Background: Angiotensin II-induced regulator of G-protein signaling-2 (RGS2) expression is an important negative feedback loop in blood pressure homeostasis.
Results: A conserved cAMP-response element-binding protein (CREB) binding site is found in the RGS2 promoter and is associated with three SNPs identified in hypertensive patients.
Conclusion: CREB links the angiotensin II/PKC/iPLA2β/PKA signaling and RGS2 transcription.
Significance: This study delineates a negative feedback loop with potential importance in the pathogenesis of human hypertension.
Keywords: Angiotensin II, CREB, G Proteins, Gene Regulation, Phospholipase, RGS2, VSMC, iPLA2
Abstract
Mice deficient in regulator of G-protein signaling-2 (RGS2) have severe hypertension, and RGS2 genetic variations occur in hypertensive humans. A potentially important negative feedback loop in blood pressure homeostasis is that angiotensin II (Ang II) increases vascular smooth muscle cell (VSMC) RGS2 expression. We reported that Group VIA phospholipase A2 (iPLA2β) is required for this response (Xie, Z., Gong, M. C., Su, W., Turk, J., and Guo, Z. (2007) J. Biol. Chem. 282, 25278–25289), but the specific molecular causes and consequences of iPLA2β activation are not known. Here we demonstrate that both protein kinases C (PKC) and A (PKA) participate in Ang II-induced VSMC RGS2 mRNA up-regulation, and that actions of PKC and PKA precede and follow iPLA2β activation, respectively. Moreover, we identified a conserved cAMP-response element (CRE) in the murine RGS2 promoter that is critical for cAMP-response element-binding protein (CREB) binding and RGS2 promoter activation. Forskolin-stimulated RGS2 mRNA up-regulation is inhibited by CREB sequestration or specific disruption of the CREB-RGS2 promoter interaction, and Ang II-induced CREB phosphorylation and nuclear localization are blocked by iPLA2β pharmacologic inhibition or genetic ablation. Ang II-induced intracellular cyclic AMP accumulation precedes CREB phosphorylation and is diminished by inhibiting iPLA2, cyclooxygenase, or lipoxygenase. Moreover, three single nucleotide polymorphisms identified in hypertensive patients are located in the human RGS2 promoter CREB binding site. Point mutations corresponding to these single nucleotide polymorphisms interfere with stimulation of human RGS2 promoter activity by forskolin. Our studies thus delineate a negative feedback loop to attenuate Ang II signaling in VSMC with potential importance in blood pressure homeostasis and the pathogenesis of human essential hypertension.
Introduction
Regulator of G-protein signaling-2 (RGS2)5 is a member of the RGS protein superfamily, which is comprised of at least 25 members in mammals (1). RGS2 is expressed ubiquitously, and its major biological function is to serve as a GTPase-activating protein that preferentially attenuates heterotrimeric G protein Gαq/11 GTPase activity (2). Several independent studies have demonstrated that Rgs2 knock-out mice have severe hypertension (3–7), suggesting that RGS2 plays a critical role in blood pressure homeostasis. Interestingly, heterozygous Rgs2 knock-out mice are also hypertensive, and their blood pressures are virtually identical to homozygous Rgs2 knock-out mice (3), indicating that two functional Rgs2 alleles are required for normal blood pressure homeostasis.
More than 30 RGS2 gene polymorphisms have been identified in human hypertensive patients (8–11), and most of them have been found in noncoding regions of the RGS2 gene, including the promoter and intronic and 3′-untranslated regions (8–11). Such polymorphisms would not alter the RGS2 protein sequence but might affect RGS2 mRNA and protein expression levels, and reductions in RGS2 expression have been observed in cells from hypertensive patients with RGS2 gene polymorphisms (10, 12). In contrast, increased RGS2 expression has been observed in cells from hypotensive patients with Bartter/Gitelman syndrome (13).
Ang II is a vasoconstrictor peptide that is important for blood pressure homeostasis (14). Ang II binds to VSMC AT1 receptors, which results in G protein Gq/11 activation, smooth muscle contraction, and increased blood pressure (14). Ang II also initiates a negative feedback loop simultaneously to restrain its vasopressor effects (2), and elements of this loop include Ang II-induced increases in RGS2 expression. The importance of this feedback loop in blood pressure homeostasis is reflected by the fact that infusion of Ang II results in an exaggerated hypertensive response in Rgs2-null mice compared with their wild-type (WT) littermates (7).
Grant et al. (15) first reported that incubation of cultured VSMC with Ang II induces a selective, dynamic increase in RGS2 mRNA expression via interaction with AT1 receptors, and this observation has been confirmed by several laboratories (16–18). Increased expression of VSMC RGS2 mRNA by Ang II results in increased levels of RGS2 protein (16, 18) and results in negative regulations of Ang II signaling (16, 18). That these in vitro observations have physiologic significance is suggested by in vivo studies that indicate that Rgs2-null mice are more sensitive to Ang II-induced hypertension than their WT littermates (7). The molecular details of the signal transduction pathway that links the Ang II-AT1 receptor interaction to increased expression of RGS2 mRNA, however, are largely unknown.
The Group VIA phospholipase A2 (iPLA2β) is a member of the PLA2 superfamily of enzymes that hydrolyze phospholipid substrates to yield a free fatty acid (e.g. arachidonic acid, AA) and a 2-lysophospholipid (e.g. 2-lysophosphatidylcholine, LPC). Both AA and LPC have intrinsic second messenger activities and are also precursors of bioactive metabolites that include eicosanoids and platelet activating factor, inter alia (19), and among the pleiotropic effects of these mediators are gene transcriptional regulatory activities (16, 20, 21). The iPLA2β resides predominantly in the cytosol of resting cells, associates with membranes upon activation, does not require Ca2+ for catalytic activity, and is inhibited by the compound bromoenol lactone (BEL) at concentrations that have little effect on members of the sPLA2 or cPLA2 families (22).
Recently, we demonstrated that iPLA2β plays an essential role in Ang II-induced RGS2 mRNA up-regulation in VSMC (16), these studies involved pharmacologic inhibition of iPLA2β activity with BEL, suppression of iPLA2β expression with antisense oligodeoxynucleotides, and genetic deletion of iPLA2β in mice by homologous recombination (16). In addition, we demonstrated that expressing iPLA2β by adenovirus-mediated gene transfer in iPLA2β-null VSMC restored the ability of Ang II to increase RGS2 mRNA expression in a concentration-dependent manner (16).
Nonetheless, the train of events that link the Ang II-AT1 receptor interaction to iPLA2β activation is largely unknown, as are the consequent events that link iPLA2β activation to increased expression of RGS2 mRNA, including the identities of transcription factor(s) that participate in these events. The current study addresses these issues in Ang II-stimulated cultured VSMC.
EXPERIMENTAL PROCEDURES
Materials and Oligodeoxynucleotide
The antibodies against total CREB and phosphorylated CREB (Ser133) were purchased from Cell Signaling (Danvers, MA). The antibody against total CREB used for supershift assay and SQ22536 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA). GF109203X and forskolin were purchased from Biomol (Plymouth Meeting, PA). H-89 was purchased from Alexis (San Diego, CA). BEL, AA, and LPC were purchased from Cayman (Ann Arbor, MI). Other chemicals were purchased from Sigma. [γ-32P]ATP (3,000 Ci/mmol) was purchased from PerkinElmer Life Sciences.
Oligodeoxynucleotides used in the inhibition studies were phosphorothioate-modified and purchased from IDT (Coralville, IA). The sequences of rat iPLA2β antisense and sense oligodeoxynucleotides were previously described (16). The sequences of cAMP regulatory element (CRE)-decoy and mismatch controls were described by Park et al. (23). The mouse RGS2-CRE1 oligodeoxynucleotide sequences are as follows: 30-mer RGS2-CRE, 5′-ATGCGGCGCCTACGTCAACAGCGCCCTCAC-3′; 30-mer RGS2-CRE mutant control, 5′-ATGCGGCGCCTACCAGTACAGCGCCCTCAC-3′.
Primary VSMC Culture
Procedures for isolating and culturing primary VSMC from rabbits, SD rats, iPLA2β knock-out, and WT littermate mice have been described (16, 24–27).
Real-time PCR
Primers for RGS2 and 18S RNA and the procedure for real-time PCR were previously described (16, 25, 28, 29).
Subcloning Murine and Human RGS2 Promoters
A bacterial artificial chromosome clone (RP23–132C16) containing the mouse RGS2 promoter was purchased from Invitrogen and used as PCR template. The forward PCR primer sequences used to amplify mouse RGS2 promoters were: 1) 5′-CCGACGCGTGAGTGAGGCAATAAAATCAAGAGTTG-3′ for −4367 bp promoter; 2) 5′-AATACGCGTACCGTGAATGACTCATGAGGA-3′ for −867 bp promoter; 3) 5′-AATACGCGTCGCACACGAGTAAAAGGTACG-3′ for −367 bp promoter; 4) 5′-AATACGCGTCGCTCTCTTGGGGCGT-3′ for the −117 bp promoter. The common reverse PCR primer sequence is 5′-AAACTCGAGTTCTCAGACTCCCGCGGC-3′. A −47 bp mouse RGS2 promoter and a −388 bp human RGS2 promoter were synthesized by IDT (Coralville, IA). All PCR products including synthetic promoters were subcloned into PGL3 basic vector (Promega, Madison, WI) to generate RGS2-Luc reporters.
