Background: The switch between autophagy and apoptosis is an important and complicated process that is not well understood.
Results: Doxorubicin treatment switches protective autophagy in sphingosine-1-phosphate phosphohydrolase-1 (SPP1)-depleted cells to apoptosis, increasing ceramide synthesis that enhances calpain activation and cleavage of pro-autophagic Atg5, generating a pro-apoptotic fragment.
Conclusion: Depletion of SPP1 sensitizes cells to doxorubicin-induced apoptosis.
Significance: Sphingolipid metabolites are involved in the cross-talk between autophagy and apoptosis.
Keywords: Apoptosis, Autophagy, Calpain, ER Stress, Sphingolipid, SPP1, Ceramide, Doxorubicin, Sphingosine 1-phosphate
Abstract
Sphingosine 1-phosphate (S1P) and ceramide have been implicated in both autophagy and apoptosis. However, the roles of these sphingolipid metabolites in the links between these two processes are not completely understood. Depletion of S1P phosphohydrolase-1 (SPP1), which degrades intracellular S1P, induces the unfolded protein response and endoplasmic reticulum stress-induced autophagy (Lépine, S., Allegood, J. C., Park, M., Dent, P., Milstien, S., and Spiegel, S. (2011) Cell Death Differ. 18, 350–361). Surprisingly, however, treatment with doxorubicin, which by itself also induced autophagy, markedly reduced the extent of autophagy mediated by depletion of SPP1. Concomitantly, doxorubicin-induced apoptosis was greatly enhanced by down-regulation of SPP1. Autophagy and apoptosis seemed to be sequentially linked because inhibiting autophagy with 3-methyladenine also markedly attenuated apoptosis. Moreover, silencing Atg5 or the three sensors of the unfolded protein response, IRE1α, ATF6, and PKR-like eIF2α kinase (PERK), significantly decreased both autophagy and apoptosis. Doxorubicin stimulated calpain activity and Atg5 cleavage, which were significantly enhanced in SPP1-depleted cells. Inhibition or depletion of calpain not only suppressed Atg5 cleavage, it also markedly decreased the robust apoptosis induced by doxorubicin in SPP1-deficient cells. Importantly, doxorubicin also increased de novo synthesis of the pro-apoptotic sphingolipid metabolite ceramide. Elevation of ceramide in turn stimulated calpain; conversely, inhibiting ceramide formation suppressed Atg5 cleavage and apoptosis. Hence, doxorubicin switches protective autophagy in SPP1-depleted cells to apoptosis by calpain-mediated Atg5 cleavage.
Introduction
Autophagy and apoptosis are important and interconnected stress-response mechanisms. Although apoptotic cell death, known as type I programmed cell death, is a universal process leading to self-killing, autophagy (“self-eating”) is a major protective pathway that usually helps cells to survive various stresses. It is generally considered that autophagy and apoptosis are antagonistic and tend to inhibit each other (1). For example, autophagy is induced to protect cells when apoptosis is blocked, whereas autophagy blockade triggers or enhances apoptosis (1). However, increased formation of autophagosomes can be associated with autophagic cell death, termed type II programmed cell death, and there is still some controversy about the functional role of autophagy in life and death outcomes (2). This functional duality is also apparent during the unfolded protein response (UPR),2 the major ER stress pathway (3), which is a potent stimulus of autophagy. Although the UPR initially improves folding and degradation of misfolded proteins, if the UPR is overwhelmed, leading to chronic ER stress, apoptosis can be initiated. Hence, multiple connections must exist between ER stress, autophagy, and apoptosis, and the molecular interplay and functional relationship between their pathways are now receiving considerable attention (1).
The sphingolipid metabolites, sphingosine 1-phosphate (S1P) and its precursor ceramide, have long been implicated as critical regulators of cell fate decisions (4). Although ceramide is generally pro-apoptotic and activates both intrinsic and extrinsic apoptotic pathways (reviewed in Ref. 5), S1P has been linked to cell survival and suppression of apoptosis (reviewed in Refs. 6 and 7). Recent studies have indicated that these sphingolipid metabolites are also important regulators of ER stress and autophagy. Elevation of ceramide, either artificially by treatment with short-chain ceramides or with the anti-cancer drug tamoxifen, leads to autophagic cell death triggered by Beclin 1 or by inhibition of Akt phosphorylation (8). Similarly, ceramide sensitized multiple cancer cell lines to apoptosis induced by the commonly used anti-cancer drug doxorubicin by promoting AMP kinase activation and mTORC1 inhibition (9). In contrast, increasing S1P by overexpression of sphingosine kinase 1 (10) or by deletion of the ER-resident S1P-degrading enzyme, S1P phosphohydrolase 1 (SPP1) (11), induced protective autophagy. Thus, it was of interest to examine the possibility that these counteracting sphingolipid metabolites, S1P and ceramide, may be involved in the link between autophagy and apoptosis.
The autophagy-related gene 5 (Atg5) product, which is required for the formation of autophagosomes, has been suggested to be a molecular link between autophagy and apoptosis as its cleavage by calpain destroys its pro-autophagic function (12) and produces a pro-apoptotic mitochondrion-permeabilizing fragment (13). In this study, we found that deficiency of SPP1 enhances doxorubicin-induced, calpain-mediated cleavage of Atg5. Ceramide, whose synthesis is increased by doxorubicin, also stimulated calpain; moreover, inhibition of ceramide formation suppressed Atg5 cleavage as well as the robust apoptosis induced by doxorubicin in SPP1-deficient cells. Thus, doxorubicin switches autophagy in SPP1-depleted cells to apoptosis by calpain-mediated Atg5 cleavage.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids
Cell culture medium and fetal bovine serum were from Invitrogen. Doxorubicin, l-3-carboxy-trans-2,3-epoxypropionyl-leu-amido-(4-guanidino-)butane-ethylester (E64d), and 3-methyladenine (3MA) were purchased from Sigma. Fumonisin B1 and myriocin were from Enzo Life Sciences (Plymouth Meeting, PA). C6:0-ceramide (d18:1/6:0, N-hexanoyl-d-erythro-sphingosine) was from Avanti Polar Lipids (Alabaster, AL). The following antibodies were used for immunoblotting: rabbit polyclonal anti-LC3 (Novus Biologicals, Littleton, CO); anti-CHOP, anti-actin, anti-p70S6K, and anti-GRP78 (Santa Cruz Biotechnology, Santa Cruz, CA); anti-β-tubulin, anti-phospho-p70S6K (Thr389), anti-cleaved poly(ADP-ribose) polymerase (PARP), anti-cleaved caspase 7, anti-phospho-eIF2α (Ser51), anti-Akt, and anti-phospho-Akt (Ser473) (Cell Signaling, Danvers, MA); anti-Atg5 (Abgent, San Diego, CA); and anti-beclin (BD Biosciences). GFP-LC3 was provided by T. Yoshimori (National Institute of Genetics, Mishima, Japan) and K. Kirkegaard (Stanford University School of Medicine). Myc-tagged dominant negative PERK (dnPERK) was from J. A. Diehl (University of Pennsylvania).
