Background: SecA has been viewed as ATPase helping precursors across SecYEG channels.
Results: SecA alone could promote protein translocation and ion channel activity, but loses specificity and efficiency, which can be restored by SecYEG.
Conclusion: SecA plays important structural roles and can function as low affinity protein conducting channels in membranes.
Significance: Establishing SecA as channels is crucial for understanding diverse mechanisms and evolution of bacterial translocation pathways.
Keywords: Ion Channels, Liposomes, Patch Clamp, Protein Secretion, Protein Translocation, SecA, SecYEG
Abstract
SecA is an essential component of the Sec-dependent protein translocation pathway across cytoplasmic membranes in bacteria. Escherichia coli SecA binds to cytoplasmic membranes at SecYEG high affinity sites and at phospholipid low affinity sites. It has been widely viewed that SecYEG functions as the essential protein-conducting channel through which precursors cross the membranes in bacterial Sec-dependent pathways, and that SecA functions as a motor to hydrolyze ATP in translocating precursors through SecYEG channels. We have now found that SecA alone can promote precursor translocation into phospholiposomes. Moreover, SecA-liposomes elicit ionic currents in Xenopus oocytes. Patch-clamp recordings further show that SecA alone promotes signal peptide- or precursor-dependent single channel activity. These activities were observed with the functional SecA at about 1–2 μm. The results show that SecA alone is sufficient to promote protein translocation into liposomes and to elicit ionic channel activity at the phospholipids low affinity binding sites, thus indicating that SecA is able to form the protein-conducting channels. Even so, such SecA-liposomes are less efficient than those with a full complement of Sec proteins, and lose the signal-peptide proofreading function, resembling the effects of PrlA mutations. Addition of purified SecYEG restores the signal peptide specificity and increases protein translocation and ion channel activities. These data show that SecA can promote protein translocation and ion channel activities both when it is bound to lipids at low affinity sites and when it is bound to SecYEG with high affinity. The latter of the two interactions confers high efficiency and specificity.
Introduction
Most proteins destined to traverse cytoplasmic membranes through the Sec-dependent pathway carry a transient N-terminal hydrophobic signal peptide that is cleaved during, or shortly after, translocation. In bacteria both biochemical and genetic approaches have provided important and complementary insight into the mechanism of this Sec secretion pathway. The current and prevailing model for Sec-dependent translocation depicts the bacterial SecYEG complex as providing the essential, protein-conducting channel, with SecA acting as a peripheral component of the translocase hydrolyzing ATP (1, 2), as it cycles on and off the cytoplasmic membrane during protein translocation (3, 4). This model of SecYEG as being the essential protein-conducting channel arises in part from the homology of SecY to Sec61p, which has been proposed to be a component of the translocation channel in yeast and mammals (5–8). The Sec61 complex is assumed to form 2 nm pores, which further assemble into 4–6 nm pores, through which nascent peptides in the ribosomes pass through the endoplasmic reticulum (5–7). The x-ray structure of an archea SecYEG complex shows similar pore size. The Escherichia coli SecYEG translocation complex (9) is significantly smaller than both the yeast Sec61 complex and the more complete SecA/SecYEGEc, as defined by more recent x-ray structural analysis (10).
However, Blobel and co-workers (11) have presented strong evidence that reconstituted E. coli membranes, lacking detectable SecY, efficiently translocate proLamB. They suggested that SecY is not the only receptor for SecA and that it may not be essential for protein translocation. Our previous work has confirmed these observations with other precursors, and showed further that both SecE-deficient or SecY-deficient membranes, derived from genetically manipulated cells, are also capable of efficiently to translocating certain precursors in vitro (12, 13). Indeed, a set of proteins (mostly inner and outer membrane proteins) were only minimally affected in E. coli cells upon depletion of SecE (14), indicating a differential requirement of SecYEG. Thus, neither SecE nor SecY is apparently essential for the translocation of all proteins. Moreover, SecA, often referred as a peripheral subunit of pre-protein translocase with ATPase activity (3, 4), has been found to integrate into membranes (15, 16). It does not cycle on and off the membrane during protein translocation (15), and it is an integral part of the protein-conducting channel (15, 17). Indeed, it has been found that SecA can bind to membranes at SecYEG high affinity sites and at phospholipids low affinity sites (15, 16). We have also found that, upon binding to anionic phospholipids, SecA undergoes conformational changes to form ring-like structures, and that these may form the core of the protein-conducting channels (18–20). These observations suggest that there may be a Sec pathway in which SecA plays a channel-forming structural role. In this study, we show that SecA alone can indeed promote protein translocation and elicit ion channel activity in liposomes, and that SecYEG restores the lost efficiency and specificity.
EXPERIMENTAL PROCEDURES
Bacterial Strains and Plasmids
E. coli K12 strain MC4100 (F−araD139 Δ(argF-lac) U169 rpsL150 relA1 deoC1rbsRfthD5301 fruA25 λ−) was from J. Beckwith (21), and a derivative of BA13 (MC4100 supFtstrpamsecA13amzch::Tn10) was from D. Oliver (22) and SecY suppression mutants, PrlA4, from T. Silhavy (21). The no-plug SecY mutants were from T. Rapoport (23). D10–1 (rna-10 relA1 spoT1 metB1 ompT::Km RNase- thi-), BL21.19 harboring plasmids pET5a/secAA (19) or pET20b/secAL43P are lab stocks. Plasmids containing secAL43P and secAR509K were from H. Takamatsu (24) and D. Oliver (25), respectively. Plasmid encoding SecYEhisG was from F. Duong (26, 27).