Site-directed Mutagenesis
CRE1 and CRE2 mutations of RGS2 promoters were performed by using QuikChange II Site-directed Mutagenesis Kit (Agilent Technologies, Inc., Santa Clara, CA) following the manufacturer's instructions. Mutations were confirmed by DNA sequencing (Davis Sequencing, Davis, CA).
RGS2 Promoter Activity Analysis
Rabbit aortic VSMC were grown in 12-well cell culture plates. When they achieved 70–80% confluence, cells were co-transfected with an RGS2-Luc reporter and pRL-SV40 Vector (Promega) using Lipofectamine Plus reagent. RGS2 promoter activity was analyzed by a dual luciferase enzyme assay system (30) in a microplate luminometer (LB960 Centro, Berthold USA, Oak Ridge, TN).
Electrophoretic Mobility Shift Assay (EMSA)
Nuclear extracts were prepared and EMSA was performed as described (31, 32). Briefly, nuclear extracts (2 μg of protein) were incubated (30 min on ice) with 30-mer 32P-labeled double-stranded WT or mutant RGS2-CRE1 probes in a mixture (15 μl total volume) consisting of 15 mm HEPES, pH 7.9, 3 mm Tris-HCl, pH 7.9, 60 mm KCl, 1 mm EDTA, 1 mm PMSF, 1 mm dithiothreitol, 4.5 μg of BSA, 2 μg of poly(dI-dC), and 15% glycerol. Reaction mixtures were analyzed on 4% nondenaturing polyacrylamide gels that were allowed to dry and were examined by autoradiography.
Western Blot Analyses
Immunofluorescence
Cultured VSMCs were grown on glass coverslips in 6-well culture plates and starved in FBS-free medium for 24 h. After Ang II stimulation, cells were fixed with 4% formaldehyde in 150 mm sodium phosphate buffer, pH 7.4, for 30 min, and then quenched and permeabilized with 1 m glycine and 0.2% Triton X-100 in PBS for 20 min. Cells were blocked in 16% normal goat serum in PBS (30 min), and then sequentially incubated with rabbit anti-phosphorylated CREB (Ser133) antibody in blocking solution (overnight, 4 °C), Alexa Fluor® 594 goat anti-rabbit IgG (Invitrogen) in blocking solution (1 h), and 300 nm DAPI in PBS (3 min). Cells were mounted in VECTASHIELD and photographed with an Olympus IX70 microscope equipped with Olympus DP70 digital camera.
Quantitation of cAMP
Rat aortic VSMC at 80% confluence were incubated in serum-free medium (24 h). Cells were stimulated with Ang II (100 nm) or isoproterenol (2 nm) for various intervals. When inhibitors were used, they were incubated with cells before stimulation (30 min). At the end of the stimulation interval, culture medium was aspirated, 0.1 n HCl was added to cells, and cellular cAMP content was assayed by Direct ELISA kit (Assay Designs) according to the manufacturer's instructions.
Statistical Analysis
Data are expressed as mean ± S.E. Statistical analyses were performed by using an unpaired t test or one-way analysis of variance (GraphPad, Prism 4) and appropriate post hoc analyses.
RESULTS
PKC Is Upstream of PKA in Ang II-induced RGS2 mRNA Up-regulation
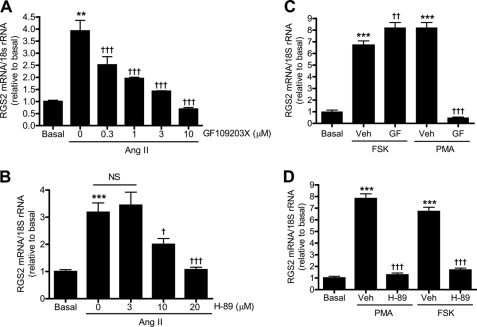
Protein Kinases A and C (PKA and PKC) can be activated by second messengers, e.g. cyclic AMP (cAMP) and 1,2-diacylglyerol, respectively, and are involved in a variety of signal transduction pathways. To examine the potential involvement of these kinases in VSMC signaling, we investigated whether inhibiting PKC or PKA affects Ang II-induced RGS2 mRNA accumulation. We found that inhibiting PKC or PKA with GF109203X (Fig. 1A) or H-89 (Fig. 1B), respectively, suppressed Ang II-induced RGS2 mRNA accumulation in a concentration-dependent manner. This suggests that both kinases participate in the signaling pathways that link the interaction of Ang II with its receptor in VSMC to increases in RGS2 mRNA and that activation of these kinases might be expected to enhance RGS2 mRNA accumulation in this setting.
FIGURE 1.
PKC is upstream of PKA in Ang II-induced RGS2 mRNA accumulation in rat aortic VSMC. A, pretreatment (30 min) of cells with various concentrations of GF109203X inhibited Ang II (100 nm, 1 h)-induced RGS2 mRNA accumulation in a concentration-dependent manner. B, pretreatment of cells with various concentrations of H-89 for 30 min inhibited Ang II-induced RGS2 mRNA accumulation in a concentration-dependent manner. C, pretreatment of cells with GF109203X (3 μm, 30 min) inhibited PMA- (100 nm, 1 h) but not FSK- (10 μm, 1 h) induced RGS2 mRNA accumulation. D, pretreatment of cells with H-89 (20 μm, 30 min) inhibited both PMA- and FSK-induced RGS2 mRNA accumulation. Veh, vehicle (Me2SO2). Data are represented as mean ± S.E. of at least three independent experiments. **, p < 0.01; ***, p < 0.001 versus basal. NS, not significant; †, p < 0.05; ††; p < 0.01; †††, p < 0.001 versus Ang II stimulation in (A and B), and PMA or FSK stimulation (Veh) in C and D.
To test these possibilities, VSMC were treated with the adenylyl cyclase activator forskolin (FSK) to stimulate PKA activity or with phorbol myristate acetate (PMA) to stimulate PKC activity. Both FSK and PMA were found to stimulate RGS2 mRNA accumulation in the absence of Ang II (Fig. 1C). This effect of FSK was not suppressed by the PKC inhibitor GF109203X (GF) (Fig. 1C) but was suppressed by PKA inhibitor H-89 (Fig. 1D). In contrast, RGS2 mRNA accumulation stimulated by PMA was suppressed by the PKC inhibitor GF109203X (Fig. 1C) but also by the PKA inhibitor H-89 (Fig. 1D). These observations indicate that RGS2 mRNA accumulation can be stimulated by activation of either PKC or PKA and that the response to either is blocked by inhibiting PKA. The response to PKA activation is not blocked by inhibiting PKC, however, which implies that activation of PKC is upstream of PKA in the signaling pathway that leads to stimulation of RGS2 mRNA accumulation.
iPLA2β Links PKC and PKA in Ang II-induced RGS2 mRNA Up-regulation
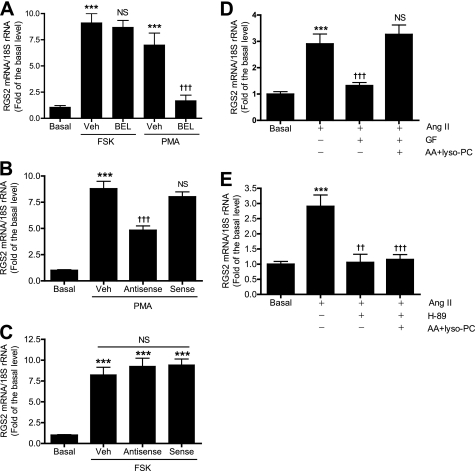
Three independent approaches were used to examine the participation of iPLA2β in RGS2 mRNA accumulation in VSMC induced by activating PKC and PKA. We first examined pharmacologic inhibition of iPLA2β activity with the compound BEL, which is an iPLA2β suicide substrate. BEL irreversibly inactivates iPLA2β, and we have previously demonstrated that BEL prevents Ang II-induced RGS2 mRNA accumulation in VSMC (16). As illustrated in Fig. 2A, BEL was found to prevent RGS2 mRNA accumulation in response to the PKC activator PMA but had little effect on the response to the adenylyl cyclase activator FSK.
