Background: Transferrin receptors are critical for survival of important Gram-negative bacterial pathogens.
Results: The anchoring peptide of TbpB mediates interaction with TbpA.
Conclusion: The anchor peptide mediates the process by which TbpB captures transferrin and delivers it to TbpA.
Significance: TbpB is critical for survival of important bacterial pathogens and this study provides insights to its role in acquiring iron.
Keywords: Bacteria, Iron, Membrane, Receptors, Transport Metals, Gram-negative, Outer Membrane, Pathogen, Surface Receptor, Transferrin
Abstract
Gram-negative bacterial pathogens belonging to the Pasteurellaceae, Moraxellaceae, and Neisseriaceae families rely on an iron acquisition system that acquires iron directly from host transferrin (Tf). The process is mediated by a surface receptor composed of transferrin-binding proteins A and B (TbpA and TbpB). TbpA is an integral outer membrane protein that functions as a gated channel for the passage of iron into the periplasm. TbpB is a surface-exposed lipoprotein that facilitates the iron uptake process. In this study, we demonstrate that the region encompassing amino acids 7–40 of Actinobacillus pleuropneumoniae TbpB is required for forming a complex with TbpA and that the formation of the complex requires the presence of porcine Tf. These results are consistent with a model in which TbpB is responsible for the initial capture of iron-loaded Tf and subsequently interacts with TbpA through the anchor peptide. We propose that TonB binding to TbpA initiates the formation of the TbpB-TbpA complex and transfer of Tf to TbpA.
Introduction
Gram-negative pathogenic bacteria in the Neisseriaceae, Moraxellaceae, and Pasteurellaceae families are major causes of important infections of humans (meningitis, pneumonia, otitis media, gonorrhea) and food production animals (shipping fever in cattle, pneumonia in pigs). They primarily reside in the respiratory and genitourinary tract of their hosts and rely on a specialized iron uptake system that acquires iron from serum transferrin (Tf).2 The surface receptor responsible for capturing Tf is highly specific for host Tf (1, 2), limiting the host range of these bacterial species. The bacterial Tf receptor is composed of a surface-exposed lipoprotein, Tf-binding protein B (TbpB), and a TonB-dependent integral outer membrane protein, TbpA (3–6). The ability of strains deficient in either TbpA or TbpB to bind Tf indicates that both receptor proteins are capable of binding to Tf, and specifically, to the C-terminal lobe of Tf (7). The interaction of Tf with the surface receptor results in removal and transport of iron across the outer membrane using energy provided by the TonB complex (8). TbpB-deficient bacteria are capable of growth on Tf (4, 6, 9), indicating that TbpA is capable of mediating the entire process of binding Tf, iron removal, and transport of iron across the outer membrane under laboratory growth conditions.
Although growth on Tf as a sole iron source is possible in the absence of TbpB, there are varying degrees of growth impairment of TbpB-deficient strains in vitro with Tf as the sole source of iron (4–6, 9, 10). Studies with a TbpB-deficient strain of the porcine pathogen Actinobacillus pleuropneumoniae demonstrated that although growth with porcine Tf as the iron source was possible in vitro, the strain was avirulent and unable to colonize the host, suggesting that TbpB is essential for iron acquisition in vivo (10). It has been proposed that TbpB may play a role in the efficient capture of the iron-loaded form of Tf (11) because TbpB, but not TbpA, has a strong preference for binding to the iron-loaded form of Tf (12, 13).
The relative susceptibility of the receptor proteins in wild-type and mutant gonococci to protease treatment has led to the conclusion that TbpA and TbpB interact in the outer membrane (14). The protection from proteolysis of TbpB by TbpA is dependent upon interaction with TonB (15), suggesting that the TbpB-TbpA interaction is linked to the iron acquisition process. Similarly, the conclusion that TbpA and TbpB “work together” during the association phase of binding radiolabeled Tf (16) implies that there is an intimate association between the receptors during the Tf binding step. However, experiments with gold-labeled antibodies in electron micrograph experiments demonstrated that TbpA and TbpB are not co-localized in the absence of added Tf (17), suggesting that they do not normally constitute a preformed receptor complex in the membrane. Thus the physical association of the receptor proteins may largely be limited to receptors actively involved in the iron acquisition process.
Recent structural studies with TbpB demonstrate that the C-terminal and N-terminal lobes have similar structural features and that the relatively unstructured N-terminal anchor peptide is associated with the C-terminal lobe. These studies have also revealed that the binding interface for Tf on TbpB is localized to a “Cap region” of the N-terminal lobe, on the opposite surface from the start of the anchor peptide region (18, 19). There are no indications of Tf interacting with the TbpB C-terminal lobe region (20, 21). Because the anchor peptide protrudes from the opposite side of the binding interface on the TbpB N-terminal lobe and associates with the C-terminal lobe, it is unlikely to directly participate in interactions with Tf (18, 19). However, the location of the anchor peptide between the Tf-binding N-lobe and the surface of the outer membrane suggests that it might play a role in modulating the position of the TbpB N-lobe relative to the outer membrane surface.
There is a large domain movement observed in structures of the holo (iron-loaded) and apo (iron-free) forms of Tf (22), from a “closed” to an “open” conformation. The interaction of TbpB with holoTf involves regions on both the C1 and the C2 domains of the C-lobe (20, 21) maintaining Tf in the closed conformation, thus limiting the release of iron (20, 23). It is unclear how TbpA is capable of binding both the holo and the apo forms of Tf (12), but it might suggest that TbpA binds both forms of Tf in an intermediate conformation. The proposal that iron removal involves separation of the two Tf domains (C1 and C2) that flank the iron-binding site (24) implies that TbpA binds to both domains. However, the only direct evidence for this is with a TbpA homologue, lactoferrin-binding protein A (25). Thus there are many unanswered questions regarding the role of TbpB and the process by which TbpB delivers Tf to TbpA. This study was initiated to investigate the TbpB-TbpA interaction and gain further insights into the role of TbpB in the iron acquisition process.
