Background: Neurodegenerative diseases are associated with intracellular protein aggregation and free radical damage.
Results: Protein aggregation of polyglutamine-containing proteins directly causes free radical production in vitro and within cells.
Conclusion: Protein aggregation during polyglutamine diseases could be targeted to prevent oxidative stress.
Significance: Intracellular protein aggregation during chronic neurodegeneration is closely linked to abnormal production of free radicals.
Keywords: Neurodegeneration, Neurodegenerative Diseases, Polyglutamine, Polyglutamine Disease, Protein Aggregation, Protein Misfolding
Abstract
Neurodegenerative diseases are characterized by intra- and/or extracellular protein aggregation and oxidative stress. Intense attention has been paid to whether protein aggregation itself contributes to abnormal production of free radicals and ensuing cellular oxidative damage. Although this question has been investigated in the context of extracellular protein aggregation, it remains unclear whether protein aggregation inside cells alters the redox homeostasis. To address this, we have used in vitro and in vivo (cellular) models of Huntington disease, one of nine polyglutamine (poly(Q)) disorders, and examined the causal relationship among intracellular protein aggregation, reactive oxygen species (ROS) production, and toxicity. Live imaging of cells expressing a fragment of huntingtin (httExon1) with a poly(Q) expansion shows increased ROS production preceding cell death. ROS production is poly(Q) length-dependent and not due to the httExon 1 flanking sequence. Aggregation inhibition by the MW7 intrabody and Pgl-135 treatment abolishes ROS production, showing that increased ROS is caused by poly(Q) aggregation itself. To examine this hypothesis further, we determined whether aggregation of poly(Q) peptides in vitro generated free radicals. Monitoring poly(Q) protein aggregation using atomic force microscopy and hydrogen peroxide (H2O2) production over time in parallel we show that oligomerization of httEx1Q53 results in early generation of H2O2. Inhibition of poly(Q) oligomerization by the single chain antibody MW7 abrogates H2O2 formation. These results demonstrate that intracellular protein aggregation directly causes free radical production, and targeting potentially toxic poly(Q) oligomers may constitute a therapeutic target to counteract oxidative stress in poly(Q) diseases.
Introduction
Neurodegenerative disorders, such as Huntington disease (HD),3 Alzheimer disease, Parkinson disease, and other proteinopathies, which involve misfolding of cellular proteins resulting in the formation of intra- or extracellular protein aggregates, have all been associated with an abnormal redox homeostasis and oxidative damage due to increased production of reactive oxygen species (ROS) (1, 2). Although the maintenance of ROS at low levels is critical to normal cell functions (3), a prolonged increase in ROS due to an impairment of the oxidative metabolism can be highly damaging to macromolecules such as DNA, proteins, and lipids (4). Although various cellular mechanisms of abnormal ROS generation have been hypothesized to occur during disease progression of proteinopathies, protein misfolding and ensuing protein aggregation reactions in the presence of metal ions themselves were suggested to participate in the production of free radicals (5–7). This hypothesis is supported by in vitro experiments using several amyloid-forming and redox-active proteins and peptides (Aβ, α-synuclein, prion-, amylin-, and British dementia (ABri) peptides) (for review, see Ref. 8) and cell studies of extracellular protein aggregation such as Aβ (5, 9). However, it is unknown whether intracellular aggregation causes abnormal ROS production.
We have used existing and novel models of polyglutamine (poly(Q)) misfolding to investigate the causal relationships between intracellular protein aggregation, ROS production and cellular toxicity. By altering the length of the poly(Q) stretch within a protein the magnitude and kinetics of protein aggregation in vitro and in vivo can be achieved. As a model we used N-terminal fragments of the huntingtin (htt) protein including the first exon (httEx1) with expanded poly(Q) stretches because these are aggregation-prone cleavage products found to aggregate within cells in the HD brain (10) and N-terminal or full-length HD mouse models (11, 12). Expression of poly(Q)-expanded htt has also been associated with oxidative stress in several cell and animals models (13–19) and the HD brain (20–23), but the mechanisms by which the cellular redox homeostasis is altered in HD remain unclear.
Given that httEx1 oligomerization and amyloid-like fibril formation can be modeled in vitro (in the test tube), we show here that both in vitro and in vivo (using cellular HD models) httEx1 aggregation is sufficient to cause an increased, detrimental poly(Q) length-dependent production of free radicals. Because increased ROS strongly coincides with the formation of oligomeric poly(Q) protein species that when suppressed also decreases ROS, our data suggest that targeting poly(Q) oligomerization could be an important avenue to prevent the abnormal redox homeostasis occurring in HD and indeed other disorders associated with intracellular protein aggregation.
EXPERIMENTAL PROCEDURES
Plasmids, Cell Culture, and Antibodies
All chemicals were purchased from Sigma unless otherwise stated. pcDNA3.1 plasmids containing httEx1 with 25, 47, 72, or 97 glutamines fused to enhanced green fluorescent protein (EGFP) at the C terminus were described previously (13). Identical httEx1 plasmids, but fused to monomeric red fluorescent protein (mRFP), were produced by excising EGFP using BamHI and XbaI restriction enzymes (Promega) and ligating mRFP that was PCR-amplified from mRFP of pRSETB (a gift from R. Tsien, University of California San Diego) using primers flanked by BamHI and XbaI sites. pCDNA3.1 plasmids encoding stretches of 15 or 81 glutamines fused to GFP were obtained from W. Strittmatter (Duke University Medical Center, Durham, NC). The MW7 intrabody was a gift from A. Khoshnan (Caltech, Pasadena, CA). Plasmid DNA preparations were sequenced after each preparation using an endonuclease-free Maxi kit (Qiagen). HeLa cells were grown in DMEM with 2 mm l-glutamine, 10% fetal bovine serum (FBS), and 100 units/ml penicillin with 100 μg/ml streptomycin at 37 °C, 10% CO2. PC12 cells were grown in RPMI 1640 medium with 2 mm l-glutamine, 10% horse serum, 5% FBS, 4.5 g/liter glucose, 10 mm Hepes, 1 mm sodium pyruvate at 37 °C, 5% CO2. The PC12 httEx1Q25/103-EGFP tebufenozide inducible cell line was a gift from E. Schweitzer (24), and the ponasterone A-inducible 14.1A PC12 cell line, originally described in Ref. 25, was cultured in DMEM with 5 mm Hepes, 5% FBS, 5% horse serum, 2 mm l-glucose, 100 units/ml penicillin with 100 μg/ml streptomycin and G418 (0.5 mg/ml) at 37 °C, 5% CO2. 1 μm tebufenozide or 5 μm ponasterone A was added to induce expression of httEx1. For all PC12 cell experiments surfaces were precoated with poly-l-lysine. 24 h after plating, cells were exposed to the appropriate DNA construct and Lipofectamine (Invitrogen) for 5 h in serum-free medium (Opti-MEM; Invitrogen) as described previously (13, 26, 27), after which 2× FBS-containing medium was added. Where appropriate, prior to transfection cells were treated with Pgl-135, Trolox, N-acetyl-l-cysteine (l-NAC), or N(G)-nitro-l-arginine methyl ester (l-NAME). For co-expression of intrabody MW7, cells were first transfected for 24 h followed by transfection of httEx1 constructs. Antibodies used were: S830 (polyclonal sheep anti-httEx1 antibody, gift from G. Bates, London), EGFP (Abcam), MW7 (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA), anti-FLAG (Sigma), cytochrome c (BD Biosciences), active caspase-3 (Promega), H2A.X-Ser(P)-139 (Upstate Biotech).