Cell Culture and Transfection
Human breast cancer MCF7 and MB-MDA-231 cells were maintained at 37 °C in 5% CO2 in phenol red-free improved minimum essential medium, supplemented with 10% heat-inactivated fetal bovine serum (FBS) and 0.25% glucose as described (11). SPP1, inositol requiring enzyme 1 (IRE1α), and calpain small subunit were down-regulated with ON-TARGETplus SMARTpool siRNA (Dharmacon, Lafayette, CO) exactly as described (11). Where indicated, 24 h after siRNA transfection, cells were transfected with expression plasmids.
Quantitative RT-PCR
Cells were disrupted in TRIzol reagent (Invitrogen), and RNA was isolated and reverse-transcribed with SuperScript II (Invitrogen). For real-time PCR, premixed primer probe sets were purchased from Applied Biosystems (Carlsbad, CA), and cDNA was amplified with ABI 7900HT.
Autophagy Assay
Autophagy was determined as described previously (11). Briefly, after transfection with GFP-LC3 and treatments as described in the legends for Figs. 1, 2, 4, and 7, cells were washed with PBS and fixed with 4% paraformaldehyde. Coverslips were examined with a Zeiss LSM 510 laser confocal microscope (Thornwood, NJ). Cells with five or more intense GFP-LC3 puncta were considered autophagic, whereas those with diffuse cytoplasmic GFP-LC3 staining were considered non-autophagic. Autophagy was quantified in at least 100 cells in three independent experiments in a double-blind manner.
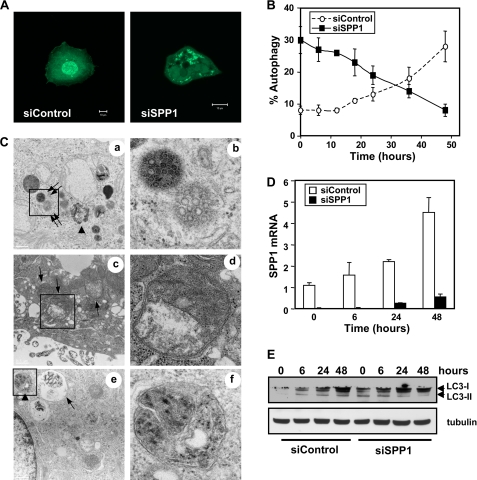
FIGURE 1.
Effect of doxorubicin on autophagy induced by SPP1 depletion. A and B, MCF7 cells were transfected with control scrambled siRNA (siControl) or siRNA targeted to SPP1 (siSPP1) and GFP-LC3 as described under “Experimental Procedures.” Cells were then treated with doxorubicin (1 μg/ml) for 6 h (A) or the indicated times (B and D), and localization of LC3 was examined by confocal microscopy. A, representative images are shown. Scale bars, 10 μm. B, autophagy was quantified. Data are expressed as the percentage of cells showing GFP-LC3 fluorescence in puncta (autophagosome formation) and are means ± S.D. of three independent experiments. At least 100 GFP-LC3-transfected cells were quantified. C–E, MCF7 cells transfected with siControl or siSPP1 were treated with doxorubicin for 6 h (C) or the indicated times (D and E). C, cells were examined by transmission electron microscopy. Typical autophagosomes (arrows), autolysosomes (arrowheads), and multivesicular bodies (double arrows) are boxed and shown at higher magnification. Scale bar, 2 μm (panels a, c, and e) and 0.5 μm (panels b, d, and f). D, SPP1 mRNA levels were determined by quantitative real-time PCR and normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA. E, cell lysates were analyzed by Western blotting using an anti-LC3 antibody. Blots were reprobed with tubulin to ensure equal loading and transfer.
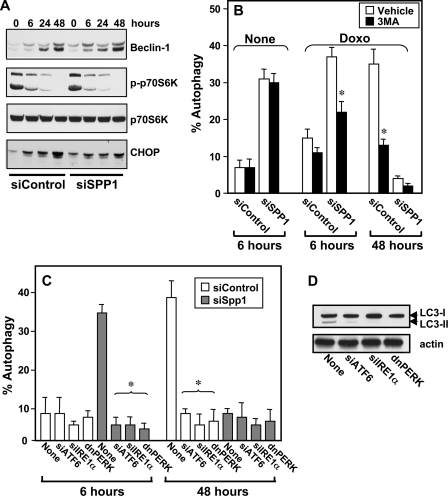
FIGURE 2.
Doxorubicin and ER stress-mediated autophagy. A, MCF7 cells transfected with siControl or siSPP1 were treated with doxorubicin (1 μg/ml) for the indicated times. Equal amounts of lysates were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. Blots were reprobed for total p70S6K to ensure equal loading and transfer. p-p70S6K, phospho-p70S6K. B, MCF7 cells were transfected with siControl or siSPP1 and GFP-LC3. Cells were treated without (None) or with doxorubicin (Doxo) for 6 or 24 h in the absence or presence of 10 mm 3MA, and autophagy was quantified by confocal microscopy. Data are means ± S.D. of three independent experiments. *, p < 0.01 as compared with cells not treated with 3MA. C, MCF7 cells were co-transfected with siControl, siSPP1, siIRE1α, siATF6, or dnPERK and GFP-LC3, as indicated. After treatment with doxorubicin for 6 or 48 h, autophagy was quantified by confocal microscopy. Values are the means ± S.D. of three independent experiments. *, p < 0.01 as compared with None. D, MCF7 cells were co-transfected with siSPP1 and siIRE1α, siATF6, or dnPERK, as indicated, and treated with doxorubicin for 6 h. Cell lysates were analyzed by Western blotting using an anti-LC3 antibody. Blots were reprobed with actin to ensure equal loading and transfer.
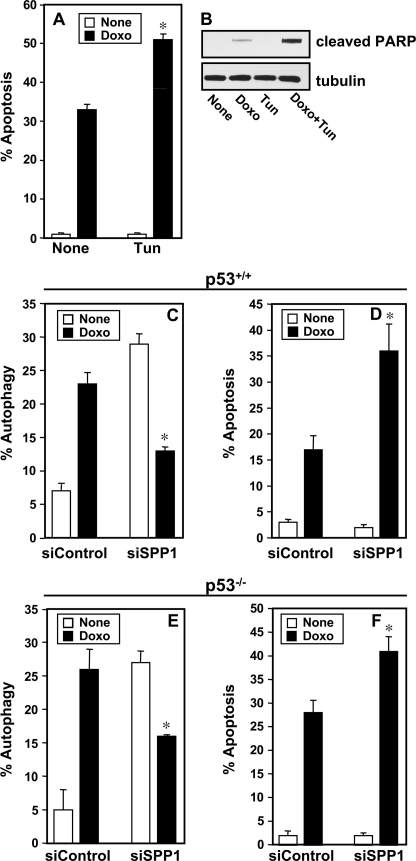
FIGURE 4.
Down-regulation of SPP1 enhances doxorubicin-induced apoptosis independently of p53. A and B, MCF7 cells were treated without (None) or with doxorubicin (Doxo, 1 μg/ml) in the absence or presence of tunicamycin (Tun, 2.5 μm) for 48 h. A, apoptosis was quantified. *, p < 0.01 as compared with cells treated only with doxorubicin. B, PARP cleavage was determined by Western blotting. C–F, wild-type HCT116 cells (C and D) or p53−/− HCT116 (E and F) were transfected with siControl or siSPP1 without or with GFP-LC3 and treated with doxorubicin (1 μg/ml) for 24 and 48 h, respectively, and autophagy (C and E) and apoptosis (D and F) were determined. *, p < 0.01 as compared with siControl treated with doxorubicin. Error bars in panels A and C–F indicate means ± S.D.
