Background: Snf1, the AMP-activated protein kinase of yeast, is regulated at the level of dephosphorylation.
Results: Ligand-mediated protection of Snf1 requires all three subunits and is specific for adenosine nucleotides.
Conclusion: ADP is the metabolic signaling molecule for Snf1 kinase.
Significance: Adenylate-mediated regulation of Snf1 connects kinase activity to energy availability.
Keywords: Adenosine, AMP-activated Kinase (AMPK), Glucose Metabolism, Phosphorylation, Protein Kinases, Signal Transduction, Snf1, Dephosphorylation
Abstract
Members of the AMP-activated protein kinase (AMPK) family are activated by phosphorylation at a conserved threonine residue in the activation loop of the kinase domain. Mammalian AMPK adopts a phosphatase-resistant conformation that is stabilized by binding low energy adenylate molecules. Similarly, binding of ADP to the Snf1 complex, yeast AMPK, protects the kinase from dephosphorylation. Here, we determined the nucleotide specificity of the ligand-mediated protection from dephosphorylation and demonstrate the subunit and domain requirements for this reaction. Protection from dephosphorylation was highly specific for adenine nucleotides, with ADP being the most effective ligand for mediating protection. The full-length α subunit (Snf1) was not competent for ADP-mediated protection, confirming the requirement for the regulatory β and γ subunits. However, Snf1 heterotrimeric complexes that lacked either the glycogen-binding domain of Gal83 or the linker region of the α subunit were competent for ADP-mediated protection. In contrast, adenylate-mediated protection of recombinant human AMPK was abolished when a portion of the linker region containing the α-hook domain was deleted. Therefore, the exact means by which the different adenylate nucleotides are distinguished by the Snf1 enzyme may differ compared with its mammalian ortholog.
Introduction
The AMP-activated protein kinase (AMPK)2 family is a conserved family of serine/threonine protein kinases present in essentially all eukaryotic cells. Under conditions of nutrient limitation or energy stress, the kinase becomes activated and plays a critical role in reorganizing metabolism and gene expression. Cellular processes that consume ATP are largely inhibited by AMPK, whereas those that produce ATP and help restore energy balance are activated (1). AMPK enzymes function as heterotrimers with a single catalytic subunit (α) and two regulatory subunits (β and γ). A number of recent studies have made great strides in understanding how these subunits interact with each other and with nucleotide ligands to respond to changes in cellular energy balance (2–4). The N-terminal half of the α subunit contains a typical kinase domain whose activity requires phosphorylation of its activation loop (5, 6). The C terminus of the α subunit is required to form a complex with the β and γ subunits. The N- and C-terminal domains of the α subunit are joined by a flexible linker that is not visible in most structural models of AMPK. Some have proposed that this linker region contains an autoinhibitory domain (7, 8), whereas a more recent report proposes that the linker plays a role in adenylate nucleotide sensing (4). The C terminus of the β subunit forms the interface between the α and γ subunits (9). The structure of the N terminus of the β subunits has not been solved, but this region plays a role in substrate specification and subcellular localization (10, 11). Most interestingly, the γ subunit is composed of four tandem cystathionine β-synthase domains, each of which forms a nucleotide-binding site. It is the cystathionine β-synthase domains in the γ subunit that bind AMP and provide the activation for which this enzyme is named.
Initial studies of AMPK showed that the addition of AMP produces large increases in kinase activity (12). More careful analyses with highly purified recombinant proteins have now shown that the allosteric activation of AMPK caused by AMP binding is relatively modest along the lines of a 2-fold stimulation (6, 13). The much larger effect of AMP is to stabilize the active form of the enzyme by making it resistant to dephosphorylation. The regulation mechanism of protection from dephosphorylation is conserved between AMPK enzymes in mammals and yeast (13, 14). The exact mechanism by which AMP binding to the γ subunit confers a phosphatase resistance to the α subunit has been a subject of great interest. Structural studies of the γ subunit bound to AMP, ATP, or no ligand found very limited changes in γ subunit conformation (15) and no obvious explanations for the change in phosphatase sensitivity. However, most structural models of AMPK were derived from crystals containing either the kinase domain or the heterotrimeric core. In a recent study, Gamblin and co-workers (4) solved the structure of the active AMPK enzyme. This new structure shows the kinase domain bound to the heterotrimeric core, with the activation loop forming a large portion of the interaction surface. The structure of the active AMPK enzyme explains the resistance to phosphatases. Furthermore, this new model proposes that the flexible linker of the α subunit wraps around the γ subunit and reaches into one of the γ subunit adenylate-binding sites. This portion of the flexible linker is referred to as the α-hook.
Yeast AMPK is known as Snf1. Regulation of Snf1 in response to changes in carbon source shares many features with the mammalian enzyme. Both yeast and mammalian AMPK enzymes are regulated at the dephosphorylation step, and both enzymes can form a phosphatase-resistant conformation. Recently, we collaborated with the members of the Carling and Gamblin laboratories and found that the Snf1 enzyme binds to ADP and becomes phosphatase-resistant in vitro (16). In this study, we optimized and more fully characterized the in vitro dephosphorylation reaction. We examined the nucleotide specificity of this reaction as well as the subunit and domain requirements for the ligand-mediated protection of Snf1 from dephosphorylation. Our results support the idea that ADP binding to the regulatory core of the enzyme stabilizes the phosphatase-resistant conformation.
EXPERIMENTAL PROCEDURES
Yeast Strains and Genetic Methods
The yeast strains used in this study were all derivatives of S228C. Wild-type Snf1 complexes were purified from FY1193 (MATa ura3-52 leu2Δ1 his3Δ200 trp1Δ63 snf1Δ10) transformed with pSNF1-TAP (17). The lithium acetate method (18) was used for transformation of yeast strains. Cells were grown at 30 °C using standard medium (19).
Protein Purifications
Snf1 kinase complexes containing primarily Snf1, Snf4, and Gal83 proteins were purified by tandem affinity purification (TAP) (20) from yeast cells lacking endogenous Snf1. The GST-tagged Snf1 kinase domain was purified from bacterial cells by affinity chromatography (21). The cDNA for human protein phosphatase (PP) 2Cα was purchased from Open Biosystems (IHS1382-8646531) and inserted into the bacterial expression plasmid pET14b (Novagen). Bacterial cells were induced with 1 mm isopropyl β-d-thiogalactopyranoside for 2.5 h at 26 °C. Extracts were prepared by sonication, and His-tagged PP2Cα was purified using nickel-nitrilotriacetic acid-agarose (Qiagen). Rabbit PP1 was purchased from Sigma (P7937). Yeast Glc7 was TAP-purified as described (21). Human AMPK composed of the α1, β1, and γ1 subunits was purified as described (22). The DNA encoding the α-hook domain (amino acids 377–411) was deleted and replaced with the codons for five alanine residues using oligonucleotide-directed mutagenesis. The resulting plasmid was confirmed by DNA sequencing. AMPK enzyme lacking the α-hook domain was purified from BL21 cells as described for the wild-type enzyme (22).
