Background: PDZ domains mediate protein interaction but may also sense lipid signaling.
Results: We characterized a new phosphatidylinositol 4,5-bisphosphate-interacting PDZ domain by in vitro, in vivo, and in silico approaches.
Conclusion: Membrane binding and subcellular localization of this domain is achieved by a combination of peptide and lipid interactions.
Significance: PDZ domains might support cooperative detection of peptide and lipid in cell signaling.
Keywords: Membrane Lipids, Membrane Proteins, Phosphatidylinositol Signaling, Protein Complexes, Scaffold Proteins, PDZ Domain, Phosphoinositides, Screen, Coincident Detection, Membrane
Abstract
PDZ domains are well known protein-protein interaction modules that, as part of multidomain proteins, assemble molecular complexes. Some PDZ domains have been reported to interact with membrane lipids, in particular phosphatidylinositol phosphates, but few studies have been aimed at elucidating the prevalence or the molecular details of such interactions. We screened 46 Drosophila PDZ domains for phosphoinositide-dependent cellular localization and discovered that the second PDZ domain of polychaetoid (Pyd PDZ2) interacts with phosphatidylinositol 4,5-bisphosphate (PtdIns(4,5)P2) at the plasma membrane. Surface plasmon resonance binding experiments with recombinant protein established that Pyd PDZ2 interacts with phosphatidylinositol phosphates with apparent affinities in the micromolar range. Electrostatic interactions involving an extended positively charged surface of Pyd PDZ2 are crucial for the PtdIns(4,5)P2-dependent membrane interactions as shown by a combination of three-dimensional modeling, mutagenesis, binding, and localization studies. In vivo localization studies further suggested that both lipid and peptide binding contribute to membrane localization. We identified the transmembrane protein Crumbs as a Pyd PDZ2 ligand and probed the relation between peptide and PtdIns(4,5)P2 binding. Contrary to the prevalent view on PDZ/peptide/lipid binding, we did not find competition between peptide and lipid ligands. Instead, preloading the protein with the 10-mer Crb3 peptide increased the apparent affinity of Pyd PDZ2 for PtdIns(4,5)P2 6-fold. Our results suggest that membrane localization of Pyd PDZ2 may be driven by a combination of peptide and PtdIns(4,5)P2 binding, which raises the intriguing possibility that the domain may coordinate protein- and phospholipid-mediated signals.
Introduction
PDZ domains are among the most common protein modules in multicellular organisms. They consist of 80–90 amino acids and have a typical fold of five to six β-strands flanked by two or three α-helices. They are generally found in multidomain proteins involved in the assembly and subcellular targeting of signaling complexes (1, 2). PDZ domains are well known as protein-interacting modules, predominantly recognizing C-termini of target proteins, but some of them have also been shown to interact with membrane phospholipids (3).
Phospholipids are major constituents of mammalian cell membranes. From a signaling perspective, phosphatidylinositol (PtdIns)4 is particularly interesting because it can be reversibly phosphorylated in a combinatorial manner on 1–3 of the hydroxyl groups of its inositol headgroup (the 3′-, 4′-, and/or 5′-positions) by an intricate system of kinases and phosphatases. The resulting seven biologically relevant PtdInsPs have distinct subcellular compartmentalization and function both as precursors for second messengers and as membrane docking sites for cytosolic proteins (4–6). For example, PtdIns(4,5)P2 is enriched at the plasma membrane in quiescent cells, where it controls, among other processes, the dynamics of the actin cytoskeleton and the early steps of endosome formation (7).
A direct link between PDZ domains and PtdInsPs was first made for the PDZ tandems (i.e. two PDZ domains connected by a short linker) of syntenin-1 (8, 9) and syntenin-2 (10), which added PDZ domains to the list of PtdInsP binding modules together with more well known examples, such as PH, FYVE, PX, ENTH, CALM, PTB, and FERM domains (5, 6). Despite the potential importance of PDZ-PtdInsP binding, few studies have addressed the particulars of these interactions, such as prevalence, binding determinants, and interplay between PtdInsPs and peptide binding. The best studied examples besides the syntenin-1 and syntenin-2 PDZ tandems are the second PDZ domain of Par-3, PICK1 PDZ, and the second PDZ domains of the zonulins (ZOs) 1 and 2 (11–13).
To identify additional PtdInsPs-interacting PDZ domains, we developed a cell-based screen with the read-out being PtdInsPs-dependent membrane localization. This is reminiscent of approaches taken to elucidate the prevalence of PtdInsP interactions among PX and PH domains from Saccharomyces cerevisiae (14, 15). We focused on the Drosophila proteome because it has fewer PDZ domains compared with the overwhelming number of human PDZ variants. We discovered that the second PDZ domain of polychaetoid (Pyd PDZ2) is targeted to the plasma membrane in a PtdIns(4,5)P2-dependent manner.
Pyd is the unique Drosophila homolog of the human ZOs proteins. These MAGUK (membrane-associated guanylate kinase) proteins are composed of three PDZ domains, one Src homology 3 domain, and one guanylate kinase domain. Pyd localizes at the sites of cell junctions (16) and has been proposed to act in dynamic remodeling of the cytoskeleton (17), in modulating the strength of adherence junctions (18), and in regulating the accumulation of various adherence junction-localized receptors (19). Relevantly, a study by Pinal et al. (20) on Drosophila photoreceptors suggested that PtdIns(4,5)P2 is enriched at these junctions.
We characterized the interactions between Pyd PDZ2 and PtdInsPs by SPR experiments and probed the molecular determinants of the membrane targeting by a combination of in silico approaches, extensive mutagenesis, binding, and in vivo localization studies. We established that Pyd PDZ2 interacts with the C-terminal peptide of the transmembrane receptor Crb3, which is an important apical determinant for maintenance of epithelial polarity (21), and used it to investigate the relation between peptide and PtdIns(4,5)P2 binding.
EXPERIMENTAL PROCEDURES
Molecular Biology
Vectors for the eukaryotic expression of eYFP fusion proteins were obtained by subcloning PDZ domains amplified by PCR from appropriate cDNA libraries in the peYFP-C2 vector (Invitrogen). The precise sequence of each construct can be obtained on demand. The Pyd PDZ2 fragment with flanking regions (residues 129–271, numbering according to Pyd isoform F) subcloned into peYFP-C2 was amplified by PCR using oligonucleotides carrying the restriction sites for NcoI and EcoRI (Roche Applied Science), digested and ligated into the pETM-11 plasmid (EMBL Heidelberg) for the expression of N-terminally His6-tagged protein. The expressed and purified protein was found to be partially degraded, and a shorter construct encoding residues 170–271 was made, allowing purification of homogeneous protein. Site-directed mutagenesis was performed using QuikChange (Stratagene) on the Pyd PDZ2 pETM-11 and the eYFP-Pyd PDZ2 templates. All constructs were verified by DNA sequencing.