Protein Purification
E. coli SecA, its derivatives and homologs, were purified as described (19, 28). Purified proOmpA, proPhoA, wild-type LamB signal peptides, and LamB deletion mutant (LamB DM) signal peptides were prepared as described (2, 15). Protein concentration was determined by A280/260 ratio or Bradford assay. SecAL43P and SecAR509K were laboratory stocks purified by L. Yu and Y. Huang, respectively.
Liposome Preparation
E. coli total lipids extracts (Avanti Polar Lipid, Inc.) were dried in the Thermo Savant vacuum and resuspended in specific buffer (150 mm KCl for oocytes injection and patch clamp, and TAK buffer containing 50 mm Tris-HCl, pH 7.6, 20 mm NH4Cl and 25 mm KCl for in vitro translocation). The suspension was subjected to sonication (Fisher Scientific Sonic Dismembrator Model 500) at the amplitude of 70% for two to three times with the pause between consecutive times of 2 min each at 0 °C water bath. The size distribution of opalescent liposomes was measured by a Beckman Coulter N5 submicron particle size analyzer and showed a normal distribution with a peak around 130 nm. The liposomes were aliquoted, stored at −80 °C, and thawed only once for experiments. SecYEG-liposomes were prepared by mixing and vortexing SecYEG with liposomes in the indicated amounts.
Membrane Vesicles and SecYEG Preparation
Inside-out membrane vesicles (IMV) of wild-type MC4100, PrlA4 and SecY plug-domain mutant were prepared as described (23, 29). SecA-depleted membrane vesicles from the secAamber mutant BA13 were prepared as described (30). SecYEG-His-tagged membranes were prepared from a C43 strain carrying a pBAD/secEhisYG plasmid and SecYEG was purified by affinity to SecE-His as described (26, 27) followed by the monoQ column using buffer containing 0.2% C12E9. The purified SecYEG was stored in 0.2% C12E9, and used in liposome preparation at least 30-fold dilution.
Protein Translocation into Liposomes
The in vitro translocation of purified proOmpA or OmpA was determined by its inaccessibility to proteinase K. Translocation mixtures per ml contained translocation buffer, 2 mm Mg(OAc)2, and an ATP-regenerating system (30), and unless indicated contained 100 μg SecA or derivatives, 1.2 μg of proOmpA and 1.2 mg of liposomes prepared in TAK buffer containing 50 mm Tris-HCl, pH 7.6, 20 mm NH4Cl and 25 mm KCl. After incubation at 30 °C for 30 min, samples were kept on ice and treated with proteinase K (0.4 mg/ml) for 25 min to remove untranslocated protein. The reaction was stopped by 2 mm PMSF as described, and the liposomes containing protease-protected proOmpA or OmpA was pelleted by centrifugation (17) in a TLA 100.3 Beckman rotor at 90,000 rpm for 25 min at 4 °C, resuspended in the sample buffer and loaded onto SDS-PAGE. The amount of translocated protein was analyzed by Western blotting followed by quantification.
Xenopus Oocyte Preparation and Voltage Clamp Measurement
The preparation of oocytes and voltage clamp measurement are as described previously (31). The 50 nl sample mixture was injected into the dark pole site of oocytes (average volume 500 nl). The current was recorded after 3 h of incubation at 23 °C. Unless otherwise noted, the amounts for membrane vesicles injection into oocytes were 60 ng membranes, 60 ng SecA, 9 ng LamB signal peptides, or 14 ng precursors (proOmpA, proPhoA) or mature proteins (OmpA, PhoA). For SecA-liposomes, the amount for injection is 120 ng liposomes and 120 ng SecA, 9 ng LamB signal peptides, or 14 ng precursors or mature proteins. For signal peptide specificity experiments, all the injections were carried out with 4 mm puromycin to release the endogenous oocytes signal peptides (31). For SecYEG proofread experiments, the purified 30 ng SecYEG was mixed with SecA-liposomes and injected with ATP, Mg, and proOmpA into oocytes.
Liposome Patch-clamp Preparation and Measurement
Currents flowing through ionic channels formed in liposomes were monitored by inside-out patch clamp techniques (32). Liposomes prepared from total E. coli lipids extracts were dehydrated on the poly-lysine-coated glasses at room temperature. After dehydration, the dried liposomes were rehydrated in the solution containing 150 mm KCl, pH 7.4 at 4 °C for 48 h. The glass micropipette, filled with 150 mm KCl, pH 7.4, was pressed against the re-hydrated liposomes and suction was applied to assist patching sealing. Once the seal was formed, the pipette was withdrawn to obtain inside-out patch and the intracellular surface of liposome was exposed to the bath solutions in 0.1 ml containing where indicated 3 mm ATP, 1 mm MgCl, 1.8 μg of LamB signal peptides, or 2.8 μg of proOmpA and 24 μg of SecA (Activation bath). After incubation in the Activation bath, the active patch where indicated was moved into Inhibition bath with 10 mm EDTA, pH 8.0.
The recordings were initiated after 15–20 min incubation in each bath solution. In the ramp recording protocol, a command of −100 mV to +100 mV was given, whereas a steady potential of −80 mV was applied for the continuous protocol. The total recording time for the continuous protocol was 1 min.
