Background: Cyclotides are plant-derived cyclic peptides that are divided into Möbius and bracelet subfamilies. Currently, only four linear variants of the Möbius subfamily have been isolated.
Results: We discovered hedyotide B2 as the first linear representative of the bracelet subfamily.
Conclusion: Hedyotide B2 shares the same connectivity as conventional cyclotides. Its linear structure is genetically predetermined.
Significance: Our study broadens our knowledge of linear cyclotides.
Keywords: Antimicrobial Peptides, Disulfide, Plant Defense, Protein Expression, Proteomics, Cyclotide, Disulfide Connectivity, Hedyotide, Top-down, Uncyclotide
Abstract
Cyclotides are heat-stable macrocyclic peptides from plants that display a wide range of biological activities. They can be divided into two subfamilies: Möbius or bracelet, based on the presence or absence of a cis-proline residue in loop 5, respectively. Currently, over 150 cyclotides have been discovered, but only four linear variants of the Möbius subfamily have been hitherto isolated. In this study, we report the discovery of two novel cyclotides, hedyotide B1 and hedyotide B2, from the aerial parts of Hedyotis biflora. Hedyotide B1 has a cyclic cystine knot structure typical of cyclotides. Interestingly, hedyotide B2 possesses a linear backbone and is the first linear representative of the bracelet subfamily. Disulfide mapping of hedyotide B2 by a top-down MS/MS approach showed that it shares the same knotted disulfide arrangement as conventional cyclotides. Its unfolding pathway also showed that the penetrating disulfide bond Cys III–VI is the most stable disulfide linkage. Cloning of the gene encoding hedyotide B2 revealed a nonsense mutation that introduces a premature stop codon at the conserved Asn residue position, which is essential for an end-to-end backbone ligation. Biophysical characterization showed that hedyotide B2 was more susceptible to exopeptidase degradation as compared with hedyotide B1. Hedyotide B2 was also inactive against all four tested bacterial strains, whereas hedyotide B1 was bactericidal to Escherichia coli and Streptococcus salivarius at low micromolar concentration. Our results provide a deeper understanding of the structures, functions, and biosynthetic processing of cyclotides and uncyclotides in plants.
Introduction
Cyclotides are plant-derived cyclic peptides containing 28–37 residues (1). They can be considered to be mini-proteins because of their well defined three-dimensional structures with an end-to-end circular peptide backbone cross-braced by three disulfide (SS)2 bonds in a cystine-knot arrangement of cysteine I–IV, II–V, and III–VI (1, 2). The extraordinary stability of cyclotides against proteolytic, thermal, or chemical degradation can be attributed to the absence of both termini and the presence of a knotted SS arrangement in a cyclic structure (3). These defining features make the cyclotide scaffold an attractive template for engineering biologic drug candidates (4). The first cyclotide, kalata B1, was discovered by a Norwegian doctor, Lorents Gran, in the early 1970s (5). During his Red Cross mission in Congo, he observed that the local women consumed a decoction called “Kalata-Kalata” to accelerate childbirth. The decoction was made from the medicinal plant Oldenlandia affinis (synonym Hedyotis affinis), which belongs to the Oldenlandia/Hedyotis genus of the Rubiaceae (coffee family). It was later identified that the peptide kalata B1 was responsible for the decoction's oxytocic activity. The NMR structure and disulfide pairings of kalata B1 were revealed by Saether et al. (6) and confirmed by chemical synthesis (7) in the 1990s, some 20 years after its discovery. Since then, approximately 150 cyclotide sequences (2) have been found in the Rubiaceae (8), Violaceae (9), Cucurbitaceae (10), and Fabaceae (11, 12) families. Cyclotides likely play a role in host defense in plants and are known to exhibit a diverse range of biological activities such as anti-HIV (13), antimicrobial (7), hemolytic (14), uterotonic (15), insecticidal (16), and nematicidal (17) activities.
With a circular structure, cyclotides can be conveniently divided into six individual loops bound by successive Cys residues (Cys I–VI). NMR studies have shown that these cyclotide loops are often solvent-exposed because of the occupancy of the interior space by the cystine knot. Topologically, cyclotides fall into two subfamilies: Möbius or bracelet, based on the presence or absence of a cis-Pro peptide bond in loop 5 (18). The cis-Pro bond causes a twist in the circular backbone of the Möbius cyclotides, a unique structural feature that is absent in the bracelet cyclotides. Of the 150 sequences found in the Cybase database (19), the majority of cyclotides (>67%) belong to the bracelet subfamily (20).
Recently, a few linear variants of cyclotides have been discovered (21, 22). Their occurrences appear to be rare, with only two naturally occurring linear variants having been reported: violacin A from Viola odorata (21) and psyle C from Psychotria leptothyrsa (22). Two other linear cyclotides have been described from O. affinis but could have been linearized from their cyclic forms by the chemical degradation of the Asn-Gly or Asp-Gly bond of loop 6 through the aspartimide formation and subsequent ring opening during the isolation procedure or the decomposition of dried plant samples (23). Interestingly, all four linear cyclotides are confined to the Möbius subfamily. Bracelet cyclotides account for approximately two-thirds of cyclotide sequences in the database (20), but their linear variants are yet to be reported.
The linear forms of the Möbius cyclotides share homologous sequences to their cyclic forms and display similar structures as determined by NMR (21, 24). A recent report shows that the formation of linear cyclotides is predetermined at the gene level (21). The cDNA encoding the precursor of the linear cyclotide violacin A contains a nonsense mutation that introduces a premature stop codon located one residue prior to the C-terminal Asn/Asp (21). This mutation prevents the translation of the C-terminal Asn/Asp, a residue essential for the backbone cyclization (25). A similar mutation may have happened in psyle C because the vital Asn/Asp residue is also absent in its primary sequence. Because the linear, uncyclized form of a cyclotide has a biosynthetic origin and its descriptive term as a linear cyclotide is an oxymoron and confusing, we propose to refer a linear cyclotide appropriately as an “uncyclotide” (uncyclized cyclotide).
In this study, we examined seven different species belonging to the Spermacoceae tribes (Rubiaceae) and identified Hedyotis biflora as a cyclotide-producing plant. H. biflora is a small annual herb traditionally used to treat body pain in fever and malaria (26). Two novel peptides, hedyotide B1 and B2, were isolated from the aerial parts of H. biflora. Hedyotide B1 was found to be a bracelet cyclotide and hedyotide B2 a bracelet uncyclotide. Interestingly, hedyotide B2 represents the first example of an uncyclotide in the bracelet subfamily. This exciting finding has prompted our further characterization of their genetic sequences, disulfide patterns, tissue-specific distribution, biodegradation pathways, and antimicrobial activities. Our results provide a deeper understanding of the structures, functions, and biosynthetic processing of cyclotides and uncyclotides in plants.
EXPERIMENTAL PROCEDURES
Screening Procedure for the Occurrence of Cyclotides in Plants
500 mg of material from each plant species was macerated and extracted with 2 ml of 50% ethanol. The extracts were diluted 5-fold and subjected to C18 solid phase extraction (SPE) columns. The SPE columns were washed with 20% acetonitrile (ACN) and eluted with 80% ACN. The eluted fractions were subjected to MALDI-TOF MS to scan for mass signals in the 2–4-kDa range. Plant extracts showing positive signal in the desired mass range were subjected further for S-reduction and S-alkylation to verify the disulfide content.
