Background: Chromogranin B (CGB) is a buffer for calcium and a functional binding partner of the inositol 1,4,5-trisphosphate receptor (InsP3R).
Results: The domains of CGB differentially regulate initiation and duration of intracellular calcium signaling.
Conclusion: C-CGB activates intracellular calcium signaling and N-CGB binds to InsP3Rs.
Significance: This study demonstrates the importance of CGB in the modulation of InsP3Rs.
Keywords: Calcium Imaging; Calcium Intracellular Release; Inositol Phosphates; Membrane Bilayer; Vesicles; 1,4,5-Trisphosphate Receptor; Calcium Signaling; Chromogranin; Dense Core Vesicles; Intracellular Calcium Channels
Abstract
The versatility of intracellular calcium as a second messenger is seen in its ability to mediate opposing events such as neuronal cell growth and apoptosis. A leading hypothesis used to explain how calcium regulates such divergent signaling pathways is that molecules responsible for maintaining calcium homeostasis have multiple roles. For example, chromogranin B (CGB), a calcium binding protein found in secretory granules and in the lumen of the endoplasmic reticulum, buffers calcium and also binds to and amplifies the activity of the inositol 1,4,5-trisphosphate receptor (InsP3R). Previous studies have identified two conserved domains of CGB, an N-terminal domain (N-CGB) and a C-terminal domain (C-CGB). N-CGB binds to the third intraluminal loop of the InsP3R and inhibits binding of full-length CGB. This displacement of CGB decreases InsP3R-dependent calcium release and alters normal signaling patterns. In the present study, we further characterized the role of N-CGB and identified roles for C-CGB. The effect of N-CGB on calcium release depended upon endogenous levels of cellular CGB, whereas the regulatory effect of C-CGB was apparent regardless of endogenous levels of CGB. When either full-length CGB or C-CGB was expressed in cells, calcium transients were increased. Additionally, the calcium signal initiation site was altered upon C-CGB expression in neuronally differentiated PC12 and SHSY5Y cells. These results show that CGB has numerous regulatory roles and that CGB is a critical component in modulating InsP3R-dependent calcium signaling.
Introduction
Chromogranin B (CGB)5 belongs to the granin family and is a low affinity, high capacity calcium binding protein found in hormone storing organelles, the nucleus, and the endoplasmic reticulum (ER) of excitable and non-excitable cells (1). Expression of CGB induces de novo secretory granule biogenesis (2), regulates transcription of many genes, and binds to and modulates the activity of the inositol 1,4,5-trisphosphate receptor (InsP3R) (1, 3, 4).
In the ER, CGB modulates the release of calcium from intracellular stores via interaction with the third intraluminal loop of the InsP3R, which resides in the lumen of the ER (4, 5). Addition of CGB to the luminal side of the InsP3R increases the open probability of channel currents 10-fold, making CGB one of the most potent coactivators of the InsP3R identified to date (4). Binding of CGB to the InsP3R is modulated by pH and is insensitive to changes in calcium (6). Unlike chromogranin A, which binds to the InsP3R at an acidic pH and not at a neutral pH, CGB has the ability to bind to the InsP3R at both an acidic and a neutral pH (6). Therefore, although both chromogranin A and CGB can modulate InsP3R function in an acidic environment, such as in secretory vesicles, only CGB will bind to the InsP3R in the ER, where the luminal pH is close to neutral.
Comparison of chromogranin A and CGB identified two domains with significant amino acid sequence homology: one at the N terminus and the other at the C terminus (1). The N-terminal domain of CGB (N-CGB) contains a stretch of 20 amino acids that bind to the third intraluminal loop of the InsP3R (1). Although this interaction has only been shown directly for InsP3R type I, it is likely that the other two InsP3R isoforms also bind CGB because the intraluminal loop regions are highly conserved among InsP3R isoforms (7). In measurements of InsP3R channel activity in lipid bilayers, addition of N-CGB to the luminal side of the channel had no effect on the open probability of the channel. However, when N-CGB was added in the presence of full-length CGB, the interaction between the InsP3R and full-length CGB was disrupted, and the loss of full-length CGB binding abrogated the increase in channel activity normally observed upon CGB and InsP3R interaction (5).
In neuronally differentiated PC12 cells, CGB is concentrated in the neurites/growth cones (8, 9). In untransfected cells, the calcium signal initiation site normally coincides with the area in which CGB is most highly expressed. Expression of N-CGB leads to a decrease in peak calcium transients after addition of extracellular agonists (9), showing that the interaction between CGB and the InsP3R at the single channel level is functionally relevant in intact cells. More importantly, attenuation in signal magnitude was accompanied by a shift in the calcium signal initiation site, from the neurites/growth cones to the soma. These findings show that N-CGB acts as a competitive inhibitor (5) and thus prevents full-length CGB from binding effectively to the InsP3R.
In this work, we examined the second homologous domain of CGB, the 23-amino acid domain of the C-terminal region (C-CGB). First, we confirmed that NIH3T3 cells do not express CGB and would therefore be a good model system for examining the effect of CGB on cell function (supplemental Fig. 1A). We then tested the expression of full-length CGB, N-CGB, C-CGB, N-deficient CGB (N-def-CGB), and C-deficient CGB (C-def-CGB) on calcium signaling. As expected, expression of N-CGB in NIH3T3 cells led to no changes in intracellular calcium signaling when compared with control cells. However, expression of C-CGB was able to mimic the effects of full-length CGB on calcium signaling. Specifically, after exogenous expression of either CGB or C-CGB in neuronally differentiated SHSY5Y cells, there was a uniform increase in calcium signaling throughout the cell, and both full-length CGB and C-CGB altered the signal initiation site from neurites/growth cones to the soma. These results show that the N and C termini of CGB have important but distinct roles in modulating calcium signaling and signal initiation.