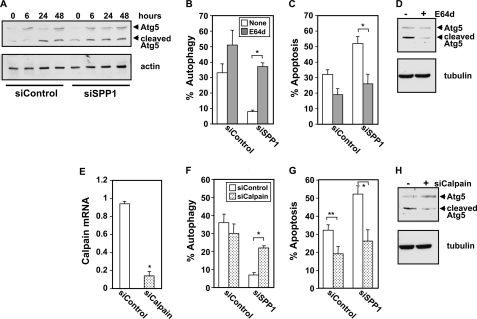
FIGURE 7.
Atg5 cleavage by calpain is involved in switching ER stress-mediated autophagy to apoptosis in SPP1-depleted cells. A, MCF7 cells transfected with siControl or siSPP1 were treated with doxorubicin (1 μg/ml) for the indicated times. Equal amounts of cell lysates were analyzed by Western blotting with an anti-Atg5 antibody. Blots were reprobed with tubulin to ensure equal loading and transfer. B and C, MCF7 cells transfected with siControl or siSPP1 without or with GFP-LC3 were treated with doxorubicin in the absence (None) or presence of E64d (10 μg/ml) for 48 h, and autophagy (B) and apoptosis (C) were determined. D, MCF7 cells transfected with siSPP1 were treated with doxorubicin (1 μg/ml) for 48 h in the absence or presence of E64d (10 μg/ml), and cleavage of Atg5 was determined by Western blotting. E, MCF7 cells were transfected with siControl or siCalpain, and calpain expression was quantified by quantitative PCR and normalized to GAPDH expression. F–G, MCF7 cells transfected with siControl or siSPP1 without or with siCalpain were treated with doxorubicin for 48 h, and autophagy (F) and apoptosis (G) were determined. H, MCF7 cells transfected with siSPP1 without or with siCalpain were treated with doxorubicin for 48 h, and cleavage of Atg5 was determined by Western blotting. *, p < 0.01, **, p < 0.05. Error bars in panels B–G indicate means ± S.D.
Electron Microscopy
For transmission electron microscopy, MCF7 cells were processed and analyzed by a transmission electron microscope (JEOL JEM-1230, Tokyo, Japan) equipped with a Gatan UltraScan 4000SP charge-coupled device camera (Pleasanton, CA) as described (11).
Apoptosis Assay
Apoptotic cells were quantified by fluorescence microscopy after labeling with Hoechst 33342 (10 μg/ml) (11). Cells exhibiting blue condensed or fragmented nuclei were considered apoptotic. At least 300 cells in three independent experiments were quantified with a Nikon Eclipse TE300 fluorescence microscope (Melville, NY). Cell viability was also determined by trypan blue exclusion.
Western Blot Analysis
Cells were lysed in buffer containing 20 mm Tris (pH 7.5), 150 mm NaCl, 1% Triton X-100, 2.5 mm sodium pyrophosphate, 1 mm β-glycerophosphate, 1 mm EDTA, 1 mm sodium orthovanadate, and 1:500 protease inhibitor mixture (Sigma). Equal amounts of proteins were separated by SDS-PAGE and transblotted to nitrocellulose, blocked with 5% nonfat dry milk for 1 h at room temperature, and then incubated overnight with the appropriate primary antibodies at 4 °C. The appropriate horseradish peroxidase-conjugated secondary antibodies were added in TBS containing 2.5% nonfat milk. Immunoreactive signals were visualized by enhanced chemiluminescence (Pierce Chemical Co).
Sphingolipid Quantitation
Lipids were extracted, and sphingolipid metabolites were quantified by liquid chromatography, electrospray ionization-tandem mass spectrometry (LC-ESI-MS/MS, 4000 QTRAP, Applied Biosystems) as described (14).
Calpain Activity
Calpain activity was determined with the Calpain-Glo protease assay kit using a luminogenic succinyl calpain substrate following the manufacturer's protocol (Promega, Madison, WI). Luminescence was measured using a Victor X4 plate reader (PerkinElmer Life Sciences).
Statistical Analysis
Experiments were repeated at least three times. Results were analyzed for statistical significance with the Student's t test for unpaired samples, and p < 0.05 was considered significant.
RESULTS
Doxorubicin Decreases ER Stress-induced Autophagy Resulting from SPP1 Depletion
We have recently demonstrated that depletion of SPP1 leads to activation of the UPR and ER stress-mediated autophagy in various cell types (11). This autophagy has unique characteristics as compared with those of other inducers of ER stress, such as tunicamycin or thapsigargin, as it did not involve p53 and did not trigger apoptosis, even after prolonged periods of autophagy or after inhibition of autophagy. Thus, it was of interest to examine whether this protective autophagy would influence the effects of the chemotherapeutic drug doxorubicin. The high level of autophagy observed in MCF7 human breast carcinoma cells depleted of SPP1, determined by the presence of characteristic punctate fluorescent GFP-LC3 patterns indicating recruitment of LC3 during autophagosome formation, was not altered by treatment with doxorubicin for 6 h (Fig. 1, A and B). Electron micrograph images of SPP1-depleted cells 6 h after doxorubicin treatment confirmed the presence of membrane-bound electron dense structures sequestering cellular components, a distinctive feature of double-membrane autophagosomes, single-membrane autolysosomes, and multivesicular bodies (Fig. 1C). However, after treatment of control siRNA transfectants with doxorubicin for 6 h, GFP-LC3 was observed predominantly as diffuse green fluorescence in the cytoplasm (Fig. 1A). Consistent with previous studies (15, 16), prolonged treatment of these cells with doxorubicin induced autophagy (Fig. 1B). Surprisingly, however, doxorubicin decreased autophagy in SPP1-depleted cells in a time-dependent manner (Fig. 1B). Notably, doxorubicin up-regulated expression of SPP1 in a time-dependent manner (Fig. 1D). However, in siSPP1-transfected cells, its expression was still markedly down-regulated even after 48 h (Fig. 1D). Immunoblots of doxorubicin-treated control scrambled siRNA (siControl) cells for the autophagy marker LC3 indicated a progressive conversion of the free LC3-I form over 48 h to its lipidated, autophagosome-associated LC3-II form, with a concomitant induction in total LC3 (Fig. 1E). In contrast, conversion to LC3-II in SPP1 down-regulated cells decreased after treatment with doxorubicin (Fig. 1E). These data suggest that autophagy induced by silencing SPP1 in MCF7 cells was not potentiated by doxorubicin, but rather tended to disappear with time. On the other hand, doxorubicin did not annihilate the autophagic process as it was able to induce autophagy in control cells within the same time frame.
Doxorubicin Also Induces ER stress-mediated Autophagy
It has previously been demonstrated that autophagy induced by doxorubicin involves the canonical pathway in which Beclin-1 (also known as Atg6) initiates the generation of autophagosomes by forming a multiprotein complex with class III PI3K and repression of the mammalian target of rapamycin (mTOR) (16). In agreement, doxorubicin increased Beclin-1 expression and suppressed mTOR activity as demonstrated by inhibition of phosphorylation of its substrate p70s6 kinase (Fig. 2A). The effect of 3MA, a class III PI3K inhibitor, on autophagy was next examined. Interestingly, although 3MA did not inhibit autophagy induced by silencing SPP1, in agreement with our previous findings (11), it markedly reduced autophagy induced by doxorubicin in the presence or absence of SPP1 expression (Fig. 2B).