Analysis of Cellular Poly(Q) Aggregation, Toxicity, and Immunocytochemistry
Cells on coverslips were washed with 1× PBS, fixed with 4% paraformaldehyde in 1× PBS for 20 min, and then processed for immunocytochemistry or mounted in Fluoromount G medium (SouthernBiotech) supplemented with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI) to allow visualization of nuclear morphology. Immunocytochemistry was performed as described (26). For quantification of histone-2A.X phosphorylation at serine 139 (H2A.X-Ser-139) 10–30 cells transfected with the respective mRFP constructs were imaged by confocal microscopy followed by analysis of the fluorescence intensity using Metamorph software (Molecular Devices). For this purpose, the pixel intensity within a circle overlaying the nucleus compared with the cytoplasmic staining (background) was determined using three randomly chosen areas within each compartment and the mean ratio for each cell was calculated. Microscopic analysis was done using an inverted Zeiss Axioplan2 epifluorescent microscope. Cells were counted as aggregate-positive if one/several inclusion bodies (IBs) were visible. We counted 200–300 mRFP/EGFP-positive cells in multiple random visual fields/coverslip in duplicate for independent experiments. Poly(Q) aggregation was also monitored by the filter trap assay (27, 28). For native gels, cell lysates were prepared in a nonreducing lysis buffer, and samples were run on 7.5% nonreducing Tris/glycine gels at 30 mA for 2–4 h and blotted overnight at 4 °C at 30 V.
For Western blotting, epitopes were detected by ECL (Amersham Biosciences) using Kodak Scientific Imaging film. Cell death was monitored by scoring the proportion of httEx1-expressing cells with fragmented and/or pyknotic nuclei as described (26), release of cytochrome c or activation of caspase-3 as described in Ref. 29 or using the MTS assay (Promega). Cell death end points were analyzed by scoring the proportion of httEx1mRFP/EGFP-expressing cells with fragmented or pyknotic nuclei, as described previously (26), showing that both apoptotic and nonapoptotic types of death can be detected. To monitor the release of cytochrome c or activation of caspase-3, cells were processed via immunocytochemistry (see above) with the respective antibodies and the httEx1-expressing cells with a clear signal for active caspase-3 antibody or homogeneous or no cytochrome c staining noted (as opposed to mitochondrially localized staining when cytochrome c is not released).
Cellular and in Vitro Free Radical Measurements
Dihydroethidium (DHE, 5 μm) and 5-(-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester (CM-H2DCF, 8 μm) were used to measure ROS. Reactive nitrogen species were measured by 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate (DAF-FM diacetate, 5 μm). All dyes (Invitrogen) were freshly suspended for each experiment. At each time point after transfection, adherent cells were washed in the 35-mm plastic dishes (used for transfection) once with DMEM without serum, and medium was replaced with 1 ml of nonsupplemented DMEM containing dyes and incubated for 20 min at 37 °C, washed 3–5 times with nonsupplemented DMEM, and multiple images of cells were taken using an excitation wavelength of 488 nm (DCF/EGFP) and 543 nm (DHE/mRFP) sequentially by confocal microscopy. The integrated morphology function in Metamorph software was employed to measure average pixel intensity of each transfected cell normalized by area (40–60 cells per experiment and condition). A -fold change comparing mean pixel intensity of cells expressing httEx1Q97 compared with mean pixel intensity of cells expressing httEx1Q25 for each experiment was calculated. For the Amplex Red assays, used to monitor in vitro H2O2 production, a working solution consisting of 10-acetyl-3;7-dihydroxyphenoxazine (Cambridge Bioscience) in dimethyl sulfoxide and horseradish peroxidase (HRP) in PBS was produced as described in Ref. 6. Standard curves were obtained using H2O2 solutions. A 15-μl sample of standard H2O2 solution or test sample of httEx1 was added to each well of a black, flat-bottomed, 384-well microtiter plate (Nunc), in triplicate, with subsequent addition of Amplex Red working solution. After a 30-min incubation at room temperature, fluorescence of resorufin was read on a plate reader (BMG Labtech) with λEx = 544 nm and λEm = 590 nm. Solutions containing no H2O2 were used as a measure of background fluorescence, which was subtracted from all values. As a control, 0.1 unit of catalase was added prior to performance of the Amplex Red assay with httEx1Q53.
Protein Purification, in Vitro Aggregation, Atomic Force Microscopy (AFM) Analysis, and Western Blotting
Expression of GST-httEx1-Q20/Q53 plasmids was performed as described (28). The protein was purified using FPLC with a 5-ml GST affinity column (GE Healthcare), and the integrity of the protein was evaluated using SDS-PAGE. The protein was dialyzed in 20 mm Tris-HCl, pH 8, 150 mm NaCl, 0.1 mm EDTA, 5% glycerol. Freshly prepared protein samples were used for each experiment. Before each experiment the GSThttEx1Q20/53 was centrifuged at 20,000 × g for 30 min to remove any preformed aggregates. GST cleavage and aggregation were initiated by addition of PreScission protease (2 units/100 μg of protein) (GE Healthcare) to 50 μm GST-httEx1Q20/53. At indicated times a volume corresponding to 12 μg of protein of the reaction was spotted onto a freshly cleaved mica disc (Agar Scientific), incubated for 2 min, and then rinsed with 200 μl of ultrapure water and dried with compressed air. Samples were imaged by AFM in air with an uncoated silicon cantilevers (FM-W, Nanoworld Innovative Technologies, Switzerland, nominal spring constant 2.8 N/m) operating in tapping mode. Western blotting was performed as described (27). Fluorescent secondary antibody-labeled membranes were scanned with a Li-COR scanner at 700 or 800 nm and quantified by using Odyssey version 1.2 software. The total number of fibrils per area was calculated by counting the number of fibrils in each 10-μm2 image with at least five random images obtained from several experiments. A fibril was defined as an elongated structure of greater than 200 nm in length. Oligomer sizes were determined by cross-sectional height analysis of individual globular aggregates from at least five AFM 2-μm2 images. Fibrils were excluded from this analysis. Lyophilized Aβ1–40 or Aβ1–42 peptide (rPeptide) was dissolved in 1 ml of 0.001% ammonium hydroxide (pH 10) and sonicated four times for 30 s in an ice beaker, with vortexing in between the sonication steps. After distribution into aliquots, ammonium hydroxide was removed using a speed vacuum, and each aliquot was stored at −20 °C until use. For aggregation experiments the peptide was resuspended in 1 mg/ml hexafluoroisopropyl alcohol, sonicated four times for 30 s in an ice beaker, and vortexed over a period of 30 min. Hexafluoroisopropyl alcohol was removed using a speed vacuum. The peptide was then resolubilized in 10 mm PBS to a final concentration of 50 μm and sonicated four times for 30 s in an ice beaker, with vortexing between sonication. Following resuspension, Aβ peptides were incubated at 37 °C for 24–72 h before imaging with AFM or performing the Amplex Red assay as described above.