These distinct properties of the C and N termini of CGB also suggest that they could have differential effects on secretory granule biogenesis (1, 2). We found that expression of full-length CGB and N-CGB was associated with an increase in the number of secretory granules. However, expression of fragments lacking the N terminus did not support granule synthesis suggesting that InsP3R-dependent calcium signaling is not required for granule synthesis, but binding of CGB or N-CGB to the InsP3R is critical.
EXPERIMENTAL PROCEDURES
Antibodies
The following antibodies were used for immunocytochemistry: CGB (BD Biosciences (1:250) or (Santa Cruz Biotechnology (Santa Cruz, CA (1:200)); c-Myc (Santa Cruz Biotechnology (1:200)); calnexin (Stressgen Bioreagents, Ann Arbor, MI (1:200)); InsP3R type I, affinity purified from rabbit polyclonal antiserum directed against the 19 C-terminal residues of the mouse InsP3R type I, custom made (Research Genetics, Huntsville, AL (1:300)); and Alexa Fluor (Invitrogen (1:500 to 1:1000)). The following antibodies were used for Western blotting: CGB (QED Biosciences, San Diego, CA (1:100)); anti-rabbit antibodies (Bio-Rad Laboratories (1:50,000)); and β-actin (Abcam, Cambridge, MA (1:10,000)).
Plasmids
Murine full-length CGB was kindly provided by Makoto Urushitani (Faculty of Medicine, Laval University). The N-deficient, and C-deficient fragments of CGB were truncated from full-length CGB. The N-terminal peptide (IIEVLSNALSKSSAPPITPE), C-terminal peptide (ELENLAAMDLELQK IAEKFSQRG), and a scrambled peptide containing the same amino acids as the C-terminal fragment (RLQSALNQDEGEIMA LKFLEKAE) were generated by annealing complementary pairs of oligonucleotides corresponding to these peptides (Integrated DNA Technologies) in vitro and cloning them into the pShooter-pCMV/Myc/ER vector (Invitrogen).
Cell Culture
NIH3T3 cells were grown in DMEM high glucose (4.5 g/liter) supplemented with 10% fetal bovine serum, penicillin, and streptomycin. PC12 cells were grown in DMEM high glucose (4.5 g/liter) supplemented with 10% heat-inactivated horse serum, 5% heat-inactivated fetal bovine serum, penicillin, and streptomycin. SHSY5Y cells were grown in 44% Eagle's minimum essential medium, 44% Ham's F12 medium, 10% fetal bovine serum, 1% non-essential amino acids (100×), penicillin and streptomycin. Cells were cultured in 5% CO2 at 37 °C. All reagents were obtained from Invitrogen.
Transfection
NIH3T3, SHSY5Y, and PC12 cells were transfected using Lipofectamine 2000 (Invitrogen) with either full-length, N-deficient, C-deficient, N-terminal, C-terminal, and C-terminal scrambled CGB. To confirm expression, cells were co-transfected with DsRed2 with a ratio of target cDNA to fluorescent protein vector of 3:1. NIH3T3 cells were plated in Lab-TeK II eight-well chambers (Nunc/Thermo Fisher Scientific), SHSY5Y in six-well plates (BD Biosciences) on poly-l-lysine coated coverslips (Sigma-Aldrich), and PC12 on collagen I-coated coverslips (BD Biosciences). Cells were used 48 h after transfection.
Neuronal Differentiation
Neurons were differentiated 48 h after transfection in antibiotic free media. Neuronal differentiation of PC12 cells was induced by supplementing growth media with 100 ng/ml nerve growth factor (mNGF 7S, Alomone Laboratories) for 7 days. SHSY5Y cells were differentiated neuronally by supplementing growth media with 25 ng/ml recombinant human nerve growth factor (βNGF, Alomone Laboratories) and 10 μm all-trans retinoic acid (Sigma-Aldrich) for 8 days. Growth medium was changed every other day.
Western Blot Analysis
Cells were lysed with M-PER (Thermo Fisher Scientific) and centrifuged at 4 °C at 10,000 × g. Protein concentration was determined using the BCA protein assay kit (Thermo Fisher Scientific). Equal amounts of protein were loaded onto Tris-HCl gels (Bio-Rad) and transferred onto PVDF membranes. Membranes were blocked with 5% milk and 0.1% Tween 20 in PBS for 1 h, followed by incubation with antibodies diluted in blocking buffer. Visualization was performed by chemiluminescence (Amersham Biosciences).
Light Microscopy
An Olympus BX60 microscope with a magnification of 100× was used to obtain light microscopic images of transfected NIH3T3 cells with the fragments introduced above.
Immunocytochemistry
Cells were fixed in 3.5% paraformaldehyde, permeabilized with 0.1% Triton X, quenched with 0.1% sodium borohydride in PBS, and blocked in 1% bovine serum albumin and 10% goat serum. Cells were incubated with primary antibody overnight and with secondary antibodies for 1 h. Coverslips were mounted with ProLong Antifade (Molecular Probes/Invitrogen). A confocal 510 LSM microscope (Zeiss, Oberkochen, Germany) was used to visualize the cells.