FIGURE 2.
iPLA2β links PKC and PKA in Ang II-induced RGS2 mRNA accumulation in rat aortic VSMC. A, pretreatment of cells with BEL (1 μm, 30 min) inhibited PMA- but not FSK-induced RGS2 mRNA up-regulation. B and C, suppression of iPLA2β expression with iPLA2β antisense oligodeoxynucleotides but not sense oligodeoxynucleotides, as described (16), inhibited PMA- but not FSK-induced RGS2 mRNA accumulation. D and E, cells were pretreated (30 min) with GF (3 μm) or H-89 (20 μm) and then stimulated with Ang II (100 nm, 1 h) in the presence or absence of 30 μm AA and LPC as described (16). Data are represented as mean ± S.E. of at least three independent experiments. ***, p < 0.001 versus basal. ††, p < 0.01; †††, p < 0.001; nonsignificant (NS) versus PMA or FSK stimulation (Veh) in A-C, and Ang II stimulation in D and E.
Because pharmacologic inhibitors including BEL can have off-target effects, we also determined effects of suppressing iPLA2β expression with an antisense oligodeoxynucleotide on RGS2 mRNA accumulation in VSMC (Fig. 2, B and C). We have previously demonstrated that an antisense oligodeoxynucleotide directed against iPLA2β effectively reduces iPLA2β protein levels and inhibits Ang II-induced RGS2 mRNA accumulation in VSMC (16). The antisense oligodeoxynucleotide was also found to significantly reduce RGS2 mRNA accumulation induced by the PKC activator PMA (Fig. 2B) but not to affect that induced by the adenylyl cyclase activator FSK (Fig. 2C). The control sense oligodeoxynucleotide did not affect the response either to PMA or FSK.
As a third approach to examine the participation of iPLA2β in RGS2 mRNA accumulation in VSMC induced by Ang II and activation of PKA and PKC, we determined whether iPLA2β reaction products could reverse the inhibition of RGS2 mRNA accumulation by inhibitors of PKA and PKC. We have previously demonstrated that both AA and LPC can induce RGS2 mRNA accumulation in VSMC in a concentration-dependent manner (16) and thus mimic effects of iPLA2β activation. We found that suppression of Ang II-induced RGS2 mRNA accumulation caused by the PKC inhibitor GF109203X was completely reversed by adding exogenous AA and LPC (Fig. 2D). In contrast, those compounds failed to reverse suppression of Ang II-induced RGS2 mRNA accumulation caused by the PKA inhibitor H-89 (Fig. 2E). These results suggest that information transfer in the signaling pathway linking Ang II receptor activation to RGS2 mRNA accumulation in VSMC proceeds via sequential activation of PKC, iPLA2β, and PKA in that order.
Isolation and Characterization of Mouse RGS2 Promoter
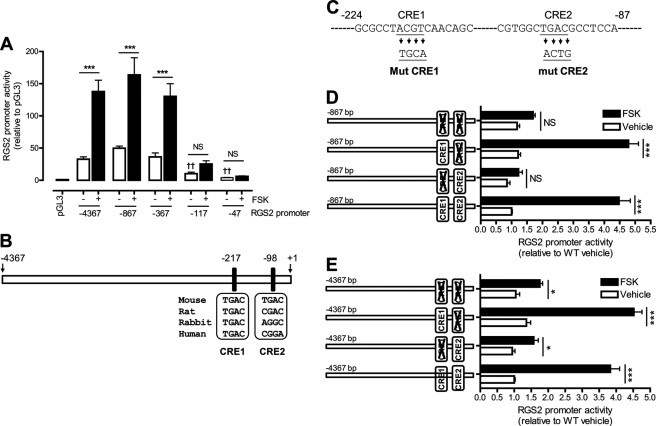
To characterize the promoter region of the Rgs2 gene and examine the interaction of potential regulatory transcription factors with the promoter, we amplified a 4,368-bp genomic DNA fragment that corresponds to the 5′-flanking sequence of the mouse Rgs2 gene by PCR from a bacterial artificial chromosome clone. This fragment includes the sequence from −4367 to +1 bp relative to the translational initiation site (33). To identify cis-regulatory elements in the RGS2 promoter, a series of 5′-end-truncated RGS2 promoter-luciferase constructs were generated from the isolated 4,368-bp fragment. The RGS2 promoter activity of these constructs was determined by dual luciferase assays in rabbit aortic VSMC.
Fig. 3A illustrates that the complete −4,367 bp RGS2 promoter fragment increased transcription by 32-fold over that achieved with the promoterless control construct, which contained only the pGL3 basic vector. 5′-Truncations of the RGS2 promoter fragment from −4,367 to −367 bp had little effect on promoter activity, but further truncation to −117 bp resulted in substantial diminution of promoter activity. This suggests that the −250 bp region between −367 and −117 bp of the RGS2 promoter contains cis-regulatory element(s) that are important for basal promoter activity.
FIGURE 3.
A highly conserved CRE is critical for RGS2 promoter activity. PGL3 basic luciferase reporter vectors containing −4367, −867, −367, −117, and −47-bp RGS2 promoters (A), −867 bp WT or mutant RGS2 promoters (D), and −4367 bp WT or mutant RGS2 promoters (E) were co-transfected with pRL-SV40 vector in rabbit aortic VSMC. Cells were then incubated (6 h) with FSK (10 μm) or vehicle (Veh) (Me2SO2). RGS2 promoter activity was analyzed by dual-luciferase assay and then normalized to pGL3 vector (A) or to WT basal (D and E). Two putative CREB binding sites, CRE1 and CRE2, were predicted in the −4367 bp mouse RGS2 promoter (B). Four key nucleotides in the RGS2 CRE1 and CRE2 sequences were subjected to site-directed mutagenesis (C). Data are represented as mean ± S.E. of at least six independent experiments. ***, p < 0.001 versus pGL3. ††, p < 0.01 versus −4367 bp basal.
To identify such elements that might transmit information from PKA activation to regulation of Rgs2 gene transcription, we examined effects of activating PKA with FSK on RGS2 promoter activity. FSK was found to strongly stimulate transcription from the full-length −4,367 bp isolated RGS2 promoter fragment and from the 5-truncation fragments from −867 and −367 bp (Fig. 3A). Stimulation by FSK declined substantially with further 5′-truncation, as with the fragment from −117 bp. This suggests that the 250 bp region between −367 and −117 bp of the RGS2 promoter contains cis-regulatory elements that are important for both basal and FSK-stimulated promoter activity.
A Highly Conserved CRE Is Critical for RGS2 Promoter Activity
The RGS2 promoter sequence from −4,367 bp was next analyzed with Genomatix MatInspector software to obtain additional information about potential cis-regulatory elements. The resultant analysis predicted a 671-bp mouse RGS2 promoter region from −554 to +116 bp that contains 128 transcription factor binding sites, including two potential CREs (Fig. 3B). The first (CRE1 from −223 to −202 bp) but not the second (CRE2 from −104 to −83 bp) CRE is located in the 250 bp between −367 bp and −117 bp of the RGS2 promoter that was demonstrated to be important for basal and FSK-stimulated RGS2 promoter activity (Fig. 3A). Moreover, the core sequence (TGAC) of CRE1 is highly conserved among mouse, rat, rabbit, and human RGS2 promoter regions (Fig. 3B), although CRE2 is not. This suggests that CRE1 may be a more important regulator of RGS2 promoter activity than CRE2.
To determine whether CRE1 and/or CRE2 affect RGS2 promoter activity under basal conditions and/or upon activation of PKA, we prepared a series of mutant −867 bp RGS2 promoter fragments that contained site-directed mutations in the sequences of CRE1 or CRE2 or both (Fig. 3C). We found that basal promoter activity of the −867 bp RGS2 promoter fragment was unaffected by mutation of either CRE1 or CRE2 (Fig. 3D). In contrast, mutation of CRE1 abolished the stimulatory effect of FSK on promoter activity without or with concomitant mutation of CRE2, although mutation of CRE2 alone had no effect (Fig. 3D). These findings indicate that CRE1 is important in mediating the effects of PKA activation on RGS2 promoter activity but that CRE2 is not.
A similar series of mutants of the −4,349 bp RGS2 promoter fragment was prepared (Fig. 3E). Again we found that basal promoter activity was unaffected by mutation of either CRE1 or CRE2 but that mutation of CRE1 abolished the stimulatory effect of FSK on promoter activity without or with concomitant mutation of CRE2 (Fig. 3E). Moreover, mutation of CRE2 alone had no effect on stimulation of promoter activity by FSK (Fig. 3E). These findings confirm that CRE1 is important in mediating effects of PKA activation on RGS2 promoter activity but that CRE2 is not.