RESULTS
Poly(Q) Expanded httEx1 Causes Increased ROS Coinciding with Aggregation but Preceding Cell Death
Previous studies showed that expression of poly(Q) expanded proteins are associated with toxicity and simultaneous ROS production that invariably occurs during cell death processes (13, 14, 30). We sought to separate ROS potentially due to protein aggregation and ROS caused by cell death. Therefore, using transient transfection of httEx1 constructs with different poly(Q) expansions (Gln-25, Gln-47, Gln-72, and Gln-97) fused to either mRFP or EGFP, we monitored the kinetics of ROS, IB formation, SDS-insoluble poly(Q) protein, and cell death in several httEx1 cell systems. The use of two fluorescent httEx1 proteins with different absorption/emission spectra allowed employment of two oxidation-sensitive molecules to determine intracellular ROS. Although httEx1Q25 did not show any IBs or formation of SDS-insoluble material (filter trap assay), aggregation of httEx1 with longer poly(Q) stretches followed a poly(Q) length- and time-dependent aggregation process as described previously (26, 30), mainly in the cytoplasm (Fig. 1, A and B, and supplemental Fig. S1, A and B). No significant differences in transgene expression levels, IB formation, and the production of SDS-insoluble protein of mRFP versus EGFP control and mutant constructs was observed (supplemental Fig. S1, C–F). Although httEx1Q97 aggregation was detectable at 24 h after transfection (Fig. 1B and supplemental Fig. S1F), a small increase in toxicity compared with httEx1Q25 expression was only detectable at 48 h that became statistically significant after 72 h (Fig. 1A). Hence, poly(Q) aggregation in this cell system occurred significantly earlier compared with nuclear abnormalities that correlated with release of cytochrome c and caspase-3 activation (supplemental Fig. S1, G and H).
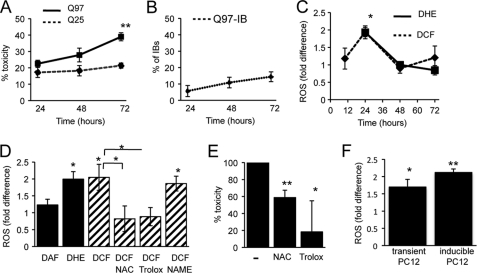
FIGURE 1.
Poly(Q) expanded httEx1 induces ROS before toxicity occurs. A, percentage of HeLa cells expressing httEx1Q25- or httEx1Q97mRFP with nuclear abnormalities. B, httEx1Q97mRFP-expressing cells with IBs (no IBs are formed by htEx1Q25mRFP). Means ± S.D. (error bars) are shown. C, -fold change of CM-H2DCF (DCF) or DHE oxidation in HeLa cells expressing httEx1Q97mRFP (DCF curve) or httEx1Q97EGFP (DHE curve) compared with httEx1Q25mRFP/EGFP. Means ± S.E. (error bars) are shown. D, -fold change in ROS at 24 h after transfection as in C (DAF used with mRFP constructs). l-NAC (1 mm), Trolox (0.1 mm), but not the NO inhibitor l-NAME (5 mm) suppressed httEx1Q97mRFP0-induced ROS. Means± S.E. are shown. E, percentage toxicity reduction 72 h after transfection with httEx1Q97mRFP by l-NAC (1 mm) or Trolox (0.1 mm). 100% toxicity is average difference in toxicity between httEx1Q97mRFP- and httEx1Q25mRFP-expressing cells with nuclear abnormalities. Means ± S.D. are shown. F, ROS induction at 24 h in httEx1Q97/Q25mRFP transiently transfected (transient PC12, DCF) or httEx1Q103/Q25EGFP tebufenozide-inducible cells (inducible PC12, DHE). Means ± S.D. are shown. A–F, n = 4–6 independent experiments. Student's t test (unpaired); *, p < 0.05; **, p < 0.01.
We next measured ROS within single, living cells by confocal microscopy using the fluorescein derivative CM-H2DCF in conjunction with mRFP constructs or DHE in combination with EGFP constructs (see supplemental Fig. S2, A and B). The mean -fold fluorescence intensity for both CM-H2DCF and DHE (as calculated by pixel intensities of cells expressing httEx1Q97 compared with httEx1Q25) showed a 2-fold increase and hence a significant ROS elevation due to httEx1Q97 expression at 24 h that decreased afterward (Fig. 1C). Comparison with untransfected (nonfluorescent) cells of the same culture dish or parallel culture dishes of untransfected cells, both httEx1Q25- and Q97mRFP-expressing cells showed an increase in CM-H2DCF oxidation (supplemental Fig. S2C). This confirms that the ROS increase in httEx1Q97 cells was not due to lower values obtained by ROS suppression via httEx1Q25 expression. We next repeated our analysis using DAF-FM diacetate to monitor NO production but did not detect any difference in oxidation between httEx1Q25- versus -Q97-expressing cells (Fig. 1D), suggesting that the response measured by CM-H2DCF and DHE is specific for ROS. Increased ROS at 24 h was suppressed by the antioxidants l-NAC and the vitamin E analog Trolox, but not the NO inhibitor l-NAME, again suggesting that ROS production was not linked to NO production (Fig. 1D). Toxicity of cells expressing httEx1Q97 treated with both l-NAC and Trolox could be significantly reduced (Fig. 1E). Therefore, the early increase in ROS when SDS-insoluble httEx1 protein was already detectable (supplemental Fig. S1F) appears to be linked to cellular toxicity occurring later, despite ROS levels having decreased by that time. An early increase in ROS due to httEx1Q97mRFP expression was also found in PC12 cells (Fig. 1F).