Western Blotting
Snf1-HA was detected with a 1:2000 dilution of HA probe (Santa Cruz Biotechnology). DyLight 680-conjugated goat anti-mouse IgG (1:5000 dilution; Thermo Scientific) was used as the secondary antibody. For detection of phosphorylated Snf1, phospho-AMPKα (Thr-172) antibody (1:1000 dilution; Cell Signaling Technology) was used. IRDye 800CW-conjugated goat anti-rabbit IgG (1:5000 dilution; LI-COR Biosciences) was used as the secondary antibody. Blots were processed using the SNAP i.d.® system (Millipore) and scanned using an Odyssey scanner (LI-COR Biosciences). Integrated intensity values of bands were quantified using Odyssey scanning software. To detect Snf1 activation loop (Thr-210) phosphorylation in vivo, cells were harvested following the addition of NaOH to 0.1 m, suspended in SDS sample buffer (62 mm Tris-Cl (pH 6.8), 10% glycerol, 5% β-mercaptoethanol, and 3% SDS), and subjected to overnight dialysis against 2 liters of radioimmunoprecipitation buffer. Protein extracts (800 μg) were immunoprecipitated in radioimmunoprecipitation buffer supplemented with protease and phosphatase inhibitors using 20 μl of HA probe-agarose conjugate (Santa Cruz Biotechnology). Bound proteins were eluted in SDS sample buffer and resolved on SDS gels. Blots were treated with Odyssey blocking buffer (LI-COR Biosciences).
Nucleotides
ATP, ADP, AMP, and AMP-PNP were purchased from Sigma (A2383, A2754, A1752, and A2647, respectively). Manufacturer claims of nucleotide purity exceeding 99% (ATP and AMP) and 95% (ADP) were confirmed by HPLC analysis (supplemental Fig. S1).
Dephosphorylation Assays
Dephosphorylation reactions (10 μl) contained purified Snf1 proteins (∼50 ng) in reaction buffer (20 mm HEPES (pH 7.0), 0.5 mm EDTA, 0.5 mm dithiothreitol, and 5 mm magnesium acetate). Nucleotides (Sigma) were dissolved in 10 mm Tris (pH 8) and 1 mm EDTA, adjusted to neutral pH, and added to reactions at a final concentration of 0.8 mm unless indicated otherwise. Titrations of purified PP2C were performed to determine the appropriate dilution needed to remove 80–90% of the Snf1 phosphorylation. Purified PP2C was diluted in reaction buffer and added to the reactions, which were then incubated at 37 °C for 10 min. Reactions were stopped by the addition of SDS sample buffer. Total and phosphorylated Snf1 proteins were analyzed by quantitative Western blotting. Phosphatase assays using the chromogenic substrate p-nitrophenyl phosphate (pNPP) were conducted in 50-μl reactions containing 20 mm HEPES (pH 8.0), 0.5 mm EDTA, 0.5 mm dithiothreitol, 2 mm MnCl2, 0.1 m NaCl, 5% (v/v) glycerol, and 20 mm pNPP. Reactions were incubated at 37 °C for 10 min and stopped by the addition of 1 ml of 1 m KPO4 (pH 8). The absorbance at 405 nm was read, and the moles of pNPP hydrolyzed were calculated using a molar extinction coefficient of 1.78 × 104 m−1 cm−1.
Statistical Analysis
For all bar plots, mean values of a minimum of three independent measurements are plotted, with error bars representing 1 S.E. Statistical significance was determined using Student's t test for unpaired variables with equal variance.
RESULTS
Adenylate-mediated Protection from Dephosphorylation
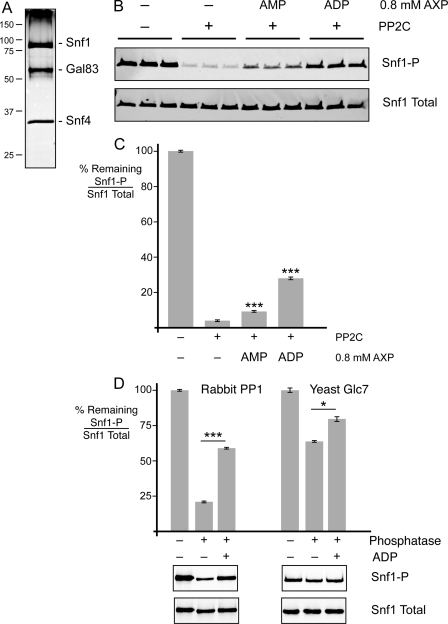
To study the dephosphorylation of Snf1 in vitro, we purified the Snf1 kinase complex from yeast cells lacking the chromosomal copy of the SNF1 gene that had been transformed with a plasmid expressing Snf1-TAP (20). Cells were grown in medium containing sucrose as the carbon source to ensure that the purified Snf1 would be phosphorylated at Thr-210. The Snf1 complex was an intact heterotrimer as judged by the abundance of the Snf1, Snf4, and Gal83 proteins on silver-stained protein gels (Fig. 1A). The complex was treated with purified recombinant phosphatase PP2C, and the reduction in phosphorylation at Thr-210 was readily detected by quantitative Western blotting using antibodies directed against phosphorylated and total Snf1 proteins (Fig. 1B). The addition of low energy adenylate molecules (0.8 mm AMP or ADP) inhibited the dephosphorylation reaction, with ADP showing a much greater level of protection from dephosphorylation. Reactions were performed in triplicate, and the levels of phosphorylated and total Snf1 proteins were determined. The ratio of phospho-Snf1 divided by total Snf1 in the absence of added PP2C was defined as 100% phosphorylated. The mean value for the percentage of phosphorylation remaining after PP2C treatment is plotted in Fig. 1C. Both AMP and ADP showed statistically significant protection of Snf1 from dephosphorylation. We were concerned that the low level of protection conferred by AMP might be due to contamination of that nucleotide with ADP. The purity of our AMP, ADP, and ATP stocks was analyzed by HPLC (supplemental Fig. S1). The AMP stock did not contain any detectable ADP. Thus, the low level of protection afforded by the addition of AMP was due to AMP itself. We concluded that low energy adenylate molecules (AMP and ADP) stabilize the phosphatase-resistant conformation of Snf1, with ADP being the most effective ligand.