Fura-2 AM Calcium Imaging and Data Analysis
Cells were transfected as described. NIH3T3 were loaded with 10 μm Fura 2-AM (Molecular Probes, Invitrogen) at 37 °C for 30 min. Ratiometric calcium imaging was performed in calcium-free buffer for no longer than 8 min per experiment. Intracellular calcium concentrations ([Ca2+]i) were derived from background-subtracted F340/F380 fluorescent ratios (R) after in situ calibration using [Ca2+]i (nm) = Kd × β × (R − Rmin)/(Rmax − R), where Kd is the dissociation constant of Fura-2 for calcium at 37 °C (225 nm), and Rmin and Rmax were determined experimentally. Calcium release was determined by subtracting the baseline calcium of an individual cell from its maximal calcium response at each time point, with the data then plotted over time. Only cells that were transfected, red fluorescent cells, were included in calculations, and all nonviable cells were excluded from calculations. The area under the curve from the time point where the agonist was introduced to the point where the cell reached a stable baseline calcium before addition of thapsigargin, indicates the total calcium release. A macro in SigmaPlot (version 10) was programmed and used to calculate total calcium release by the method described. Imaging (sampled at 1 Hz) was performed on a Zeiss Axiovert 100 microscope (Zeiss, Oberkochen, Germany).
Fluo-4 AM Calcium Imaging and Data Analysis
Experiments were done as described previously (9). Briefly, transfected PC12 or SHSY5Y cells were loaded with 10 μm Fluo-4 AM (Molecular Probes/Invitrogen) in 20% Pluronic F127 (Sigma-Aldrich) in dimethyl sulfoxide for 30 min at 37 °C. Cells were de-esterified for 15 min and imaged on a Zeiss LSM 510 NLO confocal microscope. Increases in [Ca2+]i were expressed as a ratio of the fluorescence intensity of the calcium imaging dye Fluo-4 over baseline fluorescence (F/F0) and corrected for background fluorescence Fb ((F − Fb)/(F0 − Fb)) (9). To determine regional calcium signaling, cells were divided into equal regions of interest from the growth cone to soma. The signal was determined as the time point at which F/F0 rose constantly above 10% of the interval between F0 and Fmax for the first time. Nonviable cells, defined as cells that do not respond to addition of 10 μm thapsigargin, were excluded from the analysis.
Data and Statistical Analysis
SigmaPlot was used to calculate the area under the curve to determine total calcium release and SigmaStat (Systat Software, San Jose, CA) was used for statistical analysis. Western blots were analyzed with Un-Scan-It Gel Software (Silk Scientific, Orem, UT). Raw imaging data were handled by Workbench imaging software (version 5, Indec BioSystems, Santa Clara, CA) for Fura-2 experiments and LSM imaging software (Zeiss, Oberkochen, Germany) for Fluo-4 experiments. Statistical analysis was performed using the t test for two group comparisons or one-way analysis of variance (Holm-Sidak method) for multiple group comparisons. A p value of ≤ 0.05 was considered to be statistically significant.
RESULTS
Expression of CGB and Fragments in NIH3T3 Cells
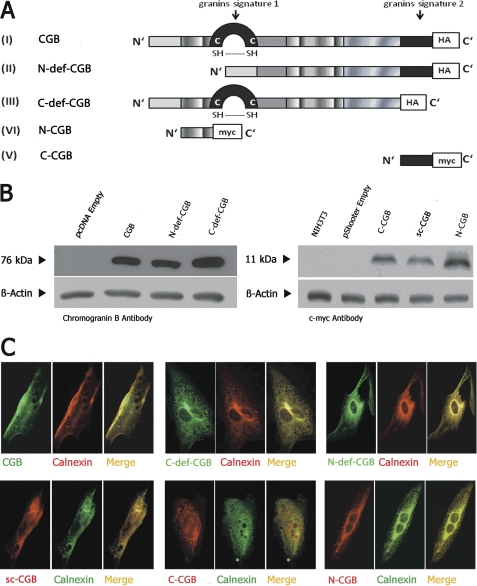
N-deficient CGB (N-def-CGB), C-deficient CGB (C-def-CGB), N-terminal CGB (N-CGB), and C-terminal CGB fragments (C-CGB) (Fig. 1A) were constructed as described under “Experimental Procedures.” Full-length CGB and its fragments were expressed in NIH3T3 cells, a mouse fibroblast cell line without endogenous CGB (10). Expression and localization were determined via Western blotting (Fig. 1B) and immunocytochemistry (Fig. 1C). CGB and its fragments were expressed (Fig. 1B) and found to co-localize with calnexin, an ER marker (Fig. 1C). Resting cytosolic calcium levels (supplemental Fig. 2A) and resting levels of ER calcium, defined as thapsigargin-releasable calcium (supplemental Fig. 2B), were unchanged by expression of full-length CGB or its fragments.
FIGURE 1.
Expression of CGB fragments in NIH3T3 cells. A, diagram of CGB constructs. B, CGB fragments were expressed in NIH3T3 cells and subjected to Western blotting. C, CGB fragments co-localized with calnexin by immunocytochemistry. A was modified from Ref. 22.
