Background: Photosystem I is a multiprotein complex essential for the photosynthetic process.
Results: Photosystem I of Chlamydomonas reinhardtii contains nine Lhca complexes arranged on one side of the core.
Conclusion: A model of the subunits organization in the Photosystem I supercomplex is presented.
Significance: The sequence of the system (dis)assembly relates to the function of the subunits.
Keywords: Chlamydomonas, Electron Microscopy (EM), Fluorescence, Photosynthesis, Protein Assembly, Photosystem I, Light Harvesting Complexes, Supramolecular Organization
Abstract
In this work we have purified the Photosystem I (PSI) complex of Chlamydomonas reinhardtii to homogeneity. Biochemical, proteomic, spectroscopic, and structural analyses reveal the main properties of this PSI-LHCI supercomplex. The data show that the largest purified complex is composed of one core complex and nine Lhca antennas and that it contains all Lhca gene products. A projection map at 15 Å resolution obtained by electron microscopy reveals that the Lhcas are organized on one side of the core in a double half-ring arrangement, in contrast with previous suggestions. A series of stable disassembled PSI-LHCI intermediates was purified. The analysis of these complexes suggests the sequence of the assembly/disassembly process. It is shown that PSI-LHCI of C. reinhardtii is larger but far less stable than the complex from higher plants. Lhca2 and Lhca9 (the red-most antenna complexes), although present in the largest complex in 1:1 ratio with the core, are only loosely associated with it. This can explain the large variation in antenna composition of PSI-LHCI from C. reinhardtii found in the literature. The analysis of several subcomplexes with reduced antenna size allows determination of the position of Lhca2 and Lhca9 and leads to a proposal for a model of the organization of the Lhcas within the PSI-LHCI supercomplex.
Introduction
The light reactions of photosynthesis are carried out by four large protein complexes: Photosystem II (PSII),2 cytochrome b6/f, Photosystem I (PSI), and ATP synthase. Like the other complexes, PSI is located in the thylakoid membrane of cyanobacteria, algae, and plants (1, 2). It catalyzes light-driven electron transport from plastocyanin, which is present in the thylakoid lumen, to ferredoxin, which is located in the stroma. With almost 100% quantum efficiency, PSI is considered to be the most efficient light-capturing and energy conversion apparatus found in nature (3, 4).
PSI evolved well over 1 billion years ago, probably over 2 billion years ago. It is thought to represent an elaboration of the simple homodimeric type I reaction centers of primordial organisms into the sophisticated machinery that we know from plants and algae (3). During evolution, different groups of photosynthetic organisms colonized diverse ecological niches. As a consequence of adaptation to different environmental conditions, PSI design underwent structural rearrangements, most having to do with the peripheral antenna system. In the lineage that produced green algae and plants, this led to the addition of several specific complexes that enhance the light-harvesting capacity of the system (5–7). In cyanobacteria PSI exists as a trimer (8), whereas in plants and green algae, PSI is monomeric and is composed of two moieties: the PSI core complex and an outer antenna system composed of light harvesting complexes (LHCI) (9).
The PSI core is responsible for light-driven charge separation and electron transfer. It coordinates around 100 chlorophylls and 20 β-carotene molecules. Its primary and tertiary structures are highly conserved among green algae and plants; 14 subunits are present in both types of organisms (PsaA-PsaL and PsaN-PsaO), whereas PsaP is present in plants but so far seems to be absent in algae (4, 10). The central part of the core complex is composed of two large proteins, PsaA and PsaB, which binds all of the cofactors of the electron transfer chain (11), except for the last 2 Fe4S4 clusters (FA and FB). These are bound to the peripheral subunit PsaC, which together with PsaD and PsaE forms the docking site for ferredoxin on the stromal side of the membrane (12, 13). PsaF and PsaN are important for electron transfer from plastocyanin to P700 (14, 15). PsaJ is a hydrophobic protein located close to PsaF and plays a role in the stabilization of this subunit conformation (16). PsaH, PsaI, PsaL, and PsaO form a cluster of integral membrane proteins, placed on one side of the core, where they are involved in interactions with LHCII during state transitions (17, 18). PsaG and PsaK are located near PsaA and PsaB, respectively (12, 13), and have been proposed to be important for the association of the outer antenna with the core (10, 19, 20). The outer antenna extends the light-harvesting capacity and ensures photoprotection (20). The genes encoding the antenna complexes of PSI of plants and green algae are members of the Lhc multigene family, which also includes the antenna complexes of PSII. These complexes show structural homology in that each Lhc polypeptide has three transmembrane α-helices and coordinates Chls a, Chls b, and different carotenoid molecules (21).
The structure of PSI-LHCI in land plants has been determined to 3.3 Å of resolution and reveals the location of four Lhca proteins (encoded by the lhca1–4 genes) on one side of the core (13, 20, 22). Two additional Lhca genes, lhca5 and lhca6, were identified in Arabidopsis thaliana (A.t.), but their products are present at much lower levels compared with the other Lhca complexes (23–25). The structure of the PSI-LHCI complex of green algae is not available, but biochemical and structural (via electron microscopy) analyses have suggested that PSI-LHCI of Chlamydomonas reinhardtii differs in size and antenna composition from its counterpart in plants (26, 27). Indeed, nine lhca genes have been identified in C. reinhardtii (28). All of them are expressed in normal conditions (29), and all gene products were shown to coordinate pigments (30). Based on their content of long wavelength Chls (“red forms”) and on their fluorescence emission maxima, the Lhca complexes were divided into three subclasses: “blue Lhca” (Lhca1, Lhca3, and Lhca7) with emission maxima at 682.5–683.5 nm, “intermediate Lhca” (Lhca5, Lhca6, and Lhca8) with maxima between 694.5 and 697.5, and “red Lhca” (Lhca2, Lhca4, and Lhca9) with maxima between 707 and 715 nm (30).
Based upon biochemical characterization, it was suggested that C. reinhardtii PSI-LHCI (C.r.PSI-LHCI) contains between 6 and 14 Lhca subunits (31–34). Recently the Lhca/core stoichiometry was estimated using mass spectrometry, suggesting the presence of 7.5 ± 1.4 Lhca subunits per PSI core (35). The Lhca complexes were categorized into three groups; that is, those present in a substoichiometric amount with respect to the core (Lhca2, Lhca5, Lhca6, Lhca8, and Lhca9), those present in a 1:1 ratio with the core (Lhca1, Lhca4, and Lhca7), and Lhca3 present at a 2:1 ratio (35), suggesting a heterogeneous composition of the PSI-LHCI complex. It was also shown that six Lhca subunits (Lhca1, Lhca4–8) may even assemble in the absence of PSI core complex (33, 36).
Three structural models of C.r.PSI-LHCI complex have been proposed based on single particle electron microscopy analysis (26, 27, 37). When compared with higher plant PSI, the C.r.PSI-LHCI particles displayed additional densities, suggesting a larger antenna size. Germano et al. (26) suggested that about 14 Lhca complexes are associated with the core. Most of them are present on the PsaJ/PsaF side, whereas 2–3 Lhca bind to the PsaH subunit. Kargul et al. (27) proposed that 11 Lhcas are arranged on the PsaG/F/J/K side, but they later revised their model, suggesting that LHCI is made up of six Lhca subunits, four of which are located at the same position as the four Lhcas in higher plants, and two of which are at the PsaH side (37).