EXPERIMENTAL PROCEDURES
Production of Transferrins
Commercial preparations of porcine transferrin (pTf) were deglycosylated with a set of recombinant deglycosylation enzymes essentially as described previously (21). Recombinant non-glycosylated pTf was produced by Pichia pastoris using the pPICZα A vector from Invitrogen. The mutant pTf gene with an N156D mutation to eliminate the asparagine involved in glycosylation was amplified with a forward primer containing the Kex2 site immediately preceding the amino-terminal amino acids of the mature protein. The recombinant non-glycosylated pTf was produced in P. pastoris culture supernatants with minor modifications of the methods described in the user manual (pPICZα, Invitrogen). Non-glycosylated pTf was precipitated from culture supernatants by ammonium sulfate and then purified by anion-exchange chromatography.
Production of Recombinant Tbp Proteins
The recombinant TbpB proteins and other anchor peptide-containing proteins and polypeptides were produced by modifications of the expression vector system used previously for production of TbpB and the N-lobe subfragment from A. pleuropneumoniae (18). All of the expression constructs included a BamHI restriction site after the tobacco etch virus (TEV) cleavage site so that the resulting proteins all had N-terminal glycine and serine residues. Thus recombinant proteins with an intact anchor peptide (2–40) had the N-terminal sequence GSSGGK (see Fig. 1), and the tagged versions had the sequence GSHHHHHHHHSGGK. The N-terminal truncations of the anchor peptide had the sequences GSFDLE (8–528), GSDLE (9–528), GSE (11–528), and so on. The untagged versions of the proteins were preceded by a polyhistidine-tagged maltose-binding protein (Mbp) and a TEV cleavage site. The tagged versions of the proteins were preceded by untagged Mbp and a TEV cleavage site. The recombinant proteins could thus be isolated by a combination of Ni-NTA chromatography, TEV cleavage, and a second round of Ni-NTA chromatography.
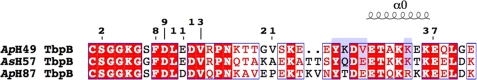
FIGURE 1.
Anchor peptide alignment of ApH49, AsH57, and ApH87 TbpBs. Identical residues and conserved residues are highlighted and boxed in red. The anchor peptide secondary structure element from ApH87 TbpB (Protein Data Bank (PDB) entry 3PQS) is shown above the sequence. Amino acids from ApH49 and AsH57 TbpBs (PDB entry 3HOL and 3PQU) that interact with the TbpB C-lobe are highlighted in blue. Numbering above the sequence illustrates the sites of N-terminal truncation of recombinant ApH49 TbpB constructs.
Recombinant TbpA from A. pleuropneumoniae strain H49 was produced using modified pET-20b plasmids (Novagen) with a BamHI restriction site immediately after a pelB signal sequence, N-terminal polyhistidine tag, and TEV cleavage site. The recombinant plasmids were transformed into Escherichia coli strain C41 (DE3) (26), and the resulting colonies were tested for expression by solid-phase binding assays with HRP-conjugated Tf or affinity capture assays with Tf-Sepharose (27).
For large scale production, 20 ml of an overnight culture was used to inoculate 1.5 liters of Terrific Broth (Sigma) supplemented with ampicillin (100 μg/ml). After 4 h, the culture was induced with 0.5 mm isopropyl-1-thio-β-d-galactopyranoside (Invitrogen) and harvested by centrifugation after a 20-h incubation. The cells were resuspended and washed with cold 50 mm Tris-HCl, 1 m NaCl, pH 8.0, buffer, resuspended in one-fifth volume of buffer containing 2% (v/v) ELUGENT (Calbiochem), and mixed thoroughly for 18 h at 4 °C. The bacterial debris was removed by centrifugation at 10,000 × g for 1 h, and the supernatant was added to Ni-NTA-agarose (Qiagen) resin (25 ml supernatant/ml resin) and mixed overnight at 4 °C. The resin was collected in a chromatography column (Bio-Rad) and washed with 10 column volumes of buffer containing 0.25% ELUGENT and 20 mm imidazole, and the bound protein was eluted with buffer containing 250 mm imidazole. The fractions containing recombinant tagged TbpA were pooled and dialyzed against 10 mm HEPES, pH 8.5, 100 mm NaCl buffer (containing 0.25% ELUGENT) and applied to a 20-ml Q-Sepharose column equilibrated with the same buffer. The flow-through fractions were collected, stored at 4 °C, and analyzed by SDS-PAGE.
Non-tagged versions of recombinant TbpB and TbpA were obtained by digestion with a recombinant His-tagged TEV enzyme (10 units/mg of recombinant TbpB, 20 units/mg of recombinant TbpA) at room temperature overnight. Recombinant TbpB was digested in 50 mm Tris-HCl, 0.5 mm EDTA, pH 8, buffer, and recombinant TbpA was digested in 50 mm Tris-HCl, 1 m NaCl, 0.25% ELUGENT. Non-tagged TbpA obtained in the flow-through of an Ni-NTA column and non-tagged TbpB obtained from Q-Sepharose chromatography were concentrated by ultrafiltration.
Affinity Capture Experiments
Capture of TbpA by Anchor Peptide-containing Proteins and Polypeptides
His-tagged versions of TbpB, TbpB derivatives, or other anchor peptide-containing polypeptides were applied to an Ni-NTA resin and incubated overnight at 4 °C (see Figs. 3 and 6). The mixtures contained a ratio of 200 μg of protein per 20 μl of Ni-NTA resin in 100 μl of 300 mm NaCl, 5 mm imidazole, 50 mm NaH2PO4, pH 8.0, buffer. The resin was collected by centrifugation, washed three times, and resuspended in buffer containing pTf (300 μg/20 μl of resin). The mixture was incubated for another 16 h, collected by centrifugation, and washed three times. 100 μl of a preparation of non-tagged TbpA or control buffer (50 mm Tris-HCl, 200 mm NaCl, pH 8.5, 0.25% ELUGENT) was added to the resin (800 μg of TbpA/20 μl of resin), and the mixture was incubated overnight 4 °C. The mixtures were centrifuged and washed three times, and SDS-PAGE sample buffer was added to the pellets to elute bound protein (60 μl of buffer/20 μl of resin). The resin was centrifuged, and 20 μl of the supernatant was applied to a 10% SDS-PAGE for analysis.