Effects of Expression of CGB and CGB Fragments on Calcium Signaling in NIH3T3 Cells
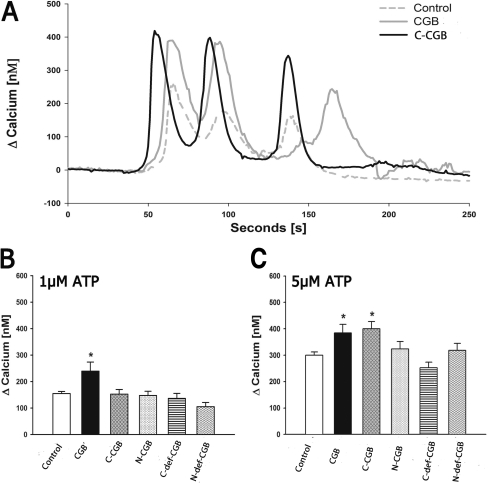
Expression of full-length CGB and C-CGB in NIH3T3 cells increased peak calcium and total calcium release in response to the addition of extracellular agonists (Fig. 2A). When stimulated with 1 μm ATP, the number of responding cells increased from 35 to 53% in cells expressing full-length CGB. On average, peak calcium release increased by 55% in cells expressing CGB (240 ± 34 nm; p ≤ 0.001) compared with control cells (154 ± 8 nm) (Fig. 2B), and the duration of the calcium response was lengthened by 121% (full-length CGB: 62 ± 7 s, p ≤ 0.0001; control: 28 ± 2 s). In terms of total calcium release, full-length CGB-transfected cells released about seven times more calcium (22,483 ± 4228 AU for CGB, p ≤ 0.0001 for CGB; 3121 ± 416 AU for control cells) when stimulated with 1 μm ATP. This increase is comparable with the change in InsP3R channel activity determined in vitro when assessing single InsP3R channels incorporated into lipid bilayers (4). All other fragments failed to induce a significant effect at 1 μm ATP.
FIGURE 2.
Expression of C-CGB increases signal duration, peak calcium release, and total calcium release. A, C-CGB increased peak and total calcium release (area under the curve) when stimulated with 5 μm ATP. B, peak calcium release increased in CGB expressing cells when stimulated with 1 μm ATP. C, peak calcium release increased in both CGB and C-CGB expressing cells when stimulated with 5 μm ATP. Data are mean ± S.E.; *, p ≤ 0.05.
When stimulated with 5 μm ATP, 66% of CGB expressing cells responded, and 59% of C-CGB cells responded. Cells expressing full-length CGB or C-CGB increased the peak calcium release by ∼30% compared with control cells (400 ± 32 nm, p ≤ 0.001 for C-CGB, 384 ± 32 nm, p ≤ 0.01 for full-length CGB-expressing cells, 300 ± 13 nm in the control cells; Fig. 2C). In terms of total calcium release when compared with control cells, full-length CGB and C-CGB expressing cells released ∼50% more calcium (16,438 ± 2376 AU, p ≤ 0.01 for full-length CGB; 15,750 ± 1783 AU, p ≤ 0.01 for C-CGB; 10735 ± 729 for control cells). All other fragments failed to induce a significant effect at 5 μm ATP.
When stimulated with 30 μm ATP, the duration of calcium release was lengthened by 23% in cells expressing C-CGB compared with control cells (95 ± 4 s for C-CGB, p ≤ 0.0001; 77 + 2 s for control cells). Even though the peak calcium release was similar, likely due to saturation of the measurable response, the change in duration shows that there is an increase of at least 32% of the total calcium release for cells expressing C-CGB in comparison to the control cells (29,128 ± 2611 AU, p ≤ 0.01 for C-CGB; 21,972 ± 1072 AU for control cells).
Overall, when cells were stimulated with 1 μm ATP, a low agonist concentration, only full-length CGB had an effect on peak calcium release and duration of the response. At 5 μm ATP, a medium agonist concentration, both C-CGB and full-length CGB were equally as efficient in increasing peak calcium and total calcium release. At high agonist concentrations of 30 μm ATP, only the expression of C-CGB was sufficiently effective to produce a further increase in duration and total calcium release. As expected, due to the removal of the InsP3R binding region, cells expressing N-def-CGB responded in a manner similar to control cells. Specifically, N-CGB expression did not have an effect in cells with no endogenous CGB because there was no CGB to displace from the binding site on the InsP3R. The C-def-CGB expressing cells also did not show a significant difference in response, supporting our hypothesis that the C-terminal region is critical for the function of CGB. It is unclear why expression of the C-terminal region had no effect at the lowest agonist concentration tested. It is possible that C-CGB associates with low affinity without the N-terminal binding domain, which means that at low agonist concentrations the enhancement of signaling would be modest.
Effects of CGB and C-terminal Fragment on Calcium Signaling and Signal Initiation in Neuronally Differentiated PC12 Cells
CGB was shown previously to be concentrated in the growth cones and neuritic branching points of neuronally differentiated PC12 cells (8, 9). This localization of CGB determines the calcium signaling initiation site (9). Upon stimulation with carbachol, an extracellular agonist, intracellular calcium signals began in the growth cones and neurites of PC12 cells, followed by a response in the soma (9). When N-CGB was localized to the lumen of the ER, this signaling pattern was disrupted (9) presumably because N-CGB inhibited proper interaction between CGB and the InsP3R.