FIGURE 1.
Adenylate-mediated protection of Snf1 dephosphorylation in vitro. A, the purity of the Snf1 complex was analyzed on an SDS-polyacrylamide gel stained with silver nitrate. The mobility of protein standards of known mass (kilodaltons) is indicated on the left. B, Snf1 complexes purified from yeast were subjected to in vitro dephosphorylation with purified recombinant PP2C in the presence of 0.8 mm AMP or ADP as shown. Triplicate reactions were assayed by quantitative Western blotting with antisera directed against total Snf1 or Snf1 phosphorylated at Thr-210 (Snf1-P). C, the mean ratio of phosphorylated Snf1 to total Snf1 remaining after phosphatase treatment was calculated from triplicate reactions. D, dephosphorylation reactions using either rabbit PP1 or yeast Glc7 were conducted with and without 0.8 mm ADP as indicated. Reactions were conducted in triplicate, and the mean values are plotted. Representative Western blots are shown below. *, p < 0.05; ***, p < 0.001.
The use of PP2C as the phosphatase in these experiments was greatly influenced by our ability to obtain large quantities of a highly active and stable protein phosphatase. The phosphatase that acts on Snf1 in vivo is Glc7, a member of the PP1 family of protein phosphatases (5, 23). ADP mediated significant protection of Snf1 from dephosphorylation when treated with commercial rabbit PP1 and with our own Glc7 preparation affinity-purified from yeast (Fig. 1D). Furthermore, ADP-mediated protection of Snf1 from Glc7 has been observed in a dose-responsive manner (16). Therefore, the ADP-mediated protection of Snf1 from dephosphorylation is not specific to PP2C but is observed with other phosphatases, including the members of the PP1 family. Although Glc7 is the cognate phosphatase acting on Snf1 in vivo, our preparations of Glc7 from yeast were less active and less stable than our preparations of recombinant PP2C. For this reason, we used human PP2C for the remaining studies presented here. We were concerned that the purified enzymes (PP2C and Snf1-TAP) might contain an activity capable of hydrolyzing adenylate nucleotides. We examined the stability of ADP after incubation with PP2C and Snf1-TAP and found that the ADP was stable and not hydrolyzed (supplemental Fig. S2).
Nucleotide Specificity of Protection from Dephosphorylation
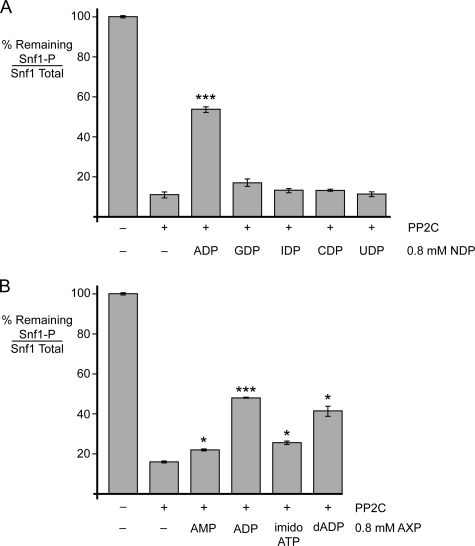
Because we detected both AMP- and ADP-mediated protection of Snf1 from dephosphorylation, we sought to determine the specificity of this reaction. We tested five nucleoside diphosphates (adenosine, guanosine, inosine, cytosine, and uridine) for the ability to mediate protection of Snf1 kinase from dephosphorylation when present at a concentration of 0.8 mm. We used a concentration of 0.8 mm in these studies because this level provides reproducible protection from dephosphorylation and is likely to be in the physiological range of adenylate nucleotides in vivo (24). Reactions were performed in triplicate, and the mean value of Thr-210 phosphorylation remaining after phosphatase treatment is plotted in Fig. 2A. Only ADP was able to mediate statistically significant protection from dephosphorylation. Therefore, the ligand-mediated protection of Snf1 from dephosphorylation is highly specific for the adenine base. We next tested the specificity of this reaction for the phosphate groups and the ribose (Fig. 2B). Again, we detected statistically significant protection mediated by both AMP and ADP, with ADP showing much greater efficacy. We were unable to test ATP directly in this system because TAP-purified Snf1 contains low levels of the Snf1-activating kinase (21). However, we tested the ability of a non-hydrolyzable ATP analog, AMP-PNP, to mediate protection and found that the addition of AMP-PNP caused a slight but statistically significant level of protection. Finally, we tested dADP and also found statistically significant protection. We conclude that the adenylate nucleotide-mediated protection of Snf1 from dephosphorylation is highly specific for the adenine base, shows a strong preference for the diphosphate, and is much less specific in differentiating between ribose and deoxyribose.
FIGURE 2.
Nucleotide specificity. Snf1 heterotrimers purified from yeast were subjected to in vitro dephosphorylation with purified recombinant PP2C in the presence or absence of 0.8 mm nucleoside diphosphates (NDP) as indicated (A) or with other adenosine nucleotides (AXP; B). Samples were assayed in triplicate by quantitative Western blotting with antisera directed against total or phosphorylated Snf1. The mean ratio of phosphorylated Snf1 to total Snf1 is plotted as the percentage remaining after phosphatase treatment. T210, Thr-210; imido-ATP, AMP-PNP. *, p < 0.05; ***, p < 0.001.