RESULTS
SecA Alone Can Promote Translocation of proOmpA into Phopholiposomes
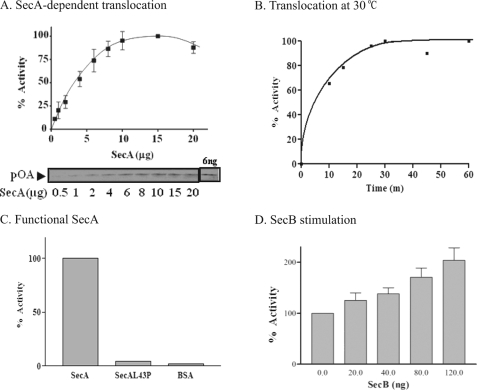
Using the traditional in vitro protein translocation assays of a precursor protein acquiring resistance to protease digestion (15, 17) we now find that SecA alone can promote translocation of proOmpA into E. coli phospholiposomes (Fig. 1). The translocation requires an increased amount of SecA (Fig. 1A). It occurs efficiently from 30–37 °C, but is more stable at 30 °C (Fig. 1B). There is little translocation (Fig. 1C) with the inactive mutant protein SecAL43P (24). Under the conditions used, the optimal amount of SecA needed to promote the maximal translocation is about 1–2 μm, which is ∼10 times higher than that required for the SecYEG-containing membranes used for in vitro translocation assays (data not shown). This higher level is close to the concentration of SecA found in bacterial cells (33–35). These results show that SecA alone in liposomes can promote protein translocation even though it is at a low level (about 10% of the input). The molecular chaperone SecB further stimulates the SecA-liposomes translocation of proOmpA (Fig. 1D).
FIGURE 1.
SecA-dependent proOmpA translocation with phospholipids liposomes. ProOmpA translocation (in 50 μl reaction mixtures) as a function of (A) SecA conc. (n = 4), and (B) incubation time at 30 °C. The highest amount of translocated proOmpA is 6.6 ng defined as 100%. C, lack of translocation of proOmpA with non-functional SecAL43P and BSA. The highest activity is 5.2 ng, defined as 100%. D, stimulation by SecB in SecA-liposomes. The translocation activity without SecB is defined as 100%.
SecA and Liposomes Are Necessary and Sufficient for Eliciting Ion Channel Activity in Oocytes
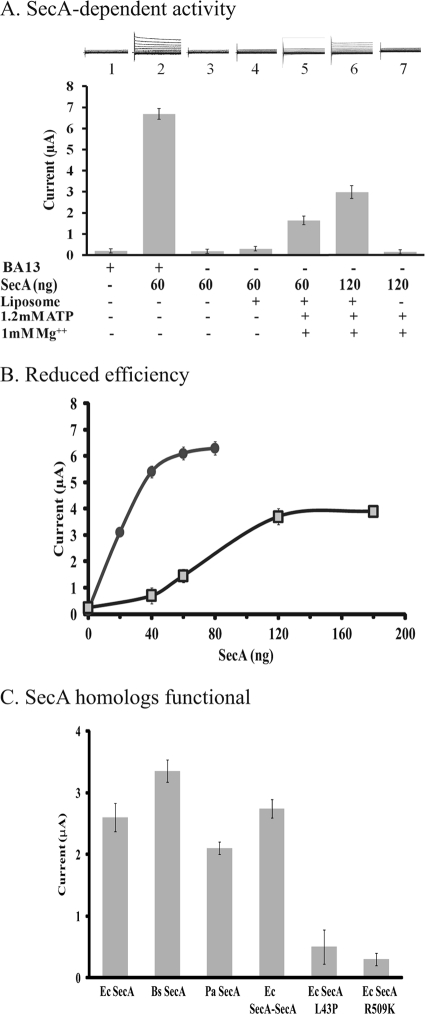
We previously developed a semi-physiological assay in Xenopus oocytes to measure bacterial SecA-dependent ion channel activities in an effort to complement the in vitro protein translocation assay (31). Because of findings that SecA-liposomes can promote protein translocation (Fig. 1), we expected similar results for the ion channel activity in the oocyte system. However, no significant activity was observed, unless ATP-Mg2+ was added together with SecA-liposomes (Fig. 2A), even though the oocytes contain sufficient ATP to promote ion channel activity with regular BA13 membranes (Fig. 2A, bar 2; Ref. 25). Optimization of the system revealed that more ATP-Mg2+ and SecA are required by SecA-liposomes (Fig. 2A and supplemental Fig. S1) than by BA13 membranes to elicit the ion channel activity, but only at a level of about 50% (Fig. 2B). The amount of SecA needed to reach the maximal activity is about 1–2 μm, which is similar to the amounts needed for in vitro protein translocation system (Fig. 1A), and in the bacterial cells as noted above. The localized concentration of SecA that forms the channels in the membrane might be even higher (33). These data indicate that SecA can elicit ion channel activity, albeit with lower efficiency.
FIGURE 2.
SecA and liposomes alone are capable for promoting ion channel activity in the oocytes. A, additional SecA and ATP-Mg2+ were required for achieving optimal channel activity with liposomes than BA13 membranes containing SecYEG. B, additional SecA is needed for channel activity with SecA-liposomes (■) as compared with BA13 with SecYEG (●). C, homologous SecAs but not non-functional E. coli SecAs can promote the ion channel activity in the oocytes. Various SecAs, 120 ng each, liposomes mixtures were injected together with proOmpA, ATP, and Mg2+ into oocytes. Ec: E. coli, EcSecA-SecA, EcSecAL43P, EcSecAR509K; Pa: Pseudomonas aeruginosa PAO1; Bs: Bacillus subtilis 168.
We next examined whether other bacterial SecA homologs can promote ion channel activity in oocytes. We found that purified SecA's of Bacillus subtilis and Pseudomonas aeruginosa, as well as the genetically constructed E. coli tandem SecA-SecA (19), were all active in eliciting ion channel activities to about the same extent (Fig. 2C). Neither of the inactive SecA L43P (36, 37), or SecA R509K (25), however, demonstrated any ion channel activity of note (Fig. 2C). These data provide further evidence that functional SecAs alone can elicit ion channel activities in liposomes.