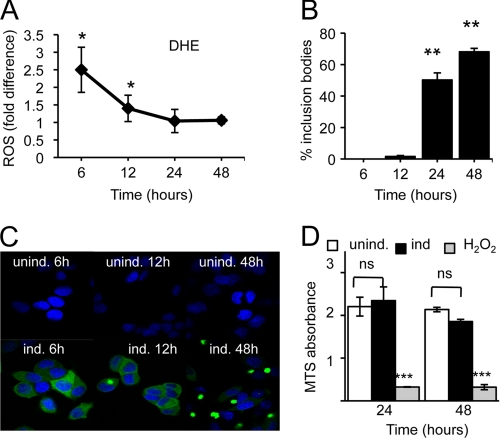
We next measured ROS and IB kinetics in a tebufenozide-inducible PC12 cell system expressing httEx1Q25- or Q103EGFP and observed again an early increase in ROS (10–24 h) that preceded cell toxicity (Fig. 1F and supplemental Fig. S3). In an uninduced state (no tebufenozide treatment) both httEx1Q25- and Q103EGFP cells showed similar levels of basal ROS levels that only increased in the httEx1Q103 cell line upon treatment with tebufenozide, excluding confounding effects due to clonal differences between these two cell lines (data not shown). l-NAC and Trolox treatment also prevented toxicity due to httEx1Q103EGFP expression in this cell system (supplemental Fig. S3). However, to verify increased ROS production due to aggregation-prone httEx1 expression independently we established a plate reader assay and performed cell population measurements. For this purpose we chose a ponasterone A httEx1Q72EGFP-inducible cell line (25) showing a rapid induction of mutant poly(Q) expression that allowed us to separate IB formation from early ROS measurements, as we suspected that oligomeric/fibrillar IB precursors could be responsible for the observed ROS elevation. A significant increase in ROS occurred at 6–12 h after transgene induction that preceded IBs in the absence of cell toxicity comparing the httEx1Q72 line in its uninduced with an induced state (Fig. 2). This early ROS induction coincided with the appearance of small oligomeric species detectable on a native gel and as SDS-insoluble material using a filter trap assay (supplemental Fig. S4). Together, these results indicate that increased ROS may be due to poly(Q) aggregation independent of the presence of IBs and toxicity.
FIGURE 2.
Increased ROS production in the absence of IB formation and toxicity in PC12 ponasterone A-inducible cells. A, -fold change in DHE oxidation due to httEx1Q72EGFP expression (induced versus uninduced). Means ± S.D. (error bars) are shown. B, percentage of cells containing one or several IBs. Means ± S.D. are shown. C, images of ponasterone A-induced versus uninduced PC12 cells showing cytoplasmic poly(Q) aggregation (IBs). D, cell toxicity (MTS absorbance) of cell populations grown under conditions as in A. H2O2 (10 mm) = positive control, two-way repeated-measure ANOVA and Bonferroni post-test, ± S.E. For A–D, n = 3 independent experiments, Student's t test (unpaired) unless otherwise stated. *, p < 0.05; **, p < 0.01; ***, p < 0.001.
ROS Production Is Poly(Q) Length-dependent and Independent of httEx1
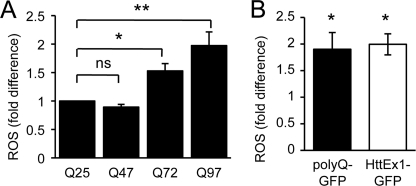
We next tested whether early ROS production was poly(Q) length-dependent given that poly(Q) aggregation kinetics strongly correlates with the length of the poly(Q) stretch. Fig. 3A shows that although no difference in ROS between httEx1Q25- and -Q47-expressing cells was detectable, there was a significant poly(Q) length-dependent increase in ROS between httEx1Q72- and -Q97-expressing cells. Expanded poly(Q) stretches alone within nondisease proteins have previously been shown to result in cell toxicity (31). Therefore we used constructs containing 19 or 81 glutamines fused to GFP (Q19/81-GFP) to investigate whether an increase in ROS could be solely due to a poly(Q) stretch. HeLa cells expressing Q81GFP produced an early increase in ROS (24 h) that was remarkably similar to cells expressing httEx1Q97EGFP (Fig. 3B) showing that ROS elevation due to httEx1Q97 is likely due to the poly(Q) stretch and not httEx1-flanking regions of the poly(Q) stretch. The aggregation kinetics (IB formation) and toxicity due to Q81-GFP compared with httEx1Q97EGFP were equivalent and increased toxicity for both constructs compared with wild-type construct expression was only detectable at 72 h after transfection (supplemental Fig. S5). Together, these results suggested a link between poly(Q) aggregation and increased ROS production.
FIGURE 3.
Mutant httEx1 ROS induction is poly(Q) length-dependent and occurs due to an expanded poly(Q) stretch only. A, DCF oxidation in HeLa cells expressing httEx1Q47-Q72 or -Q97mRFP compared with httEx1Q25mRFP (set at 1). B, ROS production due to expression of poly(Q)81-GFP constructs in HeLa cells at 24 h equivalent to that seen for httEx1Q97EGFP. For A and B, means ± S.E., n = 4 independent experiments, Student's t test (unpaired), *, p < 0.05.
Inhibition of Poly(Q) Aggregation Suppresses ROS Production
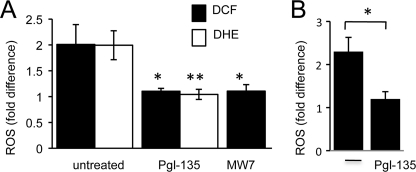
To test whether poly(Q) aggregation directly causes increased ROS we used two approaches of aggregation inhibition. HeLa cells were pretreated with Pgl-135, a benzothioazole known to reduce aggregation of httEx1 (32). Given that Pgl-135 dose-dependently suppressed httEx1Q97 IB- and SDS-insoluble protein leveling off at 50 μm (supplemental Fig. S6, A and B), HeLa cells were preincubated at this concentration with subsequent transfection with either EGFP or mRFP control and mutant httEx1 constructs, and ROS was measured after 24 h using DCF or DHE. Pgl-135 did not affect the expression levels of poly(Q) constructs (data not shown), but it suppressed httEx1Q97EGFP- or mRFP-induced ROS to levels observed in cells expressing httEx1Q25EGFP-or mRFP (Fig. 4A). Pgl-135 did not act as an antioxidant itself because it did not reduce basal ROS in httEx1Q25EGFP-expressing cells (supplemental Fig. S6C) and did not protect against oxidative stress-induced cell death due to varying concentrations of hydrogen peroxide (H2O2) (supplemental Fig. S6D). We next tested whether Pgl-135 was also able to suppress the early mutant httEx1 induced ROS in the ponasterone-inducible cell system (Fig. 2A). Plate reader assays showed that ROS was indeed significantly reduced by Pgl-135 (Fig. 4B). Pgl-135 also dose-dependently reduced IB formation in these cells (supplemental Fig. S6E). Hence, inhibition of poly(Q) aggregation using Pgl-135 inhibited the ROS increase associated with expression of poly(Q) expanded httEx1 in several cell systems.
FIGURE 4.
Poly(Q) aggregation inhibition via Pgl-135 or MW7 expression suppresses mutant httEx1-induced ROS. A, change in DCF or DHE oxidation in HeLa cells expressing httEx1Q97mRFP (DCF) or httEx1Q97EGFP (DHE) compared with httEx1Q25mRFP/EGFP with/without Pgl-135 (50 μm) treatment or co-expression of the intrabody MW7. n = 4 independent experiments, means ± S.E. (error bars). B, change in DHE oxidation in httEx1Q72EGFP ponasterone A cells induced for 6 h with/without Pgl-135 (50 μm) treatment, n = 3 independent experiments, means ± S.D. Student's t test (unpaired), *, p < 0.05; **, p < 0.01.