ADP-mediated Protection of Snf1 Requires Heterotrimeric Core Regulatory Domain
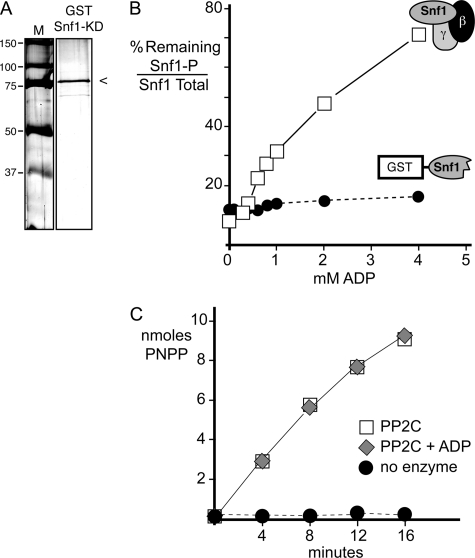
Structural studies of the Snf1 kinase have shown that the kinase and regulatory core domains can be crystallized separately (2, 25). The heterotrimeric core domain contains the entire γ subunit (Snf4), the C terminus of the β subunit (Sip2), and the C terminus of the α subunit (Snf1). Recently, a structure of the human ortholog of Snf1 was reported that shows how the N-terminal kinase domain in the α subunit binds to the heterotrimeric core domain to create a conformation that would be phosphatase-resistant (4). We have proposed a similar model for the Snf1 heterotrimer to explain the mechanism for ADP-mediated protection (16). Here, we tested this model by examining the subunit requirements for ADP-mediated protection. If ADP is in fact binding to the γ subunit (Snf4) and stabilizing the phosphatase-resistant conformation, then we would predict that the heterotrimeric core regulatory domain would be absolutely required for ADP-mediated protection. We purified the Snf1 kinase domain (residues 1–392) expressed in bacteria as a fusion to the GST protein (Fig. 3A). The Snf1 kinase domain was phosphorylated in vitro with purified Snf1-activating kinase (21) and separated from residual ATP by gel filtration. We treated the native Snf1 heterotrimer purified from yeast and the Snf1 kinase domain purified from bacteria with PP2C in the presence of increasing concentrations of ADP. Samples were analyzed by Western blotting for total and phosphorylated Snf1 proteins. The percentage of phosphorylated Snf1 remaining after phosphatase treatment is plotted in Fig. 3B. The native Snf1 heterotrimer showed strong protection from dephosphorylation that was dependent on the concentration of ADP. In contrast, the Snf1 kinase domain failed to show any ADP-mediated protection from dephosphorylation. We also assayed the ability of PP2C to hydrolyze the synthetic substrate pNPP in the presence or absence of 0.8 mm ADP (Fig. 3C). The addition of ADP had no effect on the enzymatic activity of PP2C for the synthetic substrate pNPP. We concluded that ADP-mediated protection of Snf1 requires the heterotrimeric core regulatory proteins and that ADP itself has no direct effect on the phosphatase activity of PP2C.
FIGURE 3.
Snf1 kinase domain is not sufficient for ADP-mediated protection. A, an SDS protein gel stained with Coomassie Blue was used to assess the purity of the GST-tagged Snf1 kinase domain (KD) used in this experiment. The mobility of molecular mass markers (M) is indicated in kilodaltons on the left. B, in vitro dephosphorylation assays were conducted in the presence of increasing concentrations of ADP using the Snf1 heterotrimeric complex purified from yeast (□) or the Snf1 kinase domain (residues 1–392) purified from bacteria as a GST fusion protein (●). The GST-tagged Snf1 kinase domain was phosphorylated in vitro with purified Sak1 kinase prior to the dephosphorylation assay. The mean ratio of phosphorylated Snf1 (Snf1-P) to total Snf1 is plotted as the percentage remaining after phosphatase treatment. C, the phosphatase assay was used to measure the hydrolysis of 20 mm pNPP in vitro in the absence or presence of ADP (0.8 mm) and PP2C as indicated.
Full-length α Subunit Is Not Sufficient for ADP-mediated Protection
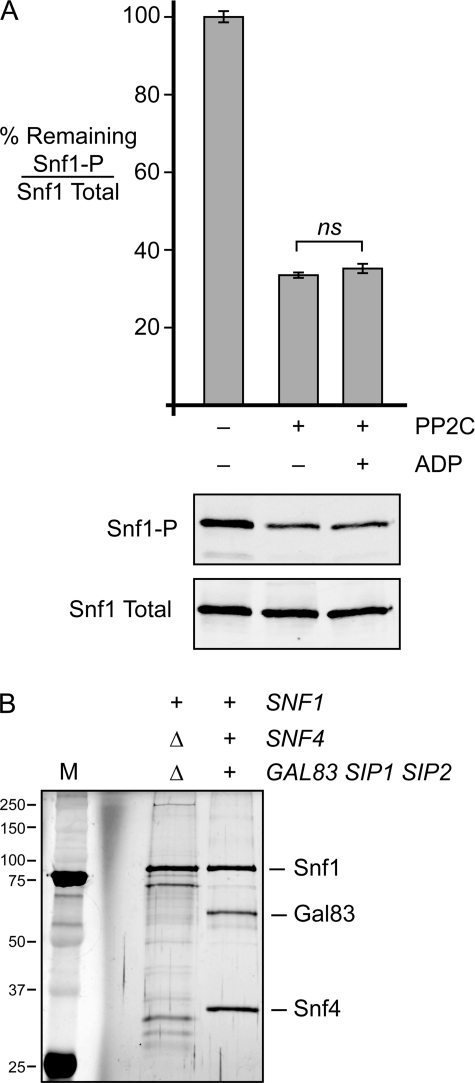
We next sought to further define the subunit requirements for ADP-mediated protection. We TAP-purified the Snf1 protein from cells that lacked either the gene for the γ subunit or all three genes for the β subunits. Consistent with previous studies (20), we found that in the absence of the γ subunit, the Snf1 protein contained substoichiometric amounts of β subunits. In the absence of β subunits, Snf1 lacked any detectable γ subunit. Therefore, the Snf1 heterotrimer requires all three subunits for stable assembly, and we were not able to isolate αβ or αγ dimers. To eliminate any uncertainty arising from trace amounts of regulatory subunits, we purified Snf1 from cells lacking the genes for the γ subunit and all three β subunit genes. The purified Snf1 protein was found to be phosphorylated and full-length (Fig. 4). Treatment of Snf1 with phosphatase reduced the phosphorylation of Thr-210. The addition of ADP offered no significant protection from dephosphorylation. Therefore, the C-terminal domain of the α subunit is not sufficient for ADP-mediated protection from dephosphorylation.
FIGURE 4.
β and γ subunits are required for ADP-mediated protection. A, phosphatase protection assays were performed in triplicate using full-length Snf1 protein purified from cells lacking the genes for the γ subunit and all three β subunits (snf4Δ sip1Δ sip2Δ gal83Δ). The mean ratio of phosphorylated Snf1 (Snf1-P) to total Snf1 is plotted as the percentage remaining after phosphatase treatment. Representative blots are shown below. ns (not significant), p > 0.05. B, an SDS protein gel stained with silver nitrate was used to assess the purity and integrity of the Snf1 protein used in this experiment. The Snf1 complex purified from wild-type cells was loaded for comparison. The mobility of molecular mass markers (M) is indicated in kilodaltons on the left.