Cell Culture, Transfections, and Microscopic Analysis
MCF-7 cells originating from the American Type Culture Collection (Manassas, VA) were grown in DMEM/F-12 medium (Invitrogen) supplemented with 10% fetal bovine serum (HyClone). For microscopy experiments, cells were plated on 8-well chamber slides (Nalge Nunc International, Roche Applied Science), transfected using FuGENE transfection reagent (Roche Applied Science), and fixed with 4% paraformaldehyde the day after transfection. For time lapse experiments, fixation was omitted. For ionomycin treatment, cells were washed with Krebs-Ringer buffer (22) before being incubated with 10 μm ionomycin (Sigma) in the same buffer. For rapamycin experiments, rapamycin (Calbiochem) was added to a final concentration of 200 nm, and the decrease in plasma membrane enrichment of eYFP-S1 PDZ1-Pyd PDZ2 was quantified by measuring confocal fluorescent intensity profiles along a line taken over cells before and after rapamycin treatment. The intensity of eYFP-S1 PDZ1-Pyd PDZ2 at sites of cell-cell contacts was normalized using the intensity of the palmitoylated FKB12-rapamycin binding domain (eCFP-FRB-PM), which served as a membrane marker. The decrease in the eYFP-S1 PDZ1-Pyd PDZ2 membrane enrichment was quantified for a total number of eight cell-cell contacts in three independent experiments and expressed as the difference (percentage) between the intensity ratio before and after the rapamycin treatment.
The enrichment of eYFP-Pyd PDZ2 at sites of cell-cell contacts was scored in living cells by wide field microscopy, alone or co-transfected with expression vector for N-terminally Myc-tagged Crb3 or Myc-Crb3ΔERLI, which lacks the last four amino acids (23). Each experiment was repeated three times, counting at the minimum 30 cell-cell contacts each time. In cases of co-transfections with Myc-Crb3, we counted only cell-cell contacts enriched in the Myc-tagged receptors, which were visualized using anti-Myc antibodies (3 μg/ml 9E10, Sigma) directly labeled with the zenon Alexa Fluor 647 mouse IgG1 labeling kit (Invitrogen). The p values were calculated using the two-tailed t test. Confocal micrographs were obtained with an Olympus Fluoview 1000 (Olympus), and wide field and time lapse micrographs were obtained with a LEICA AS-MDW working station (LEICA) using appropriate filter sets.
Protein Purifications
Escherichia coli ER2566 cells carrying the pETM-11 expression vector with wild-type or mutant Pyd PDZ2 inserts were inoculated in TYB medium containing 30 μg/ml kanamycin and grown overnight at 37 °C. Overnight cultures were diluted 100-fold into TYB medium containing 2% glucose and were grown at 37 °C to an ocular density at 600 nm of 0.6–0.8. Expression of N-terminally His-tagged proteins was induced by the addition of 1 mm isoproyl 1-thio-β-d-galactopyranoside. Cells were harvested by centrifugation after 4 h. The pellets were frozen at −20 °C overnight and then resuspended in 20 ml of buffer A (50 mm potassium phosphate buffer, pH 7.4, 20 mm imidazole, 300 mm NaCl, and 1 mm β-mercaptoethanol) with a mixture of protease inhibitors (Complete EDTA-free, Roche Applied Science) and 50 units of DNase I (Roche Applied Science). Cells were lysed by lyzosyme treatment, and the debris was pelleted by centrifugation (30 min, 15,000 rpm). Proteins were bound to a 1-ml His-trap column (GE Healthcare) pre-equilibrated with buffer A and eluted by an imidazole gradient (20 mm to 1 m in buffer A) using an Akta Explorer system (GE Healthcare). The purity of the protein samples was confirmed by SDS-PAGE with Coomassie Brilliant Blue staining. The proteins were dialyzed to appropriate buffers.
Size Exclusion Chromatography
Analytical size exclusion chromatography was performed using an Akta Explorer system and a Superdex 75 10/300 GL column (GE Healthcare). The column was equilibrated with buffer B (25 mm HEPES, 150 mm NaCl, pH 7.2, and 1 mm β-mercaptoethanol) and calibrated using a set of molecular weight standards (albumin, carbonic anhydrase, cytochrome c, aprotinin; Sigma-Aldrich). The void volume of the system was determined using blue dextran.
Equilibrium Unfolding Experiments
Urea-induced equilibrium experiments were performed in buffer B at 25 °C using a Cary Eclipse Spectrofluorimeter (Varian). Protein samples were excited at 290 nm, and the emission spectra were recorded between 320 and 380 nm.
SPR Experiments
SPR experiments were performed in a Biacore T100 (GE Healthcare). Proteins were freshly prepared, dialyzed against their respective running buffers, and typically injected at seven different concentrations. The flow was 30 μl/min, the temperature was 25 °C, and the injection time was 120–240 s, long enough for the sensorgrams to reach equilibrium. Sensorgrams were double reference-subtracted, and the responses observed 5 s before injection stop were plotted as a function of protein concentrations. Apparent KD values were determined by fitting the data to a 1:1 Langmuir binding isotherm (GraphPad Prism). Previous studies have shown this to be the most robust way for determining apparent KD values for protein-PtdInsP interactions (24).
C6-PtdIns(4,5)P2 Binding Experiments
Biotinylated glycerophosphoinositol and biotinylated C6-PtdIns(4,5)P2 (Echelon Biosciences) were immobilized (40 RUs) in the reference and sample channels of an SA chip, respectively. The buffer was 10 mm HEPES, 100 mm NaCl, 0.005% Tween 20, pH 7.2, and 1 mm β-mercaptoethanol, and the regeneration solution was 1 m NaCl, 50 mm NaOH.
Liposome Binding Experiments
Bulk phospholipids (phosphatidylcholine (PC), phosphatidylserine (PS), phosphatidylethanolamine (PE), and 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC)) were from Sigma-Aldrich. PtdIns and PtdInsPs were from Echelon Biosciences. Composite liposomes (30% PC, 20% PS, 40% PE, 5% PtdIns, and 5% PtdInsPs or 10% PtdIns for background reference) or DOPC liposomes (95% DOPC, 5% PtdInsPs or 5% PtdIns for background reference or 80% DOPC, 20% PS with 100% DOPC for background reference) were prepared and immobilized to typically 5000–6000 RUs on L1 chips as described previously (10) using buffer B. The surface was regenerated between runs by two 30-s pulses of 50 mm NaOH. The experiments were routinely repeated using different batches of liposomes and proteins.
Peptide Binding Experiments
Peptide binding experiments were performed in buffer B in a Cary Eclipse Spectrofluorimeter (Varian) at 25 °C. Synthetic peptides (LKLPPEERLI, LIRLEPELKP, and dansyl-PEERLI) were from GeneCust and of high purity (>95%). Pyd PDZ2 F194W (3 μm) was titrated with increasing peptide concentrations (10–1500 μm), and the change in intrinsic tryptophan fluorescence was recorded (excitation at 290 nm, emission at 320–375 nm). Alternatively, dansyl-PEERLI-COO− (3 μm) was titrated with increasing concentrations of Pyd PDZ2, and the change in dansyl fluorescence was recorded (excitation at 345 nm, emission at 520–580 nm). The data were fitted by a simple binding isotherm.