In this work we have purified several PSI (sub)complexes that differ in protein composition, and we have analyzed them via a combination of biochemical, spectroscopic, and electron microscopy (EM) measurements, with the aim of revealing the functional architecture of this complex.
EXPERIMENTAL PROCEDURES
Strain and Growth Conditions
A C. reinhardtii strain (JVD-1B[pGG1]) in which a hexahistidine tag had been added to the N terminus of PsaA (38) was grown in liquid Tris-acetate-phosphate medium (39). Cells were cultured at room temperature (25 °C) on a rotary shaker under an illumination flux of 10 μmol of photons photosynthetically active radiation m−2 s−1.
Thylakoids Preparations
Cells were harvested by centrifugation (4000 rpm, 6 min, 4 °C) in the mid-logarithmic phase (A750 nm ≈ 0.7), and thylakoids were prepared as in Fischer et al. (40) with a few modifications. Cells were disrupted by sonication (60 watt power in 10 cycles of 10 s on/30 s off) and centrifuged (15,000 rpm, 20 min, 4 °C). Thylakoid membranes were separated on a discontinuous gradient (24,000 rpm, 1 h, 4 °C) in a SW41 swinging bucket rotor as described (40).
Purification of the His-tagged PSI Complex
Thylakoid membranes were washed 3 times first with 10 mm Hepes (pH 7.5) buffer, then with 5 mm EDTA, and finally with 10 mm Hepes (pH 7.5). Membranes were resuspended in solubilization buffer (20 mm Hepes (pH 7.5), 0.2 m NaCl) to a chlorophyll concentration of 1 mg/ml, and an equal volume of 0.9% α-dodecylmaltoside (α-DM) was added. Samples were vortexed for few seconds and then centrifuged (15,000 rpm, 10 min, 4 °C) to remove unsolubilized material. The supernatant was loaded onto a HisTrap HP Column (GE Healthcare). The column was equilibrated with 20 mm Hepes (pH 7.5), 0.2 m NaCl, 0.03% α-DM, and washed with the same buffer. Elution was carried out with 20 mm Hepes (pH 7.5), 0.25 m imidazole, and 0.03% α-DM. The eluted fraction was loaded on a sucrose density gradient (made by freezing and thawing 0.65 m sucrose, 10 mm Tricine (pH 7.8), 0.03% α-DM buffer) and purified to homogeneity by ultracentrifugation (41,000 rpm, 17 h, 4 °C). The purification of PSI from A.t. was performed under the same conditions used for the purification of the Chlamydomonas PSI, with the difference that upon solubilization the thylakoid membranes were directly loaded on the sucrose gradient.
Gel Electrophoresis
Proteins were analyzed by a SDS-PAGE in a Tris-sulfate buffer system prepared as in Bassi (41). 3 μg of sample (in Chls) was loaded into each well. Non-denaturing gels were prepared as in Peter and Thornber (42) with a few modifications. The resolving gel contained a 4–10% acrylamide gradient, and 0.01% SDS was added to the upper running buffer. Some of the PSI-LHCI samples were solubilized with 1% β-DM and 0.4% ZWITTERGENT (at a Chl concentration of 0.4 mg ml−1).
In-gel Tryptic Digestion
For mass spectrometry-based protein identification, the green bands were excised from the gel and treated with 10 mm DTT followed by 55 mm iodoacetamide in 50 mm NH4HCO3 to reduce and alkylate cysteine residues and subsequently dehydrated by incubation for 5 min in 100% acetonitrile. The gel slices were rehydrated in 10 μl of trypsin solution (Trypsin Gold, mass spectrometry grade, Promega 10 ng/μl in 25 mm NH4HCO3) and incubated for 2 h at 37 °C. Subsequently 10 μl of 25 mm NH4HCO3 was added to prevent drying, and the incubation was prolonged overnight at 37 °C. The tryptic peptides were recovered by three subsequent extractions with 50 μl of 35%, 50, and 70% acetonitrile in 0.1% TFA. The extracted peptides were pooled and concentrated under vacuum.
For in-solution tryptic digestion, samples containing around 10 μg of protein were diluted to 20 μl in 100 mm NH4HCO3. For reduction and alkylation, the samples were incubated for 60 min at 55 °C in the presence of 10 mm DTT followed by the addition of iodoacetamide (final concentration of 50 mm) and incubation at room temperature for 30 min. After the addition of 0.5 μg of trypsin, the samples were incubated over night at 37 °C.
Liquid Chromatography-Mass Spectrometry (LC-MS)
Fractions of the peptide mixtures from in-gel trypsin digestions were diluted in 5% formic acid, passed through a pre-column (EASY-Column C18, 100 μm × 20 mm, 5-μm particle size, Thermo Scientific, Bremen, Germany), and separated on a capillary column (C18 PepMap 300, 75 μm × 250 mm, 3-μm particle size, Dionex, Amsterdam, The Netherlands) mounted on a Proxeon Easy-LC system (Proxeon Biosystems, Odense, Denmark). Solutions of 0.1% formic acid in water and a 0.1% formic acid in 100% acetonitrile were used as the mobile phases. A gradient from 4 to 40% acetonitrile was performed in 140 min at a flow rate of 300 nl/min. Eluted peptides were analyzed using a linear ion trap-Orbitrap hybrid mass spectrometer (LTQ-Orbitrap, Thermo Scientific). MS scans were acquired in the Orbitrap in the range from 400 to 1800 m/z, with a resolution of 60,000 (full-width at half-maximum). The 7 most intense ions per scan were submitted to MS/MS fragmentation (35% Normalized Collision EnergyTM) and detected in the linear ion trap.
Protein Identification
The MS raw data were submitted to Mascot (Version 2.1, Matrix Science, London, UK) using the Proteome Discoverer 1.0 analysis platform (Thermo Scientific) and searched against the C. reinhardtii proteome. Peptide tolerance was set to 10 ppm and 0.9 Da for intact peptides and fragment ions, respectively, using semi-trypsin as protease specificity and allowing for up to 2 missed cleavages. Oxidation of methionine residues, deamidation of asparagine and glutamine, and carboamidomethylation of cysteines were specified as variable modifications. The MS/MS-based peptide and protein identifications were further validated with the program Scaffold (Version Scaffold_3.0, Proteome Software Inc., Portland, OR). Protein identifications based on at least 2 unique peptides identified by MS/MS, each with a confidence of identification probability higher than 95%, were accepted. All assigned spectra used for the spectral count analysis were derived from at least two experiments with the exception of the Lhca content in fraction A4.
Spectroscopic Analysis
Room temperature absorption spectra were recorded with a Cary 4000 spectrophotometer (Varian). Room temperature and 77 K fluorescence emission spectra were recorded using a Fluorolog 3.22 spectrofluorimeter (Jobin Yvon-Spex). The excitation wavelengths were 440, 475, and 500 nm, and excitation and emission slits were set to 3.5 nm. For low temperature measurements, samples were in 66.7% glycerol (w/v), 10 mm Tricine (pH 7.8), 0.03% α-DM.