β Subunit Glycogen-binding Domain Is Not Required for ADP-mediated Protection
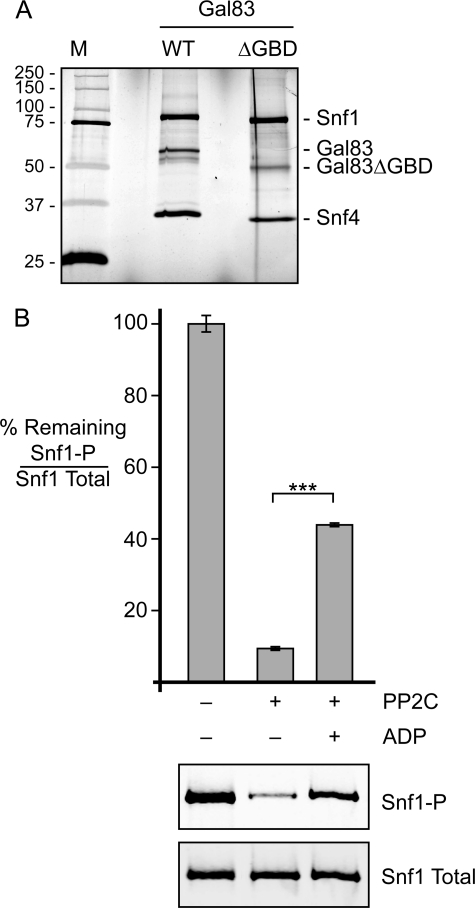
Two of the yeast β subunits, Gal83 and Sip2, have a conserved domain that is known as the glycogen-binding domain (GBD). Deletion of this domain from Gal83 results in a hyperactive Snf1 kinase (26, 27). We next asked whether this domain is required for ADP-mediated protection from dephosphorylation. Cells from which all three β subunit genes had been deleted were transformed with a low copy number plasmid expressing either wild-type Gal83 or mutant Gal83 from which the entire GBD (residues 152–244) had been deleted (Gal83ΔGBD). Snf1 heterotrimers were purified (Fig. 5A) and subjected to in vitro dephosphorylation in the presence or absence of 0.8 mm ADP (Fig. 5B). The Snf1 enzyme lacking the GBD exhibited robust ADP-mediated protection from dephosphorylation. Therefore, the GBD is not required for this regulatory mechanism.
FIGURE 5.
Gal83 GBD is not required for ADP-mediated protection. A, an SDS protein gel stained with silver nitrate was used to assess the purity and integrity of the Snf1 complex used in this experiment. The Snf1 complex purified from wild-type cells was loaded for comparison. The mobility of molecular mass markers (M) is indicated in kilodaltons on the left. B, phosphatase protection assays were performed in triplicate using the Snf1 heterotrimer purified from cells lacking the genes for all three β subunits (sip1Δ sip2Δ gal83Δ) and transformed with a plasmid expressing Gal83ΔGBD. The mean ratio of phosphorylated Snf1 (Snf1-P) to total Snf1 is plotted as the percentage remaining after phosphatase treatment. Representative Western blots are shown below. ***, p < 0.001.
Snf1 Linker Domain Is Not Required for Snf1 Regulation in Vivo
The Snf1 kinase domain and its C terminus are connected by ∼150 amino acids that are poorly conserved between species and that are predicted to be unstructured (28). Indeed, most of this linker domain could not be crystallized and is not visible in the structural studies of the Snf1 kinase domain or its heterotrimeric core (Fig. 6A). In a recent study, Gamblin and co-workers (4) were successful in crystallizing the active form of AMPK. In this new structure, the kinase domain takes on the folding of an active kinase with the alignment of the hydrophobic spine (29) and, more importantly, has the activation loop with the phosphorylated threonine nestled against the heterotrimeric core in what is likely the phosphatase-resistant conformation. In this structure, the linker between the kinase domain and its C terminus is draped over the surface of the γ subunit, with a portion known as the α-hook reaching into adenylate site 3 of γ subunit. Gamblin and co-workers proposed that the α-hook interrogates the bound adenylate and discriminates between high energy (ATP) and low energy (ADP or AMP) adenylate molecules. When ATP is bound at site 3, the α-hook cannot stably interact with site 3. Without this interaction, the phosphatase-resistant conformation becomes destabilized and vulnerable to dephosphorylation.
FIGURE 6.
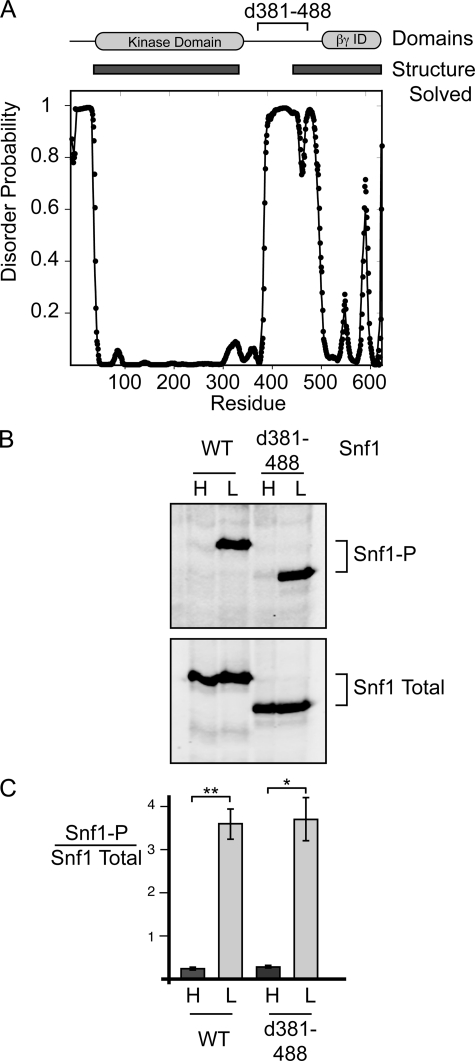
Snf1 linker region is not required for regulation of Snf1 phosphorylation in vivo. A, disorder probability (28) is plotted versus the Snf1 primary sequence. The locations of the Snf1 kinase domain, the βγ interaction domain (βγ ID), and the deletion of amino acids 381–488 (d381–488) are shown at the top. Regions of Snf1 that have had their structure solved by crystallography (1, 22) are indicated by rectangles. B, shown are Western blots of yeast extracts prepared from cells grown in high glucose (H) or 30 min after shifting to low glucose (L). Cells expressed either full-length WT Snf1 or Snf1Δ381–488 (d381–488) as indicated. Blots were probed with antibodies that react with phosphorylated (Snf1-P) or total Snf1. C, shown is the quantitation of Western blots from three independent transformants. Mean values of the ratio of phosphorylated Snf1 to total Snf1 are plotted, with error bars representing 1 S.E. *, p < 0.05; **, p < 0.01.