Electrostatic Potential Calculations and Visualization
The structure of Pyd PDZ2 was modeled based on the structure of human ZO-1 PDZ2 (PDB code 2RCZ) using the program Nest (25), and unconserved side chains were remodeled using Scap (26). Hydrogen atoms were added to the protein structures with the program CHARMM, and the structure was subjected to conjugate gradient minimization with a harmonic restraint force of 50 kcal/mol/Å applied to the heavy atoms located at the original crystallographic coordinates. Electrostatic potentials were calculated with the DelPhi program (27) using CHARMM partial charges and radii. Three-dimensional potential maps were visualized by mapping the potential values onto the molecular surface of the protein or by drawing equipotential contour meshes that connect all of the points in space having a specific potential value (here ±1 kT/e, equal to ±25 mV).
RESULTS
Identification of Pyd PDZ2 as a PtdIns(4,5)P2-interacting Module
We designed a cell-based screening assay to identify PtdInsP-interacting PDZ domains by their enrichment in/at PtdInsP-containing cellular compartments. PtdInsP binding domains often act in combination with other domains (6) because their affinities are frequently too low to confer specific localization by themselves. We therefore engineered a screening vector combining an enhanced yellow fluorescent protein and the first PDZ domain of syntenin-1 (eYFP-S1 PDZ1; Fig. 1A). eYFP provides a fluorescent probe, and S1 PDZ1 gives a low initial affinity for PtdInsPs, which on its own is not sufficient for targeting eYFP to any PtdInsPs-rich compartments (Fig. 1A, upper image). The fluorescent probe is targeted to PtdInsPs pools only when a second PtdInsP binding module is linked to the construct. The assay was validated by linking eYFP-S1 PDZ1 to the second PDZ domain of human syntenin-2, previously shown to interact with PtdIns(4,5)P2 (10). Whereas eYFP-syntenin-2 PDZ2 distributed diffusely in the cell (10), the tandem eYFP-S1 PDZ1-syntenin-2 PDZ2 was enriched at the plasma membrane and in PtdIns(4,5)P2-rich subnuclear organelles (Fig. 1A). We tested four different linkers (KIL, KILRPFQ, KIPEFVMVVRDRPFQ, and KISSSSFEFVMVVRDRPFQ) between eYFP-S1 PDZ1 and the target PDZ without observing any significant differences; the first and the last examples are shown in Fig. 1A.
FIGURE 1.
Screen for PtdInsP-interacting PDZ domains. A, the screen was based on a fluorescence microscopy assay, where we evaluated the ability of the tested PDZ domain to assist syntenin-1 PDZ1 fused to eYFP (eYFP-S1 PDZ1) in targeting PtdIns(4,5)P2-rich subcellular regions of MCF-7 cells (i.e. the plasma membrane, nuclear speckles, and/or nucleoli). Neither the individual syntenin-1 PDZ1 (top) nor syntenin-2 PDZ2 (S2 PDZ2) (10) was able to target eYFP to these regions. However, eYFP-S1 PDZ1-S2 PDZ2 localized to PtdIns(4,5)P2-rich pools (middle and bottom). Note that the length of the linker region had no impact on the subcellular enrichment. B, confocal micrographs of selected Drosophila PDZ domains fused to eYFP-S1 PDZ1 in MCF-7 cells. C, Confocal micrographs showing the enrichment of eYFP-PH PLCδ and eYFP-S1 PDZ1-Pyd PDZ2 around the PtdIns(4,5)P2-rich membranes of macropinosomes. The vesicular structures were induced by co-expression of a constitutively active Arf6-Q67L mutant (28). D, time lapse wide-field micrographs showing the delocalization of eYFP-S1 PDZ1-Pyd PDZ2 from the plasma membrane upon ionomycin-induced PtdIns(4,5)P2 breakdown. E, time lapse confocal micrographs of MCF-7 cells co-expressing eYFP-S1 PDZ1-Pyd PDZ2, mRFP-FKBP12-PtdInsPs 5′-phosphatase, and palmitoylated eCFP-FRB-PM (PM-target). eYFP-S1 PDZ1-Pyd PDZ is enriched at the plasma membrane and at cell-cell contacts. Note the reduction of this enrichment 5 min after the addition of rapamycin, which induces plasma membrane translocation of the 5′-phosphatase (right).
We screened 46 single PDZ domains and one PDZ tandem (CG31342-PDZ1-2, the Drosophila PDZ tandem most similar to syntenin) listed in supplemental Table 1. The eYFP-S1 PDZ1 fusions were expressed in MCF-7 cells, and their subcellular distribution was analyzed by confocal microscopy (supplemental Table 1 and Fig. 1B). In the majority of the cases, the fluorescent protein distributed diffusely in the cytosol. A subgroup of eight showed localization to inclusion bodies. In live imaging, the inclusions were inert, which suggests that they result from protein aggregation, and these variants were not investigated further. The eYFP-S1 PDZ1-Canoe PDZ protein concentrated at sites of cell-cell contacts (Fig. 1B) but only in a minority of the cells. The eYFP-S1 PDZ1 fused to the tandem construct of the syntenin-like CG31342-PDZ1–2 was enriched at the membrane, but the isolated PDZ domains were distributed diffusely. The most striking membrane localization was observed for Pyd PDZ2, which displayed a strong plasma membrane enrichment (Fig. 1B).
We used different cellular assays to establish if the membrane localization of eYFP-S1 PDZ1-Pyd PDZ2 was PtdIns(4,5)P2-dependent. First, we made use of a constitutively active form (Q67L) of ADP-ribosylation factor 6 (Arf6) (28), which induces the formation of intracellular macropinosomes in MCF-7 cells. These large endocytic vesicles are surrounded by PtdIns(4,5)P2-rich membranes that can be labeled with eYFP-PH-PLCδ, a well characterized probe for plasma membrane-associated PtdIns(4,5)P2 (22, 29). When eYFP-S1 PDZ1-Pyd PDZ2 was co-expressed with Arf6-Q67L, it concentrated at the macropinosomal membranes, consistent with PtdIns(4,5)P2-dependent membrane interaction (Fig. 1C).
We next investigated if the eYFP-S1 PDZ1-Pyd PDZ2 localization responded to ionomycin treatment, an assay commonly used to induce the breakdown of PtdIns(4,5)P2 (22). Supporting a PtdIns(4,5)P2-dependent membrane targeting, the addition of ionomycin to cells expressing eYFP-S1 PDZ1-Pyd PDZ2 resulted in translocation of the construct from the membrane to the cytoplasm (Fig. 1D). Finally, we used an assay, developed by Varnai et al. (30), where rapamycin treatment induces plasma membrane recruitment of a PtdInsPs 5′-phosphatase by its heterodimerization with a palmitoylated FKB12-rapamycin binding domain (eCFP-FRB-PM). The translocated phosphatase cleaves the 5′-phosphate, which results in rapid decrease of plasma membrane PtdIns(4,5)P2. We observed a decrease in the membrane enrichment of eYFP-S1 PDZ1-Pyd PDZ2 upon translocation of the enzyme (Fig. 1E). Using fluorescent intensity measurements, we found on average a 40 ± 15% decrease in the plasma membrane localization of eYFP-S1 PDZ1-Pyd PDZ2 upon translocation of the 5′-phosphatase.