Isolation and Purification of Hedyotide B1 and B2
Fresh aerial material (1 kg) of H. biflora were macerated with 5 liters of 50% ethanol and partitioned with 2.5 liters of dichloromethane for defatting. After removal of plant debris, the ethanol/water fraction was dried in vacuum and dissolved in 200 ml of 10% ethanol. The concentrated extract was subjected to flash column packed with 100 g of C18 media (Grace Davison). The column was washed with 20% ACN and eluted with 80% ACN to obtain the cyclotide-enriched fraction containing mainly hedyotide B1 and B2. Isolation of individual peptides was then achieved by repetitive RP-HPLC using a Shimadzu system. The approximate yields were ∼0.2 mg for hedyotide B1 and 0.5 mg for hedyotide B2.
S-Reduction and S-Alkylation
20 μg of each peptide was dissolved in 100 μl of NH4HCO3 buffer (100 mm, pH 7.8) containing 10 mm DTT and incubated for 1 h at 37 °C. A 2-fold excess of iodoacetamide (IAA) over the total thiol was added and incubated for 1 h at 37 °C. S-Alkylated peptides were purified by reversed phase (RP)-HPLC.
Enzymatic Digestion and Sequence Determination
Lyophilized S-alkylated peptides were digested with endoproteinase Glu-C, trypsin, or chymotrypsin and sequenced by MALDI-MS/MS as described previously (11). Assignments of isobaric residues Ile/Leu and Lys/Gln were based on cDNA sequences.
Cloning of Hedyotide Genes
RNA was prepared from fresh leaves and converted to single-stranded cDNA. Partial encoding genes of hedyotide B1 and B2 were amplified by 3′-RACE PCR (Invitrogen; catalogue number 18373-019) using two degenerate forward primers: 5′-CGATCGATTGYGGIGARAGTTGY-3′ encoding CGESC sequence and 5′-GGGGATCCTGYGGIGARACITG-3′ encoding CGETC sequence. The remaining encoding genes were obtained by 5′-RACE PCR (Invitrogen; catalogue number 18374-058) using reverse primers based on the cDNA sequences obtained from 3′-RACE PCR. Primers used for 5′-RACE PCR of hedyotide B1 and B2 were: 5′-TTCGCTCTCAGATGCAGCAGC-3′ encoding AAASESE sequence and 5′-TAACTGGAACATATTTTGTTT-3′ encoding KNKICSS sequence.
To identify intron locations, genomic DNA was extracted from fresh leaves. PCRs on DNA templates were then conducted with specific primers designed against 5′- and 3′-untranslated regions of each cyclotide.
Top-down Disulfide Mapping
Hedyotide B2 (0.2 mg) was partially reduced in 500 μl of 100 mm citrate buffer, pH 3.0, 20 mm tris(2-carboxyethyl)phosphine at 37 °C for 35 min. Subsequently, N-ethylmaleimide (NEM) powder was added directly to a final concentration of 50 mm and incubated at 37 °C for another 15 min. The reaction was quenched by immediate injection of samples into a Vydac C18 column (250 × 4.6 mm) at a flow rate of 1 ml/min. Intermediate species were separated with a linear gradient of 0.3% min−1 of 10–60% buffer B and analyzed with MALDI-TOF MS to verify the number of NEM-alkylated cysteines. NEM-alkylated intermediate species were then fully reduced with 20 mm DTT and incubated at 37 °C for 60 min. The reduced peptides were S-alkylated with 40 mm IAA and incubated at 37 °C for 30 min before stopping the reaction by injection into HPLC. S-Alkylated peptides were sequenced directly by MS/MS.
Phylogenetic Tree Construction
A phylogenetic tree was constructed using translated precursors of eight cyclotides including hedyotide B1 and B2 (H. biflora), kalata B1 (GenBankTM accession number AAL05477.1), kalata B2 (GenBankTM accession number P58454.1), kalata B3/6 (GenBankTM accession number AAL05478.1), kalata B7 (GenBankTM accession number AAL05478.1), hcf-1 (GenBankTM accession number CB083237.1), and cycloviolacin O8 (GenBankTM accession number FJ211181.1). Cycloviolacin O8 was used as an outgroup to root the phylogenetic tree. The precursor protein sequences were aligned using ClustalW (27). Phylogenetic tree was visualized by Tree View software (28).
Antibacterial Assay
Four bacterial strains from the ATCC were used including Staphylococcus aureus ATCC 12600, Streptococcus salivarius ATCC 13419, Streptococcus epidermidis ATCC 14990, and Escherichia coli ATCC 25922. All the strains were cultured in trypticase soy broth. The antimicrobial activities of hedyotide B1 and B2 were examined using the radial diffusion assay as described by Lehrer et al. (29) under a low salt condition (10 mm sodium phosphate). D4R was used as the positive control (30). D4R, (RLYR)4-[K2K], is a dendrimeric antimicrobial peptide containing four copies of a tetrapeptide RLYR on a K2K core.
Homology Modeling
Computer model of hedyotide B2 was built by MODELLER 9v7 (31) using circulin A (Protein Data Bank code 1BH4) as a template structure (32). The generated computer models were evaluated by DOPE, MolPDF, and GA341 (31). The structure with the lowest energy was used. The structures were analyzed and represented by PyMOL.
RESULTS
Screening for Novel Cyclotide-producing Plants
Seven Spermacoceae species comprising of Borreria laevicaulis, Borreria latifolia, H. biflora, Hygrophila corymbosa, Mitracarpus hirtus, Spermacoceae exilis, and Spermacoceae auricularia were screened for the presence of cyclotides. Their occurrence was determined using three criteria (8): hydrophobicity (desorbed from C18 at 25–55% ACN), mass range (2500–4000 Da), and disulfide content (three cystine bonds). Approximately 500 mg of fresh material of each plant species was homogenized in 2 ml of 50% ethanol. Each plant extract was diluted 5-fold and subjected to a C18 SPE column. Small molecules and polar substances that potentially interfered with the mass spectrometry signals were removed by washing the SPE column with 20% ACN. Cyclotides typically do not elute under this condition because of their hydrophobicity and usually desorb from SPE column between 25 and 55% ACN. To ensure complete desorption, the putative cyclotide-containing fractions were eluted with 80% ACN. These fractions were profiled and fingerprinted by MALDI-TOF MS.