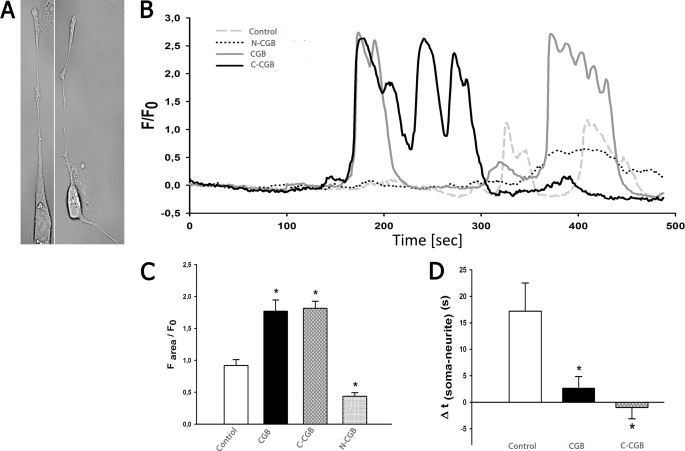
We have extended the previous findings described above by testing the effect of a second agonist, bradykinin, on the ability to evoke calcium release in PC12 cells expressing various CGB fragments. Expression of CGB or its fragments in PC12 cells did not follow the same expression pattern as that observed in cells expressing only endogenous CGB. We found that when either full-length CGB or C-CGB were expressed, CGB levels in the soma were elevated (Fig. 3A). As described earlier, expression of N-CGB also led to an even distribution of CGB between the growth cones and the soma (9). The peak calcium response was increased when full-length CGB was expressed and decreased when N-CGB was expressed (5, 9). Increased expression of full-length CGB increased InsP3-dependent signaling, but expression of N-CGB displaced full-length CGB from the InsP3R to decrease signaling. In the present series of experiments, when monitoring intracellular calcium changes with bradykinin as the agonist, we observed that expression of either full-length CGB or C-CGB increased peak calcium release (Fig. 3B, left panel). The response to bradykinin and carbachol in full-length CGB-expressing cells is similar (compare Fig. 3B and Ref. 9). The response to stimulation in C-CGB expressing cells is opposite to that observed previously in N-CGB-expressing cells. These results suggest that mechanisms downstream to InsP3 production, namely the signaling microdomains formed by the interaction between the InsP3R and CGB, are important for the amount of calcium released from intracellular stores rather than the specific agonist used to produce an increase in InsP3.
FIGURE 3.
The extent of calcium release is increased and signal initation sites are altered upon expression of CGB and C-CGB in PC12 cells. A, in control cells, endogenous CGB is mostly expressed in the growth cones and neuritic branching points. Overexpressed CGB is located throughout the cell but with the highest concentrations in the soma, whereas C-CGB is distributed and elevated throughout the cell. B, stimulation of cells expressing CGB and C-CGB with bradykinin increased the extent of calcium release. C, in cells transfected with scrambled C-CGB (control), calcium signaling was initiated in the growth cones followed by the soma. Overexpression of CGB shifted the signal initiation site to the soma, and expression of C-CGB led to calcium signaling occurring simultaneously at the soma and the growth cones. Data are mean ± S.E.; *, p ≤ 0.05.
In our previous studies, expression of N-CGB altered the signal initiation site (9). This was determined by noting the delay between agonist addition and an elevation in intracellular calcium. The signal initiation time at the neurites was subtracted from the signal initiation site at the soma and any positive value indicated signal initiation in the growth cones. In non-treated cells, intracellular calcium first rose in the growth cone, with the signal appearing in the soma after a delay of 2 s; this order was reversed in cells expressing N-CGB (9). In the present study, we expressed full-length CGB, C-CGB, or a scrambled C-terminal CGB fragment in neuronally differentiated PC12 cells. In cells that were either untransfected or were transfected with a scrambled C-terminal CGB fragment, stimulation with 2 μm bradykinin initiated calcium signaling in the growth cones followed by a response in the soma 2.3 + 0.4 s later (Fig. 3C, right panel). When full-length CGB was expressed, the signal initiation site began in the soma and was followed by a response in the growth cones 1.2 + 0.8 s later (p ≤ 0.01). When C-CGB-transfected cells were stimulated with bradykinin, the signal initiation site appeared in both the growth cones and soma at the same time; + 0.6 s (p ≤ 0.05) (Fig. 3B, right panel).
This series of experiments show that CGB is involved in defining the magnitude and initiation site of calcium signals. By changing the distribution and expression levels of CGB, calcium signaling can be altered. In addition, these experiments show that the function of full-length CGB in neuronally differentiated PC12 cells can be mimicked by C-CGB, a 23-amino acid long domain in the C-terminal of CGB.
Effects of CGB and Its C- and N-terminal Fragments on Calcium Signaling and Signal Initiation in Neuronally Differentiated SHSY5Y Cells
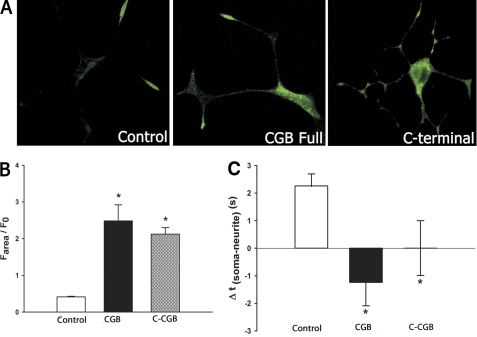
The previously described experiments were done using neuronally differentiated PC12 cells, a model commonly used to study neuronal processes. Although many studies have shown that these cells have signal transduction properties similar to that of primary neurons (11), PC12 cells have their limitations. We therefore used a second model, neuronally differentiated SHSY5Y cells, a human neuroblastoma cell line. SHSY5Y cells were successfully differentiated, which can be observed morphologically by the extensions of the neurites (Fig. 4A) and by the increased CGB levels in differentiated SHSY5Y cells (supplemental Fig. 1A).
FIGURE 4.