Additionally, we tested the PtdIns(4,5)P2 dependence of Pyd PDZ2 fused only to an eYFP tag (eYFP-Pyd PDZ2). Also, this protein was enriched at the plasma membrane although predominantly at sites of cell-cell contacts (Fig. 2A) and at the PtdIns(4,5)P2-rich membranes of Arf6-Q67L-induced macropinosomes (Fig. 2A, right image). The plasma membrane enrichment of eYFP-Pyd PDZ2 decreased upon PtdIns(4,5)P2 breakdown (Fig. 2B). Taken together, these data establish that Pyd PDZ2 is targeted to the plasma membrane in a PtdIns(4,5)P2-dependent manner.
FIGURE 2.
Pyd PDZ2 interacts with PtdIns(4,5)P2in vivo and in vitro. A, Pyd PDZ2 fused to eYFP is enriched at the plasma membrane mainly at sites of cell-cell contacts (left) and at PtdIns(4,5)P2-rich membranes of macropinosomes induced by Arf6-Q67L (right) as shown by confocal micrographs. B, the membrane enrichment of eYFP-Pyd PDZ2 is lost upon ionomycin-induced PtdIns(4,5)P2 breakdown (wide field micrographs). C, recombinant Pyd PDZ2 (50 μm) elutes as a dimer (30 kDa) from an analytical size exclusion column. D, urea-induced unfolding of Pyd PDZ2 F194W, as followed by fluorescence at 10 μm protein concentration. The observed transition was fitted to a standard two-state model. E, double reference-subtracted sensorgrams of Pyd PDZ2 perfused over biotin-C6-PtdIns(4,5)P2 (40 RU) at different concentrations (as indicated) (also see Table 1). F, the interaction between Pyd PDZ2 (12 μm) and biotin-PtdIns(4,5)P2 can be competed by the lipid headgroup (IP3) with a Hill slope of −1.5, as determined by fitting the dose-response dependence. G, Pyd PDZ2 interacts with all seven PtdInsPs species in vitro (5% PtdInsPs in DOPC liposomes) with the highest apparent affinity for PtdIns(3,4,5)P3 (black dotted line and black triangles) and the lowest for the monophosphorylated species (gray lines and symbols). H, binding isotherms of Pyd PDZ2 with PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in the background of composite liposomes intended to mimic the lipid composition of the plasma membrane (30% PC, 20% PS, 40% PE, 5% PtdIns, and 5% PtdInsPs). The binding isotherm of Pyd PDZ2 to 20% PS in DOPC liposomes is also indicated for comparison.
In Vitro PtdInsPs Binding Characteristics of Pyd PDZ2
We purified recombinant Pyd PDZ2 and determined by analytical size exclusion chromatography that it forms a dimeric structure (Fig. 2C) like its human ZOs 1–3 PDZ2 counterparts (31, 32). To ensure the stability of the recombinant protein, we engineered a F194W variant (pseudo-wild type) to be used for urea-induced unfolding experiments. Corresponding Phe to Trp mutations have been introduced in other PDZ scaffolds and have been shown not to influence the characteristics of the proteins but to provide useful fluorescence probes for unfolding and peptide binding studies (33, 34). We performed urea-induced equilibrium unfolding experiments, followed by tryptophan fluorescence, at two different protein concentrations (1 and 10 μm, the latter shown in Fig. 2D). The observed transition was fitted to a standard two-state model (35), and the stability of the protein was calculated to be 3.6 ± 0.1 kcal/mol independently of the protein concentration. We concluded that the protein forms a stable, dimeric structure that unfolds in a cooperative manner.
We determined the PtdInsPs binding characteristics of Pyd PDZ2 in a series of SPR equilibrium binding experiments, where purified recombinant protein was injected over immobilized PtdInsPs. The PtdInsPs were directly immobilized or incorporated in liposomes, and glycerophosphoinositol- or PtdIns-containing liposomes served as respective background references. The sensorgrams were corrected for binding to the references and for buffer effects (double reference-subtracted) before further data processing.
First, we established that Pyd PDZ2 binds to C6-PtdIns(4,5)P2 (Fig. 2E). The observed equilibrium responses were plotted as a function of protein concentration, and the data were fitted to a 1:1 binding isotherm, yielding an apparent affinity of 26 ± 5 μm (see Fig. 4D). We then investigated if inositol 1,4,5-trisphosphate (IP3), the PtdIns(4,5)P2 headgroup, could compete for the interaction. In these experiments, a fixed concentration of Pyd PDZ2 (12 μm) was mixed with increasing IP3 concentrations (0–1000 μm) before being injected over immobilized C6-PtdIns(4,5)P2. Consistent with competition between the IP3 in the solution and PtdIns(4,5)P2 on the surface, we observed a decrease in binding with increasing IP3 concentrations (Fig. 2F). The IC50 value for IP3 was ∼250 μm, as determined from the dose-response dependence, and the inhibition constant Ki was ∼160 μm, as calculated using the Cheng-Prusoff equation (36). Pyd PDZ2 thus interacts directly with PtdIns(4,5)P2, and the lipid headgroup is sufficient for competing with the interaction.
FIGURE 4.