The UPR and ER stress-mediated autophagy induced by SPP1 depletion involves the three canonical branches of the UPR: PKR-like eIF2α kinase (PERK), activating transcription factor-6 (ATF6), and IRE1α (11). As the role of these branches in autophagy induced by doxorubicin is still not clear (17–19), their contributions to autophagy induction by doxorubicin were examined. Doxorubicin enhanced expression of CHOP, a transcriptional regulator that is induced by all three arms of the UPR (Fig. 2A). To examine the involvement of ER stress transducers in doxorubicin-mediated autophagy, cells were transfected with specific siRNAs targeting IRE1α and ATF6, which reduced their expression by more than 90%, or dominant-negative PERK, which blocked phosphorylation of eIF2α (11). Both markedly reduced autophagy induced by doxorubicin in the presence or absence of SPP1 expression (Fig. 2C) and decreased conversion of LC3 to LC3-II (Fig. 2D).
Doxorubicin Sensitizes SPP1-depleted cells to Cell Death
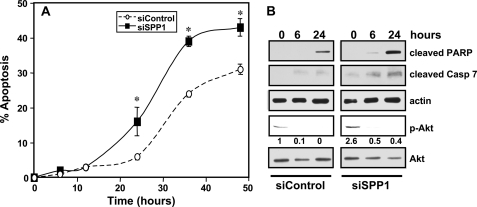
Altogether, these data demonstrate that although either depletion of SPP1 or treatment with doxorubicin induces ER stress-mediated autophagy, when combined, autophagy was markedly decreased rather than enhanced. Because ER stress-mediated autophagy has been shown to protect against cell death in some conditions (20, 21) and to promote cell killing in others (22), the possibility that doxorubicin influences the susceptibility of SPP1-depleted cells to undergo stress-mediated death was next examined. In agreement with other studies (23, 24), treatment with doxorubicin induced apoptosis in a time-dependent manner (Fig. 3A). This was accompanied by increased cleavage of PARP (Fig. 3B), a substrate for caspase-mediated proteolysis, and increased activation of caspase 7 (Fig. 3B), the main effector caspase in MCF7 cells (23), as shown by the appearance of cleaved caspase 7. Although apoptosis was undetectable in SPP1-silenced MCF7 cells (11), in the presence of doxorubicin, there was marked enhancement of apoptosis (Fig. 3A) as well as cleavage of PARP and caspase 7 (Fig. 3B). Thus, depletion of SPP1 sensitized the cells to doxorubicin-induced apoptosis. We previously showed that increased activation of the survival signal Akt in autophagy triggered by down-regulation of SPP1 protected cells from apoptosis (11). Indeed, the increased phosphorylation of Akt induced by SPP1 depletion was markedly reduced by doxorubicin (Fig. 3B), consistent with our previous observation that inactivation of Akt induced apoptosis of SPP1-depleted cells (11).
FIGURE 3.
SPP1 depletion sensitizes cells to doxorubicin-induced apoptosis. A and B, MCF7 cells transfected with siControl or siSPP1 were treated with doxorubicin (1 μg/ml) for the indicated times. A, nuclei were stained with Hoechst 33342, and apoptosis was determined by scoring the percentage of cells displaying fragmented, condensed nuclei. At least three fields were analyzed, scoring a minimum of 300 cells. Data are means ± S.D. of three independent experiments. *, p < 0.01 as compared with siControl. B, equal amounts of lysates from duplicate cultures were resolved by SDS-PAGE and immunoblotted with antibodies that recognize cleaved PARP, cleaved caspase 7 (Casp 7), and phospho-Akt (p-Akt). Numbers indicate -fold changes determined by densitometry. Blots were reprobed with anti-actin or anti-Akt to ensure equal loading and transfer.
Although down-regulation of S1P lyase, another ER-localized enzyme that cleaves S1P irreversibly, also significantly induced autophagy (10 ± 1.3% to 48 ± 10%), it did not sensitize the cells to doxorubicin-induced apoptosis. However, similar to SPP1 depletion, treatment with the ER stress inducer tunicamycin, an N-linked glycosylation inhibitor, which also by itself did not induce apoptosis, significantly enhanced apoptosis (Fig. 4A) and PARP cleavage (Fig. 4B) when combined with doxorubicin. It has been suggested that autophagy can become a death-promoting mechanism when the caspase 3-dependent pathway is blocked (25). Because MCF7 cells are devoid of caspase 3 due to functional deletion of the CASP3 gene (26), it was of interest to examine whether SPP1 depletion-mediated autophagy can be switched to cell death by doxorubicin in other types of cancer cells expressing caspase 3, such as HCT116 colon carcinoma cells. Decreasing endogenous levels of SPP1 in these cells also caused autophagy, which was also markedly reduced by doxorubicin (Fig. 4C), as a result of the concomitant increase in apoptotic cells (Fig. 4D). In this regard, the tumor suppressor p53, which is up-regulated by doxorubicin, has been shown to trigger apoptosis while inhibiting autophagy (27, 28). However, p53 is not involved, as decreased autophagy and increased apoptosis were also observed in SPP1-depleted p53 null HCT116 cells (Fig. 4, E and F). These results suggest that SPP1 plays a general role in determining sensitivity to doxorubicin-induced apoptosis.
Apoptosis and Autophagy Are Sequentially Linked
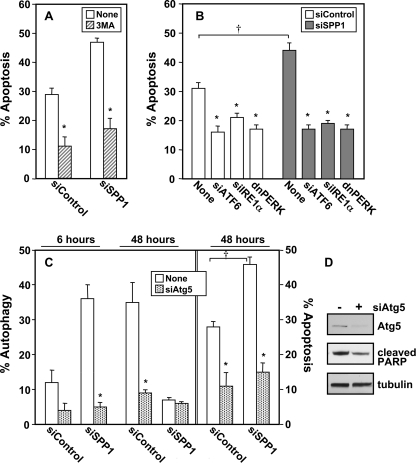
Several approaches were next utilized to further examine the link between autophagy and apoptosis. First, the effect on cell death of 3MA, which suppressed autophagy (Fig. 2B), was evaluated. 3MA did not accelerate apoptosis induced by doxorubicin in SPP1-depleted cells, but rather significantly reduced it (Fig. 5A). Moreover, down-regulation of the ER stress transducers ATF6 and IRE1α or expression of dnPERK also significantly suppressed doxorubicin-induced apoptosis in the absence or presence of SPP1 expression to similar levels (Fig. 5B). Finally, silencing Atg5, which is required in the early stages of autophagosome formation, not only suppressed autophagy (Fig. 5C), it also significantly decreased the robust apoptosis induced by doxorubicin in SPP1-depleted cells (Fig. 5C) and reduced PARP cleavage (Fig. 5D).
FIGURE 5.