SecA Alone Promotes Single Channel Activity with Liposome Patch-clamp Recording
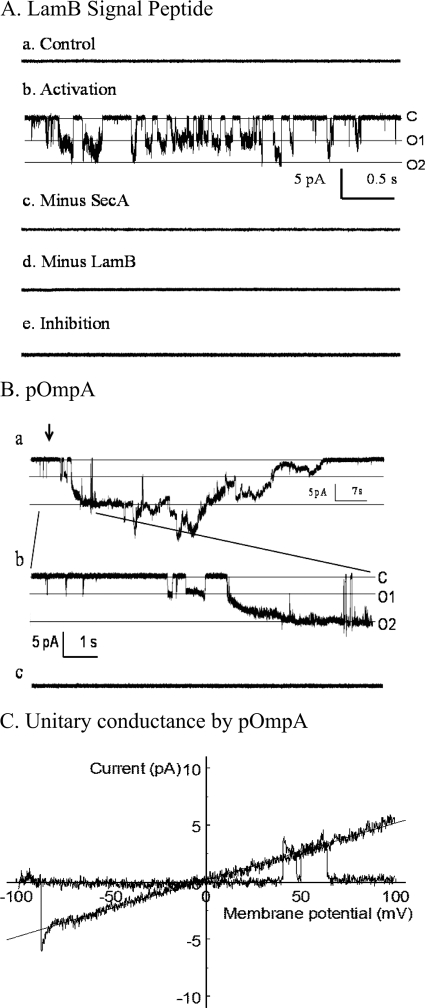
Recordings of the ionic current in the oocytes reflect the activities of a population of several hundred thousand channels (31). To explore and characterize the requirements of SecA-liposome channel activity and to determine the specificity of precursor or signal peptides for opening the channels, we developed a liposome patch-clamp recording system. We found that SecA-induced channel activities were activated by ATP-Mg2+ in the presence of LamB signal peptides (Fig. 3A). These channel activities (Fig. 3, A.b) were strictly dependent on the presence of both SecA (Fig. 3, A.c) and signal peptides (Fig. 3, A.d), whereas they were inhibited by EDTA (Fig. 3, A.e). Similar results were obtained with proOmpA (Fig. 3B). The SecA-dependent channels mediated by the signal peptide often opened and closed rapidly (Fig. 3, A.b), while those mediated by proOmpA appeared to have extensively “open states” (Fig. 3, B.a, b). The sizes of the single channels varied from 40 to 120 pS, a range similar to those reported for membrane-liposome fusions (38). However, the majority of channels with SecA-liposomes appeared to be around 50 pS (Fig. 3C). These results further provide strong evidence that in the presence of signal peptides or precursors SecA alone can elicit ionic channel activities in the liposomes.
FIGURE 3.
Patch clamp measurements of ionic channel activity in SecA-liposomes. The intracellular side of the inside-out patch was exposed to various solutions (0.1 ml). (C, closed baseline, O1, single opening, O2, double channels opening.) A, channel activity was observed only in the presence of LamB, SecA, ATP, and Mg2+. The channels opened and closed rapidly, and two channels were active, both with the same conductance of about 50 pS. The time showed is about 4 s. B, activation of SecA channels by proOmpA. Some channels opened by the Activation solution showed long opening with a large conductance (∼120 pS), while no channel activity was seen in Minus SecA solution. The arrow zoom-in area in B.c. is 10 s. C, single channel conductance by proOmpA stimulation. Single channel conductance was measured in activation treatment from Fig. 3B. The ramp voltage was from −100 mV to +100 mV. An active channel was observed, and the slope indicates the single channel conductance is 50 pS.
SecA-liposomes Lose Signal Peptide Specificity in Protein Translocation and in the Opening of Ion Channel
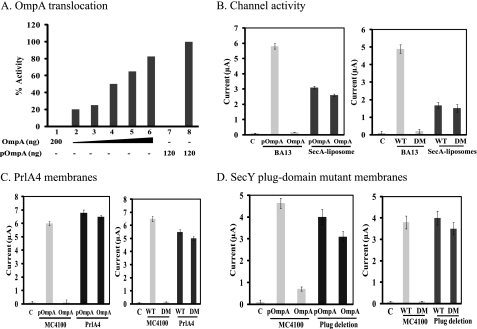
In addition to the reduction in efficiency and channel activity, these SecA-liposomes also lost the signal peptide specificity in the translocation of OmpA. Thus, while SecYEG-containing membranes could not translocate the unfolded, mature OmpA without signal peptides as expected (data not shown), the SecA-liposomes translocated OmpA efficiently in a dose-dependent manner at 30 °C, but not at 0 °C (Fig. 4A, bars 1 and 7). The translocation efficiency of the unfolded OmpA is lower than that of proOmpA (Fig. 4A).
FIGURE 4.