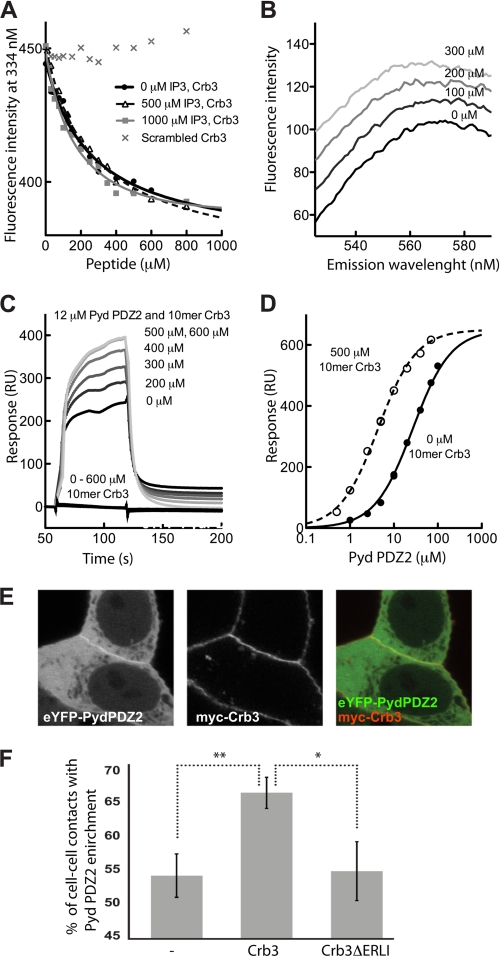
Interplay between peptide and PtdIns(4,5)P2 binding. A, equilibrium peptide binding of Pyd PDZ2 F194W and the 10-mer Crb3 peptide in the absence and presence of 500 and 1000 μm IP3 was followed by changes in intrinsic tryptophan fluorescence. Titration of the protein with a scrambled Crb3 peptide as a negative control is also indicated. B, equilibrium titration of dansyl-PEERLI-COO− with increasing concentrations of Pyd PDZ2 wild type. Note the change in fluorescence upon increasing protein concentration. C, double reference-subtracted sensorgrams of Pyd PDZ2 (12 μm) binding to biotinylated C6-PtdIns(4,5)P2 (40 RU) in the absence (black lines) and presence of increasing concentrations of LKLPPEERLI-COO− (gray lines). Shown are also control sensorgrams of different concentrations of peptide injected over the surface. D, equilibrium binding isotherms of Pyd PDZ2 to biotinylated C6-PtdIns(4,5)P2 in the absence (filled circles) and presence of 500 μm LKLPPEERLI-COO− (open circles); the apparent KD values were ∼26 and ∼4 μm, respectively. E, confocal micrographs illustrating the co-localization of eYFP-Pyd PDZ2 and Myc-Crb3 at sites of cell-cell contacts. F, bar graph of the mean percentage of cells ± S.D. (error bars), where eYFP-Pyd PDZ2 was enriched at cell-cell contacts when expressed in MCF7 cells alone, with Crb3 wild type or Crb3 with deleted PDZ binding motif (Crb3ΔERLI; *, p < 0.05; **, p < 0.01).
We next addressed the selectivity of the interaction by presenting the seven biologically relevant PtdInsP species to the protein in the background of neutral DOPC liposomes (Fig. 2G and Table 1). We found that the protein interacts in vitro with all seven PtdInsP species. The lowest apparent affinity was found for the monophosphorylated PtdInsPs, and the highest affinity was found for the most phosphorylated PtdIns(3,4,5)P3; the affinity for PtdIns(3,4,5)P3 was almost 7 times higher than for PtdIns(4,5)P2. Given the high in vitro affinity for PtdIns(3,4,5)P3, we investigated whether it might contribute to the plasma membrane targeting of Pyd PDZ2 in vivo by (i) decreasing the plasma membrane level of PtdIns(3,4,5)P3 by inhibition of the PtdIns 3-kinase using wortmannin and (ii) inducing transient plasma membrane enrichment of PtdIns(3,4,5)P3 by serum stimulation. Both treatments failed to affect the plasma membrane localization of eYFP-S1 PDZ1-Pyd PDZ2 and eYFP-Pyd PDZ2 but had clear effects on the plasma membrane enrichment of the PH domain of Akt (data not shown), a well established probe for PtdIns(3,4,5)P3 (37). Because the abundance of PtdIns(4,5)P2 at the plasma membrane is 10–20 times higher than that of PtdIns(3,4,5)P3 (6), it is not surprising that our attempts to increase or decrease the cellular PtdIns(3,4,5)P3 levels did not affect the localization of eYFP-Pyd PDZ2. However, we cannot exclude the possibility that in the context of the full protein and/or in a particular molecular environment, the interaction with other PtdInsPs may be biologically relevant.
TABLE 1.
Apparent affinities of recombinant Pyd PDZ2 for different PtdInsP species as determined by equilibrium SPR analysis of 5% PtdInsPs in the background of DOPC liposomes or composite liposomes (30% PC, 20% PS, 40% PE, 5% PtdIns)
|
KDapp |
|||||
|---|---|---|---|---|---|
| PtdIns3P, PtdIns4P, PtdIns5P | PtdIns(3,4)P2 | PtdIns(3,5)P2 | PtdIns(4,5)P2 | PtdIns(3,4,5)P3 | |
| μm | |||||
| DOPC liposomes | >30 | 18 ± 2 | 9.5 ± 0.6 | 15 ± 5 | 2.2 ± 0.2 |
| Composite liposomes | NDa | ND | ND | 0.7 ± 0.1 | 0.7 ± 0.2 |
a ND, not determined.
To tackle the apparent discrepancy between in vitro and in vivo data, we performed SPR experiments where PtdIns(4,5)P2 and PtdIns(3,4,5)P3 were presented to the protein in a background of composite liposomes (30% PC, 20% PS, 40% PE, 5% PtdIns), intended to mimic the plasma membrane composition. The apparent affinities for both PtdInsP species were significantly higher in this background, and importantly, there was no significant difference in the apparent affinities for the two PtdInsPs species (Fig. 2H and Table 1). The higher apparent affinity for PtdIns(4,5)P2 and PtdIns(3,4,5)P3 in composite liposomes suggests that additional interactions between the positively charged Pyd PDZ2 (isoelectric point 9.1) and, for example, the negatively charged PS might reinforce the interactions. We therefore tested for a direct interaction between Pyd PDZ2 and PS (20% PS in DOPC liposomes subtracted for 100% DOPC) and found that the protein associated with PS with an apparent KD of 20 ± 2 μm (Fig. 2H).
The Pyd PDZ2 Interaction with PtdIns(4,5)P2-rich Membranes Involves Multiple Electrostatic Interactions
To elucidate the molecular determinants of the interactions between Pyd PDZ2 and negatively charged membranes, we modeled the structure of Pyd PDZ2 based on the crystal structure of the homologous human ZO1 PDZ2 (Fig. 3A) and calculated its electrostatic potential (Fig. 3B). Three-dimensional visualization of the electrostatic potential of Pyd PDZ2 using equipotential contour maps revealed that Pyd PDZ2 has a strong positive potential that extends out significantly beyond the molecular surface of the protein along one face of the molecule. Such an extended positive potential predicts that, in agreement with our experimental data, the protein will have a significant affinity for negatively charged lipids and suggests that this side of the protein will face the lipid membrane. Accordingly, mutation of positively charged residues in this patch would be expected to have a significant effect on the positive potential and hence reduce the affinity for PtdIns(4,5)P2-containing membranes.
FIGURE 3.
Electrostatic interactions are important for the Pyd PDZ2-membrane interaction. A, homology model of dimeric Pyd PDZ2 (subunits in green and cyan ribbons) with residues probed by mutagenesis shown in a blue stick representation (see Table 2). Residues numbered in black indicate mutations that resulted in significant loss of lipid binding and membrane localization, whereas residues marked in gray indicate mutations with only minor effects. Asterisks indicate mutations that also caused loss of peptide binding. B, equipotential contour plot of the calculated electrostatic potential of Pyd PDZ2, visualized by drawing meshes for the values +1 kT/e (25 mV, blue mesh) and −1 kT/e (−25 mV, red mesh) outside the molecular surface of the protein. C, fluorescent confocal micrographs of eYFP-Pyd PDZ2 mutants in MCF-7 cells. Note that all mutations except R201A and R244A resulted in complete loss of membrane targeting.