In these next experiments, we investigated whether the Snf1 linker domain is required for the regulation of Snf1 phosphorylation in vivo and for ADP-mediated protection from dephosphorylation in vitro. We employed a Snf1 protein from which most of the linker domain, including the region that best aligns with the mammalian α-hook domain, was deleted (supplemental Fig. S3) but that is known from previous studies to be active (30). This Snf1 deletion mutant lacks residues 381–488. Although there is little sequence conservation between yeast and humans in this region, deletion of these 108 residues removes most of the linker sequence that connects the kinase domain with the structured C terminus. Cells expressing Snf1Δ381–488 were phenotypically Snf+ as judged by their ability to grow on raffinose medium and non-fermentable carbon sources (data not shown). The phosphorylation of the kinase activation loop was regulated in vivo in response to the carbon source in a manner indistinguishable from wild-type Snf1 (Fig. 6, B and C). Therefore, the flexible linker connecting the Snf1 kinase domain with the C terminus present in the heterotrimeric core is not required for the regulation of Snf1 phosphorylation in vivo.
Snf1 Linker Domain Is Not Required for ADP-mediated Protection or Adenylate Energy Discrimination
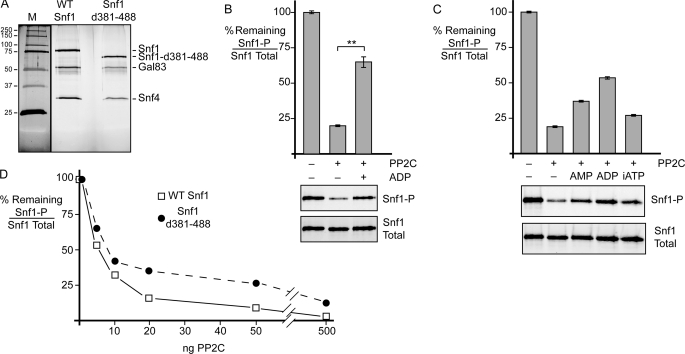
The Snf1 complex containing the deletion of amino acids 381–488 was TAP-purified from yeast cells and assayed in the in vitro dephosphorylation assay. The Δ381–488 enzyme was purified as an intact heterotrimer as judged by co-purification of the Snf4 and Gal83 proteins visible on a silver-stained SDS gel (Fig. 7A). Treatment of the Δ381–488 enzyme with phosphatase removed most of the Thr-210 phosphorylation. When ADP was present in the dephosphorylation reaction, significant protection was observed (Fig. 7B). Therefore, the linker domain of Snf1 is not required for ADP-mediation protection. The α-hook model proposes that the hook interrogates the bound adenylate nucleotide and discriminates between mono-, di-, and triphosphorylated nucleotides (4). We in asked whether the Snf1 linker is required for adenylate discrimination. Dephosphorylation reactions were performed in the absence of added nucleotide or in the presence of 0.8 mm AMP, ADP, or AMP-PNP. We used a non-hydrolyzable form of ATP to eliminate the possibility of rephosphorylation of Snf1 by trace contamination with the Sak1 kinase, a known constituent of the Snf1-TAP preparation (21). Snf1Δ381–488 was able to distinguish between the mono-, di-, and triphosphorylated adenylate nucleotides (Fig. 7C) in a manner similar to the wild-type protein (Fig. 2B). Therefore, the linker region (amino acids 381–488) between the Snf1 kinase domain and its C terminus is not required for discrimination between the high and low energy adenylate ligands. We cannot rule out the possibility that some other portion of the Snf1 protein plays the role of the α-hook by interrogating the adenylate ligand bound by the γ subunit. Deletion of the linker region did confer some phosphatase resistance to the Snf1Δ381–488 heterotrimer (Fig. 7D).
FIGURE 7.
Snf1 linker domain is not required for ligand-mediated protection in vitro or adenylate discrimination. A, SDS-polyacrylamide gel stained with silver nitrate showing the purity of the WT Snf1 heterotrimer and the heterotrimer with the linker deleted (Snf1Δ381–488 (Snf1-d381–488)). Protein size standards are shown on the left in kilodaltons (M). B, phosphatase protection assay of Snf1 heterotrimers with the linker (amino acids 381–488) deleted. Triplicate reactions were treated with PP2C with or without 0.8 mm ADP as shown. Mean values of the percentage of phosphorylated Snf1 (Snf1-P) to total Snf1 remaining are plotted, with error bars representing 1 S.E. Representative blots are shown below. C, ligand-mediated protection using the linker deletion mutant and different adenylate nucleotides present at 0.8 mm. Duplicate reactions were performed, and representative blots are shown below. Mean values are plotted, with error bars representing the range of duplicate values. iATP, non-hydrolyzable AMP-PNP. D, phosphatase resistance of wild-type Snf1 and Snf1Δ381–488. The percentage of phosphorylated Snf1 to total Snf1 remaining is plotted as a function of increasing concentrations of PP2C. **, p < 0.01.
α-Hook Domain Is Required for ADP-mediated Protection of Human AMPK in Vitro
Our finding that the linker region in the Snf1 protein is not required for ADP-mediated protection prompted us to investigate whether deletion of this region would impact the adenylate-mediated protection of the mammalian AMPK enzyme. We used the tricistronic expression plasmid developed by Dr. Dietbert Neumann to express human AMPK in bacteria (22). The DNA encoding the portion of the linker region of the human α1 protein (amino acids 377–411) containing the entire α-hook domain (Fig. 8A) was deleted and replaced with codons for five alanine residues (supplemental Fig. S3). Both wild-type AMPK and AMPKΔ377–411 were purified from Escherichia coli as intact heterotrimers in association with the human β1 and γ1 proteins (Fig. 8B). The AMPK enzymes were phosphorylated in vitro with ATP and recombinant calmodulin-dependent protein kinase kinase β, followed by gel filtration chromatography to remove residual nucleotides. The phosphorylated AMPK enzymes were assayed in the dephosphorylation assay in the absence or presence of 0.2 mm ADP (Fig. 8C). Wild-type AMPK showed significant ADP-mediated protection, consistent with a previous report (4). When the deletion form of AMPK lacking the α-hook was assayed, we found that the enzyme became more resistant to dephosphorylation (Fig. 8D) and that the adenylate-mediated protection was abolished (Fig. 8C). Therefore, the α-hook domain of human AMPK is required for adenylate-mediated protection.
FIGURE 8.