The second method of aggregation inhibition was obtained via co-expression of the intrabody MW7, specifically directed toward httEx1 (33). HeLa cells were transfected with the MW7 construct for 24 h followed by transfection with httEx1Q25- or Q97mRFP. Co-expression of MW7 did not affect httEx1 expression levels (data not shown), and co-expression of both constructs in single cells was confirmed using immunocytochemistry with an anti-FLAG antibody to detect FLAG-tagged MW7 (supplemental Fig. S7A). MW7 expression significantly reduced httEx1Q97mRFP aggregation as reported previously in cell models (33) (supplemental Fig. S7B). Cells co-transfected with MW7 and httEx1Q97mRFP produced almost identical amounts of ROS compared with cells co-transfected with httEx1Q25mRFP (Fig. 4A). Therefore, httEx1 poly(Q) aggregation inhibition by co-expression of MW7 abolished the ROS increase associated with httEx1Q97mRFP expression, similar to Pgl-135.
Free Radical Production Occurs Due to Poly(Q) Aggregation in Vitro
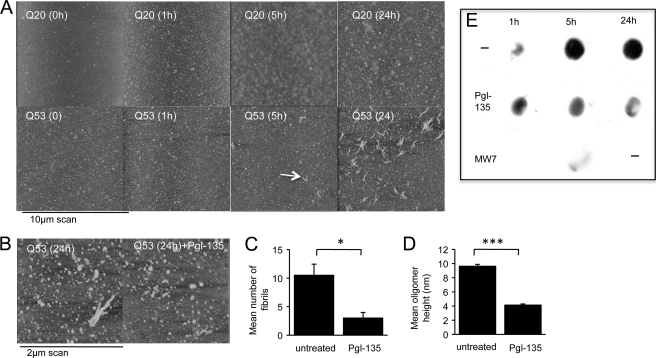
Different aggregated species of expanded poly(Q) proteins have been described, but it is unclear which of these are neurotoxic (34). To gain further insight on the potential species involved in ROS production within cells (see above) and to test whether poly(Q) aggregation itself, independent of a cellular context, could produce free radicals, we purified recombinant httEx1Q20 and -Q53 fused to GST. Cleavage of the GST tag from httEx1Q20 or -Q53 and subsequent aggregation were monitored using AFM and filter trap assays. Consistent with previous studies we found a time-dependent increase in aggregation for httEx1Q53 protein that was absent for httEx1Q20 (Fig. 5A), with oligomeric globular or annular structures and fibrils (34) (supplemental Fig. S8). We noted that some small aggregates were also present in the httEx1Q20 sample at later time points, likely due to a relatively high starting concentration enhanced by surface deposition, but at no stage were fibrils identified for httExQ20 and this aggregated material was always SDS-soluble. Given that Pgl-135 suppressed ROS in cell models (see above), it was of interest to examine whether it reduced oligomer size and fibril formation. Pgl-135 indeed reduced the appearance of fibrils, mean oligomer size, and relative numbers of small oligomers (Fig. 5, B–D, and supplemental Fig. S9) and the production of SDS-insoluble httEx1Q53 protein (Fig. 5E). Using this in vitro model of httEx1 aggregation, we next used an Amplex Red assay to measure the production of H2O2 as described previously for aggregating proteins (6). Incubation of httEx1Q53, but not -Q20, caused a rapid increase in H2O2 production after 5 h (Fig. 6A). The H2O2 formed by httEx1Q53 could be removed by addition of catalase (decomposing H2O2) before cleavage of the GST tag from httEx1Q53 (Fig. 6A). Because H2O2 production reached its maximum 5 h after httEx1Q53 cleavage when no major fibrils were present (Fig. 5A), early stages of httEx1Q53 aggregation (e.g. oligomers or protofibrils) could be responsible for H2O2 production.
FIGURE 5.
Pgl-135 reduces httEx1Q53 oligomer size and fibril formation. A, aggregation of httEx1Q53 versus httEx1Q20 analyzed by AFM. Poly(Q) fibrils are visible at 5 h. B, representative AFM images of httEx1Q53 aggregation (24 h) in the presence/absence (untreated) of Pgl-135 (50 μm). C, number of fibrillar structures/10-μm AFM scan of httEx1Q53 aggregation as in B, n = 3 independent experiments, means ± S.D. (error bars). D, httEx1Q53 oligomer size (AFM height measurements), n = 3 independent experiments, means ± S.D. E, dot blot showing that Pgl-135 (50 μm) and MW7 antibody reduce httEx1Q53 aggregation (for details, see “Experimental Procedures”). Note that MW7 24 h time point was not done.
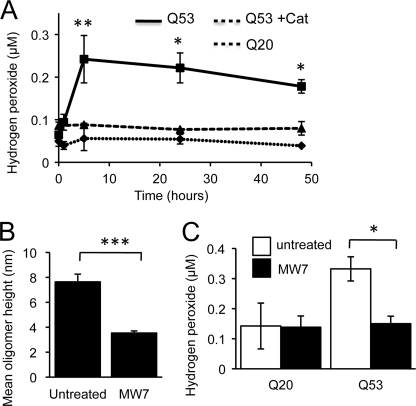
FIGURE 6.
In vitro poly(Q) aggregation is associated with hydrogen peroxide (H2O2) formation that is inhibited by the single chain antibody MW7. A, H2O2 production due to incubation of httEx1Q20/Q53. httEx1Q53 was also incubated in the presence of catalase, n = 6 independent experiments. B, addition of MW7 (1:1 molar ratio) prior to cleavage of GST from httEx1Q53 and reduction of oligomer size (5 h, AFM height measurements), n = 3 independent experiments. C, H2O2 production after 5 h of httEx1Q20/Q53 incubation with/without MW7 as in B, n = 4 independent experiments, means ± S.E. (error bars). Student's t test (unpaired), *, p < 0.05; **, p < 0.01; ***, p < 0.001.
We next asked whether inhibition of poly(Q) oligomerization was able to suppress H2O2 production. Given that the MW7 intrabody was shown to specifically inhibit httEx1 aggregation in cell models (see above; 33), MW7 antibody was added to httEx1Q20/53 reactions in a 1:1 stoichiometry before the GST tag was cleaved, and AFM samples were prepared after H2O2 levels reached its maximum (5 h). Because it was likely that early stages of aggregation caused ROS production, and only occasional fibrils were detected at this early time point (5 h), we measured mean oligomer size for httEx1Q53 reactions in parallel in the presence/absence of MW7. Fig. 6B shows that oligomer size was significantly reduced when MW7 was present. The relative number of small oligomers was similarly decreased in the presence of MW7 (supplemental Fig. S9). This finding of poly(Q) aggregation inhibition is consistent with a drastic reduction of SDS-insoluble material produced by httEx1Q53 when co-incubated with MW7 measured by the filter trap assay (Fig. 5D). httEx1Q20- and -Q53 samples were then co-incubated with MW7 and used in the Amplex Red assay to monitor H2O2 production. Although MW7 did not change basal H2O2 production measured for httEx1Q20, it reduced H2O2 in httEx1Q53 reactions to levels measured for httEx1Q20 (Fig. 6C). Therefore, reducing httEx1Q53 aggregation also suppressed httEx1Q53-mediated ROS production in vitro. Because Pgl-135 interfered with the Amplex Red assay we could not use this compound.