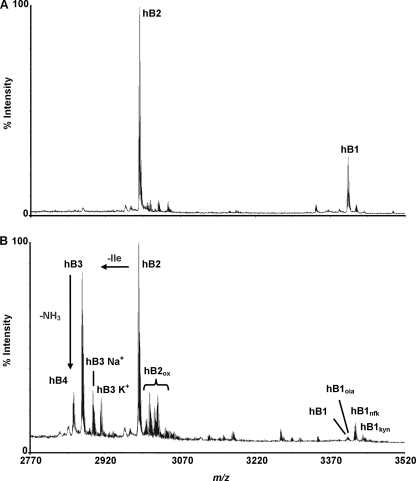
From seven plant extracts, only H. biflora showed strong positive signals in the mass range indicative of cyclotides (Fig. 1). The other six Spermacoceae species gave negative results under our screening procedure. It is possible that cyclotides in these species occur in such a low level of abundance that they escaped our detection. The disulfide content of the putative cyclotide compounds in H. biflora was then examined by comparing the mass difference before and after S-alkylation with IAA. Each S-alkylated half-cystine residue caused a mass increase of 58 Da. Most of these compounds displayed a mass shift of 348 Da, indicating the presence of three cystine bonds and suggesting the occurrence of cyclotides in H. biflora.
FIGURE 1.
MS fingerprint of cyclotides in H. biflora. Hedyotide B1 (hB1) and hedyotide B2 (hB2) are labeled at the tops of the corresponding peaks.
Isolation and Sequence Determination of Hedyotide B1 and B2
Two novel cyclotides with m/z of 3403 and 2986 Da were isolated from the aerial parts of H. biflora by C18 RP-HPLC. They were designated as hedyotide B1 and hedyotide B2, respectively. To determine their primary structures, these peptides were S-reduced with DTT and S-alkylated with IAA. The S-alkylated peptides were cleaved with trypsin, chymotrypsin, or endoproteinase Glu-C. The resulting peptide fragments were sequenced by tandem mass spectrometry.
The primary sequences of hedyotide B1 and B2 are summarized in Table 1. A database search revealed that both peptides are novel and belong to the cyclotide family. Hedyotide B1 and B2 share 58 and 85% sequence identity to cycloviolin A from Leonia cymosa and circulin A from Chassalia parvifolia, respectively. They are both categorized into the bracelet subfamily because of the absence of the cis-proline residue in loop 5. In congruency with previously reported cyclotides, hedyotide B1 possesses a typical cyclic backbone intertwined by three disulfide bridges. It is one of the most cationic cyclotides with a +3 net charge, with the only known example having six aromatic residues as opposed to the usual two to three residues found in most cyclotides. Hedyotide B2, surprisingly, has a linear structure because it lacks the essential Asn/Asp residue in the putative loop 6 of the cyclized form. Although few examples of linear cyclotides have been identified, they all belong to the Möbius subfamily (21–23). This makes hedyotide B2 the first linear variant or uncyclotide of the bracelet subfamily to be discovered. It is also worth noting that hedyotide B2 is the most abundant peptide among the cyclotides and uncyclotides expressed in the aerial tissues of H. biflora.
TABLE 1.
Novel cyclotides and uncyclotides in Hedyotis biflora
Cysteine residues are bold and underlined. pQ is pyroglutamic acid.
| Hedyotide | Sequence | AAa | Molecular weightb | Net charge |
|---|---|---|---|---|
| hB1 | GTRCGETCFVLPCWSAKFGCYCQKGFCYRN | 30 | 3402 | +3 |
| hB2c | IQCGESCVWIPCISSAWGCSCKNKICSS | 28 | 2985 | +1 |
| hB3c | QCGESCVWIPCISSAWGCSCKNKICSS | 27 | 2872 | +1 |
| hB4c | pQCGESCVWIPCISSAWGCSCKNKICSS | 27 | 2855 | +1 |
a Number of amino acids (AA).
b Molecular weights are reported as monoisotopic masses.
c Uncyclotides.
The open-ended structure of hedyotide B2 was first suspected when digestion of its S-alkylated derivative with endoproteinase Glu-C generated two fragments with m/z of 2747 and 606 Da (supplemental Fig. S1A). Because most cyclotides contain a single Glu residue, digestion with endoproteinase Glu-C will linearize the cyclic backbone, yielding a single linear fragment. There are a few examples of cyclotides containing two Glu residues, including circulin D and circulin E. In these cases, endoproteinase Glu-C treatment will generate two fragments with both terminating at a Glu residue on the C termini. De novo sequencing of the digested fragments (supplemental Fig. S1, B and C) revealed that the 2747-Da fragment ended with a Ser but not a Glu residue, suggesting the open-ended nature of hedyotide B2. In addition, its observed molecular weight was in agreement with the predicted mass of a linear form confirming the uncyclized structure of hedyotide B2.
Expression Profile of Cyclotides in H. biflora
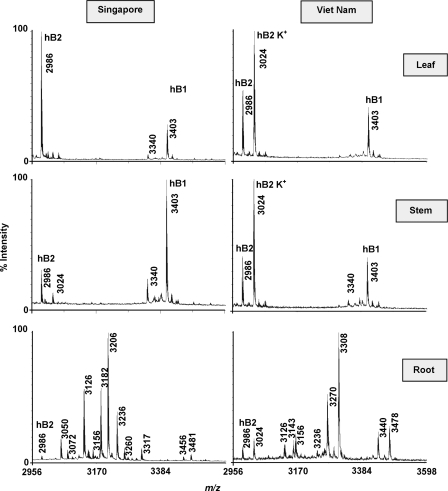
MS analysis of plant extracts from aerial parts (leaves and stems) and roots of H. biflora indicated the presence of more than 30 putative cyclotides. They displayed substantial differences in the expression patterns between the aboveground and underground parts (Fig. 2). In the aerial tissues, leaves and stems had similar profiles, with hedyotide B1 and B2 being the major cyclotide constituents. These two peptides are relatively specific to the aerial tissues with minor expression in the roots. The majority of other cyclotides were found predominantly in the underground parts. It is common that many defense molecules are found in the roots where plants have to cope with a diverse range of microbes, pests, and pathogens.
FIGURE 2.
Tissue- and region-specific distribution of cyclotides in H. biflora. Plant specimens were collected in two different countries: Singapore and Vietnam. They were divided into three different plant parts (leaf, stem, and root) and profiled separately by mass spectrometry. Peaks labeled K+ indicate the potassium adducts (+38 Da). Hedyotide B1 (hB1) and B2 (hB2) are labeled at the tops of the corresponding peaks.
Next, we compared the mass spectra of plant samples collected in Vietnam and Singapore to study the regional influence on the cyclotide compositions. We found that the cyclotide profiles of the aerial tissues did not change significantly, whereas the root tissues displayed a striking geographic variation (Fig. 2). Several cyclotides were expressed specifically in one sample but were absent in the other. The relative expression level was also different in both samples. It is interesting to note that the most abundant cyclotide in the roots appear to be region-specific. The peak with m/z of 3308 Da, for example, was the most abundant cyclotide in plants collected in Vietnam but was nearly missing in the Singapore specimen. On the other hand, the peak with m/z of 3205 Da was highly expressed only in the Singapore sample and was not detected in the Vietnam sample under our experimental conditions.