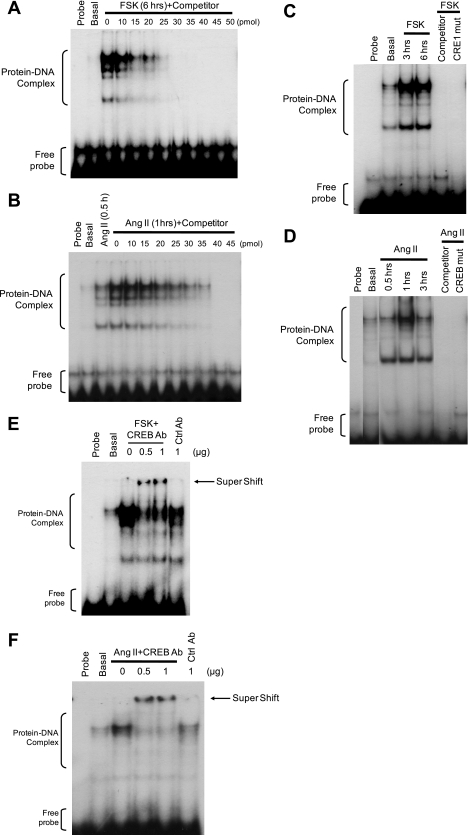
A Highly Conserved CRE Is Required for CREB Binding to RGS2 Promoter
To test whether CRE1 of the RGS2 promoter can bind CREB, nuclear extracts were prepared from rabbit VSMC that had been incubated with or without Ang II or FSK. Equivalent amounts of nuclear protein from each condition were analyzed for protein-DNA complexes using a 32P-labeled RGS2-CRE1 probe consisting of −228 to −198 bp of the mouse RGS2 promoter. FSK (Fig. 4A) and Ang II (Fig. 4B) markedly stimulated formation of protein-DNA complexes, as reflected by four discrete bands that were not observed under basal conditions (i.e. VSMC incubated without FSK or Ang II) or with only the probe (no nuclear protein). To verify that the shifted bands represented specific interactions of the labeled probe with proteins, an unlabeled (“cold”) RGS2-CRE1 oligonucleotide was used as a competitor to displace specifically bound but not nonspecifically bound probe. Radiolabeled probes in the shifted bands produced by incubating VSMC with FSK (Fig. 4A) or with Ang II (Fig. 4B) was effectively displaced by cold RGS2-CRE1 oligonucleotide competitor in a concentration-dependent manner, thereby confirming the specificity of the protein-probe interactions.
FIGURE 4.
A highly conserved CRE is required for CREB binding to the RGS2 promoter. Nuclear proteins were extracted from rabbit aortic VSMC that were quiescent or were stimulated with FSK (10 μm; A, C, and E) or Ang II (100 nm; B, D, and F) for various intervals. EMSA was performed by incubating (30 min) nuclear extract (2 μg) with 32P-labled WT CRE1 (A–F) or mutant CRE1 (C and D) probes from the mouse RGS2 promoter in the presence of various concentrations of unlabeled CRE1 probes (A and B), excess unlabeled consensus CRE probe (C and D), and an anti-CREB or control (Ctrl) antibody (E and F) as described (31, 32). Representative autoradiographs from at least three independent experiments showing protein-DNA complex, supershift (antibody-protein-DNA complex), and free probe.
To determine whether the shifted bands shown in Fig. 4, A and B, contain CREB, three independent approaches were employed. First, a consensus CRE oligonucleotide that is known to bind to CREB was used as a competitor (31). The appearance of shifted bands induced by FSK (Fig. 4C) and Ang II (Fig. 4D) was prevented when consensus CRE oligonucleotides were added. Second, protein-DNA complexes formed with a 32P-labeled mutant RGS2-CRE1 probe were examined under the same conditions used for the WT probe. This probe contained the same mutation that abolished the ability of FSK to stimulate RGS2 promoter activity in Fig. 3, C–E. Both FSK (Fig. 4C) and Ang II (Fig. 4D) failed to stimulate nuclear protein binding to the mutant probe, even though both agents stimulated protein complexation with the WT RGS2-CRE1 probe (Fig. 4, C and D). Third, a specific anti-CREB antibody induced a supershift of the uppermost of the shifted bands produced by incubating VSMC with FSK (Fig. 4E) or Ang II (Fig. 4F), although the lower bands were not supershifted and control antibodies did not affect band mobility. These findings indicate that incubating VSMC with FSK or Ang II increases CREB binding to CRE1 of the RGS2 promoter.
CREB Sequestration or Specific Disruption of CREB Binding to RGS2 Promoter Blocks Forskolin-induced RGS2 mRNA Accumulation in VSMC
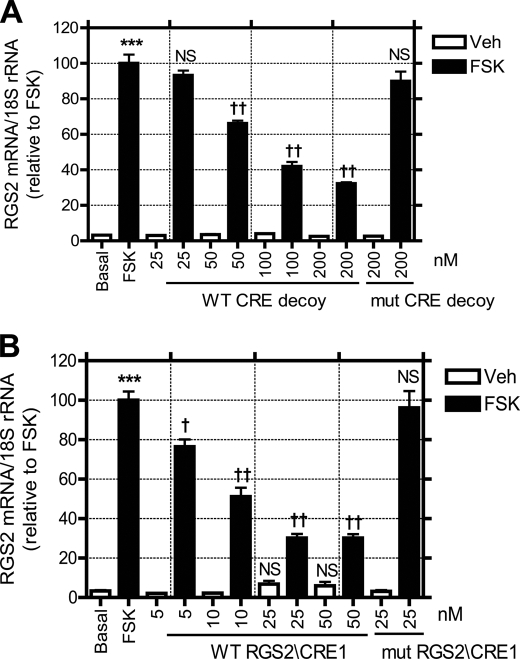
To determine what effect CREB binding to the RGS2 promoter has on RGS2 mRNA accumulation, VSMC were incubated with a 24-mer phosphorothioate oligonucleotide that contains three copies of the consensus CRE sequence. This CRE palindrome has previously been demonstrated to act as a “CRE decoy” that blocks CRE-directed transcription and thereby inhibits cancer cell growth (23). Fig. 5A illustrates that this CRE decoy significantly diminishes FSK-induced RGS2 mRNA accumulation in VSMC in a concentration-dependent manner. Maximal inhibition (67.8%) of FSK-induced RGS2 mRNA accumulation was achieved with 200 nm wild-type CRE decoy. In contrast, that concentration of a mutant phosphorothioate CRE decoy had little effect on FSK-induced RGS2 mRNA accumulation (Fig. 5A).
FIGURE 5.
Sequestration of CREB or disrupting the CREB and RGS2 promoter interaction inhibits forskolin-induced RGS2 mRNA accumulation. Rabbit aortic VSMC were transfected with various concentrations of WT or mutant (mut) CRE decoy (A) or RGS2/CRE1 oligonucleotides (B) using Lipofectamine-Plus reagent. FSK (10 μm, 1 h)-induced RGS2 mRNA up-regulation was determined by real-time PCR. Data are represented as mean ± S.E. of at least five independent experiments. ***, p < 0.001 versus, basal. †, p < 0.05; ††, p < 0.01; non-significant (NS) versus FSK stimulation.
The CRE decoy sequesters CREB and thereby blocks all CREB functions, and the results in Fig. 5A need not reflect exclusive blockade of CREB binding only to the RGS2 promoter. To examine this issue further, we incubated VSMC with a 30-mer RGS2-CRE1 phosphorothioate oligonucleotide (with a sequence identical to the probe used in Fig. 4) that contains CRE1 and flanking sequences. Fig. 5B illustrates that this RGS2-CRE1 oligonucleotide blocked FSK-induced RGS2 mRNA accumulation at much lower concentrations than those required for the CRE decoy used in Fig. 5A. Maximal inhibition (73.8%) of FSK-induced RGS2 mRNA accumulation was achieved with 25 nm RGS2-CRE1 oligonucleotide in Fig. 5B, although similar inhibition required 200 nm CRE decoy used in Fig. 5A. This indicates that the former is about 8-fold more potent than the latter. A mutant phosphorothioate RGS2-CRE1 oligonucleotide at a concentration of 25 nm had no effect on FSK-induced RGS2 mRNA up-regulation (Fig. 5B).
Ang II-induced CREB Phosphorylation and Nuclear Localization Are Blocked by Pharmacologic Inhibition or Genetic Ablation of iPLA2β Activity
Findings so far described that elucidate the signaling sequence in the Ang II/PKC/iPLA2β/PKA pathway (Figs. 1 and 2) and its coupling to the CREB/CRE1/RGS2 transcriptional machinery (Figs. 3–5) do not establish the molecular mechanisms that link all of the steps in the signaling cascade. To explore these issues further, we examined the relationship of iPLA2β activation in VSMC and Ang II-induced phosphorylation of CREB Ser133, which can be effected by PKA and other signaling kinases and is an important event in regulating transcriptional effects of CREB in a variety of cell types (34).
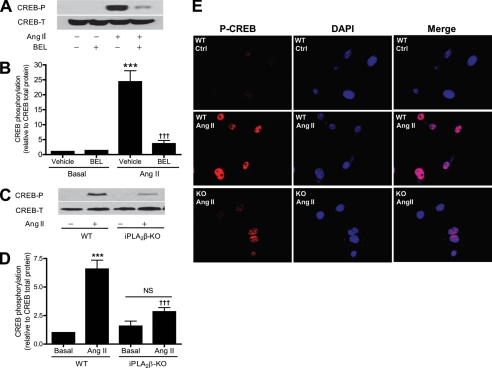
Rat aortic VSMC were incubated with BEL to inhibit iPLA2β, and CREB Ser133 phosphorylation was examined by Western blotting using an antibody that recognizes the CREB phosphopeptide that contains PO4-Ser133. Fig. 6, A and B, illustrate that Ang II strongly stimulated CREB phosphorylation in VSMC and that this effect was nearly completely prevented when iPLA2β was first inactivated with BEL.