Pigment Composition
The pigment composition of the complexes was analyzed by fitting the spectrum of the acetone extracted pigments with the spectra of the individual pigments in acetone and by HPLC as described (43). The data are the results of at least two different preparations.
Electron Microscopy and Single Particle Analysis
To improve sample purity and contrast in electron microscopy micrographs, the sucrose content of the gradient fractions was reduced by dialysis (2–4 h at 4 °C; Spectra/Por 12–14-kDa cut-off). After dialysis, specimens were prepared for negative staining with 2% uranyl acetate on glow-discharged carbon-coated copper grids. Electron microscopy was performed on a Philips CM120 electron microscope equipped with a LaB6 tip, operated at 120 kV. Images were recorded with a Gatan 4000 SP 4K slow-scan CCD camera at 133,000 magnification with a pixel size of 0.225 nm at the specimen level after binning the images. GRACE software was used for semiautomated data acquisition (44). Single particles were analyzed with the Groningen Image Processing software, including multireference and non-reference alignments, multivariate statistical analysis, and classification as previously described (45). To sharpen the final images, limited numbers of projections (500–2000) of homogeneous classes were summed, with the correlation coefficient in the final alignment step as a quality parameter.
RESULTS
Isolation and Fractionation of PSI-LHCI Complexes
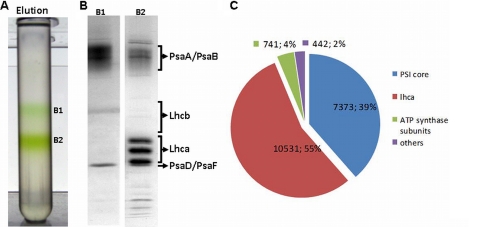
PSI-LHCI was purified in two steps from a strain carrying a hexahistidine tag at the N terminus of core subunit PsaA. Thylakoid membranes were mildly solubilized, and PSI particles were first purified on a nickel-Sepharose column. To obtain homogeneous preparations of PSI and to check for the presence of PSI complexes with different sizes, the eluted fraction was then subjected to sucrose density gradient ultracentrifugation. Two green bands were separated (B1 and B2; Fig. 1A), indicating that the elution contained two populations of complexes differing in molecular mass. These chlorophyll-protein complexes were collected and subjected to biochemical and spectroscopic analysis.
FIGURE 1.
Panel A, fractionation of PSI-LHCI eluted from the Ni(II) column by sucrose density gradient. Panel B, SDS-PAGE of green bands B1 and B2 from the gradient are shown. Panel C, shown is a Venn diagram showing the composition of the PSI-LHCI preparation (fraction B2) based on LC-MS/MS analysis of tryptic peptides. The sum of the spectra assigned to each group of proteins as well as the fraction of total spectra is indicated.
PSI-LHCI; Protein Composition
The protein compositions of the two fractions were visualized by SDS-PAGE (Fig. 1B). The polypeptide profile analysis confirmed the presence of PSI in both fractions. A broad band at high molecular mass corresponds to the PSI reaction center core subunits, PsaA and PsaB, that have an apparent molecular mass of ∼65 kDa and typically migrate as a diffuse band. Bands in the 20–27-kDa range correspond to LHCI antennae and the stromal subunits PsaD and PsaF, whereas those in the 10–20-kDa range arise from small core subunits (10, 31). No bands corresponding to PSII subunits were visible in the gel, indicating that both PSI complexes were free from PSII contamination, as seen before with the unfractionated eluate (38). Although the band pattern was very similar for the two fractions, the ratio between core subunits and Lhca complexes was higher in B1 than in B2, suggesting that the two fractions contain PSI complexes with different antenna sizes. B2 thus represents the largest complex, and in the following it is called PSI-LHCI. B1 is likely the result of a partial disassembly of PSI-LHCI during purification, as indicated by the fact that Lhca complexes were observed in the flow-through of the nickel column (supplemental Fig. S1). However, we cannot exclude that this fraction also contains the assembly intermediate recently observed by Ozawa et al. (10).
To determine in more detail the protein composition of PSI-LHCI, we performed a mass spectrometry analysis. A shotgun proteomics approach revealed the presence of all nine Lhca proteins (Lhca1–9) and all PSI core subunits, with the exception of PsaI and PsaO. These subunits are likely to be present in the complex but are unidentifiable by tryptic digestion because the resulting peptides would be too short and would not be detectable in our experimental setup. PsaN and PsaJ were not identified in all experiments and were only seen via one unique peptide, so they are not considered in the analysis.
The proteomic analysis also confirms the high purity of the preparation (Fig. 1C); 94% of the MS/MS spectra were assigned to peptides originating from PSI or LHCI, with 55% from Lhca polypeptides and 39% from PSI core subunits. Only 6% were due to impurities, of which 4% stemmed from ATP synthase subunits. No PSII or LHCII subunits were detected in the preparation.
PSI-LHCI; Pigment Composition
Pigment analysis indicated a Chl a/b ratio of 4.4 in the purified PSI-LHCI complex (Table 1). The main carotenoids were β-carotene, lutein, and violaxanthin. Loroxanthin was present in low amounts, in agreement with previous results indicating that this xanthophyll is mainly associated with PSII (46). No traces of neoxanthin could be detected, consistent with the fact that this xanthophyll is not present in PSI and that the preparation is not contaminated by PSII complexes. The Chl a/b ratio of C.r.PSI-LHCI is far lower than that of the PSI-LHCI complex of higher plants (a/b 8.5–9) (47, 48). We envision two possible origins for this difference: 1) a higher number of antenna complexes associated with C.r.PSI-LHCI as compared with plant PSI-LHCI and/or 2) a higher complement of Chl b in the Lhca complexes of C. reinhardtii as compared with the homologues of higher plants. Both effects probably contribute. Indeed, if the chlorophyll composition of the C.r.Lhca were the same as in higher plants (14 Chls per complex with a Chl a/b ratio of 3.7 (49)), then 48 Lhcas would be needed to explain a Chl a/b ratio of 4.4 in C.r.PSI-LHCI! On the other hand, to explain this value purely in terms of a different affinity for Chl a and b, the C.r.Lhcas would need to have a Chl a/b ratio of 1.2, which is far lower than observed for both native and reconstituted complexes (27, 30, 31, 33).
TABLE 1.