FIGURE 3.
The intact anchor peptide is required for capture of TbpA by a TbpB-Tf complex. Preparations of His-tagged TbpB from A. pleuropneumoniae containing the intact anchor peptide (left panel) or a truncated anchor peptide (−36TbpB) were mixed with excess non-glycosylated pTf and captured on an Ni-NTA resin, washed to remove excess pTf, and resuspended in detergent-containing buffer. Additional buffer with or without an excess of TbpA (+TbpA) was added to the resin and incubated overnight. The resins were collected by centrifugation and washed twice, and the final pellet was resuspended in SDS-PAGE buffer to elute bound proteins. The samples were applied to an SDS-PAGE gel and stained with Coomassie Blue protein stain.
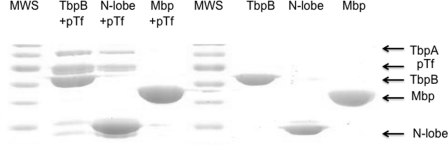
FIGURE 6.
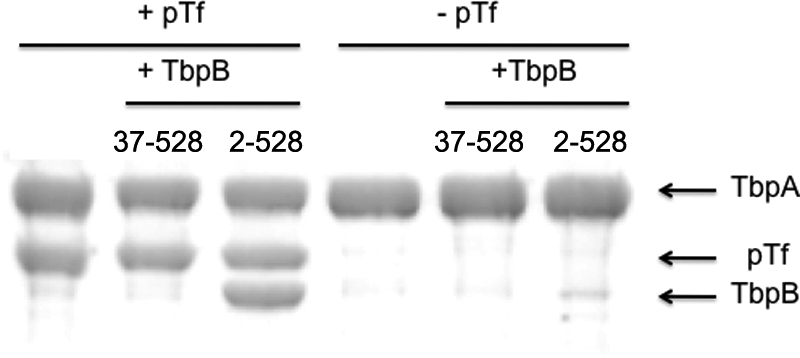
The anchor peptide is not sufficient for binding to TbpA. Preparations of His-tagged anchor peptide (amino acids 2–40) fused to intact TbpB, TbpB N-lobe, or Mbp were captured on an Ni-NTA resin. Holo (iron-loaded) pTf was added to one set of samples (+pTf) prior to the addition of untagged TbpA. After incubation overnight, the resins were collected by centrifugation and washed twice, and the final pellet was resuspended in SDS-PAGE buffer to elute bound proteins. The samples were applied to an SDS-PAGE gel and stained with Coomassie Blue protein stain. MWS, molecular weight standard.
Capture of TbpB by Tagged TbpA
1 ml of a preparation of purified, polyhistidine-tagged TbpA (200 μg) or control buffer was mixed with 20 μl of Ni-NTA resin and incubated overnight at 4 °C (see Figs. 4 and 5). Excess recombinant pTf (300 μg) was either included in the initial incubation mixture or added to the resin after incubation, washing, and resuspension in buffer for a second overnight incubation. The buffer contained 200 mm NaCl and 0.25% ELUGENT and either 50 mm Tris-HCl, pH 8 (see Fig. 4) or 10 mm HEPES, pH 8.5 (see Fig. 5). After centrifugation and washing, the resins were suspended in 1 ml of buffer containing 200 μg of recombinant, TEV-cleaved TbpB preparations and incubated overnight at 4 °C. The resin was collected by centrifugation and washed twice in buffer (containing 1 m NaCl, 20 mm imidazole, and 0.25% ELUGENT), and SDS-PAGE sample buffer was added to the pellets to elute bound protein (60 μl of buffer/20 μl of resin). After centrifugation, 20 μl of the supernatant was applied to a 10% SDS-PAGE gel for analysis.
FIGURE 4.
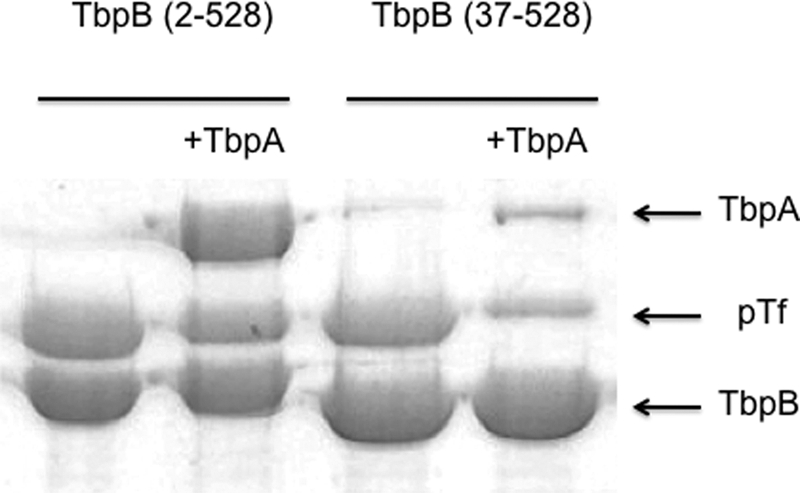
The intact anchor peptide is required for binding of TbpB to a TbpA-Tf complex. Purified, polyhistidine-tagged, recombinant TbpA from A. pleuropneumoniae strain H49 was mixed with recombinant porcine Tf (left panel) or detergent-containing buffer alone (right panel) and incubated overnight at 4 °C. Buffer without (lanes 1 and 4) or with TbpB with a truncated (37–528) or intact (2–528) N-terminal anchor peptide region was added, and the mixtures were incubated overnight at 4 °C. Ni-NTA resin was added to the incubation mixtures, collected by centrifugation, and washed twice prior to the addition of SDS-PAGE buffer to elute bound proteins.