DISCUSSION
We have determined the relationship between intracellular protein aggregation and the production of ROS using cellular models of HD. Expansions of poly(Q) repeats led to early increased ROS coinciding with poly(Q) aggregation. Inhibition of poly(Q) aggregation suppressed ROS. We then employed recombinant httEx1 protein and monitored its aggregation and ROS production in vitro. H2O2 production peaked at 1–5 h before substantial amounts of httEx1 fibrils were formed. We propose that poly(Q) aggregation intermediates are responsible for increased H2O2 production.
This finding is consistent with in vitro experiments performed with other amyloid-forming proteins (5–8). For comparison with poly(Q) experiments we have also used Aβ1–40 and Aβ1–42 peptides and found a time-dependent production of H2O2 using Amplex Red assays correlating with the aggregation kinetics of each peptide, similar to previous reports (supplemental Fig. S10). Interestingly, Tabner and colleagues (6) showed that there is a tight correlation between the time of oligomer formation and a ROS burst during such reactions, suggesting that it is the early aggregation steps and not the production of fibrillar structures that may be associated with free radical production. We tested this idea in a poly(Q) context using a single chain antibody specifically directed toward httEx1 (MW7) that altered early in vitro poly(Q) oligomerization by decreasing oligomer size (and subsequent poly(Q) aggregation steps). MW7 suppressed H2O2 and httEx1Q53 oligomerization in parallel and at an early stage (5 h), when no significant numbers of fibrils were formed (Fig. 5A). This finding strongly indicates that it is prefibrillar poly(Q) protein that may be responsible for H2O2 production. This hypothesis is also supported by the absence of a continued rise in H2O2 production in parallel with an increase in fibril formation by httEx1Q53. The mechanisms by which aggregation-prone httEx1Q53 and other aggregation-prone peptides generate H2O2, in concert with metal ions (e.g. Cu2+), is unclear and a matter of debate (35) (see below).
Our proposition of prefibrillar poly(Q) aggregates causing ROS production is supported by our httEx1 cell assays: using several cell types and ROS assays we consistently observed increased ROS production early after intracellular expression of httEx1mRFP/EGFP with poly(Q) expansions, before the formation of IBs. In the ponasterone A-inducible cell system we detected small SDS-soluble oligomeric httEx1Q72EGFP species at 6 h after transgene induction coinciding with the ROS elevation. This suggests that soluble oligomeric species of mutant huntingtin fragments may cause increased free radical production within cells (supplemental Fig. S4). Both Pgl-135 and cellular co-expression of MW7 intrabody reduced early poly(Q) aggregation and ROS in parallel, and because both MW7 and Pgl-135 acted on early poly(Q) oligomerization steps in vitro (Figs. 5, B and D, and 6B) it is likely that both treatments also impacted on cellular poly(Q) oligomerization. Therefore, our findings in cell models and the in vitro data show a direct link between poly(Q) aggregation and ROS production.
Our approach of determining ROS levels in a defined set of single, living cells when no increased mutant httEx1 toxicity was present allowed us to dissociate ROS due to poly(Q) expression from secondary redox alterations that usually occur during later stages of apoptosis (e.g. when cytochrome c release occurs) and other forms of cell death (36, 37). Basal toxicity due to mutant and wild-type httEx1 at 24 h was similar, varying from 10 to 20% in transient transfection systems, and hence the doubling of ROS in cells expressing httEx1Q97 compared with httEx1Q25 at 24 h cannot be ascribed to differences in toxicity. This finding in HeLa cells was supported by experiments performed with PC12 cells in which httEx1 transgenes were transiently transfected or stably induced with tebufenozide. An increase in ROS after only a few hours of httExQ72EGFP induction also occurred in the ponasterone A-inducible cell model in the absence of toxicity (Fig. 2). Such early induction of ROS is consistent with a recent study in Tet-Off PC12 cells (16) and shows that ROS can be induced long before the onset of cell death. The study by Bertoni et al. (16) showed a link between ROS and downstream histone protein phosphorylation, a known marker for DNA damage (16), suggesting ROS induced DNA damage. In the HeLa cell system used in the present study we also observed that mutant httEx1 expression increased H2A.X-Ser-139 phosphorylation (supplemental Fig. S11). Interestingly, the increase in phosphorylation only occurred at 24 h (supplemental Fig. S11C), mimicking the ROS kinetics measured with oxidation sensitive molecules (Fig. 1C).
The likely cellular sources of ROS, or redox imbalances due to poly(Q)-expanded proteins, are manifold. As shown in this study, poly(Q) aggregation itself causes increased ROS. Here, it is possible that aggregating structures other than oligomers (as proposed in the present report), also contribute to abnormal ROS. IBs have been suggested to be centers of oxidative reactions (38). It has been shown that oligomeric poly(Q) species are present at IBs in vivo (39), and hence it is possible that oxidative events at IBs are also driven by oligomerization reactions, likely in concert with certain metal ions. Both copper and iron ions have been proposed to be involved in amyloid-associated in vitro H2O2 production with subsequent hydroxyl radical formation via Fenton chemistry and/or the Haber-Weiss reaction (for review, see Ref. 8). Interestingly, N-terminal htt including httEx1 has been shown to be redox-active, binding copper (40). Additionally, we have demonstrated that copper increased aggregation of httEx1 with long glutamine stretches both in vitro and in vivo (cells) (41). Therefore, it is possible that copper is involved in the httEx1-driven elevation of ROS. We do not believe that poly(Q)-induced ROS itself modulates poly(Q) aggregation in a major way in the cell systems used in the present study because both l-NAC and Trolox treatment did not alter IB formation in all cell systems (supplemental Fig. S12). However, poly(Q)-induced ROS could still affect early, oligomerization reactions together with transition metals.
Poly(Q) aggregation-independent mechanisms are also likely to play significant roles in contributing to the redox imbalance in HD including transcriptional, mitochondrial, and endosomal trafficking abnormalities. Expression of PGC-1α, a key regulator of antioxidant gene expression and mitochondrial biogenesis, has been shown to be transcriptionally dysregulated in HD (42), and both full-length and httEx1 directly interact with the outer mitochondrial membrane (43, 44). Hence, transcriptional and mitochondrial abnormalities (for example alterations of the mitochondrial fusion-fission machinery) (45) are further candidate mechanisms for involvement of redox alterations in HD. Long poly(Q) stretches have also been shown to directly alter respiration of isolated mitochondria and increase ROS (46).