We probed the model experimentally by site-directed mutagenesis. We created nine mutants of Pyd PDZ2 F194W mutating arginines and lysines into alanines. We established by analytical size exclusion chromatography that the mutant proteins were dimeric (not shown). We calculated the effect of the mutations on the stability by the following,
where ΔΔGD-N is the difference between the free energies of unfolding upon mutation (Table 2). In four cases (K180A, K199A, K232A, and K236A), the mutations resulted in significant stabilization of the protein structure, whereas one mutant, R244A, had a destabilizing effect.
TABLE 2.
Binding characteristics, stability data, and in vivo localization of Pyd PDZ2 mutants
| Pyd PDZ2 | KDapp PtdIns(4,5)P2, composite liposomes | KD LKLPPEERLI | ΔΔGD-N | In vivo localizationa |
|---|---|---|---|---|
| μm | μm | kcal/mol | ||
| F194W | 0.40 ± 0.05 | 220 ± 20 | Membrane | |
| F194W/K179A | 2.9 ± 0.4 | 100 ± 30 | 0.54 ± 0.05 | Diffuse |
| F194W/K180A | 4 ± 1 | 300 ± 60 | 1.23 ± 0.04 | Diffuse |
| F194W/K199A | 2.9 ± 0.8 | 190 ± 50 | 1.59 ± 0.05 | Diffuse |
| F194W/R201A | 0.6 ± 0.1 | 231 ± 13 | −0.61 ± 0.02 | Membrane |
| F194W/K232A | 3.5 ± 0.4 | 177 ± 12 | 3.2 ± 0.3 | Diffuse |
| F194W/K235A | 2.4 ± 0.4 | 176 ± 20 | −0.16 ± 0.01 | Diffuse |
| F194W/K236A | 3.6 ± 0.6 | >1000 | 1.14 ± 0.03 | Diffuse |
| F194W/K242A | 14 ± 4 | >1000 | −0.19 ± 0.01 | Diffuse |
| F194W/R244A | 1.4 ± 0.2 | 520 ± 100 | −1.9 ± 0.2 | Membrane |
a The mutations were introduced in the background of wild-type eYFP-Pyd PDZ2.
We established the effects of the mutations on the PtdIns(4,5)P2 interactions in background of composite liposomes. Most mutations conferred significant loss in apparent affinity for PtdIns(4,5)P2 (Table 2), with K242A having the most prominent effect. The R201A and the R244A mutations had only minor effects on the lipid binding. To test the in vivo relevance of our mutagenic analysis, we introduced the same mutations in eYFP-Pyd PDZ2 and investigated the effects on the cellular localization (Fig. 3C and Table 2). We found that all but the R201A and the R244A mutations conferred complete loss of membrane localization in line with the in vitro data. The broad structural distribution of mutations affecting the PtdIns(4,5)P2 membrane binding is consistent with a model where multiple electrostatic interactions spread over an extended surface contribute to the PtdIns(4,5)P2-dependent membrane interactions.
The C-terminal Peptide of Crb3 Interacts with Pyd PDZ2
PDZ domains are known primarily as protein-protein interacting modules, and it has been shown for several PDZ domains that peptide binding may induce a small conformational change that can be followed by a change in intrinsic tryptophan fluorescence (38, 39). In the literature, there was no known peptide ligand for Pyd PDZ2, but in Drosophila, Pyd is an adherent junction protein together with several other proteins, such as the protein Crumbs. We hypothesized that the Pyd PDZ2 and Crumbs may interact directly, and we tested for binding using a fluorescence-based approach. We challenged Pyd PDZ2 F194W with the 10-mer LKLPPEERLI-COO− peptide containing the C-terminal PDZ binding motif of the Crb3, the last 7 residues being identical to the Drosophila protein Crumbs. We found a distinct change in tryptophan fluorescence with increasing concentration of the LKLPPEERLI-COO− peptide, and the transition corresponded to a KD value of ∼220 μm (Fig. 4A and Table 2). As a negative control we challenged Pyd PDZ2 with a scrambled peptide (LIRLEPELKP-COO−), and this had little effect on the intrinsic tryptophan fluorescence (Fig. 4A). We further confirmed an interaction between Pyd PDZ2 and the C-terminal of Crb3 using a dansylated 6-mer peptide (dansyl-PEERLI-COO−). This experiment served as an independent verification of the interaction between Pyd PDZ2 and the C-terminal peptide of Crb3. The rationale for the use of a shorter dansylated peptide was that such experimental set-ups have previously successfully been employed for other PDZ-peptide interactions (38). The dansyl-PEERLI-COO− peptide was titrated with increasing concentrations of wild-type Pyd PDZ2, which resulted in a distinct increase in dansyl fluorescence (Fig. 4B). Taken together, the results suggest that Pyd PDZ2 interacts with the C-terminal peptide of Crb3 in vitro, allowing its use as a model peptide.
Synergy between Peptide and PtdIns(4,5)P2 in Binding Pyd PDZ2 in Vitro
We used the 10-mer Crb3 peptide to investigate the interplay between peptide and PtdInsPs binding. The apparent KD values of the Pyd PDZ2 F194W 10-mer Crb3 interaction were determined in the absence and presence of two different IP3 concentrations (500 and 1000 μm) to be 220 ± 15 and 215 ± 15 μm, respectively, indicating that IP3 does not compete with peptide binding (Fig. 4A). Next, we investigated the relation between peptide and PtdIns(4,5)P2 binding by SPR experiments. We ensured that the 10-mer Crb3 peptide did not associate with the surface (Fig. 4C) and then incubated a fixed concentration of Pyd PDZ2 with different concentrations of LKLPPEERLI-COO− peptide before injection over C6-PtdIns(4,5)P2 (Fig. 4C). We observed a pronounced increase in response levels with increasing peptide concentrations, saturating at 500 μm peptide. We found the apparent affinity of Pyd PDZ2 for C6-PtdIns(4,5)P2 in the presence of 500 μm peptide to be 4.3 ± 0.2 μm, which is 6-fold lower than in the absence of the peptide (Fig. 4D). Importantly, the observed maximal response levels did not exceed the maximal response in the absence of peptide. The results thus suggest a synergy between peptide and PtdIns(4,5)P2 interactions.
Finally, we investigated to what extent peptide binding was affected by the nine mutations introduced to probe PtdInsP binding. Most mutations had only minor effects on the peptide binding (Table 2). However, the K236A and K242A mutations significantly affected both peptide and PtdIns(4,5)P2 interactions, possibly indicating shared structural determinants (Fig. 3).
The C Terminus of Crb3 Can Contribute to the Plasma Membrane Targeting of Pyd PDZ2 in Vivo
To establish that Crumbs represents a physiological ligand for Pyd PDZ2, we first verified that the two proteins co-localize at sites of cell-cell contacts (Fig. 4E). We then measured to what extent overexpression of Crb3 enhances the membrane localization of Pyd PDZ2. We scored, in living cells, the enrichment of eYFP-Pyd PDZ2 at sites of cell-cell contacts when expressed alone or when co-expressed with Crb3. When Pyd PDZ2 was expressed alone, it enriched at about 54% of the cell-cell contacts, and it increased to 67% upon co-expression with Crb3. As a control, we performed the same experiment using Crb3ΔERLI, which lacks the last four last amino acids that constitute the PDZ binding motif (23). The Crb3ΔERLI variant failed to increase enrichment of Pyd PDZ2 in cell-cell contacts (Fig. 4F). Taken together, the data thus identify Crumbs as a candidate physiological partner for Pyd PDZ2 and suggest that both lipid and Crumbs peptide binding contribute to plasma membrane targeting of Pyd.