Encoding cDNAs of Hedyotide B1 and B2
To determine the encoding cDNAs of hedyotide B1 and B2, degenerate primers based on the CGETC and CGESC sequences of loop 1 were used for 3′-RACE amplification. This resulted in the identification of two unique partial clones encoding for hedyotide B1 and B2, respectively. Reverse specific primers based on the cDNA sequences of each clone were then used for 5′-RACE amplification to obtain the remaining encoding sequences. Their full-length clones were designated as hbc1 and hbc2 (H. biflora clone 1 and 2 corresponding to hedyotide B1 and hedyotide B2, respectively). Overall, the hbc1 clone has a similar arrangement to the oak1 and hcf-1 clones from O. affinis (16) and Hedyotis centranthoides (8), respectively. Each contains an ER signal sequence, an N-terminal propeptide, a cyclotide domain, and a hydrophobic tail (Fig. 3). The genetic organization of hbc2 is similar to hbc1 except for the absence of the tail domain and the presence of a premature stop codon located precisely at the highly conserved C-terminal Asn/Asp position leading to a C-terminal truncated precursor that prevents translation to a cyclic structure.
FIGURE 3.
Genetic arrangements of hedyotide B1 and B2 precursors. Their primary sequences were deduced from cDNA clones and aligned with hcf-1 and kalata B1 precursors using ClustalW (27). ER signal was predicted by SignalP-HMM software (41). These precursors share an overall arrangement containing an ER signal sequence (in cyan), a pro-region (in purple), a mature cyclotide (in pink), and a short tail domain (in black). The arrow indicates the intron location. Asterisks indicate stop codons.
DNA Clones of Hedyotide Precursors
To compare the genetic structures of hedyotides at both DNA and mRNA level, genes encoding for hedyotide B1 and B2 were cloned from genomic DNA using cDNA-derived sequences as primers. In both cases, the DNA sequences revealed a single intron (104 nucleotides in length) located within the ER signal region (Fig. 3). Surprisingly, the introns are more highly conserved than the exon regions with 99% sequence identity between the two intron sequences. The high degree of conservation may reflect a possibility of functional constraints or the presence of cis-regulatory elements within the intron regions.
Phylogenetic Analysis
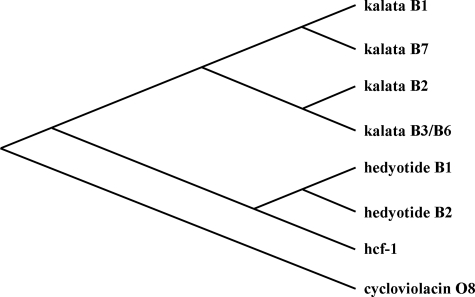
A phylogenetic tree was constructed using seven translated cyclotide precursor proteins from three Hedyotis species: hedyotide B1 and B2 (H. biflora); kalata B1, B2, B3/6, and B7 (O. affinis); and hcf-1 (H. centranthoides). As shown in Fig. 4, the hedyotide precursors have a closer evolutionary relationship to hcf-1 than to the kalata precursors. Sequence alignments showed that hedyotide B1 and hcf-1 precursors share 61% sequence identity, compared with only 27% identity between hedyotide B1 and kalata B1. This suggests that H. biflora is more closely related to H. centranthoides than to O. affinis. Our finding is consistent with the previous study by Kårehed et al. (33) using chloroplast and nuclear DNA markers to construct the phylogeny of the Rubiaceae species.
FIGURE 4.
Phylogenetic analysis of cyclotide precursors in the Hedyotis genus. The tree was constructed using eight cyclotides precursors: hedyotide B1 and B2 from H. biflora; kalata B1, B2, B3/6, and B7 from O. affinis; hcf-1 from H. centranthoides; and cycloviolacin O8 from V. odorata. Cycloviolacin O8 was used as an outgroup to root the phylogenetic tree.
Top-down Mapping of Hedyotide B2 Disulfide Connectivity
To elucidate the disulfide connectivity of hedyotide B2, we employed a top-down approach using tandem mass spectrometry. The overall strategy involved a sequential S-tagging method with two different alkylation reagents: first NEM and then IAA. The S-tagged peptides were sequenced by MALDI-collision-induced dissociation-MS/MS analysis. By optimizing the laser power and number of shots per spectrum, we were able to obtain almost complete sequence coverage across the entire backbone of the S-alkylated hedyotide B2 up to 3.6 kDa. Our approach requires no prior proteolytic digestion step as used in the conventional bottom-up strategy (34), thus allowing rapid characterization of the disulfide pattern.
Native hedyotide B2 was first partially S-reduced with tris(2-carboxyethyl)phosphine at pH 3.5. This generated a series of isoforms with one or two reduced disulfide bonds. The released thiols were immediately S-alkylated with an excess of NEM. The whole process was performed under acidic conditions at pH 3.5 to avoid the scrambling of disulfide linkages (35). The partially S-reduced and S-alkylated peptides were then purified by RP-HPLC.
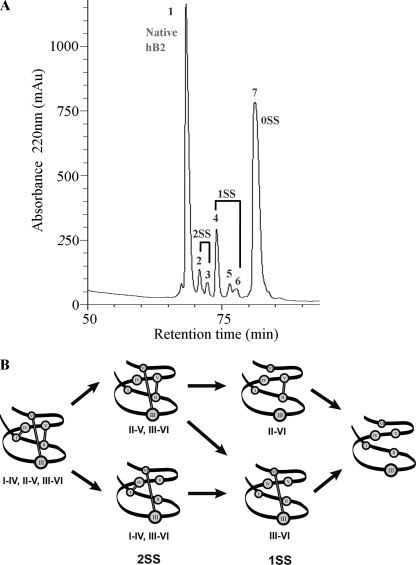
Seven HPLC-separated peaks were collected (Fig. 5A) and analyzed by MALDI-TOF MS. Because each NEM-modified cysteinyl residue caused a mass increase of 126 Da, the mass gained after the S-alkylation could be used to deduce the number of reduced disulfide bonds. Peak 1 contained the native peptide with the intact disulfide bridges. Peaks 2 and 3 had the same m/z of 3238 Da corresponding to intermediates with two NEM-labeled cysteines and two intact disulfide bonds (2SS species). Peaks 4, 5, and 6 all had m/z of 3490 Da corresponding to intermediates with four NEM-labeled cysteines and one remaining disulfide bond (1SS species). Peak 7 had all three disulfide bonds reduced and labeled by NEM (0SS species). To obtain the disulfide connectivity, 2SS and 1SS species were fully S-reduced and S-tagged with a second alkylation reagent (IAA). The doubly S-alkylated peptides were then analyzed directly by MALDI-collision-induced dissociation-MS/MS without prior proteolytic digestion.
FIGURE 5.
Disulfide mapping of hedyotide B2. A, RP-HPLC separation of hedyotide B2 derivatives after partial S-reduction and S-NEM alkylation. Peak 1 is the native peptide; peaks 2 and 3 are intermediates with two intact disulfide bonds (2SS); peaks 4, 5, and 6 are intermediates with one intact disulfide bond (1SS); and peak 7 is the fully S-NEM alkylated hedyotide B2 (0SS). B, schematic presentation of hedyotide B2 unfolding pathway. Under our experimental condition, the Cys I–IV bond is likely to be reduced first, followed by the Cys II–V bond and lastly by Cys III–VI bond.