Recently, Li et al. proposed that aberrant trafficking of the neuronal glutamate transporter EAAC1, which regulates uptake of cysteine required for glutathione (GSH) synthesis, leads to an increase in ROS in neurons expressing mutant full-length htt (15). This study showed that ROS accumulated in neurons expressing mutant Gln-140 full-length htt in knock-in mice due to an early GSH depletion. This proposed mechanism for redox alteration is likely different from the mechanism of increased oxidative stress proposed in our study. As pointed out by Li et al. (15), it is possible that the full-length model used in their study is representative of redox abnormalities at early stages of HD whereas httEx1 model systems represent a later stage of HD. However, it should be noted that the production of htt cleavage products, including fragment sizes consistent with httEx1, is an early event in HD (12), and hence httEx1 oligomerization could also contribute to alter the redox homeostasis in early HD. Another study on proteins with poly(Q) expansions argued that the lipid-localized NADPH oxidase may be an enzyme involved in abnormal redox homeostasis (16). This study showed that the relatively specific NADPH oxidase inhibitor apocynin was able to reduce ROS due to expression of httEx1 with 43 glutamines, and httEx1 interacted with gp91 (a NADPH oxidase subunit).
We propose that there are several independent sources of abnormal free radical production in HD. It is likely that on the one hand, alterations in transcription and trafficking events due to poly(Q) expansions result in increased susceptibility to oxidative stress via disruption of the glutathione and the antioxidant defense system. On the other hand poly(Q) aggregation itself and intracellular mechanisms disrupted by poly(Q) aggregation (e.g. mitochondrial functions) may contribute in parallel and/or sequentially to increased oxidative stress during poly(Q) pathology. Therefore, a beneficial approach for antioxidant therapy during chronic neurodegeneration associated with protein aggregation should involve the targeting of several mechanisms. In this context it will be important to explore the roles of non-cell autonomous mechanisms that could contribute to oxidative stress in HD. As both microglial and astrocytic cellular dysfunction due to expanded poly(Q) proteins have been reported (47, 48), glial cells may contribute to oxidative alterations in the HD brain. Although microglial cells are known for their role in the production of ROS, astrocytes regulate antioxidant control in the central nervous system. Whether increased ROS is induced due to intracellular aggregation of non-poly(Q)-containing proteins is an exciting avenue for future investigation.
Supplementary Material
Acknowledgments
We thank G. Bates for the S830 antibody, the Muchowski laboratory for plasmids, A. Koshnan for the MW7 construct, M. Cuttle and H. Schuppe for assistance with microscopy (Imaging and Microscopy Centre, Southampton), S. Moore and D. Allsop for advice with the Amplex Red assay, and V. O'Connor for fruitful discussions.
This work was supported by the Medical Research Council (to S. H. and A. W.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S12.
- HD
- Huntington disease
- AFM
- atomic force microscopy
- CM-H2DCF
- 5-(-6)-chloromethyl-2′,7′-dichlorodihydrofluorescein diacetate acetyl ester
- DAF-FM diacetate
- 4-amino-5-methylamino-2′,7′-difluorofluorescein diacetate
- DHE
- dihydroethidium
- EGFP
- enhanced GFP
- htt
- huntingtin
- httEx1
- htt exon 1
- IB
- inclusion body
- l-NAME
- N(G)-nitro-l-arginine methyl ester
- mRFP
- monomeric red fluorescent protein
- ROS
- reactive oxygen species
- poly(Q)
- polyglutamine.
REFERENCES
- 1. Barnham K. J., Masters C. L., Bush A. I. (2004) Nat. Rev. Drug Discov. 3, 205–214 [DOI] [PubMed] [Google Scholar]
- 2. Martínez A., Portero-Otin M., Pamplona R., Ferrer I. (2010) Brain Pathol. 20, 281–297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. D'Autréaux B., Toledano M. B. (2007) Nat. Rev. Mol. Cell Biol. 8, 813–824 [DOI] [PubMed] [Google Scholar]
- 4. Halliwell B., Gutteridge J. (2007) Free Radicals in Biology and Medicine, 4th Ed., Oxford University Press, New York [Google Scholar]
- 5. Opazo C., Huang X., Cherny R. A., Moir R. D., Roher A. E., White A. R., Cappai R., Masters C. L., Tanzi R. E., Inestrosa N. C., Bush A. I. (2002) J. Biol. Chem. 277, 40302–40308 [DOI] [PubMed] [Google Scholar]
- 6. Tabner B. J., El-Agnaf O. M., Turnbull S., German M. J., Paleologou K. E., Hayashi Y., Cooper L. J., Fullwood N. J., Allsop D. (2005) J. Biol. Chem. 280, 35789–35792 [DOI] [PubMed] [Google Scholar]
- 7. Turnbull S., Tabner B. J., Brown D. R., Allsop D. (2003) Biochemistry 42, 7675–7681 [DOI] [PubMed] [Google Scholar]
- 8. Allsop D., Mayes J., Moore S., Masad A., Tabner B. J. (2008) Biochem. Soc. Trans. 36, 1293–1298 [DOI] [PubMed] [Google Scholar]
- 9. Huang X., Cuajungco M. P., Atwood C. S., Hartshorn M. A., Tyndall J. D., Hanson G. R., Stokes K. C., Leopold M., Multhaup G., Goldstein L. E, Scarpa R. C., Saunders A. J., Lim J., Moir R. D., Glabe C., Bowden E. F., Masters C. L., Fairlie D. P., Tanzi R. E., Bush A. I. (1999) J. Biol. Chem. 274, 37111–37116 [DOI] [PubMed] [Google Scholar]
- 10. DiFiglia M., Sapp E., Chase K. O., Davies S. W., Bates G. P., Vonsattel J. P., Aronin N. (1997) Science 277, 1990–1993 [DOI] [PubMed] [Google Scholar]
- 11. Davies S. W., Turmaine M., Cozens B. A., DiFiglia M., Sharp A. H., Ross C. A., Scherzinger E., Wanker E. E., Mangiarini L., Bates G. P. (1997) Cell 90, 537–548 [DOI] [PubMed] [Google Scholar]
- 12. Landles C., Sathasivam K., Weiss A., Woodman B., Moffitt H., Finkbeiner S., Sun B., Gafni J., Ellerby L. M., Trottier Y., Richards W. G., Osmand A., Paganetti P., Bates G. P. (2010) J. Biol. Chem. 285, 8808–8823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wyttenbach A., Sauvageot O., Carmichael J., Diaz-Latoud C., Arrigo A. P., Rubinsztein D. C. (2002) Hum. Mol. Genet. 11, 1137–1151 [DOI] [PubMed] [Google Scholar]