FIGURE 5.
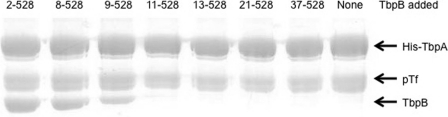
Defining the region of the anchor peptide required for binding to TbpA. Purified, polyhistidine-tagged, recombinant TbpA from A. pleuropneumoniae strain H49 was mixed with an Ni-NTA resin, and after incubation and washing, recombinant pTf was captured by the immobilized TbpA. The washed resin containing captured TbpA-pTf complex was mixed with different N-terminal truncations of TbpB. After incubation and washing, SDS-PAGE buffer was added to the resin, and the eluted proteins were analyzed by SDS-PAGE and detected by Coomassie Blue stain.
Surface Plasmon Resonance
Surface plasmon resonance experiments were performed using Biacore X (GE Healthcare) at 25 °C. Porcine transferrin was coupled to the sensor chip (CM5 research grade) via standard N-hydroxysuccinimide and N-ethyl-N-(dimethylaminopropyl) carbodiimide activation. TbpA and TbpB were diluted at various concentrations with their respective mobile phase buffer (TbpA: 20 mm Hepes, pH 7.3, 100 mm NaCl, and 0.5% C8E4 as detergent; TbpB: 10 mm Hepes, pH 7.5, 150 mm NaCl, and 0.005% surfactant P20). Samples were injected as analytes at a flow rate of 20 μl/min, and bound analytes were subsequently removed by washing with the mobile phase at 300 and 240 s after the injection of TbpA or TbpB, respectively. In the case of the TbpA experiments, the TbpA regeneration buffer (20 mm Hepes, pH 7.5, 0.5% SDS) was injected prior to each analyte injection. Kinetic constants were calculated from the sensorgrams using the simulated Biacore X evaluation software, version 4.0.1 (Biacore). The TbpA data were fitted to a 1:1 Langmuir binding model, and the TbpB data were fitted to a “heterogeneous ligand” binding model.
RESULTS
Recombinant Tbp Production
The strategy for evaluating the interaction between TbpA and TbpB was to generate tagged and non-tagged versions of each of the receptor proteins and perform affinity capture experiments to evaluate the interaction. Recombinant proteins were engineered with an N-terminal polyhistidine tag to facilitate purification. A TEV protease site was included to remove the tag and provide a non-tagged variant of the protein. The inclusion of an N-terminal Mbp fusion partner had been shown to enhance cytoplasmic expression of recombinant TbpB (18) and was incorporated into the design of the custom expression vector for TbpBs. However, to provide a polyhistidine-tagged form of TbpB without an N-terminal Mbp fusion partner, another version of the custom expression vector was designed to incorporate a polyhistidine region after the N-terminal Mbp and TEV cleavage site.
The expression vector for TbpA encoded a pelB leader peptide for export followed by the polyhistidine and TEV regions but did not encode Mbp. Preliminary analysis of expression indicated that the highest levels were achieved with TbpA from A. pleuropneumoniae, and considering the prior success with producing and crystallizing TbpBs from this species (18, 19), we decided to focus our studies on receptor proteins from this species.
To localize interaction sites with TbpA to different regions of TbpB, a variety of different derivatives of the intact TbpBs were prepared. This included a series of truncations of the N-terminal anchor peptide region (Fig. 1). For cytoplasmic expression, the N-terminal cysteine that contains covalently attached fatty acyl chains in the native protein was eliminated and replaced by a glycine and a serine residue encoded by the BamHI restriction site used for cloning. Thus these two amino acids precede all of the truncation sites.
Purified forms of recombinant TbpA expressed in the outer membrane of E. coli could be obtained by directly extracting the proteins from intact cells with buffers containing the detergents Sarkosyl or ELUGENT and 1 m NaCl followed by metal chelate chromatography and anion-exchange chromatography. Sarkosyl was shown to reduce the efficiency of the affinity capture of recombinant TbpBs; thus ELUGENT was used in all the affinity capture experiments illustrated in this study.
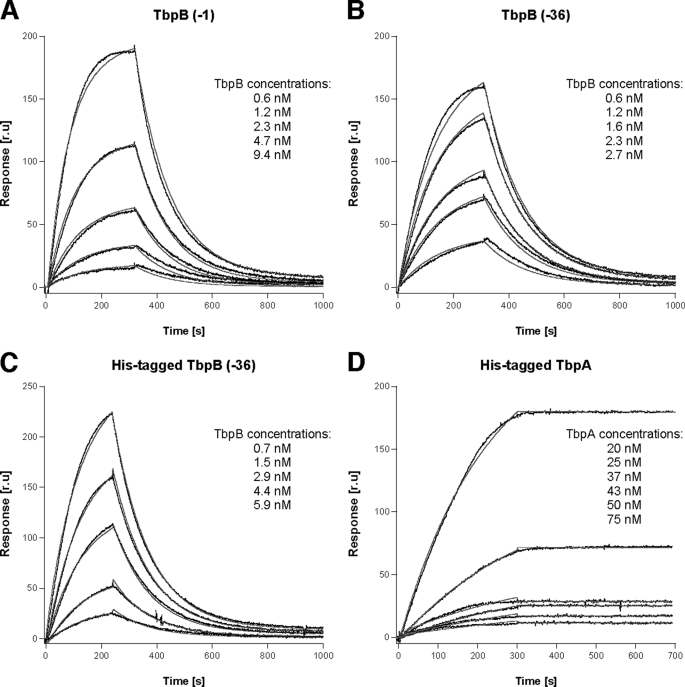
To evaluate the Tf binding properties of the recombinant proteins and determine the effect of the polyhistidine and anchor peptide regions on TbpB function, we performed surface plasmon resonance (SPR) studies using immobilized pTf. Iron-loaded pTf was covalently coupled to the chips, and TbpA or TbpB with an intact anchor peptide (amino acids 2–528) or a truncated anchor peptide (amino acids 37–528) was injected at various concentrations with the mobile phase buffer. Data from the relevant sensorgrams (Fig. 2) were fitted with an appropriate binding model. The resulting affinity constants are listed in Table 1.