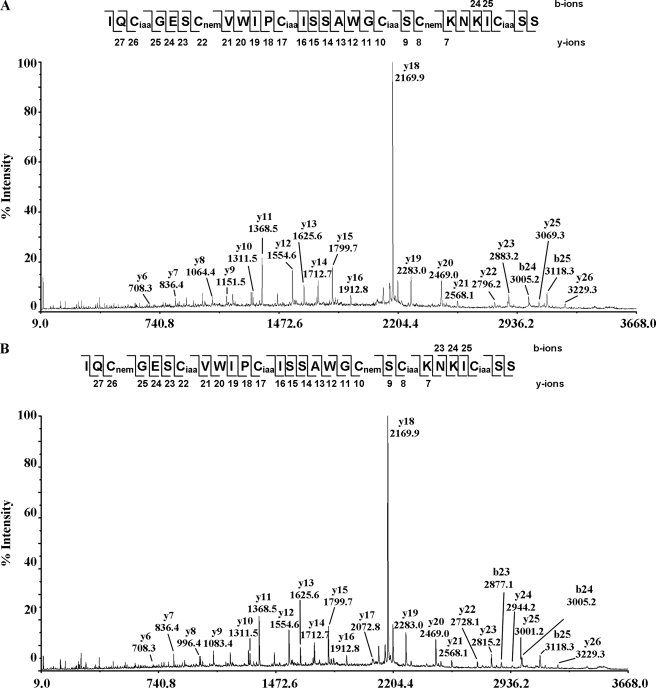
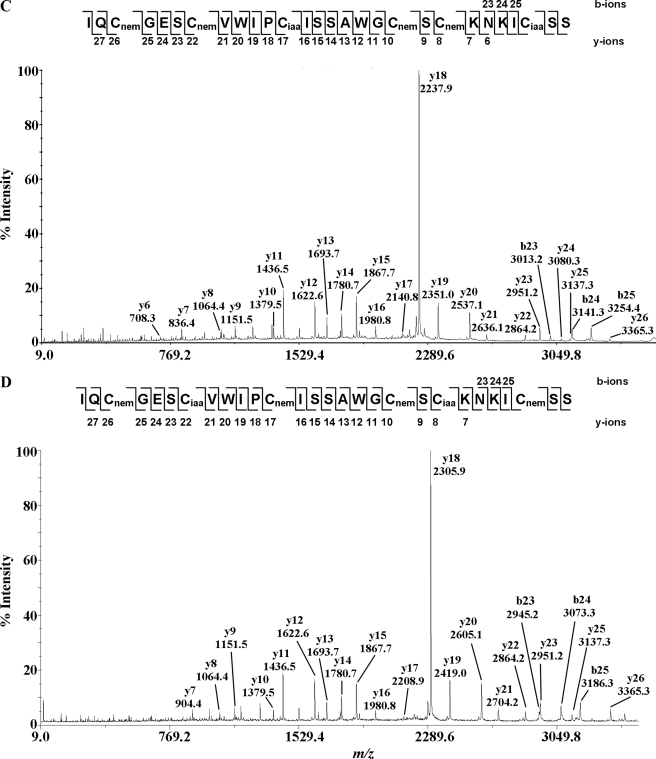
Fig. 6 shows the MS/MS spectra of the S-alkylated hedyotide B2 labeled with both NEM and IAA. Information from the differential S-tagging experiment was used to deduce the disulfide connectivity. Analysis of the S-alkylated species in peak 2 indicated S-NEM labeling on Cys II and V and S-IAA labeling on the remaining cysteines, establishing the connectivity as Cys II–V. Similarly, analyzing the MS/MS spectrum of S-alkylated species in peak 3 indicated disulfide connectivity of Cys I–IV. The disulfide patterns of 1SS species in peaks 4, 5, and 6 were characterized by a mixed labeling of four S-NEM and two S-IAA groups. Accordingly, the connectivities of the intermediates in peaks 4 and 6 were established as Cys III–VI and Cys II–V, respectively. Peak 5 contained a mixture of two 1SS intermediates with the disulfide connections of Cys III–VI and Cys II–V. The coelution of two different disulfide linkage species in peak 5 was probably due to the formation of stereoisomers resulting from the introduction of a new chiral center on the cysteinyl side chain upon S-NEM labeling (35). Our results provide unambiguous evidence that hedyotide B2, an uncyclotide, has the same cystine-knot arrangement of Cys I–IV, II–V, and III–VI as its cyclic counterparts.
FIGURE 6.
MS/MS spectra of S-alkylated hedyotide B2 doubly S-tagged with IAA and NEM. A–D show the MS/MS spectra of S-alkylated derivatives of intermediate species in peaks 2, 3, 4, and 6, respectively. Peak 5 contains isoforms of intermediate species in peaks 4 and 6 with identical MS/MS spectra and was not shown here.
Biodegradation of Cyclotides and Uncyclotides in H. biflora
To provide an understanding about the biodegradation of cyclotides, aerial materials of H. biflora were kept at room temperature for 2 weeks before extraction. MS analysis of fresh and stored materials revealed several degradation products of hedyotide B1 and B2 (Fig. 7). A number of new peptide masses corresponding to novel cyclotides were also observed. They were probably induced during the storage or degraded peptide fragments derived from large proteins because of proteolysis.
FIGURE 7.
Biodegradation of hedyotide B1 and B2. A, MS profile of freshly collected plants. B, MS profile of stored plant samples showing several degradation products of hedyotide B1 and B2. Peaks labeled Na+ and K+ indicate the sodium (+23 Da) and potassium adducts (+38 Da), respectively. hB2ox is the oxidized derivative of hedyotide B2. hB1oia, hB1nfk, and hB1kyn are the oxidized derivatives of hedyotide B1.
As shown in Fig. 7, both hedyotide B1 and B2 were prone to Trp oxidation. The indole ring of the Trp side chain was known to have three major oxidized states, namely Trpoia, Trpnfk, and Trpkyn causing a mass shift of +4, +16, and +32, respectively (23). For hedyotide B1, all three oxidized derivatives were observed, in which a significant percentage of the original peptide (>50%) was oxidized as determined by the MS ratio of the native and the oxidized products. Hedyotide B1 contains a single Trp residue located on loop 3, which is exposed on the peptide surface, making it relatively susceptible to oxidation. Similarly, hedyotide B2 also underwent oxidation, but the interpretation of the oxidative states was complicated by having two Trp residues in its primary sequence. It is possible to form up to 15 different oxidized derivatives of hedyotide B2. They can be easily recognized by RP-HPLC profiles, which display the characteristic loss of 280-nm absorbance caused by the modification of the indole ring of Trp residues. Thirteen of them were observed at molecular weight level. Their masses and possible oxidation states were summarized in supplemental Table S1.