Expression of C-CGB in SHSY5Y cells altered the signal initation site and increased peak calcium release. A, differentiation of SHSY5Y was successful, as indicated by neurite extensions. B and C, when stimulated with 15 μm carbachol, peak calcium release was increased in cells expressing CGB and C-CGB. Expression of N-CGB decreased peak calcium release. D, expression of CCB significantly decreased the delay between the first neuritic and somatic signal whereas C-CGB shifted the signal initiation site to the soma. Data are mean ± S.E.; *, p ≤ 0.05.
When full-length CGB or CGB fragments were overexpressed in SHSY5Y cells, resting cytoplasmic and ER calcium levels were unchanged (supplemental Fig. 1, B and C). InsP3R type I, the predominant isoform in neurons, was found to be distributed equally in the soma and growth cones (supplemental Fig. 3) (8), indicating that distribution of the InsP3R cannot explain the initiation site for the calcium signals. When cells were stimulated with 15 μm carbachol, the peak calcium response was nearly double in cells overexpressing full-length CGB or C-CGB, when compared with control cells (1.8 ± 0.1 AU, p ≤ 0.0001 for CGB; 1.8 ± 0.1 AU, p ≤ 0.0001 for C-CGB and 0.9 ± 0.09 AU for control cells) (Fig. 4, B and C). Expression of N-CGB decreased the peak calcium release by 47% (0.4 ± 0.05 AU, p ≤ 0.01). Cells expressing only C-CGB also responded 68% longer than the control cells, increasing the duration of calcium release from 99 + 8 s in the control cells to 167 + 12 s in cells expressing C-CGB (p ≤ 0.0001). Expression of the fragments also altered the sensitivity of InsP3-induced calcium release. Full-length CGB-transfected cells increased the number of responsive cells from 59% in the control cells to 80%, C-CGB expression increased the number of responsive cells to 86%, and N-CGB expression resulted in an 8% decrease in the percentage of cells responding.
Under basal conditions, the growth cones of neuronally differentiated SHSY5Y cells were more sensitive to InsP3-mediated calcium release than the soma. In the control cells, the calcium signal was detected in the growth cones followed by a response in the soma 17 ± 5 s later. Expression of full-length CGB decreased the delay in response to 2.6 ± 2.2 s (p ≤ 0.01), and expression of C-CGB caused a shift in the initiation site from the growth cone to the soma with a delay of 1 ± 2.1 s later (p ≤ 0.01) (Fig. 5D).
FIGURE 5.
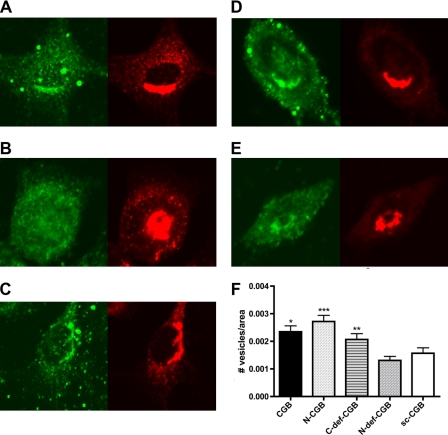
N-CGB is required for secretory vesicle biogenesis. CGB fragments were expressed in NIH3T3 cells and stained for CGB (green) and vesicles (β-COP, red). A, CGB full-length; B, N-CGB; C, C-def-CGB; D, N-def-CGB; E, scrambled C-terminal CGB fragment (sc-CGB); F, when vesicles were quantified, N-CGB, CGB full-length, and C-def-CGB were shown to induce vesicle biogenesis, whereas N-def-CGB did not increase vesicle biogenesis over control (scrambled C-terminal CGB fragment). Data are mean ± S.E.; *, p ≤ 0.05; **, p ≤ 0.03; ***, p ≤ 0.01.
In these experiments, we show that CGB can play a role in determining the magnitude and initiation site of calcium signaling. Expression of the N-CGB diminished the calcium transient. In contrast, expression of full-length CGB or C-CGB induced dramatic increases in peak calcium release and altered the initiation sites for calcium signaling.
Effects of Expression of CGB and CGB Fragments on Secretory Granule Synthesis
To examine the roles of the N- and C-terminal domains on cellular function, we decided to monitor the effect of CGB expression on secretory vesicle biogenesis. Full-length CGB, N-CGB, or C-def-CGB fragments were expressed in NIH3T3 cells, and de novo secretory granule biogenesis was induced (Fig. 5, A–C). In contrast, expression of the N-def-CGB fragment and the scrambled peptide (Fig. 5, D and E) failed to induce vesicle formation. The green staining shows the distribution of CGB, and the red corresponds to β-COP, a marker for secretory granules. Similar results were obtained when vesicles were observed by light microscopy (supplemental Fig. 4). These experiments show that expression of full-length CGB and fragments that contain the InsP3R binding portion of CGB will induce secretory granule formation. However, fragments that lack the binding region, even if they contain the portion of CGB that activates the InsP3R channel, do not induce secretory granule biogenesis.
DISCUSSION
CGB is known for its involvement in calcium signaling via interaction with the InsP3R at the luminal side of the ER. In addition, two potentially important regions of CGB have been identified, the N- and C-terminal domain (12). The interaction of these two domains with the InsP3R has been proposed to aid in regulating spatial and temporal calcium signaling patterns (9). In this study, we identified distinct roles for the N- and C-terminal domain of CGB. We found previously that the N-terminal domain is crucial for binding to the InsP3R and the C-terminal domain amplifies calcium signaling.