Autophagy and apoptosis are sequentially linked. A, MCF7 cells transfected with siControl or siSPP1 were treated with doxorubicin (1 μg/ml) in the absence (None) or presence of 10 mm 3MA for 48 h, and apoptosis was determined. B, MCF7 cells were co-transfected with siControl, siSPP1, siIRE1α, siATF6, or dnPERK, as indicated. Apoptosis was quantified after 48 h of treatment with doxorubicin. C, MCF7 cells co-transfected with siControl or siSPP1 and siAtg5 were treated with doxorubicin (1 μg/ml) for the indicated times, and autophagy and apoptosis were determined. *, p < 0.01 as compared with None. †, p < 0.01 as compared with siControl. Error bars in panels A–C indicate means ± S.D. D, MCF7 cells co-transfected with siControl or siSPP1 and siATG5 were treated with doxorubicin for 48 h. Cell lysates were analyzed by Western blotting using antibodies for Atg5 and cleaved PARP. Blots were stripped and reprobed with anti-tubulin to ensure equal loading and transfer.
Autophagy and Apoptosis Are Linked through Atg5 Cleavage by Calpain
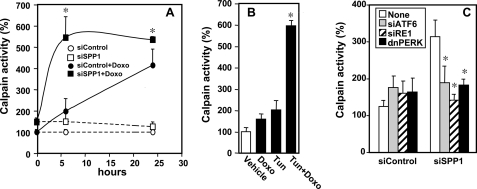
It has previously been suggested that calpain-mediated cleavage of Atg5 switches autophagy to apoptosis, whereby the N-terminal fragment of Atg5 with a molecular mass of 24 kDa is translocated to the mitochondria, inducing mitochondrial damage leading to apoptosis (13). Therefore, we first examined calpain activity by measuring cleavage of a succinyl proluminescent calpain substrate. Depletion of SPP1 itself only slightly increased calpain activity (Fig. 6A). Doxorubicin treatment increased calpain activity in a time-dependent manner, reaching a maximum at 24 h, which was dramatically accelerated in SPP1-depleted cells, and maximum activity was observed within 6 h (Fig. 6A). Similarly, treatment with the ER stress inducer tunicamycin also significantly enhanced doxorubicin-induced calpain activity (Fig. 6B). Inhibition of the three sensors of the UPR and ER stress by silencing ATF6 or IRE1α or overexpression of dnPERK completely prevented the enhanced calpain activation in SPP1-depleted cells treated with doxorubicin (Fig. 6C).
FIGURE 6.
Doxorubicin-induced calpain activity is enhanced in SPP1-depleted cells. A, MCF7 cells transfected with siControl or siSPP1 were treated without or with doxorubicin (Doxo, 1 μg/ml), and calpain activity was measured at the indicated times with a luminescence assay. *, p < 0.01 as compared with siControl treated with doxorubicin. B, MCF7 cells were treated for 6 h with vehicle, doxorubicin (1 μg/ml), tunicamycin (Tun, 2.5 μm), or both, and calpain activity measured. *, p < 0.01 as compared with vehicle. C, MCF7 cells were co-transfected with siControl, siSPP1, siIRE1α, siATF6, or dnPERK, as indicated, and treated with doxorubicin (1 μg/ml) for 6 h, and calpain activity was measured. Activities were normalized to untreated siControl cells (None) at time 0. *, p < 0.01 as compared with None. Error bars in all panels indicate means ± S.D.
Next we examined whether cleavage of Atg5 by calpain is a determinant of conversion of autophagy to apoptosis. Doxorubicin enhanced the appearance of a smaller Atg5 band with the expected molecular mass of 24 kDa detected with an antibody against the N terminus of Atg5 (Fig. 7A). Cleavage of Atg5 in cells depleted of SPP1 was accelerated as the fragment was detected within 6 h (Fig. 7A), in agreement with increased calpain activity determined at the same time point (Fig. 6A). To confirm the involvement of calpain-mediated Atg5 cleavage, the papain-like cysteine protease inhibitor (E64d) was used. E64d reversed the suppressive effect of doxorubicin on autophagy mediated by SPP1 depletion (Fig. 7B) and decreased doxorubicin-induced apoptosis in SPP1-depleted cells (Fig. 7C). Treatment with E64d, in agreement with its inhibitory effect on calpain activity, decreased cleavage of Atg5 (Fig. 7D). To further substantiate the involvement of calpain, its expression was down-regulated by more than 85% with a specific siRNA (Fig. 7E). Depletion of calpain also attenuated doxorubicin-induced switching of autophagy (Fig. 7F) to apoptosis (Fig. 7G). Doxorubicin treatment did not induce cleavage of Atg5 in these calpain-deficient MCF7 cells (Fig. 7H), further supporting the notion that calpain is the major protease responsible for this cleavage. Altogether, these results indicate that calpain-mediated Atg5 cleavage switches autophagy to apoptosis in SPP1-depleted cells treated with doxorubicin.
Involvement of Ceramide Formation in Atg5 Cleavage and Apoptosis
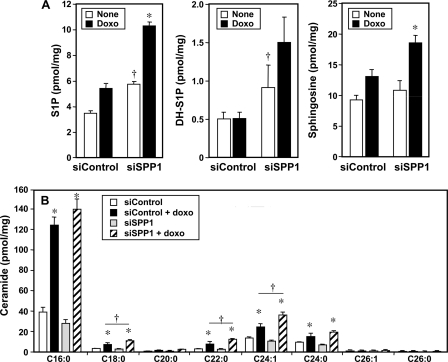
Numerous studies have indicated that increased de novo synthesis of ceramide, the precursor of sphingosine and S1P, is one factor that determines sensitivity to chemotherapeutics such as doxorubicin (5, 29–31). Because depletion of SPP1 markedly sensitizes cells to doxorubicin-induced apoptosis, we examined the effect of treatment with doxorubicin on sphingolipid metabolite levels in SPP1-depleted cells. In agreement with previous studies (11, 32, 33), we found that down-regulation of SPP1 increased intracellular levels of S1P and dihydro-S1P (Fig. 8A). Treatment with doxorubicin enhanced these increases in phosphorylated sphingoid bases. Although the changes in cellular S1P levels appear small, it should be noted that it is still not possible with current technology to determine S1P levels in the ER where SPP1 is located and that local increases in S1P could be much larger. In agreement with previous studies (5, 29, 31, 34), doxorubicin markedly increased ceramide levels in the presence or absence of SPP1 (Fig. 8B). Almost all ceramide species were increased with a predominant increase in the C16:0 species (Fig. 8B). Increases in C18:0, C22:0, and C24:1 were also more pronounced in SPP1-depleted cells (Fig. 8B).
FIGURE 8.
Effect of doxorubicin on levels of sphingolipid metabolites in SPP1-depleted cells. A and B, MCF7 cells transfected with siControl or siSPP1 were treated without (None) or with doxorubicin (Doxo, 1 μg/ml) for 24 h. Lipids were extracted, and S1P, dihydro-S1P (DH-S1P), sphingosine (A), and ceramide species (B) were determined by LC-ESI-MS/MS. Numbers indicate chain length followed by the number of double bonds in the fatty acid. Data are averages of triplicates and expressed as pmol of lipid/mg of protein. *, p < 0.01 as compared with untreated cells. †, p < 0.01 as compared with siControl. Similar results were found in three additional experiments. Error bars in both panels indicate means ± S.D.