FIGURE 2.
Kinetics of binding of TbpA and TbpB to immobilized pTf. A–D, the binding curves with increasing concentrations of TbpB (−1) (A), TbpB (−36) (B), His-tagged TbpB (−36) (C), and His-tagged TbpA (D) with pTf immobilized on an SPR biosensor as a function of time are illustrated. Black lines show the experimental data, and the red line shows fitting data. Concentrations of each Tbp are indicated on the respective sensorgrams. The calculated kinetic constants are listed in Table 1. r. u, response units.
TABLE 1.
Rate and affinity constants for binding of pTf by TbpA and TbpB
The rate and dissociation constants were calculated from the recorded sensorgrams for the binding of TbpA and TbpB to immobilized pTf on SPR biosensor. polyhis, polyhistidine; aa, amino acids.
| Protein | kon | koff | Kd |
|---|---|---|---|
| m−1s−1 | s−1 | ||
| ApH49 TbpB (aa 2–528) | 5.21 × 105 | 7.97 × 10−3 | 15.3 ± 0.1 nm |
| ApH49 TbpB (aa 37–528) | 3.75 × 105 | 6.87 × 10−3 | 18.3 ± 0.5 nm |
| ApH49 polyhisTbpB (aa 37–528) | 1.75 × 105 | 7.7 × 10−3 | 44 ± 1 nm |
| ApH49 polyhisTbpA | 5.96 × 104 | 7.84 × 10−6 | 0.13 ± 0.04 nm |
The calculated affinity constants for TbpA, −1 TbpB (amino acids 2–528), and −36 TbpB (amino acids 37–528) of 0.13, 15.3, and 18.3 nm, respectively, confirm that TbpA has a greater affinity for binding iron-loaded Tf than TbpB. The results also demonstrate that TbpB with the intact anchor peptide does not have an increased affinity for binding Tf, consistent with the expectation that the anchor peptide would not directly interact with Tf. Similarly, the inclusion of a polyhistidine region on the N terminus has minimal impact on binding of TbpB to Tf. The TbpA-Tf complex is surprisingly stable as 50% ethylene glycol or high salt (4 m NaCl) added to a non-ionic detergent (0.06% n-dodecyl β-d-maltoside) was unable to disrupt the TbpA-Tf complex. The full regeneration of the covalently coupled Tf required injection of a minimum content of 0.5% SDS (25 μl) in the mobile phase, which did not result in any loss in the binding capacity of the chip.
Anchor Peptide Is Required for TbpB-TbpA Interaction
In an attempt to emulate the in vivo situation, a polyhistidine-tagged form of TbpB was exposed to porcine Tf and then used to capture an untagged TbpA. When a form of recombinant TbpB containing a nearly complete anchor peptide (amino acids 2–528) was used, it was capable of capturing an untagged form of recombinant TbpA (Fig. 3, left panel). In contrast, when the experiment was performed with recombinant TbpB lacking the first 36 amino acids of the anchor peptide region (amino acids 37–528), little or no TbpA was captured by the preformed complex (right panel). The small amounts of TbpA and pTf observed in this figure may be due to incomplete washing. Also notable in this experiment is that the addition of untagged TbpA to the anchor peptide-deficient TbpB complexed to pTf resulted in removal of pTf from the immobilized complex (note the lower intensity of the pTf band in the +TbpA lane). This is perhaps not surprising considering that the affinity of TbpA for pTf is considerably greater than the affinity of TbpB for pTf (Fig. 2 and Table 1).
To further investigate the TbpA-TbpB interaction, a reciprocal experiment was performed in which a tagged form of TbpA was tested for the ability to capture non-tagged forms of TbpB. When iron-loaded porcine Tf was mixed with the polyhistidine-tagged form of TbpA from A. pleuropneumoniae and the resulting complex was exposed to the untagged intact TbpB (amino acids 2–528), it effectively captured this form of TbpB (Fig. 4, left panel). In contrast, when the experiment was performed with recombinant TbpB containing a truncated anchor peptide (amino acids 37–528), it was not associated with the immobilized complex. When the experiment was repeated in the absence of pTf (Fig. 4, right panel), neither form of TbpB was captured by the immobilized form of TbpA.
Region of Anchor Peptide Required for Binding to TbpA
The results in Fig. 4 clearly illustrate that a truncated form of TbpB lacking nearly the entire anchor peptide region is defective in binding to TbpA. A series of N-terminal truncations of TbpB from A. pleuropneumoniae strain H49 (Fig. 1) was prepared to determine what portion of the anchor peptide region is involved in binding to TbpA. We postulated that the six amino acids after the N-terminal cysteine (SGGKGS) likely serve as a flexible arm not involved in binding to TbpA; thus the first truncation started with the eight residue, phenylalanine. The series of truncations was focused on the region immediately after Phe-8 in anticipation that if secondary structural elements were important, this region may be important. The series of truncated proteins was mixed with a preformed TbpA-pTf complex to evaluate their ability to be captured by the complex (Fig. 5). The results illustrate that after removal of the first seven amino acids (including the N-terminal cysteine), there was relatively little impact on the capture of TbpB by the TbpA-pTf complex. However, removal of the subsequent phenylalanine (9–528) reduces the ability of the TbpA-pTf complex to capture the recombinant TbpB, and the removal of the following glutamic acid and valine residues (11–528) results in the complete loss in binding to the complex. The dramatic loss in TbpA binding by removal of these conserved residues suggests that either they are key residues involved in binding to TbpA or they are essential for the formation of secondary structural features required for the interaction.