Pigment compositions of PSI-LHCI supercomplexes from C. reinhardtii (this study) and A. thaliana (48)
The values of individual carotenoids are normalized to 100 Chls (a+b).
| Chl a/b | Chl/carotenoids | Loroxanthin | Violaxanthin | Lutein | β-Carotene | |
|---|---|---|---|---|---|---|
| C.r.PSI-LHCI | 4.4 ± 0.1 | 5.0 ± 0.2 | 2.0 ± 0.6 | 4.8 ± 0.6 | 7.0 ± 1.0 | 8.5 ± 0.3 |
| A.t.PSI-LHCI | 9.7 ± 0.3 | 4.8 ± 0.1 | 2.3 ± 0.3 | 5.5 ± 0.3 | 13.1 ± 0.3 |
PSI-LHCI; Spectroscopic Characterization
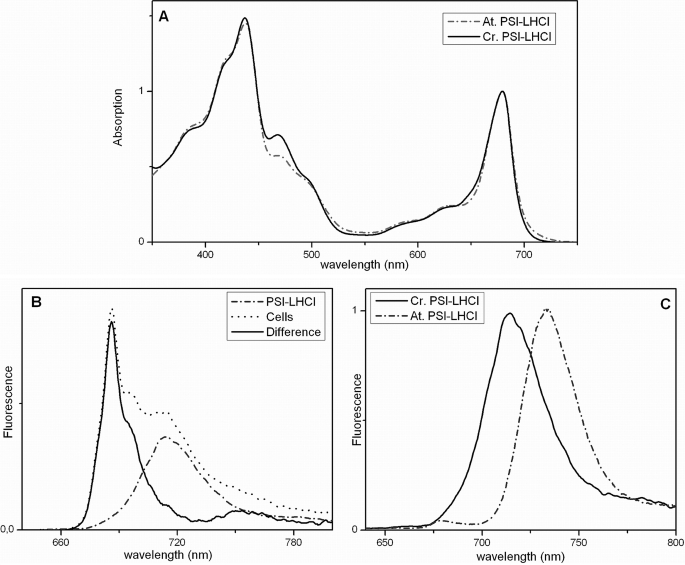
The PSI-LHCI absorption spectrum at room temperature shows a maximum at 679 nm (Fig. 2a). Compared with the complex of higher plants (Fig. 2a), C.r.PSI-LHCI is characterized by (i) an increased absorption in the Chl b region (630–660 nm), in agreement with the lower Chl a/b ratio and (ii) a decreased absorption in the region above 700 nm, indicating a lower content of red forms, as observed previously (26, 31).
FIGURE 2.
Room-temperature absorption (A) and 77 K fluorescence emission (B and C) spectra of C.r.PSI-LHCI (panels A and C, solid line; panel B, dashed line) compared with those of cells (panel B, dotted line) and A.t.PSI-LHCI (panels A and C, dashed line). Excitation used for fluorescence spectra was at 440 nm. The difference between normalized fluorescence emission spectra of cells and C.r.PSI-LHCI is shown in panel B (solid line).
The fluorescence emission spectrum of C.r.PSI-LHCI at low temperature (77 K) exhibits a maximum at 715 nm, and it is practically identical to the spectrum of PSI in the cell (Fig. 2b), indicating that the purified complex maintains its in vivo properties. Although the emission maximum of the C. reinhardtii complex is 18 nm-blue-shifted compared with the spectrum of plant PSI-LHCI (Fig. 2c and Refs. 27, 33, 38, and 50)), the two spectra have similar characteristics. They are very broad and strongly red-shifted with respect to the absorption of the complexes, suggesting a similar origin of the red forms (i.e. mixing of the lowest excitonic state with a charge transfer state (51, 52)) in the two organisms.
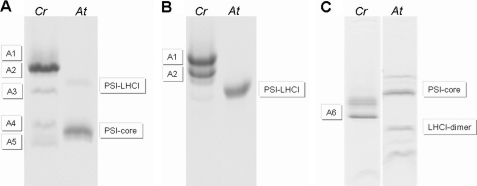
Native PAGE of PSI-LHCI Subcomplexes
To investigate the stability of PSI-LHCI and to assess the strength of the association of the individual Lhca complexes with the PSI core, PSI-LHCI was treated with a combination of Zwittergent and β-DM, and the resulting subcomplexes were separated by native PAGE. Five bands (A1–A5; Fig. 3A) were observed. Surprisingly, even a very low concentration of ionic detergent led to a partial dissociation of the complex, as indicated by the separation of PSI-LHCI into two main bands in 0.01% SDS (Fig. 3B), suggesting that part of the antenna is associated with the core in a relatively loose fashion. The molecular mass of the PSI (sub)complexes can be roughly estimated by using A.t.PSI-LHCI (600 KDa) and A.t.PSI-core (440 kDa) particles as standards. Approximate values of 770, 690, 570, 460, and 370 kDa were found for the molecular masses of bands A1–A5, respectively. Assuming an average molecular mass of 35 kDa for an Lhca complex, the A1 fraction should, therefore, contain a PSI complex with 5 Lhcas more than A.t.PSI-LHCI and, thus, 9 in total. The A2 fraction would, therefore, have lost two Lhcas compared with A1, whereas only three-four Lhcas are expected to be present in fraction A3. A4 contains a complex smaller than A.t.PSI-LHCI but larger than A.t.PSI-core, suggesting that it probably still binds one-two Lhcas. Fraction A5 migrates below the A.t.PSI-core, indicating that it had lost some of the core subunits. An additional complex, fraction A6, with an apparent molecular mass of 140 kDa, was obtained upon very mild solubilization of the B1 band.
FIGURE 3.
Native PAGE of PSI-LHCI supercomplexes of C. reinhardtii (Cr) or A. thaliana (At) solubilized with 1% β-DM and 0.4% Zwittergent-16 (A) or with 0.01% SDS (B). C, shown is a native gel of the B1 band solubilized with 0.01% SDS (Cr) compared with the PSI-LHCI (sub)complexes of A. thaliana (At).
PSI (Sub)complexes: Protein Composition
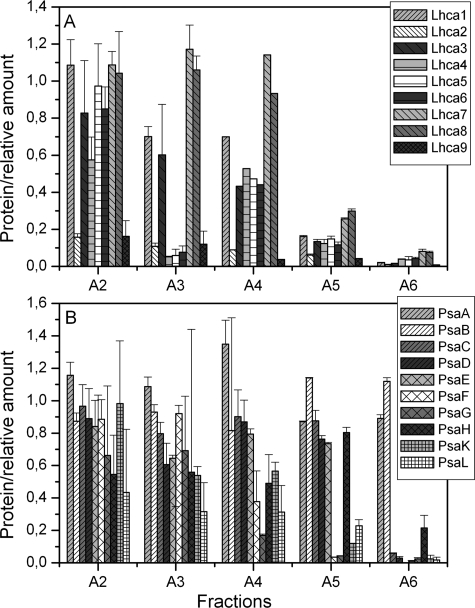
To get information about the protein composition of the PSI complexes, the A1-A6 fractions were analyzed by mass spectrometry. To determine the relative abundance of each subunit in the different fractions, spectral count normalization was applied (53).
The Lhca content from all bands is presented in Fig. 4A as relative values normalized to the abundance of the same subunit in the largest complex (A1 fraction; see supplemental Table S1 for the protein identification data). The A2 fraction shows a 90% decrease of Lhca2 and Lhca9, whereas A3 shows an additional loss of 95% of Lhca4, Lhca5, and Lhca6. Surprisingly, in fractions A4 and A5, for which the mobility on the gel suggests that they represent PSI with strongly reduced antenna size, all Lhcas were detected with the exception of Lhca2 and Lhca9. However, in fraction A4, although the amount of Lhca7 and Lhca8 is almost unchanged compared with A1, it is around 40% for all other Lhcas. In the case of fraction A5, all the Lhca polypeptides are present in similar amounts, but around 20–30% of their content is in A1. Virtually no Lhca proteins were detected in fraction A6.