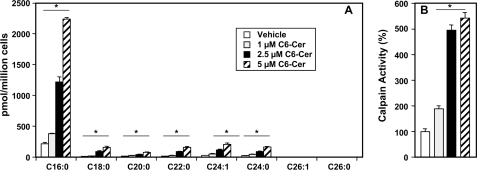
Next, we asked whether ceramide elevation leads to calpain activation. To this end, cells were treated with C6-ceramide, a short-chain analog that is readily taken up by cells and converted to endogenous long-chain ceramides (35). Mass spectrometry analysis revealed that levels of endogenous long-chain ceramide species were significantly increased, particularly C16:0, C18:0, C22:0, and C24:1, within 3 h after C6:0-ceramide treatment (Fig. 9A). Calpain activity was increased up to 5-fold concomitantly with ceramide elevation (Fig. 9B).
FIGURE 9.
C6:0-ceramide is rapidly converted to endogenous long-chain ceramides and activates calpain. A and B, MCF7 cells were treated with the indicated concentrations of C6-ceramide (C6-Cer) for 3 h. A, generation of endogenous ceramide species was analyzed by LC-ESI-MS/MS. Data are averages of triplicates and expressed as pmol of lipid per million cells. B, calpain activity was measured in duplicate cultures with a luminescence assay. *, p < 0.01 as compared with vehicle-treated cells. Error bars in both panels indicate means ± S.D.
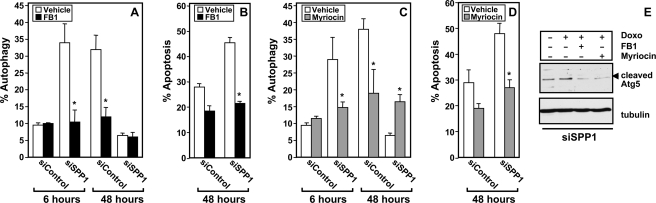
Pretreatment with fumonisin B1, an inhibitor of ceramide synthase, or myriocin, an inhibitor of serine palmitoyltransferase, the first committed step in de novo synthesis of ceramide, as expected reduced ceramide elevation in response to doxorubicin (data not shown). Importantly, fumonisin B1 (FB1) or myriocin suppressed autophagy in SPP1-depleted cells (Fig. 10, A and C). Moreover, these inhibitors drastically inhibited the robust apoptosis observed in SPP1-depleted cells treated with doxorubicin (Fig. 10, B and D). Furthermore, inhibition of ceramide generation by FB1 or myriocin suppressed Atg5 cleavage in SPP1-depleted cells treated with doxorubicin (Fig. 10E). These results suggest that ceramide elevation in response to doxorubicin contributes to Atg5 cleavage.
FIGURE 10.
Fumonisin B1 and myriocin suppress doxorubicin-mediated autophagic cell death. MCF7 cells transfected with siControl or siSPP1 were treated with doxorubicin (1 μg/ml) in the absence or presence of FB1 (25 μm) (A and B) or myriocin (10 μm) (C and D) for the indicated times, and autophagy (A and C) and apoptosis (B and D) were determined. *, p < 0.01 as compared with vehicle-treated cells. Error bars in panels A–D indicate means ± S.D. E, equal amounts of MCF7 cell lysates transfected with siSPP1 and treated with doxorubicin (Doxo) in the absence or presence of FB1 or myriocin as indicated were analyzed by Western blotting with an anti-Atg5 antibody. Blots were reprobed with tubulin.
DISCUSSION
We previously demonstrated that increasing intracellular S1P at the ER by suppressing expression of SPP1 induces ER stress and triggers p53-independent autophagy that does not lead to autophagic cell death (11). Moreover, SPP1 levels change in response to some stimuli that alter the UPR. Analysis of MCF7 breast cancer cells overexpressing XBP-1, a transcription factor that participates in the UPR and confers both estrogen independence and anti-estrogen resistance, revealed that SPP1 was among the genes that were significantly up-regulated (36). In agreement, we found that doxorubicin, similar to other stimuli that alter the UPR, up-regulates SPP1. Interestingly, a systematic analysis using an RNA interference screen across a broad range of cancers to define critical pathways of drug resistance and sensitivity identified the ceramide transport protein (CERT) and SPP1. Out of more than 50 siRNAs targeted to ceramide metabolism-related genes (the “ceramidome”), only expression of CERT and SPP1 was found to be important for sensitivity of breast cancer cells to doxorubicin (34). In the present study, we examined how depletion of SPP1 sensitizes MCF7 cells to doxorubicin-induced apoptosis.
We have now shown that treatment of SPP1-depleted cells with doxorubicin decreases the number of autophagic cells and switches them to apoptosis. As both ER stress and autophagy are self-limited processes that usually protect cells from cell death by diverse mechanisms, it is reasonable that induction of apoptosis would be coupled to inactivation of autophagy. In agreement, inhibition of ER stress by inactivation of the three sensors of the UPR to ER stress, IRE1α, ATF6, and PERK, or by attenuating autophagy with 3-methyladenine also markedly suppressed apoptosis in SPP1-depleted cells treated with doxorubicin. Moreover, in agreement with other studies, we found that doxorubicin significantly enhanced de novo biosynthesis of ceramide (5, 31, 34), and inhibition of ceramide formation also reduced apoptosis. It was initially thought that ceramide induced autophagic cell death by up-regulating Beclin 1 expression in a JNK-dependent manner (8, 37), by the cell death-inducing mitochondrial BH3-only protein, BNIP3 (38), or by down-regulation of nutrient transporters (39). Ceramide also suppresses Akt and thus interferes with mTOR signaling and other survival pathways (8, 40). However, other studies indicate that ceramide elevation may be linked to protective autophagy. Down-regulation of ceramide synthase 2 (Cers2), although decreasing very long-chain ceramides and increasing long-chain C14 and C16 ceramides, did not induce apoptotic cell death but rather induced autophagy and UPR (41). However, decreasing C16:0-ceramide by down-regulation of CerS6 induces ER stress via the ATF6/CHOP arm, which leads to cell death, whereas C18:0 ceramide generated by CerS1 has pro-apoptotic effects (42). This is reminiscent of studies in worms where ceramides with different chain lengths have opposing effects on survival; although very long-chain C24-C26 ceramides increase sensitivity to anoxia-mediated death, shorter-chain C20-C22 ceramides decrease it (43). These results suggest that ceramides with different fatty acid chain lengths in specific subcellular locations might have distinct functions in regulation of UPR, ER stress, autophagy, and apoptosis. Moreover, it has also become increasingly evident that the location of S1P formation dictates its function (44). In agreement with this notion, although decreasing degradation of S1P by depletion of S1P lyase also induced autophagy, it did not sensitize cells to doxorubicin. It should be noted that the lyase and SPP1 have distinct actions. Although dephosphorylation of S1P by SPP1 is important for the salvage pathway and formation of sphingosine and ceramide, the lyase degrades S1P irreversibly to phosphoethanolamine (precursor of phosphatidylethanolamine) and hexadecenal, a bioactive product that also induces apoptosis (45).