Anchor Peptide Is Not Sufficient for Binding to TbpA
The experiments illustrated in Figs. 3–5 do not conclusively demonstrate that the anchor peptide is directly involved in binding to TbpA, only that a nearly intact (amino acids 7–40) anchor peptide region is required for binding. To more completely address what elements of TbpB are involved in the interaction with TbpA, a series of recombinant proteins with intact anchor peptide was prepared. These include the TbpB N-lobe (lacking the C-terminal lobe) and a hybrid protein consisting of the N-terminal intact anchor peptide fused to maltose-binding protein. A polyhistidine-tagged form of the intact anchor peptide was also produced.
The tagged versions of the anchor peptide (amino acids 2–40) and the anchor peptide-containing proteins were used to capture untagged TbpA. Porcine Tf was first applied to the resins to saturate the intact TbpB and TbpB N-lobe preparations. Both the intact TbpB and the TbpB N-lobe subfragment with added pTf effectively captured the TbpA complex (Fig. 6), indicating that the TbpB C-lobe is not required for interaction with TbpA. The hybrid protein with the anchor peptide fused to Mbp did not capture the TbpA-pTf. Many different permutations of this type of experiment were performed with these proteins and with the isolated anchor peptide, including capture with tagged TbpA complexed with pTf, but we were unable to demonstrate any binding to TbpA by the isolated anchor peptide. Similarly, in various permutations of affinity capture experiments, we were unable to demonstrate binding of the anchor peptide to pTf. Collectively, these results demonstrate that the anchor peptide is not capable of binding directly to pTf or TbpA and can only mediate binding of TbpB or the TbpB N-lobe to a complex of TbpA and pTf. These results seem to indicate either that a portion of the TbpB N-lobe is involved in the interaction with TbpA or that the conformation of anchor peptide required for interaction with TbpA is influenced by the presence of the N-lobe.
DISCUSSION
The Tf receptors present in Gram-negative species in the Neisseriaceae, Pasteurellaceae, and Moraxellaceae families play a critical role in the survival of these bacteria in the iron-restricted environment of their vertebrate hosts. The original isolation of two Tf receptor proteins by affinity capture experiments (28) and the early experiments demonstrating that both proteins were required for growth on Tf as an iron source (5) led to an initial view that they formed a functional receptor complex responsible for binding Tf at the cell surface and removing iron that was transported into the cell. However, the subsequent demonstration that strains lacking the TbpB lipoprotein component were capable of growth on Tf as a sole iron source (6, 9, 29) indicated that TbpB was not essential for the iron acquisition process and begged the question, “What is the role of TbpB?”
The invariant presence of functional tbpB genes in clinical disease isolates from human and veterinary pathogens strongly suggests that TbpB plays a critical role for survival in vivo, which seems inconsistent with the ability of TbpB−ve strains to grow on transferrin in laboratory experiments. Experiments with the pig pathogen A. pleuropneumoniae (10) demonstrated that an isogenic mutant of TbpB capable of growth on exogenous Tf in the laboratory was avirulent in a pig infection model, suggesting that TbpB was required for iron acquisition under the more demanding conditions in vivo. However, it is unclear what characteristics of growth in vivo are responsible for the TbpB requirement, and currently, there are no in vitro growth systems that emulate the stringent requirements of growth in the host.
The strong preference of TbpB for binding the iron-loaded form of Tf (12, 13) and the potential for an unstructured anchor peptide to extend far from the surface of the outer membrane (18) suggest that the primary role of TbpB many be in the initial capture of iron-loaded Tf (19) (Fig. 7). Even with the C-terminal portion of the anchor peptide wrapped around the C-terminal lobe, the Tf-binding cap region of the N-terminal lobe could be positioned >60 Å from the outer membrane surface. The observation that maximal binding of labeled Tf by intact cells was attained at the earliest measured time point in a TbpB+ve, TbpA−ve strain, but not in a TbpB−ve, TbpA+ve strain (16), is consistent with a model in which TbpB is fully accessible at the cell surface. Because the intact anchor peptide does not mediate binding of TbpB to TbpA in the absence of Tf (Fig. 4), it is likely that TbpB is not part of a preformed receptor complex at this stage, consistent with observations made in prior EM studies (17).
FIGURE 7.
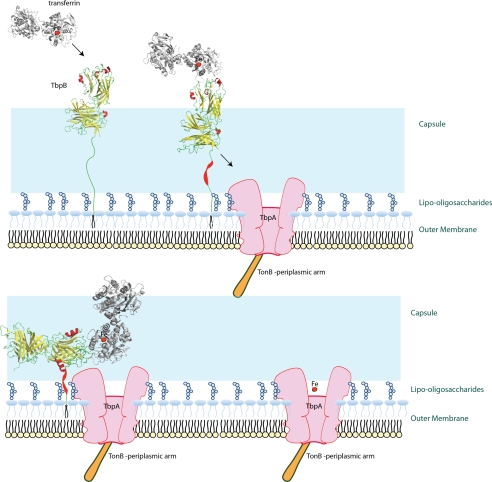
Model for capture of Tf by TbpB. Iron-loaded (holo) Tf is captured by TbpB with the N-terminal anchor region in a fully extended conformation. This places TbpB beyond the lipo oligosaccharide side chains and the polysaccharide capsule. The binding of Tf by the N-terminal lobe of TbpB displaces the anchor peptide from its interaction with the C-terminal lobe and results in conformational changes in the anchor peptide that bring the TbpB-Tf complex closer to the surface of the outer membrane and capable of interacting with “activated” TbpA (bound to TonB). Tf released by TbpB is readily captured by the activated TbpA so that the iron removal and transport process can proceed, resulting in release of apoTf.