FIGURE 4.
Quantification of the LHCI (A) and PSI core (B) polypeptides found in the PSI-LHCI subcomplexes. The number of spectra assigned to individual subunits was normalized to the number of spectra assigned to the PsaA/PsaB polypeptides in the same fraction. The values are reported relative to the values determined for each polypeptide in fraction A1.
The composition of the PSI core subunits in the different fractions was also analyzed (Fig. 4B). The data indicate a strong reduction of PsaL and PsaH in fractions A2 and A3. Interestingly, in fractions A4 and A5, PsaF together with PsaG and PsaK, which have been suggested to stabilize the binding of Lhca complexes to the core, are significantly reduced. In fraction A6 the stromal subunits PsaC, -D, and -E are also missing; it still contains PsaA and PsaB and likely represents a core heterodimer lacking all small subunits or peripheral antennae.
PSI (Sub)complexes: Absorption, Fluorescence, and Pigment Analysis
The absorption maxima of the complexes shift to the blue as one descends to less and less assembled states. Although the absorption maximum of A1 is 679 nm, it is 677 nm in A4 and A5 (Fig. 5a). A decrease in the Chl b absorption region is also observed going from A1 to A5, with the exception of A4, which shows higher absorption in this region than A3.
FIGURE 5.
Absorption (a) and low temperature fluorescence emission (b) spectra of PSI-LHCI subcomplexes shown in Fig. 3.
Fluorescence emission spectra were measured at 77 K (Fig. 5b). The emission maximum is 715 nm for A1 and A2, 711 nm for A3, and 708 nm for A4 and A5 (Table 2). The second peak around 680 nm observed in A3 is due to the presence of a pool of disconnected Chls in this fraction.
TABLE 2.
Pigment composition of PSI-LHCI subcomplexes A1-A5
The error in these measurements is below 0.2.
| Chl a/b | Chl/carotenoids | Absorbance maximum | Emission maximum | |
|---|---|---|---|---|
| nm | nm | |||
| A1 | 4.4 ± 0.1 | 5.5 ± 0.4 | 679 | 715 |
| A2 | 4.5 ± 0.1 | 5.6 ± 0.4 | 679 | 715 |
| A3 | 5.7 ± 0.3 | 6.3 ± 1.7 | 678 | 711 |
| A4 | 4.6 ± 0.2 | 6.7 ± 0.3 | 677 | 708 |
| A5 | 6.5 ± 0.5 | 7.6 ± 1.6 | 677 | 708 |
The pigment compositions of the five fractions were also analyzed and are reported in Table 2. An increase in Chl a/b ratio was observed in going from fraction A1 to A5, again with the exception of A4, which shows a lower Chl a/b ratio as compared with band A3.
Electron Microscopy and Single-particle Analysis
To determine the structural organization of PSI-LHCI, electron microscopy and single-particle analysis were performed. Specimens of PSI-LHCI yielded a relatively homogeneous preparation (see supplemental Fig. S2). Single-particle analysis of more than 50,000 projections gave two major classes that represent two types of complexes of a slightly different overall size. The larger PSI-LHCI complex (Fig. 6a) represents about 40% of the dataset, whereas the smaller type (Fig. 6b) accounts for about 55%. The main difference between the two particles is on the right side of the projection map, with a clear absence of density in the smaller complex. The difference is not a matter of a variation in tilt on the carbon support film (see supplemental Fig. S4) but is likely due to the loss of a specific subunit.
FIGURE 6.
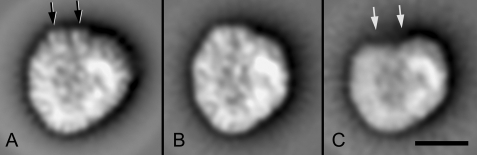
Average projection maps of C.r.PSI-LHCI (A and B) and the A2 fraction (C). The maps in panels A and B depict the two major class averages of PSI-LHCI complex, representing sums of about 2000 particles each. The larger particle (A) represents ∼40% of the population in the fraction, while the smaller one (B) represents ∼55% of the particles. Panel C shows the sum of 500 particles of the Lhca2/Lhca9-less complex from fraction A2. The black and gray arrows indicate the presence and absence of two densities of similar size and shape at the top. Space bar = 10 nm.
For a structural assignment of the PSI-LHCI projection maps, determination of their handedness is crucial. This was facilitated by the presence of a characteristic crescent-shaped density at the right side of the EM projection maps (Fig. 6), which is typical for the PSI-LHCI complex seen from the stromal side (see supplemental Fig. S3A and Ref. 48). This characteristic density facilitated a manual fitting of the x-ray model of plant PSI-LHCI (54) into the projection maps of the C.r.PSI-LHCI (Fig. 7, a and b). The fit shows an overall good agreement between x-ray and EM data, especially for the larger complex (Fig. 7b). In the case of the smaller complex, the lack of EM density on the right side of the projection map may correspond to the absence of PsaL and PsaH (Fig. 7, white arrows) rather than to the absence or presence of a larger-sized Lhca subunit. This assignment is supported by the biochemical data indicating that PsaH and PsaL are the first subunits to be lost (see above). An on-scale comparison of the C.r.PSI-LHCI projection maps with the structure of pea PSI-LHCI clearly indicates that the antenna proteins of C. reinhardtii are all bound at one side of the core (Fig. 7, a and b).
FIGURE 7.
Structural assignment of the smaller (a) and larger (b) type of C. r.PSI-LHCI complex by fitting the high resolution structure of the plant PSI-LHCI complex (PDB code 2WSC) (54). The white arrow indicates the position of two peripheral subunits PsaL and PsaH. The yellow arrow shows the best-resolved density of Lhca protein of the inner row, which corresponds to the position of Lhca1 in the plant PSI-LHCI complex; the blue arrow indicates the density of the best-resolved Lhca protein of the outer row of LHCI. Black asterisks (a) mark the high density spots of the proposed outer LHCI half-ring, which are presumed to correspond to the C helix of the Lhca polypeptide. A tentative fitting of the outer row of LHCI using the structure of the Lhca2 protein is indicated in panel b. Panel c, the PSI-LHCI-LHCII supercomplex of A. thaliana (adapted from Kouril et al. (63)) is shown at the same scale for a direct comparison of the PSI antenna size between green algae and plants.