- 14. van Roon-Mom W. M., Pepers B. A., 't Hoen P. A., Verwijmeren C. A., den Dunnen J. T., Dorsman J. C., van Ommen G. B. (2008) BMC Mol. Biol. 9, 84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Li X., Valencia A., Sapp E., Masso N., Alexander J., Reeves P., Kegel K. B., Aronin N., Difiglia M. (2010) J. Neurosci. 30, 4552–4561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bertoni A., Giuliano P., Galgani M., Rotoli D., Ulianich L., Adornetto A., Santillo M. R., Porcellini A., Avvedimento V. E. (2011) J. Biol. Chem. 286, 4727–4741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Pérez-Severiano F., Santamaría A., Pedraza-Chaverri J., Medina-Campos O. N., Ríos C., Segovia J. (2004) Neurochem. Res. 29, 729–733 [DOI] [PubMed] [Google Scholar]
- 18. Bogdanov M. B., Andreassen O. A., Dedeoglu A., Ferrante R. J., Beal M. F. (2001) J. Neurochem. 79, 1246–1249 [DOI] [PubMed] [Google Scholar]
- 19. Tabrizi S. J., Workman J., Hart P. E., Mangiarini L., Mahal A., Bates G., Cooper J. M., Schapira A. H. (2000) Ann. Neurol. 47, 80–86 [DOI] [PubMed] [Google Scholar]
- 20. Browne S. E., Bowling A. C., MacGarvey U., Baik M. J., Berger S. C., Muqit M. M., Bird E. D., Beal M. F. (1997) Ann. Neurol. 41, 646–653 [DOI] [PubMed] [Google Scholar]
- 21. Browne S. E., Ferrante R. J., Beal M. F. (1999) Brain Pathol. 9, 147–163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Polidori M. C., Mecocci P., Browne S. E., Senin U., Beal M. F. (1999) Neurosci. Lett. 272, 53–56 [DOI] [PubMed] [Google Scholar]
- 23. Tabrizi S. J., Cleeter M. W., Xuereb J., Taanman J. W., Cooper J. M., Schapira A. H. (1999) Ann. Neurol. 45, 25–32 [DOI] [PubMed] [Google Scholar]
- 24. Aiken C. T., Tobin A. J., Schweitzer E. S. (2004) Neurobiol. Dis. 16, 546–555 [DOI] [PubMed] [Google Scholar]
- 25. Apostol B. L., Kazantsev A., Raffioni S., Illes K., Pallos J., Bodai L., Slepko N., Bear J. E., Gertler F. B., Hersch S., Housman D. E., Marsh J. L., Thompson L. M. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 5950–5955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Wyttenbach A., Carmichael J., Swartz J., Furlong R. A., Narain Y., Rankin J., Rubinsztein D. C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. King M. A., Hands S., Hafiz F., Mizushima N., Tolkovsky A. M., Wyttenbach A. (2008) Mol. Pharmacol. 73, 1052–1063 [DOI] [PubMed] [Google Scholar]
- 28. Wacker J. L., Zareie M. H., Fong H., Sarikaya M., Muchowski P. J. (2004) Nat. Struct. Mol. Biol. 11, 1215–1222 [DOI] [PubMed] [Google Scholar]
- 29. Wyttenbach A., Tolkovsky A. M. (2006) J. Neurochem. 96, 1213–1226 [DOI] [PubMed] [Google Scholar]
- 30. Firdaus W. J., Wyttenbach A., Diaz-Latoud C., Currie R. W., Arrigo A. P. (2006) FEBS J. 273, 3076–3093 [DOI] [PubMed] [Google Scholar]
- 31. Marsh J. L., Walker H., Theisen H., Zhu Y. Z., Fielder T., Purcell J., Thompson L. M. (2000) Hum. Mol. Genet. 9, 13–25 [DOI] [PubMed] [Google Scholar]
- 32. Hockly E., Tse J., Barker A. L., Moolman D. L., Beunard J. L., Revington A. P., Holt K., Sunshine S., Moffitt H., Sathasivam K., Woodman B., Wanker E. E., Lowden P. A., Bates G. P. (2006) Neurobiol. Dis. 21, 228–236 [DOI] [PubMed] [Google Scholar]
- 33. Khoshnan A., Ko J., Patterson P. H. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hands S. L., Wyttenbach A. (2010) Acta Neuropathol. 120, 419–437 [DOI] [PubMed] [Google Scholar]
- 35. Nadal R. C., Rigby S. E., Viles J. H. (2008) Biochemistry 47, 11653–11664 [DOI] [PubMed] [Google Scholar]
- 36. Kirkland R. A., Franklin J. L. (2001) J. Neurosci. 21, 1949–1963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Orrenius S. (2007) Drug Metab. Rev. 39, 443–455 [DOI] [PubMed] [Google Scholar]
- 38. Firdaus W. J., Wyttenbach A., Giuliano P., Kretz-Remy C., Currie R. W., Arrigo A. P. (2006) FEBS J. 273, 5428–5441 [DOI] [PubMed] [Google Scholar]
- 39. Legleiter J., Mitchell E., Lotz G. P., Sapp E., Ng C., DiFiglia M., Thompson L. M., Muchowski P. J. (2010) J. Biol. Chem. 285, 14777–14790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Fox J. H., Kama J. A., Lieberman G., Chopra R., Dorsey K., Chopra V., Volitakis I., Cherny R. A., Bush A. I., Hersch S. (2007) PLoS One 2, e334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hands S. L., Mason R., Sajjad M. U., Giorgini F., Wyttenbach A. (2010) Biochem. Soc. Trans. 38, 552–558 [DOI] [PubMed] [Google Scholar]
- 42. Cui L., Jeong H., Borovecki F., Parkhurst C. N., Tanese N., Krainc D. (2006) Cell 127, 59–69 [DOI] [PubMed] [Google Scholar]
- 43. Choo Y. S., Johnson G. V., MacDonald M., Detloff P. J., Lesort M. (2004) Hum. Mol. Genet. 13, 1407–1420 [DOI] [PubMed] [Google Scholar]
- 44. Panov A. V., Gutekunst C. A., Leavitt B. R., Hayden M. R., Burke J. R., Strittmatter W. J., Greenamyre J. T. (2002) Nat. Neurosci. 5, 731–736 [DOI] [PubMed] [Google Scholar]
- 45. Song W., Chen J., Petrilli A., Liot G., Klinglmayr E., Zhou Y., Poquiz P., Tjong J., Pouladi M. A., Hayden M. R., Masliah E., Ellisman M., Rouiller I., Schwarzenbacher R., Bossy B., Perkins G., Bossy-Wetzel E. (2011) Nat. Med. 17, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Puranam K. L., Wu G., Strittmatter W. J., Burke J. R. (2006) Biochem. Biophys. Res. Commun. 341, 607–613 [DOI] [PubMed] [Google Scholar]
- 47. Bradford J., Shin J. Y., Roberts M., Wang C. E., Li X. J., Li S. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 22480–22485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Björkqvist M., Wild E. J., Thiele J., Silvestroni A., Andre R., Lahiri N., Raibon E., Lee R. V., Benn C. L., Soulet D., Magnusson A., Woodman B., Landles C., Pouladi M. A., Hayden M. R., Khalili-Shirazi A., Lowdell M. W., Brundin P., Bates G. P., Leavitt B. R., Möller T., Tabrizi S. J. (2008) J. Exp. Med. 205, 1869–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.