FIGURE 6.
Inhibition of iPLA2β with BEL or genetic deletion of iPLA2β alleviates Ang II-induced CREB phosphorylation and nuclear localization. A and B, quiescent rat aortic VSMC were pretreated (30 min) with BEL (3 μm) or vehicle (Veh) (Me2SO) and then stimulated (5 min) with Ang II (100 nm). C, D, and E, quiescent mouse aortic VSMC isolated from WT or iPLA2β-KO (KO) were stimulated with Ang II. Representative Western blots of total CREB and phosphorylated CREB (A and C). Data are represented as mean ± S.E. of at least three independent experiments (B and D). Representative fluorescent micrographs of three independent experiments (E). ***, p < 0.001 versus Veh or basal. †††, p < 0.001 versus Ang II stimulation.
Moreover, although Ang II stimulated CREB Ser133 phosphorylation by 6.6-fold in wild-type mouse VSMC, there was a much smaller effect (1.8-fold) in VSMC from iPLA2β-null mice (Fig. 6, C and D), reflecting about a 75% diminution of Ang II-stimulated CREB Ser133 phosphorylation in VSMC as a consequence of genetic deletion of iPLA2β. The pharmacologic and genetic evidence in Fig. 6 thus indicates that iPLA2β plays an important role in Ang II-induced CREB phosphorylation in VSMC.
It has been demonstrated previously that CREB phosphorylation results in its cytosolic to nuclear translocation in VSMC (35). To determine whether iPLA2β participates in CREB nuclear localization by affecting CREB phosphorylation, we examined effects of Ang II on the subcellular distribution of phospho-CREB (p-CREB) in VSMC from WT and iPLA2β-null mice. The subcellular location of p-CREB was visualized with a phosphopeptide-specific antibody, and nuclei were stained with DAPI. Colocalization of p-CREB and DAPI was interpreted to reflect nuclear localization of CREB. Fig. 6E illustrates that incubating WT VSMC with Ang II caused a marked increase in p-CREB nuclear staining compared with unstimulated cells, but this effect of Ang II was greatly diminished in iPLA2β-null VSMC.
Stimulated cAMP Production by Adenylyl Cyclase Contributes to Ang II-induced PKA/CREB Activation and RGS2 mRNA Up-regulation through Cyclooxygenase (COX)- and Lipoxygenase (LO)-mediated Eicosanoids
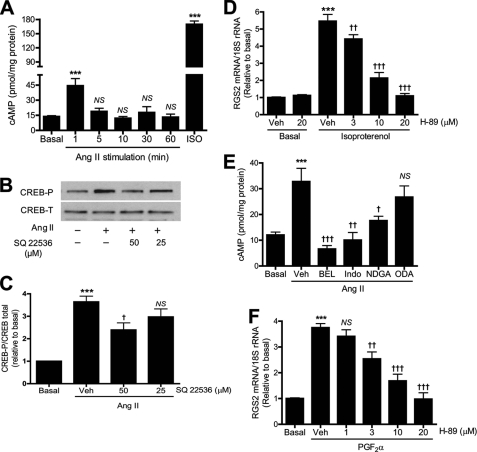
One possible signal pathway by which Ang II may activate PKA is by stimulating cAMP production by adenylyl cyclase. To examine this possibility, rat aortic VSMCs were stimulated with Ang II for various intervals and intracellular cAMP levels were measured by enzyme immunoassay. Ang II was found to stimulate cAMP accumulation, and levels achieved a peak at 1 min and thereafter returned to basal values by 10 min (Fig. 7A).
FIGURE 7.
Ang II-induced cAMP accumulation is mediated by iPLA2, COX, and LO and contributes to CREB phosphorylation and RGS2 mRNA accumulation. A, rat aortic VSMCs were stimulated with Ang II (100 nm) for various intervals or with isoproterenol (ISO, 2 nm, 1 min). Intracellular cAMP levels were measured by enzyme immunoassay. B, representative Western blots of total CREB and phosphorylated CREB. Cells were treated (30 min) with SQ22536 before Ang II stimulation (100 nm, 5 min). C, summary of data from Fig. 7B (n = 3). D, VSMCs were incubated (30 min) with H-89 before incubation (1 h) with isoproterenol (2 nm). E, VSMCs were pretreated (30 min) with BEL (10 μm), nordihydroguaiaretic acid (NDGA) (30 μm), indomethacin (50 μm) (Indo), or 17-octadecynoic acid (ODA) (10 μm), respectively, before incubation (1 h) with Ang II (100 nm). n = 4–12. F, VSMCs were incubated (30 min) with H-89 before incubation (1 h) with PGF2α (30 μm). n = 4. ***, p < 0.001 versus vehicle (Veh) (Me2SO2) or basal. †, p < 0.05; ††; p < 0.01; †††, p < 0.001; NS versus Veh in C–F and basal in A.
To determine whether this rapid and transient increase in cAMP accumulation is involved in Ang II-induced PKA/CREB activation and/or RGS2 transcription, we first examined the effect of the adenylyl cyclase inhibitor SQ22536 on Ang II-induced CREB phosphorylation in VSMC. SQ22536 partially but significantly attenuated Ang II-induced CREB phosphorylation (Fig. 7, B and C). This suggests that activation of adenylyl cyclase contributes, at least in part, to activation of CREB by Ang II.
To further explore the role of adenylyl cyclase in Ang II-induced RGS2 transcription, we examined the effect on RGS2 mRNA accumulation of the β-adrenergic receptor agonist isoproterenol, which is known to simulate adenylyl cyclase activity via Gs (36). Isoproterenol was found to strongly stimulate accumulation of both cAMP (Fig. 7A) and RGS2 mRNA (Fig. 7D). Inhibiting PKA with H-89 diminished isoproterenol-induced RGS2 mRNA accumulation in a concentration-dependent manner. These results indicate that direct activation of Gs-coupled β-adrenergic receptors is sufficient to stimulate RGS2 mRNA accumulation through a PKA-dependent mechanism in VSMC.
To determine whether iPLA2β and products of its action, e.g. arachidonic acid and/or its oxygenated metabolites, are involved in Ang II-induced cAMP accumulation, VSMCs were pretreated with pharmacologic inhibitors of relevant enzymes (16, 25). These include BEL, an iPLA2 inhibitor; nordihydroguaiaretic acid, an inhibitor of LO; indomethacin, an inhibitor of COX-1 and -2; and 17-octadecynoic acid, an inhibitor of cytochrome P450 epoxygenases. Ang II was added after VSMC had been preincubated with one of these inhibitors, and cAMP levels were measured after various intervals. Ang II-induced cAMP accumulation was found to be prevented completely by BEL or indomethacin and was partially inhibited by nordihydroguaiaretic acid, although it was not significantly affected by 17-octadecynoic acid (Fig. 7E). These data suggest that the action of iPLA2 to release AA and its subsequent oxygenation by COX and LO are required for Ang II to induce cAMP accumulation.
To determine whether activating other Gq-coupled receptors can trigger the same PKA/CREB pathway for up-regulating RGS2 transcription, we examined the effect of the PKA inhibitor H-89 on prostaglandin F2α (PGF2α)-induced RGS2 mRNA accumulation. PGFα2 is known to act via Gq, and H-89 was found to inhibit PGFα2-induced RGS2 mRNA accumulation in a concentration-dependent manner (Fig. 7F). These data are consistent with our previous report (16) and suggest that the iPLA2β/PKA/CREB might be a common pathway that links G protein-coupled receptors and RGS2 transcription.
SNPs Identified in Human Hypertensive Patients Are Located in Highly Conserved CRE of RGS2 Promoter and Affect the Ability of FSK to Stimulate Human RGS2 Promoter Activity
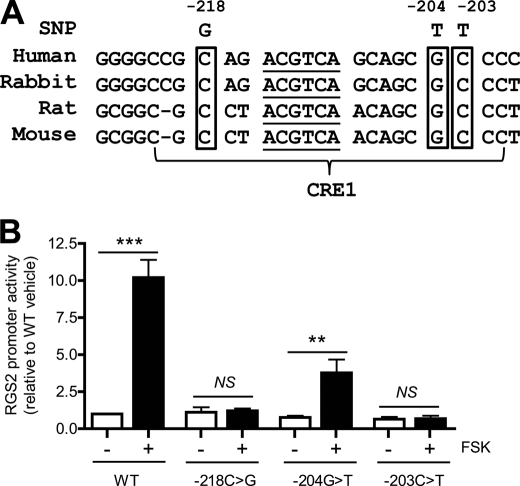
More than 30 RGS2 gene polymorphisms have so far been identified in human hypertensive patients (8–11). Interestingly, three of them (−218C>G, −204G>T, and −203C>T (8)) are located within the CRE1 sequence of the human RGS2 promoter (Fig. 8A). The identities of the nucleotides at each of these three sites are highly conserved in the RGS2 promoter sequences of humans, rabbits, rats, and mice (Fig. 8A). These observations raise the possibility that the three human RGS2 SNPs might negatively affect RGS2 mRNA expression, which might be expected to predispose to hypertension by analogy with effects of reduced RGS2 expression on blood pressure in genetically modified mice (3–7).