To delve further into the organization of the Lhca complexes, we performed a similar analysis of fraction A2, which lacks Lhca2 and Lhca9. Electron microscopy of the A2 specimen followed by image analysis of about 20,000 single-particle projections produced a projection map similar to but smaller than that of the C.r.PSI-LHCI complex (Fig. 6c). A direct comparison of the projection maps of PSI-LHCI and A2 (Fig. 6, a and c) leads to the localization of Lhca2 and Lhca9. One of them occupies the binding site where Lhca1 binds to the core in higher plants, whereas the other is located in a position that is not present in higher plants (Fig. 7, b and c). Regarding the other Lhca complexes, the improved map allows recognition of small, but strong, circular densities (Fig. 7, a and b, black asterisks) that we assume correspond to helix C of the Lhca proteins, which is roughly perpendicular to the membrane plane, thus resulting in a strong density in the projection map (see also supplemental Fig. S3A). Based on the x-ray model fitting, the inner row of Lhca proteins in C. reinhardtii comprises four Lhca proteins that can directly associate with the core, in principle in a mode similar to that of plants. However, a local mismatch in the x-ray model fitting indicates that at least one Lhca protein of the inner row, the first from the top, has a slightly different binding position/orientation in C. reinhardtii (Fig. 7, a and b, yellow arrows). The fitting of the plant PSI-LHCI structure into the C.r.PSI-LHCI map highlights an additional area of the projection map, comprising an outer row of Lhca proteins absent in the plant structure (Fig. 7a). The localization of the topmost Lhca in the outer row is evident from the well resolved EM density (Fig. 7, a and b, blue arrows). The fitting of the four remaining Lhca proteins is facilitated by the localization of their C helices (Fig. 7b), allowing determination of the position of all nine Lhca complexes in the structure of C.r.PSI-LHCI.
DISCUSSION
In contrast to higher plants, a consistent picture of the composition and organization of the antenna subunits in PSI of C. reinhardtii is not yet available. There is also no consensus about the number of Lhca copies associated with PSI, as Lhca/core stoichiometries ranging from 4 to 14 have been proposed (26, 27, 31–33, 35, 37). In this work we have purified a His6-tagged version of C.r.PSI-LHCI after mild detergent solubilization of the membranes. PSI-LHCI subcomplexes with different antenna sizes have been obtained after further solubilization of the purified complex, suggesting that the association of at least part of the antenna to the core is relatively loose, which could at least partially explain the discrepancy in the number of Lhcas that have been found associated with the core.
Lhca/Core Stoichiometry; Nine Lhca Complexes Are Associated with PSI Core
The largest PSI-LHCI complex purified was shown to be highly homogeneous (supplemental Fig. 2). Several lines of evidence suggest that the Lhca/core ratio of this complex is 9; (i) the comparison of the C.r.PSI-LHCI projections with either the A. thaliana complex or the pea PSI-LHCI crystal structure suggests the presence of 5 extra Lhcas in the former (Fig. 7); (ii) the apparent molecular mass of C.r.PSI-LHCI is 770 kDa, corresponding to a complex with 9 Lhcas; (iii) the Chl a/b ratio of 4.4 of C.r.PSI-LHCI is consistent with the presence of 9 Lhca complexes with an average Chl a/b ratio of 2.2, in agreement with the results on reconstituted Lhcas (30); (iv) a PSI subcomplex with molecular mass of 690 kDa (i.e. still far larger than the plant supercomplex) was shown (by MS and EM) to lack two Lhca subunits and some core subunits compared with the largest PSI-LHCI supercomplex.
EM analysis shows that the C.r.PSI-LHCI preparation contains only two types of particles differing by some density at the PsaH/PsaL site. This density, however, is too small to accommodate a Lhca complex. The preparation is thus homogeneous with respect to the antenna composition. Comparison of the EM maps of C.r.PSI-LHCI and the A2 subcomplex reveals the positions of Lhca2 and Lhca9 as two well defined densities. These densities are present in all particles in the PSI-LHCI fraction, implying a 1:1 stoichiometry of each polypeptide with the core.
In a recent study it was found that most of the Lhca complexes, including Lhca2 and Lhca9, are present in a substoichiometric amount with respect to the core (35). However, these data were obtained by averaging the results of three preparations with different Chl a/b ratios and thus very likely with different antenna sizes. Indeed, Stauber et al. (35) suggest that 7.5 ± 1.4 Lhca proteins are present per core complex, with the highest value in that range being in good agreement with our results. Moreover, the Lhca complexes that were suggested to be in a 1:1 ratio with the core are Lhca1, Lhca3, Lhca4, and Lhca7, which correspond (with the exception of Lhca4) to the complexes that are still present in the smaller particles (Fig. 4A) and are thus likely to be more stably associated with the core.
Disassembly of the Complex
The sequential disassembly of the C.r.PSI-LHCI complex after secondary solubilization provides information about the hierarchic organization of the complex. The first step in the disassembly process (i.e. production of subcomplex A2) corresponds to the loss of PsaL, PsaH, Lhca2, and Lhca9. The fact that these polypeptides are lost first in the presence of a very low concentration of ionic detergent indicates that they are rather loosely bound. It also explains previous reports in which Lhca2 and Lhca9 were either absent or present in substoichiometric amounts (33, 35). The facile loss of these LHCI complexes, which are known to harbor the red-most forms of the antenna pigments (30), might also explain the large discrepancies in excitation energy transfer and trapping studies of C.r.PSI-LHCI found in the literature (55–59).
The second disassembly step leads to a complex (A3) with an apparent molecular mass of 570 kDa and which lacks Lhca 4, Lhca5, and Lhca6. No loss of core subunits beyond PsaH and PsaL, which are already absent in the A2 complex, was observed. The 120-kDa difference between A2 and A3 can thus be fully attributed to the loss of 3 Lhca antenna complexes.
The next subcomplex (A4) has an apparent molecular mass of 460 kDa. It has lost PsaK, PsaG, and PsaF, but it seems to retain part of the antenna. Indeed, all Lhcas but Lhca2 and Lhca9 could be detected in this fraction. However, their level was around 40% that in the largest complex (A1), with the exception of Lhca7 and Lhca8, whose presence was unchanged. Fraction A4 also shows a lower Chl a/b ratio as compared with fraction A3, in agreement with a higher content of Lhca. The apparent discrepancy of having a complex with reduced molecular mass but with higher antenna content can be reconciled by taking into account the fact that Lhca complexes can also form a large complex (300–540 kDa) in the absence of the core (10, 33). This complex has been shown to be composed of all Lhcas but Lhca3, Lhca2, and Lhca9 and to have an emission maximum at 708 nm. These are also the characteristics of the A4 fraction (except for the presence of Lhca3), suggesting that this fraction contains a PSI subcomplex co-migrating with a Lhca-oligomer. The 460-kDa complex corresponds well to a core that has lost PsaK, PsaG, PsaF, PsaL, and PsaH but still retains two Lhcas. The best candidates for these are Lhca7 and Lhca8, which are still present at WT levels, or Lhca3, which is present in this fraction but not expected to be part of the Lhca-oligomer.
Fraction A5, with an apparent molecular mass of 370 kDa, represents a core complex that has lost the full Lhca complement in addition to the above-mentioned PSI-core subunits. The Lhca proteins detected in this fraction (all but Lhca2 and 9) are all present at low but comparable levels. This is likely due to co-migration of an antenna oligomer, which can apparently exhibit different sizes.