DISCUSSION
We developed a cell-based screening assay for the identification of PtdInsP-interacting PDZ domains and applied it to the Drosophila proteome. We found one domain, Pyd PDZ2 (of 46 tested), that displayed sufficiently high affinity to target it to plasma membrane PtdIns(4,5)P2. A rather low frequency of high affinity PtdInsP-interacting domains has been observed also for other lipid-interacting modules, such as the PH and PX domains (14, 15). However, the outcome of our screen is surprisingly low, given previous estimates suggesting that up to 20% of PDZ domains function as lipid binding modules (13). However, this estimate was from a screen based on a co-sedimentation assay with liposomes prepared from bovine brain lipid extracts, which have complex and variable composition. It is therefore not clear which lipids bound to these PDZ domains and if these interactions are of physiological relevance. However, we cannot exclude the possibility that some of the PDZ domains we tested do interact with membrane lipids, although with too low affinities to target the screening construct to PtdInsPs-rich membranes; such weak interactions might nevertheless be relevant in the context of multidomain proteins. Indeed, it is quite common that individual lipid binding modules have rather low membrane affinity and require oligomerization or combination with other modules to achieve membrane targeting (40). In this context, it is noteworthy that our positive hit, Pyd PDZ2, forms a stable dimer.
Our study indicates that the interaction of Pyd PDZ2 with PtdIns(4,5)P2-containing membranes is multivalent and contains components of nonspecific electrostatic interactions. The large extended positive patch along one side of the molecule seems crucial for the electrostatic interactions with the negatively charged cytosolic face of the membrane. Interestingly, a similar positive electrostatic patch can be observed for the homologous human ZO-1 PDZ2 (data not shown). In this case, the extension of the positive patch is smaller and would be expected to provide weaker electrostatic interactions with negatively charged lipids. Indeed, ZO-1 PDZ2 has previously been shown to be a PtdInsP binding module although it does not target the plasma membrane PtdIns(4,5)P2 (11). From the experimental data, it is not obvious if the Pyd PDZ2 structure harbors a specific docking site for the PtdIns(4,5)P2 headgroup. However, we surmise that such a site might locate in the vicinity of the peptide carboxylate binding groove, where the extended α-helices face the lipid membrane (Fig. 3, A and B), because the strongest effect of the mutations on the lipid binding was observed for this region. Two mutations (K236A and K242A) significantly affect both peptide and PtdIns(4,5)P2 liposome binding, suggesting shared structural determinants.
The apparent cooperativity between peptide and PtdIns(4,5)P2 is an unexpected finding of our study, because it stands in contrast to the predominant view, that peptide and PtdInsP binding are competitive (8, 11, 13). At least two explanations can account for the observed synergy between peptide and PtdIns(4,5)P2 binding; peptide binding may induce conformational changes that lead to a higher PtdIns(4,5)P2 binding affinity, or additional electrostatic interactions contributed by the LKLPPEERLI-COO− peptide may stabilize the interactions with immobilized PtdIns(4,5)P2. In any case, it is an exciting finding because it suggests that PtdIns(4,5)P2 and peptide binding may work in a concerted manner to recruit the protein to the membrane. Exactly how Pyd PDZ2 structurally accommodates peptide and PtdIns(4,5)P2 binding is still not clear at this point. However, studies on other small protein modules, such as the PH domain of OSBP1 (41) and the disabled-1 PTB domain (42), have demonstrated that such dual recognition can be accomplished independently by small modular protein domains.
Our finding is particularly interesting, considering that some transmembrane receptors with PDZ binding motifs colocalize with PtdIns(4,5)P2 at the cellular membrane (e.g. the TRPc4α receptor) (43), whereas other receptors (e.g. Syndecan-4) (44), directly interact with membrane PtdIns(4,5)P2. This points toward an intriguing possibility, coincident detection of peptide and PtdInsPs signaling by PDZ domains, with potentially important implications for the biology of PDZ-containing proteins. In the case of Pyd, it may be of importance for its biological function in regulating the accumulation of various receptors at the PtdIns(4,5)P2-rich adherens junctions (19, 20). The synergy between peptide and PtdIns(4,5)P2 binding shown here may be representative for other PDZ domains, which paves the way for further studies.
Supplementary Material
Acknowledgments
We are grateful to Dieter Vermeire and Dimitri Jordens for technical support and to Helga Ceulemans and Joachim Schultz for providing Drosophila cDNAs. Arf6-Q67L was a generous gift from Bernhard Wehrle-Haller (University of Geneva), the mRFP-FKBP12-PtdIns(4,5)P2 5-phosphatase and eCFP-FRB-PM were from Peter Varnai (Semmelweis University, Budapest) and Tamas Balla (National Institutes of Health, Bethesda, MD), and the Crb3 expression vectors were from Ben Margolis (University of Michigan, Ann Arbor, MI).
This work was supported by the Fund for Scientific Research-Flanders (FWO), the Concerted Actions Program of the Katholieke Universiteit Leuven, the Belgian Federation against Cancer (BFK), the Interuniversity Attraction Poles of the Prime Ministers Services (IUAP), and the EMBO young investigator program (to P. Z.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1.
- PtdIns
- phosphatidylinositol
- PtdInsP
- phosphatidylinositol phosphate
- PtdIns(4,5)P2
- phosphatidylinositol 4,5-bisphosphate
- PtdIns(3,4,5)P3
- phosphatidylinositol 3,4,5-trisphosphate
- PH
- pleckstrin homology
- eYFP
- enhanced yellow fluorescent protein
- eCFP
- enhanced cyan fluorescent protein
- PC
- phosphatidylcholine
- PS
- phosphatidylserine
- PE
- phosphatidylethanolamine
- DOPC
- 1,2-dioleoyl-sn-glycero-3-phosphocholine
- IP3
- inositol 1,4,5-trisphosphate
- RU
- response unit.