FIGURE 8.
Three SNPs identified in human hypertensive patients are located in the highly conserved CRE and implicated in FSK-induced human RGS2 promoter activity. A, a multiple sequence alignment of RGS2 promoters with highlights of the core sequence of CRE1 (underlined) and three human SNPs. B, promoter assay of WT or mutant 399-bp human RGS2 promoter containing single SNP. Data are represented as mean ± S.E. of four independent experiments. **, p < 0.01; ***, p < 0.001.
To examine this possibility, we cloned a 399-bp fragment of the human RGS2 promoter (−398 to +1 bp) and created a series of 3 point mutants that correspond to the sequence alterations observed in the three SNPs identified in the human RGS2 promoter region of hypertensive patients. As expected, FSK strongly stimulated WT human RGS2 promoter activity (Fig. 8B) in a manner similar to that observed with the WT mouse RGS2 promoter (Fig. 3). Each of the three mutant RGS2 promoter constructs that corresponded to the identified human SNPs exhibited a greatly diminished or absent response to FSK. In particular, the −218C>G and −203C>T RGS2 promoter mutants were completely unresponsive to FSK, although the −204G>T mutant exhibited a detectable but much diminished stimulatory response to FSK (Fig. 8B). None of the three RGS2 promoter mutant constructs exhibited altered basal promoter activity, and only the response to FSK was affected.
DISCUSSION
Several novel findings have been obtained in the current study that have not been previously reported to our knowledge. First, PKC activation has been demonstrated to lie upstream of the sequential activation of iPLA2β and PKA in the signaling pathway through which Ang II stimulates RGS2 mRNA accumulation. Second, a highly conserved CRE in the murine RGS2 promoter has been identified as a key cis-regulatory element that is critical for FSK- or Ang II-stimulated CREB binding to the RGS2 promoter and its consequent activation. Third, FSK-induced RGS2 mRNA accumulation has been found to be prevented by CREB sequestration or specific disruption of CREB binding to the RGS2 promoter. Fourth, Ang II-induced CREB phosphorylation and nuclear localization are demonstrated to require iPLA2β activation. Fifth, SNPs identified in human hypertensive patients have been demonstrated to reside in the highly conserved CRE of the RGS2 promoter and affect the ability of FSK to stimulate human RGS2 promoter activity.
Ang II increases transcription of the Rgs2 gene in VSMC, and this represents a potentially important negative feedback mechanism to attenuate Ang II signaling and maintain blood pressure homeostasis (2). The signaling pathways that govern the regulation of Rgs2 gene transcription and the effects of Ang II on that process have been incompletely understood. Although there are multiple RGS isoforms (including RGS2, RGS3S, RGS3L, RGS4, RGS5, RGS10, RGS11, RGS12, and GAIP), Ang II induces a rapid and selective increase only in RGS2 mRNA in cultured VSMC, and the magnitude of this effect is reduced by pharmacologic inhibition of PKC (15). The downstream effectors of PKC activation that mediate Ang II-induced RGS2 mRNA accumulation, however, had heretofore remained unidentified.
In cultured PC12 cells activation of PKA by any of several means strongly stimulates RGS2 mRNA accumulation, and these include addition of cAMP analogs dibutryl-cAMP or 8-CPT-cAMP or by stimulating endogenous cAMP production with the adenylyl cyclase activator forskolin (37). Similar effects occur in other cell types (38, 39), including human VSMC (17). The possibilities that PKA participates in Ang II-induced RGS2 mRNA accumulation and that PKC and PKA cooperate in this process have not previously been examined to our knowledge. Here, we demonstrate that the PKA inhibitor H-89 suppresses Ang II-induced RGS2 mRNA accumulation in VSMC in a concentration-dependent manner (Fig. 1B), and similar results were obtained by inhibiting PKA with (Rp)-8-bromo-cAMP (data not shown). Our findings in experiments that employed combinations of pharmacologic inhibitors and activators of PKC and the adenylyl cyclase activator forskolin indicate that activation of PKC lies upstream of PKA activation in the signaling sequence by which Ang II stimulates RGS2 mRNA accumulation in cultured VSMC, as illustrated in Fig. 1, C and D.
We recently reported that iPLA2β plays an essential role in Ang II-stimulated Rgs2 gene transcription in cultured VSMC (16), although the detailed molecular causes and consequences of iPLA2β activation were not established. In the current study, we examined the relationships among activation of iPLA2β, PKC, and PKA in the signaling cascade whereby Ang II stimulates transcription of the Rgs2 gene.
By using approaches that include pharmacologic inhibition of iPLA2β activity, molecular biologic suppression of iPLA2β expression, and reversal of signaling blockade by adding products of iPLA2β action, we have delineated events in the signaling pathway downstream from the interaction of Ang II with its receptor that involves sequential activation of PKC, iPLA2β, and PKA. Unresolved issues include the mechanism whereby PKC activates iPLA2β, although it is tempting to speculate that iPLA2β phosphorylation is involved. At present, however, there is no direct evidence that PKC can phosphorylate iPLA2β, although PKC activation is clearly involved in effecting iPLA2β translocation from the cytosol to membrane in macrophage-like P388D1 cells (40) and ventricular myocytes (41). Whether a similar mechanism operates in VSMC and participates in the signaling pathway whereby Ang II stimulates RGS2 transcription remains to be determined.
Mouse and rat RGS2 promoters have been previously characterized after their cloning from osteoblasts (42) and 3T3-L1 cells (39). Their promoter activity has been found to be stimulated by activation of cAMP signaling with parathyroid hormone and FSK (42) or 8-bromo-cAMP (39) in a concentration-dependent manner. This is consistent with several reports that PKA is involved in RGS2 transcriptional regulation (17, 37–39). No CRE sites have heretofore been identified in rat or mouse RGS2 promoter regions (42), however, and a role for CREB in regulating RGS2 promoter activity has not previously been established.
Here we provide several lines of evidence that a CRE site in the mouse RGS2 promoter that is highly conserved among several mammalian species including human serves as a key cis-regulatory element that binds CREB and thereby participates in regulating RGS2 promoter activity and its stimulation by Ang II and FSK in VSMC. Evidence supporting these possibilities derive from experiments including promoter deletion analysis (Fig. 3A), predictions based on bioinformatics analysis of the promoter sequence (Fig. 3B), site-directed mutagenesis (Fig. 3, C–E), EMSA (Fig. 4, A–F), and studies involving a CRE decoy or an RGS2/CRE1 oligonucleotide competitor (Fig. 5, A and B).
The prototypical CRE is an 8-base pair palindrome (TGACGTCA). The highly conserved CRE sequence in the promoter region of the RGS2 gene that we have identified contains only the first six nucleotides of the classical 8-bp CRE and thus represents a half-site CRE motif (34). This may account for the failure of previous studies to identify a CRE sequence in RGS2 promoters (42). A half-site motif CRE is less active than the classical 8-bp CRE motif but serves the same function (34). Of 105 genes with functional CREs so far identified, about 50% exhibit the half-site CRE motif (34). In fact, sequence variations in CRE sites of promoters is recognized to be an important mechanism for differential regulation of target genes by cAMP (34).
Our current studies demonstrate that replacing the ACGT sequence in CRE1 of the RGS2 promoter with the sequence TGCA in the RGS2 promoter fragments from −4,367 or −867 bp in the mouse promoter abolishes the ability of FSK to stimulate RGS2 promoter activity (Fig. 3, C–E). This also ablates CREB binding to the RGS2 promoter induced by incubating VSMC with FSK or Ang II (Fig. 4). Moreover, the critical importance of this half-site CRE in the RGS2 promoter is further illustrated by the findings in Fig. 5 that a 30-mer RGS2-CRE oligonucleotide that contains the WT half-site CRE blocks FSK-induced CREB binding to the RGS2 promoter (Fig. 4A) and also blocks accumulation of RGS2 mRNA (Fig. 5B). In contrast, no such blockade is produced by a mutant half-site CRE (Figs. 4, C and D, and 5B).