In the last step of the disassembly, an additional complex appears that has lost the stromal subunits PsaC, PsaD, and PsaE. It is composed of only the central subunits PsaA and PsaB and thus represents a stripped-down PsaA/B heterodimer. The implied disassembly sequence, PsaH/L > PsaG/K/F > PsaC/D/E > PsaA/B, suggests that the assembly of the PSI complex occurs in the opposite order, in partial agreement with recent results (10). Moreover, it corresponds to more and more fundamental biochemical function, with the PsaA/B heterodimer being the locus of charge separation, PsaC/D/E adding the terminal acceptors and ferredoxin-binding site, PsaF/J (the latter implied) adding the plastocyanin-binding site, and PsaG/K adding the ability to bind LHCI antenna complexes (12, 13, 16, 60). Finally, PsaL/H add the ability to bind LHCII trimers during state transitions (17, 18).
Photosystem I Organization
Although several studies have focused on the structural organization of the PSI-LHCI complex of C. reinhardtii (26, 27, 37, 61), this has not yet led to a conclusive model for the antenna organization. The discrepancy between previous models is largely due to limited resolution in the electron microscopy density maps. This made it impossible to determine the positions of individual Lhca proteins, and even the orientation and positioning of the PSI core moiety can be questioned (27). Moreover, at the time of the first studies (26, 27), the plant PSI-LHCI structure, which turned out to be crucial for assignment of the correct stoichiometry and position of the four Lhca antenna proteins (20), was not yet available. In this study we obtained a PSI-LHCI projection map at about 15 Å resolution, and the density profile enabled us to position the core complex and 9 Lhca antenna polypeptides. In particular, the assignment of five similar high density spots in the peripheral part of the LHCI antenna, assumed to correspond to the density of the C helix of the 5 outer Lhca proteins, was helpful to establish the stoichiometry. The positions of Lhca2 and Lhca9 were assigned with high confidence. Further data are necessary to determine the exact location of the other seven Lhca proteins, but based on the reasonable assumption that PSI-LHCI complexes disassemble by losing the most external antenna complexes first, we propose that Lhca4, Lhca5, and Lhca6 are located in the outer half-ring of the supercomplex, whereas Lhca1, Lhca3, Lhca7, and/or Lhca8 compose the inner half-ring, with Lhca3 located next to PsaK, based on previous results (62). It is interesting to observe that the loss of PsaG, PsaK, and PsaF leads to the dissociation of a large part of the antenna, confirming the important role of these subunits in the stable association of LHCI with PSI (10).
PSI of Chlamydomonas Versus PSI of Higher Plants
The comparison of the PSI-LHCI supercomplexes from green algae and plants reveals that they differ in several respects. (i) The outer antenna of green algal PSI-LHCI in is larger than in plants (nine Lhcas versus four). (ii) The Lhcas are organized into two half-rings on one side of the core; the outer half-ring is not present in plant supercomplexes. (iii) C.r.PSI-LHCI is less stable than the complex from higher plants and easily loses some of the Lhcas, starting with Lhca9 and Lhca2. (vi) The amount of red forms is far lower for C.r.PSI-LHCI than for the plant supercomplex; the C. reinhardtii complex has an emission maximum at 715 nm versus 735 nm for the one from A. thaliana. (v) LHCI complexes from C. reinhardtii have a higher Chl b content as compared with plant LHCI.
Supplementary Material
This work was supported by De Nederlandse Organisatie voor Wetenschappelijk Onderzoek, Earth and Life Sciences, through a Vidi grant (to R. C.). Work in the Redding laboratory was supported by National Science Foundation Grant MCB-1052573.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1–S4.
- PSI and II
- and Photosystem I and II, respectively
- LHCI
- light harvesting complexes
- A.t.
- Arabidopsis thaliana
- Chl
- chlorophyll
- C.r.
- C. reinhardtii
- α-DM
- α-dodecylmaltoside
- Tricine
- N-[2-hydroxy-1,1-bis(hydroxymethyl)ethyl]glycine.
REFERENCES
- 1. Arnon D. I. (1971) Proc. Natl. Acad. Sci. U.S.A. 68, 2883–2892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Dekker J. P., Boekema E. J. (2005) Biochim. Biophys. Acta 1706, 12–39 [DOI] [PubMed] [Google Scholar]
- 3. Amunts A., Nelson N. (2009) Structure 17, 637–650 [DOI] [PubMed] [Google Scholar]
- 4. Jensen P. E., Haldrup A., Rosgaard L., Scheller H. V. (2003) Physiol. Plant. 119, 313–321 [Google Scholar]
- 5. Amunts A., Nelson N. (2008) Plant Physiol. Biochem. 46, 228–237 [DOI] [PubMed] [Google Scholar]
- 6. Ben-Shem A., Frolow F., Nelson N. (2004) Photosynth. Res. 81, 239–250 [DOI] [PubMed] [Google Scholar]
- 7. Nelson N., Ben-Shem A. (2005) Bioessays 27, 914–922 [DOI] [PubMed] [Google Scholar]
- 8. Boekema E. J., Dekker J. P., Vanheel M. G., Rogner M., Saenger W., Witt I., Witt H. T. (1987) FEBS Lett. 217, 283–286 [Google Scholar]
- 9. Lam E., Oritz W., Mayfield S., Malkin R. (1984) Plant Physiol. 74, 650–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Ozawa S., Onishi T., Takahashi Y. (2010) J. Biol. Chem. 285, 20072–20079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jordan P., Fromme P., Witt H. T., Klukas O., Saenger W., Krauss N. (2001) Nature 411, 909–917 [DOI] [PubMed] [Google Scholar]
- 12. Jensen P. E., Bassi R., Boekema E. J., Dekker J. P., Jansson S., Leister D., Robinson C., Scheller H. V. (2007) Bba-Bioenergetics 1767, 335–352 [DOI] [PubMed] [Google Scholar]
- 13. Scheller H. V., Jensen P. E., Haldrup A., Lunde C., Knoetzel J. (2001) Bba-Bioenergetics 1507, 41–60 [DOI] [PubMed] [Google Scholar]
- 14. Farah J., Rappaport F., Choquet Y., Joliot P., Rochaix J. D. (1995) EMBO J. 14, 4976–4984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Haldrup A., Naver H., Scheller H. V. (1999) Plant J. 17, 689–698 [DOI] [PubMed] [Google Scholar]
- 16. Fischer N., Boudreau E., Hippler M., Drepper F., Haehnel W., Rochaix J. D. (1999) Biochemistry 38, 5546–5552 [DOI] [PubMed] [Google Scholar]