Although TbpB with an extended anchor peptide might be optimally localized for the capture of iron-loaded Tf at the cell surface, it would not be ideally positioned for transfer of Tf to TbpA. Thus it is tempting to speculate that binding of Tf to TbpB at the cell surface triggers a conformational change in the anchor peptide region that brings TbpB closer to the cell surface and facilitates interaction with TbpA so that efficient transfer of Tf to TbpA can occur (Fig. 7). Structural studies have shown that portions of the anchor peptide are capable of forming secondary structure (19), and the rapid loss in the ability of the anchor peptide to mediate binding to TbpA when the conserved FDL motif is removed (Fig. 5) may suggest that it is involved in the formation of secondary or tertiary structure. However, the inability of the anchor peptide fused to Mbp to mediate interaction with TbpA, whereas the TbpB N-lobe does complex with TbpA (Fig. 6), suggests that a different portion of the N-lobe either contributes to the interaction with TbpA or influences the formation of secondary structural features of the anchor peptide. Clearly, further studies are required to provide direct evidence for the proposed properties and features of the anchor peptide.
Structural studies have revealed that the C-terminal portion of the TbpB anchor peptide is intimately associated with the C-terminal lobe of TbpB, effectively wrapping around it (18, 19). The observation that a portion of the anchor peptide “released” from interacting with the C-lobe in one structure formed an α-helix (19) begs the question as to whether the interaction between anchor peptide and C-lobe is designed to modulate the anchor peptide conformation and function. This leads to an obvious hypothesis regarding TbpB function, in which binding of Tf leads to disruption of the interaction between C-lobe and anchor peptide so that the anchor peptide is free to interact with TbpA. Clearly, experiments designed to specifically address this hypothesis are needed.
The results from hydrogen-deuterium exchange coupled to mass spectrometry studies indicate that TbpB binds to regions on both domains of the C-lobe of Tf on the “lip” flanking the interdomain cleft and that the C-lobe is likely in a closed conformation (20, 21), consistent with the preference for the iron-loaded, closed form. There was no detectable release of iron from porcine Tf bound to A. pleuropneumoniae TbpB to the periplasmic iron-binding protein (FbpA) from Haemophilus influenzae using stability of unpurified proteins by rates of hydrogen-deuterium exchange (SUPREX) (20), supporting the view that Tf is maintained in the closed iron-loaded conformation. Thus holoTf in the closed conformation would be released near the membrane surface (Fig. 7) and compensate for the inability of TbpA to discriminate between the holo and apo forms of Tf.
Once TbpB is brought into close proximity to TbpA (Fig. 7), the binding kinetics (Fig. 2) would favor rapid transfer of Tf to TbpB. The unidirectional transfer of Tf from TbpB to TbpA is observed experimentally (Fig. 3, last lane). If TbpA binds to both domains of the C-lobe, as has been demonstrated with the TbpA homologue, lactoferrin-binding protein A (LbpA) (25), it implies either that there is a conformational change upon binding Tf or that TbpA captures Tf in a conformation common to both holoTF and apoTf (closed, open, intermediate). A conformational change related to Tf binding might account for the nearly irreversible binding to TbpAs (Fig. 2 and Table 1) and may relate to the observation that the efficient release of bound Tf from TbpA at the gonococcal cell surface required the presence of TonB (16). In the TbpB−ve, TonB−ve strains, there was no detectable displacement of the bound, labeled Tf by unlabeled Tf after a 40-min incubation when compared with >60% released in the TonB-proficient strain. This suggests that the provision of energy by TonB to facilitate the removal and transport of iron somehow facilitates the release of bound Tf, possibly by inducing a conformational change in TbpA. It is likely that the purified, recombinant TbpA used in the experiments illustrated in Figs. 3–6 shares the properties of TbpA in a TonB-deficient strain in that it does not readily release Tf. This certainly is consistent with prior experience in the affinity capture of the native TbpA as elution from a Tf affinity column requires the inclusion of relatively high concentrations of denaturants such as guanidine hydrochloride (27).
The inability of tagged recombinant TbpA to capture TbpB in the absence of bound Tf (Fig. 4, right panel) might imply that the capture is mediated by simultaneous binding of TbpB and TbpA to Tf. Because this does not occur in the absence of the intact anchor peptide (Fig. 4, left panel, 37–528), this interpretation requires that the anchor peptide either binds to Tf directly or modulates the binding by TbpB or TbpA to permit simultaneous binding to Tf. Recent hydrogen-deuterium exchange coupled to mass spectrometry studies in our laboratory have shown that the TbpA footprint on Tf partially overlaps the TbpB footprint such that there would be steric interference with simultaneous binding by the two receptor proteins (data not shown).
An alternate interpretation of the requirement for Tf is that binding of Tf maintains recombinant TbpA in a conformation that is favorable for complex formation, a conformation that is not favored with the detergent-solubilized recombinant TbpA. One conformation that may not be favored in the recombinant form of TbpA is one that is dependent upon or induced by interaction with TonB. In this context, it is interesting to consider that TbpB is protected from protease by TbpA in intact cells only when a functional TonB is present (15). Thus TonB interaction with TbpA may induce a conformation that is favorable for interacting with TbpB, and this conformation may be induced in our experiments by binding of Tf. The transport process would be most efficient if binding of Tf to TbpB released the anchor peptide for interaction with TbpA and the interaction was preferentially with TbpA that had interacted with TonB (Fig. 7). This would provide a ready supply of the appropriate iron-loaded Tf to TbpA that was available for transport. The model in which TonB interaction with TbpA initiates the formation of a Tbp-TbpA complex and transfer of Tf to TbpA raises many questions regarding the subsequent mechanism of iron removal and transport.