In addition to the Trp oxidation, hedyotide B2 was also susceptible to degradation by exopeptidases because of its open-ended structure. The Ile residue at the N terminus was trimmed giving rise to hedyotide B3 with a m/z of 2873 Da. Subsequently, the Gln residue of hedyotide B3 at the N terminus spontaneously cyclized to form a pyroglutamyl derivative generating hedyotide B4 with a m/z of 2856 Da. It should be noted that hedyotide B3 and B4 are also prone to oxidation, and each may generate a set of 15 different oxidation products. Hedyotide B2 thus can give rise to up to 47 different modification variants, including the formation of hedyotides B3 and B4 and their derivatives. Our results provide further understanding of the biodegradation of cyclotides and uncyclotides. The cyclic backbone provides better resistance to exopeptidase degradation as compared with its linear counterpart.
Antimicrobial Activity
Antibacterial activities of hedyotide B1 and B2 were tested against four different bacterial strains: E. coli, S. aureus, S. epidermidis, and S. salivarius (Table 2). Hedyotide B1 was active against E. coli and S. salivarius with the MIC values of 3.4 and 5.9 μm, respectively. Hedyotide B2 was inactive against all tested bacterial strains up to 80 μm in concentration. Neither peptide had inhibitory activity against S. aureus or S. epidermidis.
TABLE 2.
Antibacterial activities of hedyotide B1 and B2
| Organism | MIC |
||
|---|---|---|---|
| Hedyotide B1 | Hedyotide B2 | D4Ra | |
| μm | |||
| E. coli | 3.4 | >80 | 1.29 |
| S. salivarius | 5.9 | >80 | 0.96 |
| S. epidermidis | >80 | >80 | 0.71 |
| S. aureus | >80 | >80 | 1.9 |
a Synthetic antibacterial peptide as positive control.
DISCUSSION
In this study, we have analyzed seven species belonging to the Spermacoceae tribe and identified H. biflora as a novel cyclotide-producing plant. Two novel peptides and their oxidized derivatives were isolated from the aerial parts of H. biflora. Hedyotide B1 has a cyclic structure typical of a cyclotide, whereas hedyotide B2 is an uncyclotide possessing a linear backbone. Disulfide mapping of hedyotide B2 showed that it shares the same knotted disulfide arrangement as conventional cyclotides. The encoding genes of hedyotide B1 and B2 were subsequently cloned, revealing new insights about the biosynthesis processing of cyclotides and uncyclotides in plants.
Distribution of Cyclotides in the Hedyotis Genus
Our finding of cyclotides in H. biflora made it the fourth Hedyotis species and the first annual herb of its genus classified as a cyclotide-bearing plant. The morphologies of the other three species vary from perennial herbs (O. affinis), shrubs (H. centranthoides), and perennial woody trees (Hedyotis terminalis). In addition to hedyotides B1 and B2 and their derivatives, MS analysis of H. biflora extract revealed the presence of more than 30 unique cyclotide masses, which is in agreement with the previous study that suggested an average number of 34 cyclotides/rubiaceous plant (8).
Intriguingly, despite the discovery of cyclotides in multiple Hedyotis species, not all plants in this genus express cyclotides. Hedyotis corymbosa, for example, is a widely distributed species of the Hedyotis genus. It is closely related to O. affinis, and a detailed comparison has been studied extensively by Datta and Sen (36) and Gran et al. (15). Regardless of its close relationship with H. biflora and O. affinis, H. corymbosa has been shown to be a non-cyclotide-containing species (8). This work provides further support that H. corymbosa is not a cyclotide-producing plant. This raises the question of why cyclotides have been lost in several Hedyotis plants or more broadly in the Rubiaceae lineages during their course of evolution. Cyclotides are known to possess a wide range of defense-related functions such as antimicrobial (7), anti-HIV (13), and insecticidal (16) functions. It is intriguing to speculate how plants ensure their fitness in the absence of cyclotide-defense armory. It is possible that plants may have developed alternative protection strategies to compensate for the loss of cyclotide expression.
Top-down Mapping of Hedyotide B2 Disulfide Linkage
The discovery of hedyotide B2 as the first linear variant of bracelet subfamily has stimulated our interest in characterizing its disulfide pattern. To our knowledge, cystine connectivity of an uncyclotide has not been established by a chemical method, although it has been proposed to possess the same knotted arrangement based on an NMR study (21). Here, we mapped the connectivity of hedyotide B2 using a top-down approach instead of the conventional bottom-up strategy where proteins are subjected to proteolysis prior to MS analysis (37). Although the bottom-up approach provides a straightforward strategy for characterizing the cystine linkages, it requires extra steps of proteolytic digestion and purification that are not desirable for rapid disulfide mapping. In addition, it is common that many of the digested fragments are not observed in MS because of inappropriate charge, mass, or insolubility upon digestion (34). Valuable information may be lost making it difficult for complete characterization of a protein. Therefore, by eliminating the proteolytic digestion step and subjecting whole peptides directly to MS/MS analysis, we overcame these limitations and thus allowed rapid characterization of disulfide connectivity.
In our approach, hedyotide B2 was sequentially S-tagged with NEM and IAA followed directly by MS/MS analysis. In the first S-alkylation with NEM, five different intermediates eluted in peaks 2–6 were collected. In the previous work by Göransson and Craik (35) on the disulfide characterization of kalata B1, only two intermediate species were isolated, which led to the identification of the Cys II–V and III–VI linkages. The connectivity of the third disulfide bond Cys I–IV was obtained by deduction. MS characterizations of five intermediates species in our study provided the first chemical evidence for all three cystine linkages establishing the knotted arrangement of hedyotide B2.
In addition to the characterization of disulfide patterns, the analysis of intermediate products also provides additional information about the unfolding mechanism of hedyotide B2. A possible unfolding pathway showing intermediate species at various reduction stages was presented in Fig. 5B. The Cys III–VI linkage appeared to be the most stable disulfide bond because no 2SS species with breakage of the Cys III–VI linkage was isolated under our experimental condition. In addition, 1SS species with the intact Cys III–VI linkage was the most abundant form among the five intermediate species. This finding is consistent with our understanding of the cystine-knot arrangement because the Cys III–VI linkage is the most shielded and buried inside the cystine core. Our results are also consistent with the previous studies on cliotide T2 (11) and kalata B1 unfolding pathway (35), where the penetrating disulfide bond (Cys III–VI) appears to be the most stable and the last to be broken.
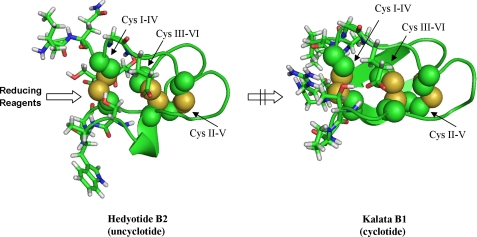
The reduction order of the two remaining disulfide bonds is more difficult to predict because both 2SS species with the breakage of Cys I–IV and II–V linkages were isolated. However, no 1SS species with the intact Cys I–IV linkage was identified, suggesting that this disulfide bond is first to be broken. This result differs from the unfolding pathway of kalata B1 (11, 35) and cliotide T2 (11), in which the Cys II–V linkage is the first disulfide bond being reduced. The difference in the reduction order is likely attributed to the difference in the cyclic and linear nature of kalata B1/cliotide T2 and hedyotide B2, respectively. A computer model of hedyotide B2 based on circulin A was used for comparing with kalata B1 structure (Fig. 8). In kalata B1, the Cys II–V linkage is most surface-exposed and thus most readily reduced (35). The Cys I–IV linkage is shielded by the cyclic backbone, limiting its access to reducing reagents. In hedyotide B2, the open-ended structure increases the accessibility of the Cys I–IV bond, making it more easily broken than the Cys II–V bond. Thus, our analysis suggests a reverse order of reduction of the Cys I–IV and II–V linkages in the linear and cyclic forms.