We have now examined the molecular events induced by doxorubicin that switch protective autophagy in SPP1-depleted cells to apoptosis and shown that calpain-mediated cleavage of Atg5, which is indispensable for autophagosome formation, is the key that connects autophagy and apoptosis. It has previously been demonstrated that proteolysis of Atg5 by the calcium-dependent cysteine protease calpain destroys its pro-autophagic function (12), while generating a pro-apoptotic mitochondrion-permeabilizing fragment (13). Several lines of evidence indicate that this truncated fragment of Atg5 is the major contributor to the switch between protective autophagy and apoptosis that we observed. First, depletion of SPP1 enhanced doxorubicin-induced calpain activity and cleavage of Atg5. Second, inhibition or down-regulation of calpain suppressed Atg5 cleavage and apoptosis induced by doxorubicin in SPP1-depleted cells. Third, down-regulation of Atg5 also suppressed doxorubicin-induced apoptosis in these cells. Fourth, ceramide, whose synthesis is enhanced by doxorubicin, also stimulated calpain, and inhibition of ceramide biosynthesis with FB1 or myriocin not only suppressed Atg5 cleavage but also the robust apoptosis induced by doxorubicin in SPP1-deficient cells. Hence, ceramide formation in response to doxorubicin activates calpain-mediated Atg5 cleavage that in turn switches autophagy in SPP1-depleted cells to apoptosis. Similarly, in prostate cancer cells, melanoma differentiation-associated gene 7 (mda-7)/interleukin-24 (IL-24) enhanced cleavage of Atg5 by calpain and facilitated a switch from autophagy to apoptosis that was also dependent on ceramide generation (46). Furthermore, it was recently reported that cis-4-methylsphingosine phosphate, a semisynthetic analog of S1P formed at the ER, induces ER stress and calcium release, leading to activation of calpain and caspases and neuronal apoptosis (47). In this regard, S1P generated in the ER membrane was shown many years ago to activate release of calcium from inositol 1,4,5-trisphosphate-sensitive calcium pools (48). It is therefore possible that S1P accumulation in the ER could also contribute to enhancement of calpain activity and Atg5 cleavage in SPP1-depleted cells. Because depletion of SPP1 activates Akt in a PERK-dependent manner that counteracts ER stress-induced apoptotic signaling (11), our finding that doxorubicin inhibits Akt activation in these cells suggests an additional mechanism for enhancement of apoptosis. As mentioned above, these effects could be attributed to ceramide elevation as ceramide has been long known to potently inhibit activation of Akt (49). This notion is in agreement with the observation that a specific Akt inhibitor also induced apoptosis in SPP1-depleted cells (11).
Taken together, our results solidify the notion that sphingolipid metabolic pathways are involved in governing the cross-talk between autophagy and apoptosis.
Acknowledgments
We are grateful to Drs. T. Yoshimori (National Institute of Genetics, Mishima, Japan), K. Kirkegaard (Stanford University School of Medicine, Stanford, CA), J. A. Diehl (University of Pennsylvania), David Ron (Skirball Institute, New York University), and Andrew Thorburn (University of Colorado) for generous gifts of reagents. Confocal microscopy and electron microscopy were supported in part by National Institutes of Health Grant P30 CA16059 to the Massey Cancer Center and NINDS Center core Grant 5P30NS047463, respectively.
This work was supported, in whole or in part, by National Institutes of Health Grant 5R37GM043880 from NIGMS (to S. S.).
- UPR
- unfolded protein response
- ATF6
- activating transcription factor 6
- Atg
- autophagy-related gene
- CHOP
- C/EBP homologous protein
- C/EBP
- CCAAT/enhancer-binding protein
- dn
- dominant-negative
- E64d
- l-3-carboxy-trans-2,3-epoxypropionyl-leu-amido-(4-guanidino-)butane-ethylester
- ER
- endoplasmic reticulum
- FB1
- fumonisin B1
- IRE1α
- inositol-requiring transmembrane kinase/endonuclease α
- LC3
- microtubule-associated protein 1 light chain 3
- 3MA
- 3-methyladenine
- mTOR
- mammalian target of rapamycin
- PARP
- poly(ADP-ribose) polymerase
- PERK
- PKR-like eIF2α kinase
- S1P
- sphingosine 1-phosphate
- SPP1
- S1P phosphohydrolase 1
- siControl
- control scrambled siRNA
- siSPP1
- siRNA targeted to SPP1.
REFERENCES
- 1. Kroemer G., Mariño G., Levine B. (2010) Mol. Cell 40, 280–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Eisenberg-Lerner A., Bialik S., Simon H. U., Kimchi A. (2009) Cell Death Differ 16, 966–975 [DOI] [PubMed] [Google Scholar]
- 3. Buchberger A., Bukau B., Sommer T. (2010) Mol. Cell 40, 238–252 [DOI] [PubMed] [Google Scholar]
- 4. Cuvillier O., Pirianov G., Kleuser B., Vanek P. G., Coso O. A., Gutkind S., Spiegel S. (1996) Nature 381, 800–803 [DOI] [PubMed] [Google Scholar]
- 5. Ogretmen B., Hannun Y. A. (2004) Nat. Rev. Cancer 4, 604–616 [DOI] [PubMed] [Google Scholar]
- 6. Alvarez S. E., Milstien S., Spiegel S. (2007) Trends Endocrinol. Metab. 18, 300–307 [DOI] [PubMed] [Google Scholar]
- 7. Pyne N. J., Pyne S. (2010) Nat. Rev. Cancer 10, 489–503 [DOI] [PubMed] [Google Scholar]
- 8. Scarlatti F., Bauvy C., Ventruti A., Sala G., Cluzeaud F., Vandewalle A., Ghidoni R., Codogno P. (2004) J. Biol. Chem. 279, 18384–18391 [DOI] [PubMed] [Google Scholar]
- 9. Ji C., Yang B., Yang Y. L., He S. H., Miao D. S., He L., Bi Z. G. (2010) Oncogene 29, 6557–6568 [DOI] [PubMed] [Google Scholar]
- 10. Lavieu G., Scarlatti F., Sala G., Carpentier S., Levade T., Ghidoni R., Botti J., Codogno P. (2006) J. Biol. Chem. 281, 8518–8527 [DOI] [PubMed] [Google Scholar]
- 11. Lépine S., Allegood J. C., Park M., Dent P., Milstien S., Spiegel S. (2011) Cell Death Differ. 18, 350–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Xia H. G., Zhang L., Chen G., Zhang T., Liu J., Jin M., Ma X., Ma D., Yuan J. (2010) Autophagy 6, 61–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yousefi S., Perozzo R., Schmid I., Ziemiecki A., Schaffner T., Scapozza L., Brunner T., Simon H. U. (2006) Nat. Cell Biol. 8, 1124–1132 [DOI] [PubMed] [Google Scholar]