- 17. Lunde C., Jensen P. E., Haldrup A., Knoetzel J., Scheller H. V. (2000) Nature 408, 613–615 [DOI] [PubMed] [Google Scholar]
- 18. Zhang S., Scheller H. V. (2004) J. Biol. Chem. 279, 3180–3187 [DOI] [PubMed] [Google Scholar]
- 19. Jensen P. E., Gilpin M., Knoetzel J., Scheller H. V. (2000) J. Biol. Chem. 275, 24701–24708 [DOI] [PubMed] [Google Scholar]
- 20. Ben-Shem A., Frolow F., Nelson N. (2003) Nature 426, 630–635 [DOI] [PubMed] [Google Scholar]
- 21. Jansson S. (1999) Trends Plant Sci. 4, 236–240 [DOI] [PubMed] [Google Scholar]
- 22. Amunts A., Drory O., Nelson N. (2007) Nature 447, 58–63 [DOI] [PubMed] [Google Scholar]
- 23. Ganeteg U., Klimmek F., Jansson S. (2004) Plant Mol. Biol. 54, 641–651 [DOI] [PubMed] [Google Scholar]
- 24. Lucinski R., Schmid V. H., Jansson S., Klimmek F. (2006) FEBS Lett. 580, 6485–6488 [DOI] [PubMed] [Google Scholar]
- 25. Storf S., Jansson S., Schmid V. H. (2005) J. Biol. Chem. 280, 5163–5168 [DOI] [PubMed] [Google Scholar]
- 26. Germano M., Yakushevska A. E., Keegstra W., van Gorkom H. J., Dekker J. P., Boekema E. J. (2002) FEBS Lett. 525, 121–125 [DOI] [PubMed] [Google Scholar]
- 27. Kargul J., Nield J., Barber J. (2003) J. Biol. Chem. 278, 16135–16141 [DOI] [PubMed] [Google Scholar]
- 28. Elrad D., Grossman A. R. (2004) Curr. Genet. 45, 61–75 [DOI] [PubMed] [Google Scholar]
- 29. Stauber E. J., Fink A., Markert C., Kruse O., Johanningmeier U., Hippler M. (2003) Eukaryot. Cell 2, 978–994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mozzo M., Mantelli M., Passarini F., Caffarri S., Croce R., Bassi R. (2010) Biochim. Biophys. Acta 1797, 212–221 [DOI] [PubMed] [Google Scholar]
- 31. Bassi R., Soen S. Y., Frank G., Zuber H., Rochaix J. D. (1992) J. Biol. Chem. 267, 25714–25721 [PubMed] [Google Scholar]
- 32. Subramanyam R., Jolley C., Brune D. C., Fromme P., Webber A. N. (2006) FEBS Lett. 580, 233–238 [DOI] [PubMed] [Google Scholar]
- 33. Takahashi Y., Yasui T. A., Stauber E. J., Hippler M. (2004) Biochemistry 43, 7816–7823 [DOI] [PubMed] [Google Scholar]
- 34. Tokutsu R., Teramoto H., Takahashi Y., Ono T. A., Minagawa J. (2004) Plant Cell Physiol. 45, 138–145 [DOI] [PubMed] [Google Scholar]
- 35. Stauber E. J., Busch A., Naumann B., Svatos A., Hippler M. (2009) Proteomics 9, 398–408 [DOI] [PubMed] [Google Scholar]
- 36. Wollman F. A., Bennoun P. (1982) Biochim. Biophys. Acta 680, 352–360 [Google Scholar]
- 37. Kargul J., Turkina M. V., Nield J., Benson S., Vener A. V., Barber J. (2005) FEBS J. 272, 4797–4806 [DOI] [PubMed] [Google Scholar]
- 38. Gulis G., Narasimhulu K. V., Fox L. N., Redding K. E. (2008) Photosynth. Res. 96, 51–60 [DOI] [PubMed] [Google Scholar]
- 39. Gorman D. S., Levine R. P. (1965) Proc. Natl. Acad. Sci. U.S.A. 54, 1665–1669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fischer N., Sétif P., Rochaix J. D. (1997) Biochemistry 36, 93–102 [DOI] [PubMed] [Google Scholar]
- 41. Bassi R. (1985) Carlsberg Research. Commun. 50, 127–143 [DOI] [PubMed] [Google Scholar]
- 42. Peter G. F., Thornber J. P. (1991) J. Biol. Chem. 266, 16745–16754 [PubMed] [Google Scholar]
- 43. Croce R., Canino G., Ros F., Bassi R. (2002) Biochemistry 41, 7334–7343 [DOI] [PubMed] [Google Scholar]
- 44. Oostergetel G. T., Keegstra W., Brisson A. (1998) Ultramicroscopy 74, 47–59 [Google Scholar]
- 45. Boekema E. J., Van Roon H., Van Breemen J. F., Dekker J. P. (1999) Eur. J. Biochem. 266, 444–452 [DOI] [PubMed] [Google Scholar]
- 46. Pineau B., Gerard-Hirne C., Selve C. (2001) Plant Physiol. Biochem. 39, 73–85 [Google Scholar]
- 47. van Oort B., Amunts A., Borst J. W., van Hoek A., Nelson N., van Amerongen H., Croce R. (2008) Biophys. J. 95, 5851–5861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Wientjes E., Oostergetel G. T., Jansson S., Boekema E. J., Croce R. (2009) J. Biol. Chem. 284, 7803–7810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wientjes E., Croce R. (2011) Biochem. J. 433, 477–485 [DOI] [PubMed] [Google Scholar]
- 50. Gibasiewicz K., Szrajner A., Ihalainen J. A., Germano M., Dekker J. P., van Grondelle R. (2005) J. Phys. Chem. B 109, 21180–21186 [DOI] [PubMed] [Google Scholar]
- 51. Ihalainen J. A., Ratsep M., Jensen P. E., Scheller H. V., Croce R., Bassi R., Korppi-Tommola J. E. I., Freiberg A. (2003) J. Phys. Chem. B 107, 9086–9093 [Google Scholar]
- 52. Romero E., Mozzo M., van Stokkum I. H., Dekker J. P., van Grondelle R., Croce R. (2009) Biophys. J. 96, L35–L37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu H., Sadygov R. G., Yates J. R., 3rd (2004) Anal Chem. 76, 4193–4201 [DOI] [PubMed] [Google Scholar]
- 54. Amunts A., Toporik H., Borovikova A., Nelson N. (2010) J. Biol. Chem. 285, 3478–3486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ihalainen J. A., van Stokkum I. H., Gibasiewicz K., Germano M., van Grondelle R., Dekker J. P. (2005) Biochim. Biophys. Acta 1706, 267–275 [DOI] [PubMed] [Google Scholar]
- 56. Owens T. G., Webb S. P., Mets L., Alberte R. S., Fleming G. R. (1989) Biophys. J. 56, 95–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Melkozernov A. N., Kargul J., Lin S., Barber J., Blankenship R. E. (2004) J. Phys. Chem. B 108, 10547–10555 [Google Scholar]
- 58. Melkozernov A. N., Kargul J., Lin S., Barber J., Blankenship R. E. (2005) Photosynth. Res. 86, 203–215 [DOI] [PubMed] [Google Scholar]
- 59. Owens T. G., Webb S. P., Alberte R. S., Mets L., Fleming G. R. (1988) Biophys. J. 53, 733–745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Fischer N., Hippler M., Sétif P., Jacquot J. P., Rochaix J. D. (1998) EMBO J. 17, 849–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Busch A., Hippler M. (2011) Biochim. Biophys. Acta 1807, 864–877 [DOI] [PubMed] [Google Scholar]
- 62. Naumann B., Stauber E. J., Busch A., Sommer F., Hippler M. (2005) J. Biol. Chem. 280, 20431–20441 [DOI] [PubMed] [Google Scholar]
- 63. Kouril R., Zygadlo A., Arteni A. A., de Wit C. D., Dekker J. P., Jensen P. E., Scheller H. V., Boekema E. J. (2005) Biochemistry 44, 10935–10940 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.