FIGURE 8.
Computer models of hedyotide B2 and kalata B1 (Protein Data Bank code 1NB1). Disulfide bonds (indicated by the arrow) are shown as spheres, and the surrounding amino acids are in stick configuration. For hedyotide B2, its open-ended structure increases the accessibility of Cys I–IV to reducing reagents, whereas for kalata B1 this bond is steric hindered by the cyclic backbone.
Biosynthesis Pathway of Hedyotide B2
Based on the sequence comparison of cyclotides in the database, the linear structure of hedyotide B2 is likely genetically predetermined and is unlikely to have resulted from ring opening or chemical degradation. Its mature sequence lacks both highly conserved residues, the N-terminal Gly and C-terminal Asn, which are required for the backbone cyclization. The cDNA encoding clone of hedyotide B2 (hbc2) reveals a nonsense mutation that introduces a premature stop codon precisely at the conserved Asn position. The Asn residue has been proposed to act as recognition site for a putative asparaginyl endopeptidase, an enzyme believed to catalyze the peptide backbone cyclization (25, 38). The open-ended nature of hedyotide B2 can be attributed to the absence of this key residue. Mutation of another residue at loop 6 has been reported in violacin A, the first identified linear cyclotide from V. odorata (21). The stop codon in this peptide, however, occurs one residue prior to the conserved Asn. Thus, an uncyclotide can be formed if a nonsense mutation occurs in loop 6 that replaces, truncates, or prevents the translation of the conserved Asp/Asn residue essential in the end-to-end ligation processing.
The hbc2 clone also revealed that the conserved Gly residue at the N terminus is indeed present in the precursor protein but is absent in the mature hedyotide B2 sequence. It is uncertain whether hedyotide B2 is synthesized through a direct cleavage of the precursor protein, resulting in the mature product without the Gly residue, or through a short-lived intermediate with the N-terminal Gly, which is subsequently removed by proteolysis (Fig. 9). The latter seems more likely because the sequence comparison with hedyotide B1 and rubiaceous cyclotides suggested that the cleavage sites are highly conserved and located between the dipeptide motif “Leu-Lys” and the conserved Gly residue. Trimming of the Gly residue is probably caused by aminopeptidases, which are ubiquitous in plants. This process appears to be highly efficient because no mature peptide with the conserved Gly residue was detected from the plant extract under our isolation procedure.
FIGURE 9.
Proposed model for the biosynthesis of hedyotide B2, B3, and B4. Processing at the N terminus occurs between Lys and Gly, followed by sequential trimming of N-terminal residues and deamidation to give rise to hedyotide B2, B3, and B4.
Biodegradation of Cyclotides in H. biflora
Characterization of stored plant samples provides an understanding about the biodegradation of cyclotides in the natural environment. Several degraded products of hedyotide B1 and B2 were formed during the prolonged storage of plant materials. They can be classified into two categories: Trp-oxidized products and truncated products. In the first category, three and thirteen putative Trp-oxidized products of hedyotide B1 and B2, respectively, were observed. The modifications of Trp have been proposed to be facilitated by exposure to sunlight (23). In our study, the plant materials were kept at room temperature away from sunlight, suggesting that other factors in addition to UV released during the plant decomposition process may accelerate the Trp oxidation.
The second category affects only hedyotide B2 because of its open-ended structure, whereas hedyotide B1 with a cyclic backbone is not affected. The N-terminal trimming of hedyotide B2 led to the formation of hedyotide B3 and B4. No detectable amount of these two peptides was found upon the prolonged storage of pure hedyotide B2 at room temperature. This suggests the involvement of exopeptidases in the trimming process. The decomposition of plant materials are known to occur through several stages. It begins with leaching of water and releasing of most water-soluble compounds (39). The trimming of hedyotide B2 is probably facilitated by aminopeptidase liberated during this leaching process. Another early event is the breakdown of the plant materials that provide nutrients and surface area for bacteria and fungi colonization (39). These microorganisms produce a great diversity of enzymes that possibly contribute to the degradation of hedyotide B2. Knowledge of these degradation pathways provides a better understanding of the half-life and potential applications of cyclotides and uncyclotides in drug development.
Antimicrobial Activity
Hedyotide B1 and B2 show a clear difference in their antimicrobial actions. Hedyotide B1 was active against both Gram-positive (S. salivarius) and Gram-negative bacteria (E. coli) with MIC values of 5.9 and 3.4 μm, respectively. A synthetic version of hedyotide B1 has been tested previously for the antimicrobial activity (40) that shows slightly different MIC values for E. coli (2 μm) and S. aureus (37 μm). For E. coli, the difference in the MIC values is less than 2-fold and within the experimental error. For S. aureus, both works show that hedyotide B1 is, at best, moderately active at high concentration of 37 μm or >80 μm. These differences can be attributed to the different strains of S. aureus used in the antimicrobial assays. Despite the small variations in the MIC values in both works, the antimicrobial activity is consistent, with hedyotide B1 being active against E. coli and only weakly active against S. aureus.
Hedyotide B2 was inactive against all tested strains under our experimental conditions. Although the endogenous functions of hedyotide B2 are still uncertain, its high expression level in the aerial tissues suggests that it may have important physiological functions in plants.
In summary, this study describes an in-depth characterization of two novel peptides, hedyotide B1 and B2, from the aerial parts of H. biflora. Elucidation of their genetic structures revealed that the presence of a premature stop codon in loop 6 prevents the translation of the conserved Asn/Asp leading to uncyclotide formation. Furthermore, we also demonstrated that the uncyclotide hedyotide B2 possesses the same knotted disulfide arrangement as conventional cyclotides. The biological activity of hedyotide B2, however, was still undetermined and would warrant further research to provide more thorough understanding of the mechanistic action and physiological functions of uncyclotides in plants.
Supplementary Material
This work was supported by Biomedical Research Council Grant BMRC 09/1/22/19/612, A*STAR, and Grant ARC21/08 from the Academic Research Fund of the Ministry of Education in Singapore.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Fig. S1.
- SS
- disulfide
- SPE
- solid phase extraction
- ACN
- acetonitrile
- IAA
- iodoacetamide
- NEM
- N-ethylmaleimide
- ER
- endoplasmic reticulum
- MIC
- minimal inhibitory concentration.