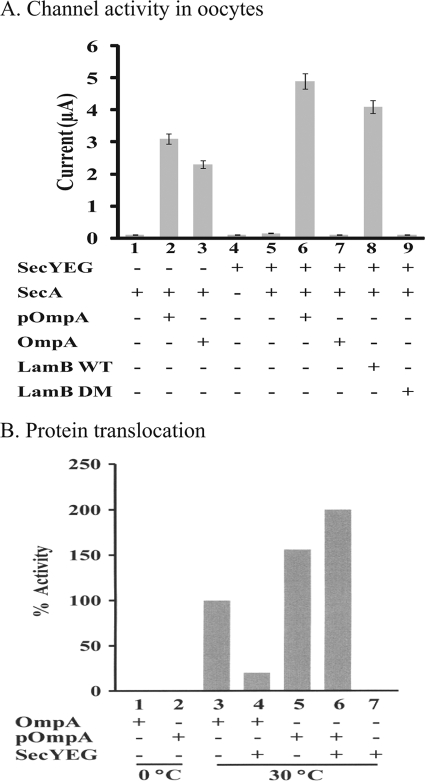
Loss of signal peptide specificity of SecA-liposomes and SecY mutant membranes. A, translocation of OmpA and proOmpA in SecA-liposomes. The reaction was carried out in 0.1 ml. Bars 1 and 7 were carried out at 0 °C as controls; bars 2–6 (30, 60, 100, 150, and 200 ng) and bar 8 were at 30 °C. B–D, ion channel activities in oocytes in the presence of 4 mm puromycin. Unfolded OmpA, proOmpA, LamB wild-type (WT), or deletion mutant (DM) signal peptides were injected with 120 ng SecA-liposomes (B), or 60 ng of PrlA4 membranes (C), or SecY plug-deletion membranes (D). c: control with puromycin to remove endogenous precursors without exogenous precursor or signal peptide.
We further characterized the features of SecA-liposomes activity in the oocytes which allow easier measurement of ionic activities with the sum of hundred-thousand channels (31). To determine the signal peptide specificity, we used puromycin to release the oocyte endogenous signal peptides such that the channel activity is strictly dependent on the addition of exogenous signal peptides or precursors. As shown in Fig. 4B, SecA-liposomes were active in generating ion channel activity with unfolded mature OmpA without signal peptides (Fig. 4B), while BA13 membranes were not active as expected (31). By comparison, proOmpA was able to elicit ion channel activities in either BA13 membranes or SecA-liposomes (Fig. 4B). Similar results were obtained with precursor protein proPhoA or mature unfolded PhoA (by urea and DTT treatment); though the mature PhoA without unfolding was not active (data not shown), indicating some limitation for activities.
Similarly, SecA-liposomes were less efficient but also lost the specificity for proofreading recognition of signal peptides (31). Thus, while BA13 membranes differentiated the wild-type from defective LamB signal peptides for channel activity (Fig. 4B, also see Ref. 25), SecA-liposomes generated similar channel activity with both LamB WT and LamB DM signal peptides, indicating that the SecA-liposomes channels were also opened by defective signal peptides (Fig. 4B).
The loss of proofreading of signal peptides recognition in SecA-liposomes resembles those of PrlA/SecY mutants (23, 29, 38, 39). We therefore tested the SecY suppressor mutant, PrlA4, which enables translocation of pre-proteins with a defective or missing signal peptides (23, 39), for the proofreading function of channel activity in the oocytes. The PrlA4 membranes showed current activities with OmpA or LamB DM (Fig. 4C), and PhoA (data not shown), while wild-type MC4100 membranes did not. As expected, proOmpA (Fig. 4C), and proPhoA (data not shown) stimulated similar ionic currents with wild-type as well as PrlA4 suppressor membranes.
Similar results were obtained with membranes isolated from SecYEG plug-mutants that also lose proofreading specificity (23). These membranes elicit similar ion channel activities with unfolded OmpA or proOmpA, and LamB wild-type or defective signal peptides (Fig. 4D). Similar results were obtained with PhoA or proPhoA protein (data not shown). These data indicate that SecA-liposomes lose the signal peptide recognition specificity, mimicking the SecY/PrlA mutants in eliciting ionic channel activities in the oocytes.
SecYEG Restores Signal Peptide Specificity, and Enhances Ion Channel and Protein Translocation Activities
Because the SecA-liposomes have the same ion channel activity characteristics as PrlA4 and SecY-plug mutants, we further tested whether purified SecYEG reconstituted into liposome could restore the proofreading. The purified SecYEG liposomes had no ion channel activity without SecA-liposomes in the oocytes (Fig. 5A, bar 4). The ion channel activities elicited by SecA-liposomes with unfolded OmpA and LamB DM were abolished in the presence of SecYEG-liposomes (Fig. 5A, bars 7, 9) while the proOmpA or LamB WT were able to stimulate the ion channel activity in SecA-liposomes and SecA/SecYEG liposomes (Fig. 5A, bar 2, 6). In the presence of SecYEG the activities were about twice as high as in its absence (Fig. 5A), and reduced the requirements of increased SecA and ATP in the oocytes (data not shown). These results indicate that SecYEG not only restores the signal peptide proofreading capabilities to discriminate among different functional signal sequences, but also enhances the ion channel activity of SecA-liposomes.
FIGURE 5.
Restoration of efficiency and signal peptide specificity by SecYEG. A, SecYEG restores signal peptide specificity in ion channel activity. Purified SecYEG (30 ng) was mixed by vortexing with SecA-liposomes (120 ng) and injected into oocytes in the presence of various precursors or signal peptides with 4 mm puromycin. LamB WT, wild-type LamB signal peptide. LamB DM, LamB deletion signal peptide. B, protein translocation. Translocation mixtures in 0.1 ml containing where indicated 120 ng SecYEG, 120 ng SecB, 10 μg SecA, 120 ng proOmpA, or 120 ng unfolded mature OmpA were incubated at 30 °C or 0 °C for 30 min.