Several protein kinases, including PKA, have been implicated in Ang II-induced CREB phosphorylation in cultured VSMC (43), but upstream signals that link the Ang II-AT1 receptor interaction to activation of PKA and CREB phosphorylation had not previously been clearly established. In macrophages (20, 21) and endothelial cells (44), iPLA2β has been found to be involved in CREB phosphorylation induced by double-stranded RNA, but whether iPLA2β plays a similar role in Ang II-induced CREB phosphorylation in VSMC had not previously been reported to our knowledge. Here we demonstrate that iPLA2β activity is required for Ang II to induce CREB phosphorylation in VSMC in experiments that involve pharmacologic inhibition of iPLA2β with BEL or genetic ablation of its expression in iPLA2β-null mice (Fig. 6).
Although the molecular mechanism through which iPLA2β regulates PKA/CREB has not yet been established, the iPLA2β reaction products AA and lysophosphatidylcholine and their metabolites are known to be capable of stimulating PKA activity and/or CREB phosphorylation (31, 32, 45, 46). The current study demonstrates that stimulating VSMC with Ang II results in rapid and transient accumulation of cAMP (Fig. 7A). Several lines of evidence suggest that this cAMP accumulation contributes to CREB phosphorylation induced by Ang II and mediated by PKA that stimulates RGS2 transcription. First, Ang II-induced cAMP accumulation peaks at 1 min (Fig. 7A) and precedes Ang II-induced CREB phosphorylation at 5 min (Fig. 6). Second, pharmacologic inhibition of iPLA2, COX, or LO attenuates Ang II-induced cAMP accumulation (Fig. 7E). These data are consistent with our previous report that inhibition of iPLA2, COX, or LO results in suppression of RGS2 transcription (16). Third, activation of adenylyl cyclase with the β-adrenergic receptor agonist isoproterenol stimulates RGS2 transcription in a PKA-dependent manner (Fig. 7D). Notably, although the cAMP level achieved after stimulation with isoproterenol is 4-fold higher than that produced by Ang II (Fig. 7A), both agents induce comparable levels of accumulation of RGS2 mRNA (see Fig. 1A versus 7D). These data imply that cAMP accumulation contributes to the stimulation of RGS2 transcription by Ang II but also suggest that additional mechanisms may be involved. It has been demonstrated previously that Ang II can stimulate PKA in a cAMP-independent manner in VSMC (47) and that the PLA2 reaction product lysoplasmenylcholine can directly stimulate purified PKA activity (45). These observations are consistent with our findings that inhibiting adenylyl cyclase results in partial attenuation of Ang II-induced CREB phosphorylation (Fig. 7, B and C) but that inhibiting iPLA2β with BEL results in complete inhibition CREB phosphorylation (Fig. 6), as does inhibiting PKA with H-89 (Fig. 1B).
It is striking that heterozygous Rgs2 knock-out mice are as hypertensive as homozygous Rgs2 knock-out mice (3), which indicates that modest reduction of RGS2 levels is sufficient to result in hypertension in mice and that the complete absence of RGS2 is not required for this effect. This and similar findings (4) highlight a potentially important role of RGS2 in the pathogenesis of hypertension and have motivated clinical studies in which more than 30 SNPs within the RGS2 gene have been identified in hypertensive patients (8–11). Although it is assumed that RGS2 SNPs associated with hypertension may alter RGS2 expression and/or activity, there is little direct evidence for this. An intriguing aspect of the studies reported here is that three human RGS2 SNPs that have been identified in hypertensive patients are demonstrated to be located at sites within the CREB binding site of human RGS2 promoter. Moreover, mutagenesis of each of these sites to produce promoter sequences corresponding to those that result from the SNPs resulted in reduction or abolition of the effect of FSK to stimulate human RGS2 promoter activity (Fig. 8).
It has been recognized that binding of CREB to CRE in certain promoters is influenced both by the core sequence (TGACGTCA) and sequences that flank the CRE palindrome (34). Indeed, each of the reported SNPs in the human RGS2 promoter occur at sites that flank the human RGS2 CRE core sequence. The mechanism(s) by which an individual RGS2 SNP preferentially diminishes the ability of FSK to stimulate human RGS2 promoter activity have not yet been elucidated, but it is tempting to suggest that different RGS2 SNPs may cause distinct DNA conformational changes that differentially affect CREB binding. Regardless of the exact molecular mechanism that is responsible for the loss of the ability of FSK to stimulate human RGS2 promoter activity associated with the SNPs, the findings reported here demonstrate that three SNPs that have been identified in the human RGS2 promoter affect regulation of promoter activity and may contribute to the pathogenesis of essential hypertension via their effects on RGS2 mRNA expression.
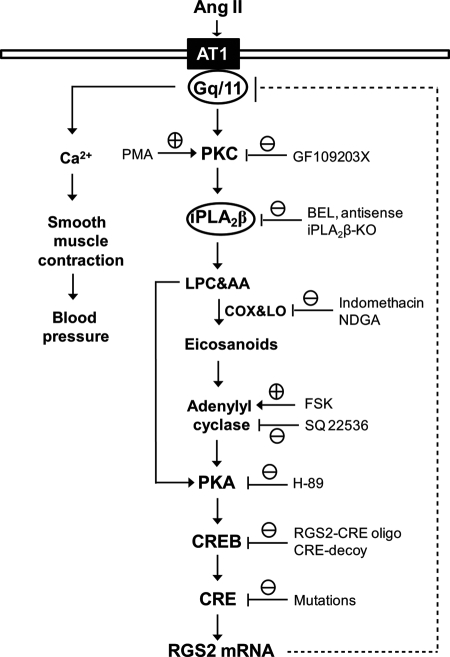
In summary, our studies have identified a highly conserved CRE in the RGS2 promoter that is a key cis-regulatory element in transcriptional regulation of the RGS2 gene that is affected by the Ang II signaling pathway in VSMC. Fig. 9 illustrates a model based on our findings that outlines the sequence of events in the Ang II signaling pathway and summarizes the experimental approaches used to characterize them. Although the effect of Ang II interaction with the AT1 receptor has long been recognized to induce vasoconstriction and a rise in blood pressure, it has only recently been appreciated that Ang II simultaneously initiates a negative feedback loop that serves to restrain its vasopressor effect (2–4). The current study delineates several steps involved in this negative feedback loop and demonstrates sequential activation of PKC, iPLA2β, adenylyl cyclase, PKA, and CREB that result in CREB binding to a CRE site in the RGS2 promoter that activates transcription of the RGS2 gene. These findings identify the interaction between CREB and the RGS2 promoter CRE as a potential therapeutic target for prevention and/or treatment of hypertension and associated cardiovascular disease.
FIGURE 9.
Model for Ang II-induced and PKC/iPLA2β/PKA/CREB-mediated RGS2 mRNA accumulation in VSMC. The pharmacological, molecular, and genetic approaches used in this study are also indicated.
Acknowledgment
We thank Dr. Ming C. Gong for valuable suggestions.
This work was supported, in whole or in part, by National Institutes of Health Grants HL088389 and HL088389-02S1 and a Scientist Development Grant from the American Heart Association (to Z. G.).
- RGS2
- regulator of G-protein signaling-2
- Ang II
- angiotensin II
- VSMC
- vascular smooth muscle cells
- iPLA2
- calcium-independent phospholipase A2
- PKC
- protein kinase C
- PKA
- protein kinase A
- BEL
- bromoenol lactone
- CRE
- cAMP-response element
- CREB
- cAMP-response element-binding protein
- SNP
- single nucleotide polymorphism
- AA
- arachidonic acid
- LPC
- 1-radyl, 2-lyso-glycerophosphocholine
- FSK
- forskolin
- GF
- GF109203X
- PMA
- phorbol myristate acetate
- COX
- cyclooxygenase
- LO
- lipoxygenase
- CYP
- cytochrome P450-dependent epoxygenase
- PGF2α
- prostaglandin F2α.