REFERENCES
- 1. Craik D. J., Daly N. L., Bond T., Waine C. (1999) J. Mol. Biol. 294, 1327–1336 [DOI] [PubMed] [Google Scholar]
- 2. Sze S. K., Wang W., Meng W., Yuan R., Guo T., Zhu Y., Tam J. P. (2009) Anal. Chem. 81, 1079–1088 [DOI] [PubMed] [Google Scholar]
- 3. Colgrave M. L., Craik D. J. (2004) Biochemistry 43, 5965–5975 [DOI] [PubMed] [Google Scholar]
- 4. Henriques S. T., Craik D. J. (2010) Drug Discov. Today 15, 57–64 [DOI] [PubMed] [Google Scholar]
- 5. Gran L. (1973) J. Nat. Prod. (Lloydia) 36, 207–208 [Google Scholar]
- 6. Saether O., Craik D. J., Campbell I. D., Sletten K., Juul J., Norman D. G. (1995) Biochemistry 34, 4147–4158 [DOI] [PubMed] [Google Scholar]
- 7. Tam J. P., Lu Y. A., Yang J. L., Chiu K. W. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 8913–8918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gruber C. W., Elliott A. G., Ireland D. C., Delprete P. G., Dessein S., Göransson U., Trabi M., Wang C. K., Kinghorn A. B., Robbrecht E., Craik D. J. (2008) Plant Cell 20, 2471–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Trabi M., Svangård E., Herrmann A., Göransson U., Claeson P., Craik D. J., Bohlin L. (2004) J. Nat. Prod. 67, 806–810 [DOI] [PubMed] [Google Scholar]
- 10. Hernandez J. F., Gagnon J., Chiche L., Nguyen T. M., Andrieu J. P., Heitz A., Trinh Hong T., Pham T. T., Le Nguyen D. (2000) Biochemistry 39, 5722–5730 [DOI] [PubMed] [Google Scholar]
- 11. Nguyen G. K., Zhang S., Nguyen N. T., Nguyen P. Q., Chiu M. S., Hardjojo A., Tam J. P. (2011) J. Biol. Chem. 286, 24275–24287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Poth A. G., Colgrave M. L., Philip R., Kerenga B., Daly N. L., Anderson M. A., Craik D. J. (2011) ACS Chem. Biol. 6, 345–355 [DOI] [PubMed] [Google Scholar]
- 13. Gustafson K. R., McKee T. C., Bokesch H. R. (2004) Curr. Protein Pept. Sci. 5, 331–340 [DOI] [PubMed] [Google Scholar]
- 14. Göransson U., Luijendijk T., Johansson S., Bohlin L., Claeson P. (1999) J. Nat. Prod. 62, 283–286 [DOI] [PubMed] [Google Scholar]
- 15. Gran L., Sandberg F., Sletten K. (2000) J. Ethnopharmacol. 70, 197–203 [DOI] [PubMed] [Google Scholar]
- 16. Jennings C., West J., Waine C., Craik D., Anderson M. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 10614–10619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Colgrave M. L., Kotze A. C., Huang Y. H., O'Grady J., Simonsen S. M., Craik D. J. (2008) Biochemistry 47, 5581–5589 [DOI] [PubMed] [Google Scholar]
- 18. Craik D. J., Cemazar M., Wang C. K., Daly N. L. (2006) Biopolymers 84, 250–266 [DOI] [PubMed] [Google Scholar]
- 19. Wang C. K., Kaas Q., Chiche L., Craik D. J. (2008) Nucleic Acids Res. 36, D206–D210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Simonsen S. M., Sando L., Ireland D. C., Colgrave M. L., Bharathi R., Göransson U., Craik D. J. (2005) Plant Cell 17, 3176–3189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ireland D. C., Colgrave M. L., Nguyencong P., Daly N. L., Craik D. J. (2006) J. Mol. Biol. 357, 1522–1535 [DOI] [PubMed] [Google Scholar]
- 22. Gerlach S. L., Burman R., Bohlin L., Mondal D., Göransson U. (2010) J. Nat. Prod. 73, 1207–1213 [DOI] [PubMed] [Google Scholar]
- 23. Plan M. R., Göransson U., Clark R. J., Daly N. L., Colgrave M. L., Craik D. J. (2007) Chembiochem. 8, 1001–1011 [DOI] [PubMed] [Google Scholar]
- 24. Barry D. G., Daly N. L., Clark R. J., Sando L., Craik D. J. (2003) Biochemistry 42, 6688–6695 [DOI] [PubMed] [Google Scholar]
- 25. Saska I., Gillon A. D., Hatsugai N., Dietzgen R. G., Hara-Nishimura I., Anderson M. A., Craik D. J. (2007) J. Biol. Chem. 282, 29721–29728 [DOI] [PubMed] [Google Scholar]
- 26. Dagar H. S., Dagar J. C. (1991) Econ. Bot. 45, 114–119 [Google Scholar]
- 27. Thompson J. D., Higgins D. G., Gibson T. J. (1994) Nucleic Acids Res. 22, 4673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Page R. D. (1996) Comput. Appl. Biosci. 12, 357–358 [DOI] [PubMed] [Google Scholar]
- 29. Lehrer R. I., Rosenman M., Harwig S. S., Jackson R., Eisenhauer P. (1991) J. Immunol. Methods 137, 167–173 [DOI] [PubMed] [Google Scholar]
- 30. Tam J. P., Lu Y. A., Yang J. L. (2002) Eur. J. Biochem. 269, 923–932 [DOI] [PubMed] [Google Scholar]
- 31. Fiser A., Sali A. (2003) Methods Enzymol. 374, 461–491 [DOI] [PubMed] [Google Scholar]
- 32. Daly N. L., Koltay A., Gustafson K. R., Boyd M. R., Casas-Finet J. R., Craik D. J. (1999) J. Mol. Biol. 285, 333–345 [DOI] [PubMed] [Google Scholar]
- 33. Kårehed J., Groeninckx I., Dessein S., Motley T. J., Bremer B. (2008) Mol. Phylogenet. Evol. 49, 843–866 [DOI] [PubMed] [Google Scholar]
- 34. Reid G. E., McLuckey S. A. (2002) J. Mass Spectrom. 37, 663–675 [DOI] [PubMed] [Google Scholar]
- 35. Göransson U., Craik D. J. (2003) J. Biol. Chem. 278, 48188–48196 [DOI] [PubMed] [Google Scholar]
- 36. Datta P. C., Sen A. (1969) Q. J. Crude Drug Res. 9, 1356–1374 [Google Scholar]
- 37. Kelleher N. L., Lin H. Y., Valaskovic G. A., Aaserud D. J., Fridriksson E. K., McLafferty F. W. (1999) J. Am. Chem. Soc. 121, 806–812 [Google Scholar]
- 38. Gillon A. D., Saska I., Jennings C. V., Guarino R. F., Craik D. J., Anderson M. A. (2008) Plant J. 53, 505–515 [DOI] [PubMed] [Google Scholar]
- 39. Gupta M. K., Shrivastava P., Singhal P. K. (1996) Hydrobiologia 335, 33–41 [Google Scholar]
- 40. Wong C. T., Taichi M., Nishio H., Nishiuchi Y., Tam J. P. (2011) Biochemistry 50, 7275–7283 [DOI] [PubMed] [Google Scholar]
- 41. Emanuelsson O., Brunak S., von Heijne G., Nielsen H. (2007) Nat. Protoc. 2, 953–971 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.