CGB and Signaling Microdomains
Because CGB is not distributed uniformly within cells, its interaction with the InsP3R creates a microdomain, which in turn increases the sensitivity of the InsP3R. In this study, we found that the localization of CGB was critical for the creation of these microdomains. Functionally, a calcium microdomain is a restricted region where an intracellular elevation of calcium occurs around the calcium release channel without necessarily leading to a rise in cytosolic calcium levels. There are a number of different compositions of signaling complexes that can determine/regulate the timing and shape of the calcium response. For example, in neuronally differentiated PC12 cells, the distribution of the InsP3R-CGB complex is crucial for determining the signal initiation site (8). In these cells, an elevation of calcium is initiated in the distal parts of the neurite where the concentration of CGB is highest. Disruption of the interaction between the InsP3R type I and CGB shifts the signal initiation to the soma (9). The interaction of the bradykinin receptor with the InsP3R via actin filaments in sympathetic ganglion cells is another example of a complex that is important for signaling. Disruption of the actin filaments allows the two receptors to separate leading to diminished calcium signaling (13). As shown in this paper, the C- and N-terminal domains of CGB may play a crucial role in regulating the development of calcium microdomains, thus aiding in the modulation of calcium responses.
The distribution of CGB contrasts with that of many other proteins crucial for intracellular calcium signaling. ATPases in the plasma membrane (plasma membrane calcium ATPase 1, PMCA1) and ER (sarcoplamic endoplasmic reticulum calcium ATPase 2), and levels of parvalbumin and other calcium buffers important for neuronal function are distributed uniformly (14, 15). From a comparison of the distribution of these proteins, it has been suggested that proteins associated with “on-mechanisms” are often distributed heterogeneously within neurons, whereas “off-mechanisms” are distributed homogeneously. Therefore, as CGB is distributed heterogeneously, it is likely that it acts as an on-mechanism and is crucial for initiating calcium release from the InsP3R.
ER Proteins That Regulate Signaling
Other ER resident proteins such as ERp44, calnexin, and junctate regulate the InsP3R as well, although in a manner different from that of CGB. Regulation of the InsP3R by other proteins enables for the development of complex signaling. In addition, similarities and differences of these proteins allow us to gain insight into other potential mechanisms by which CGB regulates the InsP3R. ERp44, a 43.9-kDa protein, mediates the retention of oxidoreductins and proteins and is thought to act as a redox sensor for the ER (16). In a manner similar to CGB, ERp44 interacts with the third intraluminal loop of the InsP3R. In contrast to CGB, which binds with its N-terminal domain to the conserved region of the third intraluminal loop (L3C), ERp44 binds to the variable part of the intraluminal loop (L3V) (17). The L3V is structurally different among subtypes of InsP3R, unlike the L3C, and as a result, ERp44 only interacts with InsP3R type I; chromogranin interacts with all three subtypes of the InsP3R (6). Additionally, the molecular interaction of ERp44 with the InsP3R is affected not only by pH, as CGB primarily is, but also by redox and calcium. Mutations of the InsP3R cysteines within L3V were found to disrupt the interaction of ERp44 with the InsP3R. Moreover, the high calcium concentration and oxidizing environment normally found in the ER also disrupts ERp44 interaction with the InsP3R (17). Therefore, it seems as if the role of ERp44 is to decrease the release of calcium, as indicated by inhibition of the InsP3R channel opening upon binding of ERp44. This is opposite to the role that CGB plays and suggests that ERp44 may act to prevent ER calcium stores from completely emptying when the calcium concentration in the ER is decreased. Interplay between CGB and ERp44 could have an important role in regulation of ER calcium stores.
The InsP3R is also regulated by another calcium binding protein of the ER, junctate. Similar to ERp44 and CGB, junctate has regulatory N- and C-terminal domains. Junctate is an integral membrane protein that binds to all three subtypes of the InsP3R via its N-terminal domain (18). Upon overexpression of full-length junctate, there is amplification of InsP3 mediated calcium release, whereas neither its C- nor N-terminal domain alters calcium release (18). Although junctate does not modulate InsP3R calcium release via its N- or C-terminal domain, junctate is similar to CGB in that it acts as a calcium buffer in the ER and is a moderate affinity, high capacity calcium binding protein. One of the primary functions of the junctate is in regulating receptor-activated and store depletion-induced calcium entry. Upon stimulation of the InsP3R and/or depletion of intracellular calcium stores, a set of receptor/store-operated calcium channels on the plasma membrane are activated, allowing entry of extracellular calcium and replenishment of intracellular calcium stores (19–21). Overexpression of the N-terminal domain of junctate induces ER proliferation and increases peripheral couplings of the ER with the plasma membrane. This interaction with the InsP3R, via the N-terminal domain of junctate, modulates receptor-activated calcium entry. In contrast, modulation of store depletion-induced calcium entry occurs only upon overexpression of full-length junctate. In this manner, junctate is similar to CGB in that its role as a modulator is domain-specific.
Conclusions
In summary, this study supports the role of CGB in the maintenance and modulation of calcium signaling, as well as in granule biogenesis. We found that the C-terminal domain is crucial for inducing calcium release from the InsP3R. We also further characterized the role of CGB and found that expression of the N-terminal domain was sufficient to induce biogenesis of vesicles, in addition to its role as a competitive inhibitor of CGB. When full-length CGB is present, the N terminus competes with full-length CGB for its binding site on the InsP3R, therefore decreasing calcium release. On the other hand, in cells lacking CGB, the competition between full-length CGB and N-terminal CGB does not occur, therefore causing calcium release to be unaltered.