Human AMPK α-hook domain is required for adenylate-mediated protection in vitro. A, structural model for human AMPK in the active conformation (31) showing the α subunit (green), with its α-hook domain (red sticks) reaching into site 3 of the γ subunit (cyan) bound to ADP (blue). B, Coomassie Blue-stained protein gel showing the purity of the recombinant wild-type AMPK α1β1γ1 complex and the same complex with the α-hook deleted (AMPK d377–411). The mobility of molecular mass markers (M) is indicated in kilodaltons on the left. C, phosphatase protection assay with AMPK heterotrimers with and without the α-hook domain. Triplicate reactions were treated with PP2C with or without 0.2 mm ADP as shown. Mean values of the percentage of phosphorylated AMPK (AMPK-P) to total AMPK remaining after phosphatase treatment are plotted. Representative blots are shown below. D, titration of PP2C using wild-type AMPK (□) or AMPKΔ377–411 (●). The percentage of phosphorylated AMPK to total AMPK remaining after phosphatase treatment is plotted as a function of added PP2C. ***, p < 0.001; ns, p > 0.05.
DISCUSSION
The discovery that the Snf1 kinase of yeast and the AMP-activated kinase of mammals are orthologs has greatly enhanced the rate of new discoveries in both fields of study. The two enzymes are so similar in structure that it was surprising when differences in regulation were found. For instance, AMP is able to allosterically activate mammalian AMPK yet has no effect on the kinase activity of yeast Snf1 (31). With the availability of purified recombinant enzymes, the allosteric effect of AMP was found to be relatively modest especially compared with the large effect that AMP has on the activity of AMPK by stabilizing the active phosphatase-resistant conformation (6, 13). However, once again, the yeast enzyme did not seem to be regulated similarly by AMP. However, this conundrum was solved when it was discovered that the regulatory ligand for Snf1 is ADP, not AMP. Binding of ADP to the Snf1 complex does in fact promote the formation of a phosphatase-resistant conformation (16), thus unifying the yeast and mammalian enzymes with a shared regulatory mechanism. This in vitro study showing adenylate-mediated protection of Snf1 is supported by in vivo studies demonstrating that the primary determinant of Snf1 phosphorylation status is the rate of dephosphorylation (14). In this study, we characterized the ligand requirements for mediating protection and determined the subunit and domain requirements of this reaction.
The addition of low energy adenylate ligands inhibits the dephosphorylation of purified Snf1 in vitro. Consistent with our previous work (16), we found that ADP is the most efficient nucleotide for protection from dephosphorylation. The addition of other nucleotide diphosphates had little effect on Snf1 dephosphorylation (Fig. 2), demonstrating that the adenine base is essential for this mode of regulation. dADP can also mediate significant protection, indicating that the oxygen of the 2′-carbon of the ribose is not a key binding determinant. This finding is consistent with mutagenesis studies that found that mutation of the aspartate residues thought to interact with the ribose moiety in the γ subunit cystathionine β-synthase domains has only modest effects on adenylate binding to the yeast γ subunit (16). Measurements of nucleotide pools in Saccharomyces cerevisiae show that ribonucleosides are 50–200 times more abundant than deoxyribonucleosides (24). Taken together, these data strongly suggest that ADP is the predominant regulatory ligand for Snf1 in vivo. It is not clear at this time why the mammalian enzyme shows equivalent protection by either AMP or ADP (4), whereas the yeast enzyme shows a strong preference for ADP (Fig. 1). The different responses to the low energy adenylate ligands could reflect different adenylate nucleotide levels in mammalian and yeast cells, or they could reflect subtle differences in the AMPK enzymes themselves.
Adenylate-mediated protection of AMPK from dephosphorylation is thought to occur when adenylate binding to the γ subunit stabilizes the active phosphatase-resistant conformation of the heterotrimer (4). We tested the subunit and domain requirements for adenylate-mediated protection of Snf1. As predicted, adenylate-mediated protection of Snf1 was not observed with the isolated Snf1 kinase domain (Fig. 3) or with the full-length α subunit (Fig. 4). The intact heterotrimer is required, consistent with the idea that the phosphatase resistance is acquired when the kinase domain binds the heterotrimeric core. The formation of the compact phosphatase-resistant structure of active Snf1/AMPK is likely conserved throughout eukaryotic evolution. The phosphorylated activation loop forms a large part of the interaction surface between the kinase domain and the heterotrimeric core (4). Furthermore, the two histidine residues present in the C terminus of the β subunit that contact the activation loop are absolutely conserved in the β subunits from all eukaryotic species, including humans, yeast, and even choanoflagellates (supplemental Fig. S4). Mutation of the conserved histidine residues destabilizes the phosphatase-resistant conformation and increases the rate of dephosphorylation for both the yeast and human AMPK enzymes (4, 16).
A key question is the molecular mechanism by which adenylate binding to the γ subunit promotes the formation of the phosphatase-resistant conformation. Crystallography studies showed negligible differences in the structure of the γ subunit bound to high or low energy adenylate molecules (3, 15), leaving unanswered the question of how the enzyme distinguishes between adenylate ligands and how that information is transmitted to the α subunit. The recently proposed α-hook model envisions that the linker region of the α subunit, which connects its kinase domain with its C-terminal domain, interacts with the γ subunit and contacts adenylate ligands bound in site 3 (4). This model is rather appealing, as it provides a rational explanation for the discrimination between adenylate ligands and for the stabilization of the phosphatase-resistant conformation. We asked whether the α-hook model also applies to regulation of the yeast Snf1 complex. Previously, we found that a large deletion of the Snf1 linker region that removed the residues that best align with the mammalian α-hook domain resulted in a functional Snf1 kinase (30). In this study, we analyzed the Snf1Δ381–488 deletion mutant in greater detail and found that this portion of the linker region was not required for regulation of Snf1 phosphorylation in vivo (Fig. 6). We purified the Snf1Δ381–488 heterotrimer and found that the mutant enzyme was proficient in adenylate-mediated protection from dephosphorylation (Fig. 7). We are tempted to conclude that the α-hook model does not apply to the yeast enzyme; however, we cannot rule out the possibility that other regions of the Snf1 linker that are not visible in the structural models and that are still present in the Δ381–488 enzyme may engage the γ subunit in a manner analogous to the mammalian α-hook. Indeed, studies of Schizosaccharomyces pombe AMPK show an ADP bound at site 2 of the γ subunit (32), a site that is not used by human AMPK and that is closer to the center of the heterotrimeric core than is site 3. Thus, fungal AMPK enzymes may use a different site for adenylate binding, which would necessitate the use of a different region of the α subunit for site interrogation.
Our studies do support the α-hook model for the human enzyme. Removal of the α-hook from human AMPK does eliminate adenylate-mediated protection (Fig. 8), as do missense mutations in the hook itself (4). Interestingly, deletion of the α-hook domain conferred significant phosphatase resistance to human AMPK (Fig. 8D). A similar but less pronounced phosphatase resistance was observed when the linker region of the yeast Snf1 enzyme was deleted (Fig. 7D). Forcing the kinase domain and the heterotrimeric core into closer proximity by reducing the size of the linker may favor the formation of the phosphatase-resistant conformation independent of ligand binding.