We also tested the effect of SecYEG on OmpA translocation activity of SecA-liposomes. As with ion channel activities, the loss of recognition specificity of OmpA translocation in SecA-liposomes was also restored by the addition of purified SecYEG such that the unfolded OmpA could no longer be translocated in the presence of SecYEG (Fig. 5B), as in the membranes containing SecYEG. Interestingly, the addition of SecYEG also increased the translocation activity of proOmpA by 2-fold for SecA-liposomes.
Thus, the SecA-liposomes are fundamentally functional to promote ion channel and protein translocation activities, but lose efficiency and signal peptide specificity, both of which can be restored by SecYEG.
DISCUSSION
SecA-mediated Protein Translocation and Ion Channel Activity at Lipid Low Affinity Binding Sites
In this study, we demonstrate that SecA alone can promote protein translocation and ion channel activities in the liposomes, indicating that SecA could function as a protein-conducting channel. The activity and signal peptide specificity of SecA-liposomes can be enhanced by the addition of SecYEG. It is known that bacterial cells contain more molecules of SecA than SecYEG (13), and that the excess SecA could integrate into the anionic phospholipids at “low-affinity” sites. Similar phenomena were observed earlier. Ben de Kruijff and co-workers (40, 41) showed SecA insertion into lipid bilayers in which two distinct pools of membrane-bound SecA were identified: one is lipid-bound and other is SecYEG-bound (42). Thus, the two membrane-bound SecAs that differ in the degree of lipid association enlighten our observation that lipid-bound SecA, in the absence of SecYEG, plays a structural role to penetrate into lipids forming protein conducting channels (20). We hypothesize, therefore, that there are potentially two kinds of SecA-dependent protein-conducting channels: One that forms when SecA binds with high affinity to the SecYEG complexes and another that forms when SecA binds, with low affinity, to anionic phospholipids (4). There are many examples of pairs of high and low affinity transport systems for particular amino acids, which presumably offer a selective advantage. Thus, the existence of two SecA-dependent protein-conducting channels in bacteria would also offer a similar selective advantage for protein translocation. The current prevailing view is that bacterial SecYEG is the essential protein-conducting channel, while SecA is the peripheral ATPase that pushes precursor peptides through these high affinity channels. Our findings indicate an alternative understanding, that SecA alone can integrate into the membrane to form a low affinity site protein-conducting channel. In support of this view, we have previously observed that SecA forms a ring-structure of 8.4 nm with a potential pore size of 2–3 nm (20, 39) upon interaction with anionic phospholipids, while (together with SecYEG) SecA forms a 10.9 nm-diameter structure with a 3.0–4.8 nm cavity (39). The latter parameters are remarkably similar to those of the Sec61 complex. Thus, our work supports the idea that SecA, in the absence of SecYEG, forms channels in phospholipids that are capable of mediating protein translocation and eliciting ion channel activity.
We have used both electrophysiological conductance and protein translocation assays to analyze activities of SecA-dependent protein conducting channels in liposomes. The two assays measure somewhat different activities. Even so, it is likely that conductance measures only opening of ion channels, not necessarily complete translocation process. Thus, LamB signal peptides and proPhoA can open the channel, but still cannot be secreted (12, 13). For proOmpA, both translocation and channel activities are mediated by SecA alone in liposomes. We have also measured ion channel activities within SecA-liposomes by both oocyte whole cell and single channel recordings with similar conclusion that provide confidence for using oocytes as a tool for measuring SecA channel activities. While single-channel recordings provide specific information about the channel characteristics, the oocyte whole cell recordings are easier to manipulate with large numbers of test samples (typically 20–30 oocytes can be measured for each experimental condition), and yield the sum of more than a hundred-thousand channel activities (31). With the oocyte recordings, we further show that inactive EcSecA has no channel activity, but that other SecA homologs yield comparable channel activities. Moreover, these same experiments show that SecYEG-liposomes alone without SecA have no activity in either ion channel or protein translocation activities (Fig. 5).
Earlier studies have shown that protein translocation can take place in the absence of SecYEG (11–14); see also Introduction). Our findings that SecA-liposomes alone can promote protein translocation and ion channel activities provide a plausible explanation and basis to reconcile with the then inconceivable observations.
Reduction of Efficiency and Loss of Signal Peptide Specificity of SecA-liposomes and the Restoration by SecYEG
The SecA-liposomes are active in promoting both protein translocation and ion channel activities, but they are less efficient and lose the specificity for signal peptides. Thus, more SecA and ATP (though still in the concentration ranges found in the bacterial cells) are required for SecA-liposomes to reach maximal activities of about 50% activities obtained from SecYEG-containing membranes, and the defective, or no signal peptides can still promote activities (Fig. 4). The loss in the ability of SecA-liposomes to differentiate specificity of signal peptides resembles that of PrlA suppressors and the “no-plug” SecY mutant (Fig. 4, C and D). In SecYEG complex, SecY serves as a “gating” mechanism to select the proper precursors with typical signal peptides for translocation (23). Removal of SecYEG plug causes the similar translocation defects as PrlA suppressor mutants which allow the translocation of proteins lacking of signal peptide such as OmpA and PhoA in the cells (29, 43), as have been observed in this study with SecA-liposomes. Taken together, though SecA-liposomes lose the proofreading function and is less efficient, they may represent the basic essential core for the Sec-dependent protein-conducting channels.