- 14. Hait N. C., Allegood J., Maceyka M., Strub G. M., Harikumar K. B., Singh S. K., Luo C., Marmorstein R., Kordula T., Milstien S., Spiegel S. (2009) Science 325, 1254–1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Akar U., Chaves-Reyez A., Barria M., Tari A., Sanguino A., Kondo Y., Kondo S., Arun B., Lopez-Berestein G., Ozpolat B. (2008) Autophagy 4, 669–679 [DOI] [PubMed] [Google Scholar]
- 16. Muñoz-Gámez J. A., Rodríguez-Vargas J. M., Quiles-Pérez R., Aguilar-Quesada R., Martín-Oliva D., de Murcia G., Menissier de Murcia J., Almendros A., Ruiz de Almodóvar M., Oliver F. J. (2009) Autophagy 5, 61–74 [DOI] [PubMed] [Google Scholar]
- 17. Kim S. J., Park K. M., Kim N., Yeom Y. I. (2006) Biochem. Biophys. Res. Commun. 339, 463–468 [DOI] [PubMed] [Google Scholar]
- 18. Lai H. C., Yeh Y. C., Ting C. T., Lee W. L., Lee H. W., Wang L. C., Wang K. Y., Lai H. C., Wu A., Liu T. J. (2010) Eur. J. Pharmacol. 644, 176–187 [DOI] [PubMed] [Google Scholar]
- 19. Bar-Joseph H., Ben-Aharon I., Rizel S., Stemmer S. M., Tzabari M., Shalgi R. (2010) Reprod. Toxicol. 30, 566–572 [DOI] [PubMed] [Google Scholar]
- 20. Ogata M., Hino S., Saito A., Morikawa K., Kondo S., Kanemoto S., Murakami T., Taniguchi M., Tanii I., Yoshinaga K., Shiosaka S., Hammarback J. A., Urano F., Imaizumi K. (2006) Mol. Cell. Biol. 26, 9220–9231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bernales S., McDonald K. L., Walter P. (2006) PLoS Biol. 4, e423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ding W. X., Ni H. M., Gao W., Hou Y. F., Melan M. A., Chen X., Stolz D. B., Shao Z. M., Yin X. M. (2007) J. Biol. Chem. 282, 4702–4710 [DOI] [PubMed] [Google Scholar]
- 23. Cuvillier O., Nava V. E., Murthy S. K., Edsall L. C., Levade T., Milstien S., Spiegel S. (2001) Cell Death Differ. 8, 162–171 [DOI] [PubMed] [Google Scholar]
- 24. Sankala H. M., Hait N. C., Paugh S. W., Shida D., Lépine S., Elmore L. W., Dent P., Milstien S., Spiegel S. (2007) Cancer Res. 67, 10466–10474 [DOI] [PubMed] [Google Scholar]
- 25. Gozuacik D., Bialik S., Raveh T., Mitou G., Shohat G., Sabanay H., Mizushima N., Yoshimori T., Kimchi A. (2008) Cell Death Differ. 15, 1875–1886 [DOI] [PubMed] [Google Scholar]
- 26. Jänicke R. U., Sprengart M. L., Wati M. R., Porter A. G. (1998) J. Biol. Chem. 273, 9357–9360 [DOI] [PubMed] [Google Scholar]
- 27. Tasdemir E., Maiuri M. C., Galluzzi L., Vitale I., Djavaheri-Mergny M., D'Amelio M., Criollo A., Morselli E., Zhu C., Harper F., Nannmark U., Samara C., Pinton P., Vicencio J. M., Carnuccio R., Moll U. M., Madeo F., Paterlini-Brechot P., Rizzuto R., Szabadkai G., Pierron G., Blomgren K., Tavernarakis N., Codogno P., Cecconi F., Kroemer G. (2008) Nat. Cell Biol. 10, 676–687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Green D. R., Kroemer G. (2009) Nature 458, 1127–1130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kolesnick R. (2002) J. Clin. Invest. 110, 3–8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kolesnick R., Altieri D., Fuks Z. (2007) Cancer Cell 11, 473–475 [DOI] [PubMed] [Google Scholar]
- 31. Liu Y. Y., Yu J. Y., Yin D., Patwardhan G. A., Gupta V., Hirabayashi Y., Holleran W. M., Giuliano A. E., Jazwinski S. M., Gouaze-Andersson V., Consoli D. P., Cabot M. C. (2008) FASEB J. 22, 2541–2551 [DOI] [PubMed] [Google Scholar]
- 32. Johnson K. R., Johnson K. Y., Becker K. P., Bielawski J., Mao C., Obeid L. M. (2003) J. Biol. Chem. 278, 34541–34547 [DOI] [PubMed] [Google Scholar]
- 33. Zhao Y., Kalari S. K., Usatyuk P. V., Gorshkova I., He D., Watkins T., Brindley D. N., Sun C., Bittman R., Garcia J. G., Berdyshev E. V., Natarajan V. (2007) J. Biol. Chem. 282, 14165–14177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Swanton C., Marani M., Pardo O., Warne P. H., Kelly G., Sahai E., Elustondo F., Chang J., Temple J., Ahmed A. A., Brenton J. D., Downward J., Nicke B. (2007) Cancer Cell 11, 498–512 [DOI] [PubMed] [Google Scholar]
- 35. Ogretmen B., Pettus B. J., Rossi M. J., Wood R., Usta J., Szulc Z., Bielawska A., Obeid L. M., Hannun Y. A. (2002) J. Biol. Chem. 277, 12960–12969 [DOI] [PubMed] [Google Scholar]
- 36. Gomez B. P., Riggins R. B., Shajahan A. N., Klimach U., Wang A., Crawford A. C., Zhu Y., Zwart A., Wang M., Clarke R. (2007) FASEB J. 21, 4013–4027 [DOI] [PubMed] [Google Scholar]
- 37. Li D. D., Wang L. L., Deng R., Tang J., Shen Y., Guo J. F., Wang Y., Xia L. P., Feng G. K., Liu Q. Q., Huang W. L., Zeng Y. X., Zhu X. F. (2009) Oncogene 28, 886–898 [DOI] [PubMed] [Google Scholar]
- 38. Daido S., Kanzawa T., Yamamoto A., Takeuchi H., Kondo Y., Kondo S. (2004) Cancer Res. 64, 4286–4293 [DOI] [PubMed] [Google Scholar]
- 39. Guenther G. G., Peralta E. R., Rosales K. R., Wong S. Y., Siskind L. J., Edinger A. L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 17402–17407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Coward J., Ambrosini G., Musi E., Truman J. P., Haimovitz-Friedman A., Allegood J. C., Wang E., Merrill A. H., Jr., Schwartz G. K. (2009) Autophagy 5, 184–193 [DOI] [PubMed] [Google Scholar]
- 41. Spassieva S. D., Mullen T. D., Townsend D. M., Obeid L. M. (2009) Biochem. J. 424, 273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Senkal C. E., Ponnusamy S., Bielawski J., Hannun Y. A., Ogretmen B. (2010) FASEB J. 24, 296–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Menuz V., Howell K. S., Gentina S., Epstein S., Riezman I., Fornallaz-Mulhauser M., Hengartner M. O., Gomez M., Riezman H., Martinou J. C. (2009) Science 324, 381–384 [DOI] [PubMed] [Google Scholar]
- 44. Spiegel S., Milstien S. (2011) Nat. Rev. Immunol. 11, 403–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kumar A., Byun H. S., Bittman R., Saba J. D. (2011) Cell Signal. 23, 1144–1152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Bhutia S. K., Dash R., Das S. K., Azab B., Su Z. Z., Lee S. G., Grant S., Yacoub A., Dent P., Curiel D. T., Sarkar D., Fisher P. B. (2010) Cancer Res. 70, 3667–3676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Hagen N., Hans M., Hartmann D., Swandulla D., van Echten-Deckert G. (2011) Cell Death Differ. 18, 1356–1365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ghosh T. K., Bian J., Gill D. L. (1990) Science 248, 1653–1656 [DOI] [PubMed] [Google Scholar]
- 49. Zhou H., Summers S. A., Birnbaum M. J., Pittman R. N. (1998) J. Biol. Chem. 273, 16568–16575 [DOI] [PubMed] [Google Scholar]