The results in this study clearly demonstrate that the presence of the N-terminal anchor peptide region on TbpB is required for interacting with TbpA, define the N-terminal residues not required, and demonstrate that the presence of Tf is required for this interaction. However, the details of the sites of interaction and conformation of the anchor peptide required for the interaction are yet to be determined. The ability to form ternary Tf-TbpB-TbpA complexes provides the potential for pursuing structural studies that could directly address some of these questions. However, a combination of biochemical and genetic approaches and mass shift analyses may provide alternate and complementary approaches to probe these interactions.
This work was supported by Grant 49603 from the Canadian Institutes of Health Research and by a Team Program grant from Alberta Innovates – Health Solutions.
This manuscript is dedicated to the memory of Xue Yang.
- Tf
- transferrin
- pTf
- porcine Tf
- Tbp
- transferrin-binding protein
- TEV
- tobacco etch virus
- Ni-NTA
- nickel-nitrilotriacetic acid
- Mbp
- maltose-binding protein.
REFERENCES
- 1. Gray-Owen S. D., Schryvers A. B. (1993) Microb. Pathog. 14, 389–398 [DOI] [PubMed] [Google Scholar]
- 2. Schryvers A. B., Gonzalez G. C. (1990) Can. J. Microbiol. 36, 145–147 [DOI] [PubMed] [Google Scholar]
- 3. Gray-Owen S. D., Schryvers A. B. (1996) Trends Microbiol. 4, 185–191 [DOI] [PubMed] [Google Scholar]
- 4. Gray-Owen S. D., Loosmore S., Schryvers A. B. (1995) Infect. Immun. 63, 1201–1210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Irwin S. W., Averil N., Cheng C. Y., Schryvers A. B. (1993) Mol. Microbiol. 8, 1125–1133 [DOI] [PubMed] [Google Scholar]
- 6. Luke N. R., Campagnari A. A. (1999) Infect. Immun. 67, 5815–5819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Alcantara J., Yu R. H., Schryvers A. B. (1993) Mol. Microbiol. 8, 1135–1143 [DOI] [PubMed] [Google Scholar]
- 8. Stojiljkovic I., Srinivasan N. (1997) J. Bacteriol. 179, 805–812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Anderson J. E., Sparling P. F., Cornelissen C. N. (1994) J. Bacteriol. 176, 3162–3170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Baltes N., Hennig-Pauka I., Gerlach G. F. (2002) FEMS Microbiol. Lett. 209, 283–287 [DOI] [PubMed] [Google Scholar]
- 11. Krell T., Renauld-Mongénie G., Nicolaï M. C., Fraysse S., Chevalier M., Bérard Y., Oakhill J., Evans R. W., Gorringe A., Lissolo L. (2003) J. Biol. Chem. 278, 14712–14722 [DOI] [PubMed] [Google Scholar]
- 12. Retzer M. D., Yu R., Zhang Y., Gonzalez G. C., Schryvers A. B. (1998) Microb. Pathog. 25, 175–180 [DOI] [PubMed] [Google Scholar]
- 13. Yu R. H., Schryvers A. B. (1993) Microb. Pathog. 15, 433–445 [DOI] [PubMed] [Google Scholar]
- 14. Cornelissen C. N., Sparling P. F. (1996) J. Bacteriol. 178, 1437–1444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cornelissen C. N., Anderson J. E., Sparling P. F. (1997) Mol. Microbiol. 26, 25–35 [DOI] [PubMed] [Google Scholar]
- 16. DeRocco A. J., Yost-Daljev M. K., Kenney C. D., Cornelissen C. N. (2009) Biometals 22, 439–451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Powell N. B., Bishop K., Palmer H. M., Ala'Aldeen D. A., Gorringe A. R., Borriello S. P. (1998) J. Med. Microbiol. 47, 257–264 [DOI] [PubMed] [Google Scholar]
- 18. Moraes T. F., Yu R. H., Strynadka N. C., Schryvers A. B. (2009) Mol. Cell 35, 523–533 [DOI] [PubMed] [Google Scholar]
- 19. Calmettes C., Yu R. H., Silva L. P., Curran D., Schriemer D. C., Schryvers A. B., Moraes T. F. (2011) J. Biol. Chem. 286, 12683–12692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Silva L. P., Yu R., Calmettes C., Yang X., Moraes T. F., Schryvers A. B., Schriemer D. C. (2011) J. Biol. Chem. 286, 21353–21360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ling J. M., Shima C. H., Schriemer D. C., Schryvers A. B. (2010) Mol. Microbiol. 77, 1301–1314 [DOI] [PubMed] [Google Scholar]
- 22. MacGillivray R. T., Moore S. A., Chen J., Anderson B. F., Baker H., Luo Y., Bewley M., Smith C. A., Murphy M. E., Wang Y., Mason A. B., Woodworth R. C., Brayer G. D., Baker E. N. (1998) Biochemistry 37, 7919–7928 [DOI] [PubMed] [Google Scholar]
- 23. Nemish U., Yu R. H., Tari L. W., Krewulak K., Schryvers A. B. (2003) Biochem. Cell Biol. 81, 275–283 [DOI] [PubMed] [Google Scholar]
- 24. Schryvers A. B., Stojiljkovic I. (1999) Mol. Microbiol. 32, 1117–1123 [DOI] [PubMed] [Google Scholar]
- 25. Wong H., Schryvers A. B. (2003) Microbiology 149, 1729–1737 [DOI] [PubMed] [Google Scholar]
- 26. Miroux B., Walker J. E. (1996) J. Mol. Biol. 260, 289–298 [DOI] [PubMed] [Google Scholar]
- 27. DeWinter L. M., Schryvers A. B. (2002) in Meningococcal Vaccines: Methods and Protocols (Pollard A. J., Maiden M. C., eds) pp. 109–120, Humana Press Inc., Totowa, NJ [Google Scholar]
- 28. Schryvers A. B., Morris L. J. (1988) Infect. Immun. 56, 1144–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gray-Owen S. D., Schryvers A. B. (1995) Infect. Immun. 63, 3809–3815 [DOI] [PMC free article] [PubMed] [Google Scholar]