Supplementary Material
Acknowledgments
We thank Drs. Faith Mayer and David Carling for the generous gift of purified PP2C, the calmodulin-dependent protein kinase kinase expression plasmid, and communication of results prior to publication. We thank Noah Gale for construction of the Snf1 linker deletion plasmid and Dr. Dietbert Neumann for the gift of the AMPK expression plasmid. We thank Drs. Lena Miller and Michael Parniak for conducting HPLC analysis of nucleotides.
This work was supported, in whole or in part, by National Institutes of Health Grant GM46443.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
- AMPK
- AMP-activated protein kinase
- TAP
- tandem affinity purification
- PP
- protein phosphatase
- AMP-PNP
- adenosine 5′-(β,γ-imido)triphosphate
- pNPP
- p-nitrophenyl phosphate
- GBD
- glycogen-binding domain.
REFERENCES
- 1. Hardie D. G. (2011) Am. J. Clin. Nutr. 93, 891S–896S [DOI] [PubMed] [Google Scholar]
- 2. Amodeo G. A., Rudolph M. J., Tong L. (2007) Nature 449, 492–495 [DOI] [PubMed] [Google Scholar]
- 3. Townley R., Shapiro L. (2007) Science 315, 1726–1729 [DOI] [PubMed] [Google Scholar]
- 4. Xiao B., Sanders M. J., Underwood E., Heath R., Mayer F. V., Carmena D., Jing C., Walker P. A., Eccleston J. F., Haire L. F., Saiu P., Howell S. A., Aasland R., Martin S. R., Carling D., Gamblin S. J. (2011) Nature 472, 230–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. McCartney R. R., Schmidt M. C. (2001) J. Biol. Chem. 276, 36460–36466 [DOI] [PubMed] [Google Scholar]
- 6. Suter M., Riek U., Tuerk R., Schlattner U., Wallimann T., Neumann D. (2006) J. Biol. Chem. 281, 32207–32216 [DOI] [PubMed] [Google Scholar]
- 7. Chen L., Jiao Z. H., Zheng L. S., Zhang Y. Y., Xie S. T., Wang Z. X., Wu J. W. (2009) Nature 459, 1146–1149 [DOI] [PubMed] [Google Scholar]
- 8. Crute B. E., Seefeld K., Gamble J., Kemp B. E., Witters L. A. (1998) J. Biol. Chem. 273, 35347–35354 [DOI] [PubMed] [Google Scholar]
- 9. Iseli T. J., Walter M., van Denderen B. J., Katsis F., Witters L. A., Kemp B. E., Michell B. J., Stapleton D. (2005) J. Biol. Chem. 280, 13395–13400 [DOI] [PubMed] [Google Scholar]
- 10. Schmidt M. C., McCartney R. R. (2000) EMBO J. 19, 4936–4943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vincent O., Townley R., Kuchin S., Carlson M. (2001) Genes Dev. 15, 1104–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Carling D., Clarke P. R., Zammit V. A., Hardie D. G. (1989) Eur. J. Biochem. 186, 129–136 [DOI] [PubMed] [Google Scholar]
- 13. Sanders M. J., Grondin P. O., Hegarty B. D., Snowden M. A., Carling D. (2007) Biochem. J. 403, 139–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Rubenstein E. M., McCartney R. R., Zhang C., Shokat K. M., Shirra M. K., Arndt K. M., Schmidt M. C. (2008) J. Biol. Chem. 283, 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Xiao B., Heath R., Saiu P., Leiper F. C., Leone P., Jing C., Walker P. A., Haire L., Eccleston J. F., Davis C. T., Martin S. R., Carling D., Gamblin S. J. (2007) Nature 449, 496–500 [DOI] [PubMed] [Google Scholar]
- 16. Mayer F., Heath R., Underwood E., Jimenez D. C., Sanders M. J., Leiper F. C., Xiao B., Jing C., Walker P. A., McCartney R. R., Martin S. R., Schmidt M. C., Gamblin S. J., Carling D. (2011) Cell Metab. 14, 707–714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Nath N., McCartney R. R., Schmidt M. C. (2002) J. Biol. Chem. 277, 50403–50408 [DOI] [PubMed] [Google Scholar]
- 18. Gietz R. D., Schiestl R. H., Willems A. R., Woods R. A. (1995) Yeast 11, 355–360 [DOI] [PubMed] [Google Scholar]
- 19. Rose M. D., Winston F., Hieter P. (eds) (1990) Methods in Yeast Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY [Google Scholar]
- 20. Elbing K., Rubenstein E. M., McCartney R. R., Schmidt M. C. (2006) J. Biol. Chem. 281, 26170–26180 [DOI] [PubMed] [Google Scholar]
- 21. Elbing K., McCartney R. R., Schmidt M. C. (2006) Biochem. J. 393, 797–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Neumann D., Woods A., Carling D., Wallimann T., Schlattner U. (2003) Protein Expr. Purif. 30, 230–237 [DOI] [PubMed] [Google Scholar]
- 23. Sanz P., Alms G. R., Haystead T. A., Carlson M. (2000) Mol. Cell. Biol. 20, 1321–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nick McElhinny S. A., Watts B. E., Kumar D., Watt D. L., Lundström E. B., Burgers P. M., Johansson E., Chabes A., Kunkel T. A. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 4949–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rudolph M. J., Amodeo G. A., Bai Y., Tong L. (2005) Biochem. Biophys. Res. Commun. 337, 1224–1228 [DOI] [PubMed] [Google Scholar]
- 26. Momcilovic M., Iram S. H., Liu Y., Carlson M. (2008) J. Biol. Chem. 283, 19521–19529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mangat S., Chandrashekarappa D., McCartney R. R., Elbing K., Schmidt M. C. (2010) Eukaryot. Cell 9, 173–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ward J. J., Sodhi J. S., McGuffin L. J., Buxton B. F., Jones D. T. (2004) J. Mol. Biol. 337, 635–645 [DOI] [PubMed] [Google Scholar]
- 29. Kornev A. P., Haste N. M., Taylor S. S., Eyck L. F. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 17783–17788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Leech A., Nath N., McCartney R. R., Schmidt M. C. (2003) Eukaryot. Cell 2, 265–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wilson W. A., Hawley S. A., Hardie D. G. (1996) Curr. Biol. 6, 1426–1434 [DOI] [PubMed] [Google Scholar]
- 32. Jin X., Townley R., Shapiro L. (2007) Structure 15, 1285–1295 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.