The fact that domains of CGB have distinct functions emphasizes the importance of regulating intracellular calcium release and vesicle biogenesis. If the C-terminal domain was altered by mutations or conformational/functional changes due to its environment, then the ability to maintain calcium release will be challenged. Similarly, vesicle biogenesis depends upon the integrity of the N-terminal domain of CGB. In this way, alterations that prevent CGB from functioning will disrupt calcium homeostasis and lead to pathophysiological conditions.
Supplementary Material
Acknowledgments
We thank Ivana Kuo, Colleen Feriod, Brenda DeGray, Christin Schulze, Jennifer Benbow, and Edward Petri for discussions and comments on the manuscript. The authors thank Dr. Makoto Urushitani (Faculty of Medicine, Laval University) for generously providing reagents used in this study.
This work was supported, in whole or in part, by National Institutes of Health Grants DK57751 and DK61747. This work was also supported by scholarships from the German National Merit Foundation (to S. S.) and the Howard Hughes Medical Institute (to M. M.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. 1–4.
- CGB
- chromogranin B
- InsP3R
- inositol 1,4,5,-trisphosphate receptor
- ER
- endoplasmic reticulum
- N-CGB
- N-terminal CGB
- C-CGB
- C-terminal CGB
- N-def-CGB
- N-terminal-deficient CGB
- C-def-CGB
- C-terminal-deficient CGB
- AU
- area units
- L3C
- conserved third-intraluminal loop of the InsP3R
- L3V
- variable third-intraluminal loop of the InsP3R.
REFERENCES
- 1. Huh Y. H., Jeon S. H., Yoo J. A., Park S. Y., Yoo S. H. (2005) Biochemistry 44, 6122–6132 [DOI] [PubMed] [Google Scholar]
- 2. Glombik M. M., Krömer A., Salm T., Huttner W. B., Gerdes H. H. (1999) EMBO J. 18, 1059–1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Thrower E. C., Park H. Y., So S. H., Yoo S. H., Ehrlich B. E. (2002) J. Biol. Chem. 277, 15801–15806 [DOI] [PubMed] [Google Scholar]
- 4. Thrower E. C., Choe C. U., So S. H., Jeon S. H., Ehrlich B. E., Yoo S. H. (2003) J. Biol. Chem. 278, 49699–49706 [DOI] [PubMed] [Google Scholar]
- 5. Choe C. U., Harrison K. D., Grant W., Ehrlich B. E. (2004) J. Biol. Chem. 279, 35551–35556 [DOI] [PubMed] [Google Scholar]
- 6. Yoo S. H., So S. H., Kweon H. S., Lee J. S., Kang M. K., Jeon C. J. (2000) J. Biol. Chem. 275, 12553–12559 [DOI] [PubMed] [Google Scholar]
- 7. Bezprozvanny I. (2005) Cell Calcium 38, 261–272 [DOI] [PubMed] [Google Scholar]
- 8. Johenning F. W., Zochowski M., Conway S. J., Holmes A. B., Koulen P., Ehrlich B. E. (2002) J. Neurosci. 22, 5344–5353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jacob S. N., Choe C. U., Uhlen P., DeGray B., Yeckel M. F., Ehrlich B. E. (2005) J. Neurosci. 25, 2853–2864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Huh Y. H., Chu S. Y., Park S. Y., Huh S. K., Yoo S. H. (2006) Biochemistry 45, 1212–1226 [DOI] [PubMed] [Google Scholar]
- 11. Greene L. A., Tischler A. S. (1982) in Advances in Cellular Neurobiology, pp. 373–414, Academic Press, New York [Google Scholar]
- 12. Yoo S. H. (2000) Trends Neurosci. 23, 424–428 [DOI] [PubMed] [Google Scholar]
- 13. Delmas P., Wanaverbecq N., Abogadie F. C., Mistry M., Brown D. A. (2002) Neuron 34, 209–220 [DOI] [PubMed] [Google Scholar]
- 14. Collin T., Chat M., Lucas M. G., Moreno H., Racay P., Schwaller B., Marty A., Llano I. (2005) J. Neurosci. 25, 96–107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Caillard O., Moreno H., Schwaller B., Llano I., Celio M. R., Marty A. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 13372–13377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Anelli T., Alessio M., Bachi A., Bergamelli L., Bertoli G., Camerini S., Mezghrani A., Ruffato E., Simmen T., Sitia R. (2003) EMBO J. 22, 5015–5022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Higo T., Hattori M., Nakamura T., Natsume T., Michikawa T., Mikoshiba K. (2005) Cell 120, 85–98 [DOI] [PubMed] [Google Scholar]
- 18. Treves S., Franzini-Armstrong C., Moccagatta L., Arnoult C., Grasso C., Schrum A., Ducreux S., Zhu M. X., Mikoshiba K., Girard T., Smida-Rezgui S., Ronjat M., Zorzato F. (2004) J. Cell Biol. 166, 537–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Irvine R. F. (1990) FEBS Lett. 263, 5–9 [DOI] [PubMed] [Google Scholar]
- 20. Zhu X., Jiang M., Peyton M., Boulay G., Hurst R., Stefani E., Birnbaumer L. (1996) Cell 85, 661–671 [DOI] [PubMed] [Google Scholar]
- 21. Putney J. W., Jr., Broad L. M., Braun F. J., Lievremont J. P., Bird G. S. (2001) J. Cell Sci. 114, 2223–2229 [DOI] [PubMed] [Google Scholar]
- 22. Urushitani M., Sik A., Sakurai T., Nukina N., Takahashi R., Julien J. P. (2006) Nat. Neurosci. 9, 108–118 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.