27. Chauhan N. B., Siegel G. J., Lee J. M. (2001) J. Chem. Neuroanat. 21, 277–288 [DOI] [PubMed] [Google Scholar]
- 28. Hunot S., Bernard V., Faucheux B., Boissière F., Leguern E., Brana C., Gautris P. P., Guérin J., Bloch B., Agid Y., Hirsch E. C. (1996) J. Neural. Transm. 103, 1043–1052 [DOI] [PubMed] [Google Scholar]
- 29. Siegel G. J., Chauhan N. B. (2000) Brain Res. Brain Res. Rev. 33, 199–227 [DOI] [PubMed] [Google Scholar]
- 30. Straten G., Eschweiler G. W., Maetzler W., Laske C., Leyhe T. (2009) J. Alzheimers Dis. 18, 331–337 [DOI] [PubMed] [Google Scholar]
- 31. Schroeter M. L., Stein T., Maslowski N., Neumann J. (2009) Neuroimage 47, 1196–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Herndon E. S., Hladik C. L., Shang P., Burns D. K., Raisanen J., White C. L., 3rd (2009) J. Neuropathol. Exp. Neurol. 68, 250–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Airavaara M., Pickens C. L., Stern A. L., Wihbey K. A., Harvey B. K., Bossert J. M., Liu Q. R., Hoffer B. J., Shaham Y. (2011) Addict. Biol. 16, 261–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Kent W. J., Baertsch R., Hinrichs A., Miller W., Haussler D. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 11484–11489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schwartz S., Kent W. J., Smit A., Zhang Z., Baertsch R., Hardison R. C., Haussler D., Miller W. (2003) Genome Res. 13, 103–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Blanchette M., Kent W. J., Riemer C., Elnitski L., Smit A. F., Roskin K. M., Baertsch R., Rosenbloom K., Clawson H., Green E. D., Haussler D., Miller W. (2004) Genome Res. 14, 708–715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Troncoso J. C., Cataldo A. M., Nixon R. A., Barnett J. L., Lee M. K., Checler F., Fowler D. R., Smialek J. E., Crain B., Martin L. J., Kawas C. H. (1998) Ann. Neurol. 43, 673–676 [DOI] [PubMed] [Google Scholar]
- 38. Bradley L. H., Fuqua J., Richardson A., Turchan-Cholewo J., Ai Y., Kelps K. A., Glass J. D., He X., Zhang Z., Grondin R., Littrell O. M., Huettl P., Pomerleau F., Gash D. M., Gerhardt G. A. (2010) PLoS One 5, e9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Immonen T., Alakuijala A., Hytönen M., Sainio K., Poteryaev D., Saarma M., Pasternack M., Sariola H. (2008) Exp. Neurol. 210, 793–796 [DOI] [PubMed] [Google Scholar]
- 40. Wang J., Carnicella S., Ahmadiantehrani S., He D. Y., Barak S., Kharazia V., Ben Hamida S., Zapata A., Shippenberg T. S., Ron D. (2010) J. Neurosci. 30, 14502–14512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lipovich L., Johnson R., Lin C. Y. (2010) Biochim. Biophys. Acta 1799, 597–615 [DOI] [PubMed] [Google Scholar]
- 42. Sillerud L. O., Larson R. S. (2005) Curr. Protein Pept. Sci. 6, 151–169 [DOI] [PubMed] [Google Scholar]
- 43. Dorus S., Vallender E. J., Evans P. D., Anderson J. R., Gilbert S. L., Mahowald M., Wyckoff G. J., Malcom C. M., Lahn B. T. (2004) Cell 119, 1027–1040 [DOI] [PubMed] [Google Scholar]
- 44. Rhead B., Karolchik D., Kuhn R. M., Hinrichs A. S., Zweig A. S., Fujita P. A., Diekhans M., Smith K. E., Rosenbloom K. R., Raney B. J., Pohl A., Pheasant M., Meyer L. R., Learned K., Hsu F., Hillman-Jackson J., Harte R. A., Giardine B., Dreszer T. R., Clawson H., Barber G. P., Haussler D., Kent W. J. (2010) Nucleic Acids Res. 38, D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Rhead B., Karolchik D., Kuhn R. M., Hinrichs A. S., Zweig A. S., Fujita P. A., Diekhans M., Smith K. E., Rosenbloom K. R., Raney B. J., Pohl A., Pheasant M., Meyer L. R., Learned K., Hsu F., Hillman-Jackson J., Harte R. A., Giardine B., Dreszer T. R., Clawson H., Barber G. P., Haussler D., Kent W. J. (2010) Nucleic Acids Res. 38, D613–D619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yang Z. (2007) Mol. Biol. Evol. 24, 1586–1591 [DOI] [PubMed] [Google Scholar]
- 47. Wu H. M., Tzeng N. S., Qian L., Wei S. J., Hu X., Chen S. H., Rawls S. M., Flood P., Hong J. S., Lu R. B. (2009) Neuropsychopharmacology 34, 2344–2357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Li C. Y., Zhang Y., Wang Z., Zhang Y., Cao C., Zhang P. W., Lu S. J., Li X. M., Yu Q., Zheng X., Du Q., Uhl G. R., Liu Q. R., Wei L. (2010) PLoS Comput. Biol. 6, e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Stan A. D., Ghose S., Gao X. M., Roberts R. C., Lewis-Amezcua K., Hatanpaa K. J., Tamminga C. A. (2006) Brain Res. 1123, 1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Ghitza U. E., Zhai H., Wu P., Airavaara M., Shaham Y., Lu L. (2010) Neurosci. Biobehav. Rev. 35, 157–171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Liu Q. R., Lu L., Zhu X. G., Gong J. P., Shaham Y., Uhl G. R. (2006) Brain Res. 1067, 1–12 [DOI] [PubMed] [Google Scholar]
- 52. Liu Q. R., Walther D., Drgon T., Polesskaya O., Lesnick T. G., Strain K. J., de Andrade M., Bower J. H., Maraganore D. M., Uhl G. R. (2005) Am. J. Med. Genet. B Neuropsychiatr. Genet. 134B, 93–103 [DOI] [PubMed] [Google Scholar]
- 53. Pruunsild P., Kazantseva A., Aid T., Palm K., Timmusk T. (2007) Genomics 90, 397–406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Li J. T., Zhang Y., Kong L., Liu Q. R., Wei L. (2008) Nucleic Acids Res. 36, 4833–4844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Zhang Y. E., Landback P., Vibranovski M. D., Long M. (2011) PLoS Biol. 9, e1001179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Oddo S., Caccamo A., Shepherd J. D., Murphy M. P., Golde T. E., Kayed R., Metherate R., Mattson M. P., Akbari Y., LaFerla F. M. (2003) Neuron 39, 409–421 [DOI] [PubMed] [Google Scholar]