In support of this idea, we show that the reduction of efficiency and loss of signal peptide specificity in SecA-liposomes can be restored by SecYEG (Fig. 5). Thus, the addition of purified SecYEG to the SecA-liposomes not only enhances both protein translocation and ion channel activities, but also regains the ability to differentiate the specificity of signal peptides. Moreover, the presence of SecYEG reduces the requirements of more SecA and ATP in the oocytes. We propose that binding of SecA at low affinity sites in the membrane constitutes the basic protein-conducting channel, while the binding at SecYEG high affinity sites confers additional efficiency and signal peptide specificity. Though SecA-liposomes resemble PrlA/SecY mutants in the loss of signal peptide specificity, whether such low affinity SecA sites are functional in the cells remains to be determined. It is worth noting that certain precursors may have different requirements: proPhoA cannot be translocated in our in vitro system in the absence of SecYEG (12, 13) and certain proteins are differentially affected during SecYEG depletion in the cells (14).
Possible Evolution of Sec Protein-conducting Channels
The general Sec secretion pathway is evolutionally conserved from prokaryotes to eukaryotes. Indeed, one of the principle arguments for the SecYEG being the essential channel is that it is conserved evolutionarily (4, 8). The SecY homolog, Sec61, is found widely, from mammals to bacteria, and the signal recognition particles (SRP)3 from elephants to E. coli (44). While SRP is an obligatory component of co-translational translocation in the endoplasmic reticulum, the bacterial SRP analog is but one of several targeting components required for only some membrane protein integration, but not for others. Yet each component of FtsY/Ffh/4.5S RNA of the bacterial SRP is essential for cell growth. Similarly, the secY and secE genes are essential for cell growth, but not for the translocation of all secreted proteins. Thus, there is no compelling reason why SecYEG should be the only core protein-conducting channel in bacteria, despite the extensive literature that is based mainly on in vitro reconstitution of proteoliposomes with SecYEG. The secretory system has undergone significant evolutionary modification. SRP is essential for eukaryotic co-translational secretion, but in bacteria it is only one of the possible components in membrane protein biogenesis systems; many proteins can be secreted without it, and most can be translocated post-translationally. Evolutionary conservation of a molecule is not proof that the molecule continues to play the same role. SecA is the only essential component of all Sec secretion pathways in bacteria, yet it does not survive the evolutionary division into eukaryotes.
Thus, while it is possible that two SecA-dependent pathways have evolved independently (one with and the other without SecYEG), a division that reflects the diverse pathways in bacterial secretion; we suggest a possible simpler evolutionary route: there is only one SecA-dependent secretion pathway. In it, SecA provides the basic protein-conducting channel, while SecYEG confers a higher affinity and specificity, much the same as RNA polymerase being the core enzyme in bacteria with sigma factors conferring specificity. SecA function is markedly energy profligate, expending more than a thousand ATPs for each precursor that passes through the membrane (3), either co- or post-translationally (1, 2). Although such costs can be tolerated across the relatively energy-rich bacterial membrane (including ATP generated from protonmotive force), selective pressure -exerted by the sub-compartmentalization of cells into organelles in eukaryotes that lack their own, proximal energy source, perhaps has intensified the needs for energy conservation, leading to the extinction of the spend-thrift SecA in higher organisms. Under such selective pressure, Sec61p might have evolved in to becoming the channel, mediating a fundamentally co-translational process that utilizes energy from protein synthesis to push the nascent chain across the channels.
Our data do not differentiate between these two evolutionary routes: whether two independent SecA-dependent secretion pathways have evolved (one at the high affinity binding site with SecYEG, and another at the low affinity binding site with phospholipids, albeit with lower efficiency and specificity) or a single SecA-dependent pathway has emerged, which has subsequently diverged into two. Whatever the mechanism of change, it is clear from the current studies that, even though SecYEG enhances the efficiency and specificity of protein translocation in bacteria, SecA alone is necessary and, by itself, can promote bacterial protein translocation and ion channel activity.
Supplementary Material
Acknowledgments
We thank J. Beckwith, T. Silhavy, T. Rapoport, D. Oliver, F Duong, and H. Takamatsu for bacterial strains/plasmids, L. Gierasch for LamB signal peptides, and C. Derby, J. Houghton, J. Ingraham and I. Weber, for comments and L.Yu and Y.Huang for SecA homologs. This article is dedicated to the memory of Bernard Davis, and to John Ingraham for his unfailing encouragement over the years. We specifically acknowledge Gunter Blobel for first suggesting in 1990 that SecY is not essential for protein translocation and is not the only receptor for SecA.
This work was supported in part by National Institutes of Health Grant GM34676 (to P. C. T.) and Chinese NNSF Grant 30830028 and NBRP Grant 2010CD833706/2010CB912400 (to S.-f. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- SRP
- signal recognition particle.