REFERENCES
- 1. Wieland T., Mittmann C. (2003) Pharmacol. Ther. 97, 95–115 [DOI] [PubMed] [Google Scholar]
- 2. Gu S., Cifelli C., Wang S., Heximer S. P. (2009) Clin. Sci. 116, 391–399 [DOI] [PubMed] [Google Scholar]
- 3. Heximer S. P., Knutsen R. H., Sun X., Kaltenbronn K. M., Rhee M. H., Peng N., Oliveira-dos-Santos A., Penninger J. M., Muslin A. J., Steinberg T. H., Wyss J. M., Mecham R. P., Blumer K. J. (2003) J. Clin. Invest. 111, 445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tang K. M., Wang G. R., Lu P., Karas R. H., Aronovitz M., Heximer S. P., Kaltenbronn K. M., Blumer K. J., Siderovski D. P., Zhu Y., Mendelsohn M. E., Tang M., Wang G. (2003) Nat. Med. 9, 1506–1512 [DOI] [PubMed] [Google Scholar]
- 5. Sun X., Kaltenbronn K. M., Steinberg T. H., Blumer K. J. (2005) Mol. Pharmacol. 67, 631–639 [DOI] [PubMed] [Google Scholar]
- 6. Gross V., Tank J., Obst M., Plehm R., Blumer K. J., Diedrich A., Jordan J., Luft F. C. (2005) Am. J. Physiol. Regul. Integr. Comp. Physiol. 288, R1134–1142 [DOI] [PubMed] [Google Scholar]
- 7. Hercule H. C., Tank J., Plehm R., Wellner M., da Costa Goncalves A. C., Gollasch M., Diedrich A., Jordan J., Luft F. C., Gross V. (2007) Exp. Physiol. 92, 1014–1022 [DOI] [PubMed] [Google Scholar]
- 8. Yang J., Kamide K., Kokubo Y., Takiuchi S., Tanaka C., Banno M., Miwa Y., Yoshii M., Horio T., Okayama A., Tomoike H., Kawano Y., Miyata T. (2005) J. Hypertens. 23, 1497–1505 [DOI] [PubMed] [Google Scholar]
- 9. Zhao Q., Wang L., Yang W., Chen S., Huang J., Fan Z., Li H., Lu X., Gu D. (2008) Pharmacogenet. Genomics 18, 459–466 [DOI] [PubMed] [Google Scholar]
- 10. Semplicini A., Lenzini L., Sartori M., Papparella I., Calò L. A., Pagnin E., Strapazzon G., Benna C., Costa R., Avogaro A., Ceolotto G., Pessina A. C. (2006) J. Hypertens. 24, 1115–1124 [DOI] [PubMed] [Google Scholar]
- 11. Riddle E. L., Rana B. K., Murthy K. K., Rao F., Eskin E., O'Connor D. T., Insel P. A. (2006) Hypertension 47, 415–420 [DOI] [PubMed] [Google Scholar]
- 12. Semplicini A., Strapazzon G., Papparella I., Sartori M., Realdi A., Macchini L., Calò L. A., Ceolotto G. (2010) J. Hypertens. 28, 1104–1108 [DOI] [PubMed] [Google Scholar]
- 13. Calò L. A., Pagnin E., Davis P. A., Sartori M., Ceolotto G., Pessina A. C., Semplicini A. (2004) J. Clin. Endocrinol. Metab. 89, 4153–4157 [DOI] [PubMed] [Google Scholar]
- 14. Touyz R. M., Schiffrin E. L. (2000) Pharmacol. Rev. 52, 639–672 [PubMed] [Google Scholar]
- 15. Grant S. L., Lassègue B., Griendling K. K., Ushio-Fukai M., Lyons P. R., Alexander R. W. (2000) Mol. Pharmacol. 57, 460–467 [DOI] [PubMed] [Google Scholar]
- 16. Xie Z., Gong M. C., Su W., Turk J., Guo Z. (2007) J. Biol. Chem. 282, 25278–25289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cho H., Harrison K., Schwartz O., Kehrl J. H. (2003) Biochem. J. 371, 973–980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Li Y., Hashim S., Anand-Srivastava M. B. (2005) Cardiovasc. Res. 66, 503–511 [DOI] [PubMed] [Google Scholar]
- 19. Ma Z., Turk J. (2001) Prog. Nucleic Acids Res. Mol. Biol. 67, 1–33 [DOI] [PubMed] [Google Scholar]
- 20. Maggi L. B., Jr., Moran J. M., Scarim A. L., Ford D. A., Yoon J. W., McHowat J., Buller R. M., Corbett J. A. (2002) J. Biol. Chem. 277, 38449–38455 [DOI] [PubMed] [Google Scholar]
- 21. Moran J. M., Buller R. M., McHowat J., Turk J., Wohltmann M., Gross R. W., Corbett J. A. (2005) J. Biol. Chem. 280, 28162–28168 [DOI] [PubMed] [Google Scholar]
- 22. Hazen S. L., Zupan L. A., Weiss R. H., Getman D. P., Gross R. W. (1991) J. Biol. Chem. 266, 7227–7232 [PubMed] [Google Scholar]
- 23. Park Y. G., Nesterova M., Agrawal S., Cho-Chung Y. S. (1999) J. Biol. Chem. 274, 1573–1580 [DOI] [PubMed] [Google Scholar]
- 24. Xie Z., Su W., Guo Z., Pang H., Post S. R., Gong M. C. (2006) Cardiovasc. Res. 69, 491–501 [DOI] [PubMed] [Google Scholar]
- 25. Xie Z., Gong M. C., Su W., Xie D., Turk J., Guo Z. (2010) J. Biol. Chem. 285, 8628–8638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pang H., Guo Z., Su W., Xie Z., Eto M., Gong M. C. (2005) Am. J. Physiol. Cell Physiol. 289, C352–360 [DOI] [PubMed] [Google Scholar]
- 27. Pang H., Guo Z., Xie Z., Su W., Gong M. C. (2006) Am. J. Physiol. Cell Physiol. 290, C892–899 [DOI] [PubMed] [Google Scholar]
- 28. Guo Z., Su W., Allen S., Pang H., Daugherty A., Smart E., Gong M. C. (2005) Cardiovasc. Res. 67, 723–735 [DOI] [PubMed] [Google Scholar]
- 29. Su W., Guo Z., Randall D. C., Cassis L., Brown D. R., Gong M. C. (2008) Am. J. Physiol. Heart Circ. Physiol. 295, H1634–1641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Dyer B. W., Ferrer F. A., Klinedinst D. K., Rodriguez R. (2000) Anal. Biochem. 282, 158–161 [DOI] [PubMed] [Google Scholar]
- 31. Dronadula N., Rizvi F., Blaskova E., Li Q., Rao G. N. (2006) J. Lipid Res. 47, 767–777 [DOI] [PubMed] [Google Scholar]
- 32. Chava K. R., Karpurapu M., Wang D., Bhanoori M., Kundumani-Sridharan V., Zhang Q., Ichiki T., Glasgow W. C., Rao G. N. (2009) Arterioscler. Thromb. Vasc. Biol. 29, 809–815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Antonarakis S. E. (1998) Hum. Mutat. 11, 1–3 [DOI] [PubMed] [Google Scholar]
- 34. Mayr B., Montminy M. (2001) Nat. Rev. Mol. Cell Biol. 2, 599–609 [DOI] [PubMed] [Google Scholar]
- 35. Stevenson A. S., Cartin L., Wellman T. L., Dick M. H., Nelson M. T., Lounsbury K. M. (2001) Exp. Cell Res. 263, 118–130 [DOI] [PubMed] [Google Scholar]
- 36. Kubalak S. W., Webb J. G. (1993) Am. J. Physiol. Heart Circ. Physiol. 264, H86–96 [DOI] [PubMed] [Google Scholar]
- 37. Pepperl D. J., Shah-Basu S., VanLeeuwen D., Granneman J. G., MacKenzie R. G. (1998) Biochem. Biophys. Res. Commun. 243, 52–55 [DOI] [PubMed] [Google Scholar]
- 38. Miles R. R., Sluka J. P., Santerre R. F., Hale L. V., Bloem L., Boguslawski G., Thirunavukkarasu K., Hock J. M., Onyia J. E. (2000) Endocrinology 141, 28–36 [DOI] [PubMed] [Google Scholar]
- 39. Cheng Y. S., Lee T. S., Hsu H. C., Kou Y. R., Wu Y. L. (2008) J. Cell. Biochem. 105, 922–930 [DOI] [PubMed] [Google Scholar]
- 40. Akiba S., Mizunaga S., Kume K., Hayama M., Sato T. (1999) J. Biol. Chem. 274, 19906–19912 [DOI] [PubMed] [Google Scholar]
- 41. Steer S. A., Wirsig K. C., Creer M. H., Ford D. A., McHowat J. (2002) Am. J. Physiol. Cell Physiol. 283, C1621–1626 [DOI] [PubMed] [Google Scholar]
- 42. Thirunavukkarasu K., Halladay D. L., Miles R. R., Geringer C. D., Onyia J. E. (2002) J. Cell. Biochem. 85, 837–850 [DOI] [PubMed] [Google Scholar]
- 43. Funakoshi Y., Ichiki T., Takeda K., Tokuno T., Iino N., Takeshita A. (2002) J. Biol. Chem. 277, 18710–18717 [DOI] [PubMed] [Google Scholar]
- 44. Martinson B. D., Albert C. J., Corbett J. A., Wysolmerski R. B., Ford D. A. (2003) J. Lipid Res. 44, 1686–1691 [DOI] [PubMed] [Google Scholar]
- 45. Williams S. D., Ford D. A. (1997) FEBS Lett. 420, 33–38 [DOI] [PubMed] [Google Scholar]
- 46. Williams S. D., Ford D. A. (2001) Am. J. Physiol. Heart Circ. Physiol. 281, H168–176 [DOI] [PubMed] [Google Scholar]
- 47. Dulin N. O., Niu J., Browning D. D., Ye R. D., Voyno-Yasenetskaya T. (2001) J. Biol. Chem. 276, 20827–20830 [DOI] [PubMed] [Google Scholar]