REFERENCES
- 1. Chen L., Tai P. C. (1985) Proc. Natl. Acad. Sci. U.S.A. 82, 4384–4388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Chen L. L., Tai P. C. (1987) Nature 328, 164–166 [DOI] [PubMed] [Google Scholar]
- 3. Wickner W., Leonard M. R. (1996) J. Biol. Chem. 271, 29514–29516 [DOI] [PubMed] [Google Scholar]
- 4. Driessen A. J., Nouwen N. (2008) Annu. Rev. Biochem. 77, 643–667 [DOI] [PubMed] [Google Scholar]
- 5. Beckmann R., Bubeck D., Grassucci R., Penczek P., Verschoor A., Blobel G., Frank J. (1997) Science 278, 2123–2126 [DOI] [PubMed] [Google Scholar]
- 6. Hamman B. D., Chen J. C., Johnson E. E., Johnson A. E. (1997) Cell 89, 535–544 [DOI] [PubMed] [Google Scholar]
- 7. Hanein D., Matlack K. E., Jungnickel B., Plath K., Kalies K. U., Miller K. R., Rapoport T. A., Akey C. W. (1996) Cell 87, 721–732 [DOI] [PubMed] [Google Scholar]
- 8. Pohlschröder M., Hartmann E., Hand N. J., Dilks K., Haddad A. (2005) Annu. Rev. Microbiol 59, 91–111 [DOI] [PubMed] [Google Scholar]
- 9. Breyton C., Haase W., Rapoport T. A., Kühlbrandt W., Collinson I. (2002) Nature 418, 662–665 [DOI] [PubMed] [Google Scholar]
- 10. Zimmer J., Nam Y., Rapoport T. A. (2008) Nature 455, 936–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Watanabe M., Nicchitta C. V., Blobel G. (1990) Proc. Natl. Acad. Sci. U.S.A. 87, 1960–1964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Yang Y. B., Lian J., Tai P. C. (1997) J. Bacteriol 179, 7386–7393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Yang Y. B., Yu N., Tai P. C. (1997) J. Biol. Chem. 272, 13660–13665 [DOI] [PubMed] [Google Scholar]
- 14. Baars L., Wagner S., Wickström D., Klepsch M., Ytterberg A. J., van Wijk K. J., de Gier J. W. (2008) J. Bacteriol 190, 3505–3525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen X., Xu H., Tai P. C. (1996) J. Biol. Chem. 271, 29698–29706 [DOI] [PubMed] [Google Scholar]
- 16. Kim Y. J., Rajapandi T., Oliver D. (1994) Cell 78, 845–853 [DOI] [PubMed] [Google Scholar]
- 17. Chen X., Brown T., Tai P. C. (1998) J. Bacteriol 180, 527–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen Y., Tai P. C., Sui S. F. (2007) J. Struct. Biol. 159, 149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang H., Na B., Yang H., Tai P. C. (2008) J. Bacteriol 190, 1413–1418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wang H. W., Chen Y., Yang H., Chen X., Duan M. X., Tai P. C., Sui S. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 4221–4226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Silhavy T. J., Beckwith J. (1983) Methods Enzymol. 97, 11–40 [DOI] [PubMed] [Google Scholar]
- 22. Oliver D. B., Beckwith J. (1982) J. Bacteriol. 150, 686–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Li W., Schulman S., Boyd D., Erlandson K., Beckwith J., Rapoport T. A. (2007) Mol. Cell 26, 511–521 [DOI] [PubMed] [Google Scholar]
- 24. Takamatsu H., Nakane A., Oguro A., Sadaie Y., Nakamura K., Yamane K. (1994) J. Biochem. 116, 1287–1294 [DOI] [PubMed] [Google Scholar]
- 25. Mitchell C., Oliver D. (1993) Mol. Microbiol. 10, 483–497 [DOI] [PubMed] [Google Scholar]
- 26. Collinson I., Breyton C., Duong F., Tziatzios C., Schubert D., Or E., Rapoport T., Kühlbrandt W. (2001) EMBO J. 20, 2462–2471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Maillard A. P., Lalani S., Silva F., Belin D., Duong F. (2007) J. Biol. Chem. 282, 1281–1287 [DOI] [PubMed] [Google Scholar]
- 28. Cabelli R. J., Chen L., Tai P. C., Oliver D. B. (1988) Cell 55, 683–692 [DOI] [PubMed] [Google Scholar]
- 29. Duong F., Wickner W. (1999) EMBO J. 18, 3263–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tai P. C., Tian G., Xu H., Lian J. P., Yu J. N. (1991) in Methods in Cell Biology (Alan M. T. ed), pp. 167–187, Academic Press; [PubMed] [Google Scholar]
- 31. Lin B. R., Gierasch L. M., Jiang C., Tai P. C. (2006) J. Membr Biol. 214, 103–113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hamill O. P., Marty A., Neher E., Sakmann B., Sigworth F. J. (1981) Pflugers Arch 391, 85–100 [DOI] [PubMed] [Google Scholar]
- 33. Das S., Stivison E., Folta-Stogniew E., Oliver D. (2008) J. Bacteriol 190, 7302–7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Or E., Rapoport T. (2007) FEBS Lett. 581, 2616–2620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Woodbury R. L., Hardy S. J., Randall L. L. (2002) Protein Sci. 11, 875–882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Oliver D. B., Beckwith J. (1981) Cell 25, 765–772 [DOI] [PubMed] [Google Scholar]
- 37. Yu L., Yang H., Ho Q., Tai P. C. (2006) Protein Expr. Purif. 50, 179–184 [DOI] [PubMed] [Google Scholar]
- 38. Simon S. M., Blobel G. (1992) Cell 69, 677–684 [DOI] [PubMed] [Google Scholar]
- 39. Tang Y., Pan X., Tai P. C., Sui S. (2010) Sci. China Life Sci. 53, 1049–1056 [DOI] [PubMed] [Google Scholar]
- 40. Breukink E., Demel R. A., de Korte-Kool G., de Kruijff B. (1992) Biochemistry 31, 1119–1124 [DOI] [PubMed] [Google Scholar]
- 41. Keller R. C., Snel M. M., de Kruijff B., Marsh D. (1995) FEBS Letts. 358, 251–254 [DOI] [PubMed] [Google Scholar]
- 42. van Voorst F., van der Does C., Brunner J., Driessen A. J., de Kruijff B. (1998) Biochemistry 37, 12261–12268 [DOI] [PubMed] [Google Scholar]
- 43. Derman A. I., Puziss J. W., Bassford P. J., Jr., Beckwith J. (1993) EMBO J. 12, 879–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wolin S. L. (1994) Cell 77, 787–790 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.