REFERENCES
- 1. Feng W., Zhang M. (2009) Nat. Rev. Neurosci. 10, 87–99 [DOI] [PubMed] [Google Scholar]
- 2. Harris B. Z., Lim W. A. (2001) J. Cell Sci. 114, 3219–3231 [DOI] [PubMed] [Google Scholar]
- 3. Gallardo R., Ivarsson Y., Schymkowitz J., Rousseau F., Zimmermann P. (2010) Chembiochem 11, 456–467 [DOI] [PubMed] [Google Scholar]
- 4. Balla T. (2005) J. Cell Sci. 118, 2093–2104 [DOI] [PubMed] [Google Scholar]
- 5. Kutateladze T. G. (2010) Nat. Chem. Biol. 6, 507–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lemmon M. A. (2008) Nat. Rev. Mol. Cell Biol. 9, 99–111 [DOI] [PubMed] [Google Scholar]
- 7. Di Paolo G., De Camilli P. (2006) Nature 443, 651–657 [DOI] [PubMed] [Google Scholar]
- 8. Zimmermann P., Meerschaert K., Reekmans G., Leenaerts I., Small J. V., Vandekerckhove J., David G., Gettemans J. (2002) Mol. Cell 9, 1215–1225 [DOI] [PubMed] [Google Scholar]
- 9. Zimmermann P., Zhang Z., Degeest G., Mortier E., Leenaerts I., Coomans C., Schulz J., N'Kuli F., Courtoy P. J., David G. (2005) Dev Cell 9, 377–388 [DOI] [PubMed] [Google Scholar]
- 10. Mortier E., Wuytens G., Leenaerts I., Hannes F., Heung M. Y., Degeest G., David G., Zimmermann P. (2005) EMBO J. 24, 2556–2565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Meerschaert K., Tun M. P., Remue E., De Ganck A., Boucherie C., Vanloo B., Degeest G., Vandekerckhove J., Zimmermann P., Bhardwaj N., Lu H., Cho W., Gettemans J. (2009) Cell Mol. Life Sci. 66, 3951–3966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Pan L., Wu H., Shen C., Shi Y., Jin W., Xia J., Zhang M. (2007) EMBO J. 26, 4576–4587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu H., Feng W., Chen J., Chan L. N., Huang S., Zhang M. (2007) Mol. Cell 28, 886–898 [DOI] [PubMed] [Google Scholar]
- 14. Yu J. W., Lemmon M. A. (2001) J. Biol. Chem. 276, 44179–44184 [DOI] [PubMed] [Google Scholar]
- 15. Yu J. W., Mendrola J. M., Audhya A., Singh S., Keleti D., DeWald D. B., Murray D., Emr S. D., Lemmon M. A. (2004) Mol. Cell 13, 677–688 [DOI] [PubMed] [Google Scholar]
- 16. Wei X., Ellis H. M. (2001) Mech. Dev. 100, 217–231 [DOI] [PubMed] [Google Scholar]
- 17. Takahashi K., Matsuo T., Katsube T., Ueda R., Yamamoto D. (1998) Mech. Dev. 78, 97–111 [DOI] [PubMed] [Google Scholar]
- 18. Seppa M. J., Johnson R. I., Bao S., Cagan R. L. (2008) Dev. Biol. 318, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Djiane A., Shimizu H., Wilkin M., Mazleyrat S., Jennings M. D., Avis J., Bray S., Baron M. (2011) J. Cell Biol. 192, 189–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Pinal N., Goberdhan D. C., Collinson L., Fujita Y., Cox I. M., Wilson C., Pichaud F. (2006) Curr. Biol. 16, 140–149 [DOI] [PubMed] [Google Scholar]
- 21. Médina E., Lemmers C., Lane-Guermonprez L., Le Bivic A. (2002) Biol. Cell 94, 305–313 [DOI] [PubMed] [Google Scholar]
- 22. Várnai P., Balla T. (1998) J. Cell Biol. 143, 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roh M. H., Fan S., Liu C. J., Margolis B. (2003) J. Cell Sci. 116, 2895–2906 [DOI] [PubMed] [Google Scholar]
- 24. Narayan K., Lemmon M. A. (2006) Methods 39, 122–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Petrey D., Xiang Z., Tang C. L., Xie L., Gimpelev M., Mitros T., Soto C. S., Goldsmith-Fischman S., Kernytsky A., Schlessinger A., Koh I. Y., Alexov E., Honig B. (2003) Proteins 53, Suppl. 6, 430–435 [DOI] [PubMed] [Google Scholar]
- 26. Xiang Z., Honig B. (2001) J. Mol. Biol. 311, 421–430 [DOI] [PubMed] [Google Scholar]
- 27. Rocchia W., Sridharan S., Nicholls A., Alexov E., Chiabrera A., Honig B. (2002) J. Comput. Chem. 23, 128–137 [DOI] [PubMed] [Google Scholar]
- 28. Peters P. J., Hsu V. W., Ooi C. E., Finazzi D., Teal S. B., Oorschot V., Donaldson J. G., Klausner R. D. (1995) J. Cell Biol. 128, 1003–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Brown F. D., Rozelle A. L., Yin H. L., Balla T., Donaldson J. G. (2001) J. Cell Biol. 154, 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Varnai P., Thyagarajan B., Rohacs T., Balla T. (2006) J. Cell Biol. 175, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen J., Pan L., Wei Z., Zhao Y., Zhang M. (2008) EMBO J. 27, 2113–2123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Fanning A. S., Lye M. F., Anderson J. M., Lavie A. (2007) J. Biol. Chem. 282, 37710–37716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chi C. N., Bach A., Engström A., Wang H., Strømgaard K., Gianni S., Jemth P. (2009) Biochemistry 48, 7089–7097 [DOI] [PubMed] [Google Scholar]
- 34. Gianni S., Ivarsson Y., De Simone A., Travaglini-Allocatelli C., Brunori M., Vendruscolo M. (2010) Nat. Struct. Mol. Biol. 17, 1431–1437 [DOI] [PubMed] [Google Scholar]
- 35. Jackson S. E., Fersht A. R. (1991) Biochemistry 30, 10428–10435 [DOI] [PubMed] [Google Scholar]
- 36. Cheng Y., Prusoff W. H. (1973) Biochem. Pharmacol. 22, 3099–3108 [DOI] [PubMed] [Google Scholar]
- 37. Watton S. J., Downward J. (1999) Curr. Biol. 9, 433–436 [DOI] [PubMed] [Google Scholar]
- 38. Gianni S., Engström A., Larsson M., Calosci N., Malatesta F., Eklund L., Ngang C. C., Travaglini-Allocatelli C., Jemth P. (2005) J. Biol. Chem. 280, 34805–34812 [DOI] [PubMed] [Google Scholar]
- 39. Ivarsson Y., Travaglini-Allocatelli C., Brunori M., Gianni S. (2008) J. Biol. Chem. 283, 8954–8960 [DOI] [PubMed] [Google Scholar]
- 40. Lemmon M. A. (2003) Traffic 4, 201–213 [DOI] [PubMed] [Google Scholar]
- 41. He J., Scott J. L., Heroux A., Roy S., Lenoir M., Overduin M., Stahelin R. V., Kutateladze T. G. (2011) J. Biol. Chem. 286, 18650–18657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stolt P. C., Jeon H., Song H. K., Herz J., Eck M. J., Blacklow S. C. (2003) Structure 11, 569–579 [DOI] [PubMed] [Google Scholar]
- 43. Otsuguro K., Tang J., Tang Y., Xiao R., Freichel M., Tsvilovskyy V., Ito S., Flockerzi V., Zhu M. X., Zholos A. V. (2008) J. Biol. Chem. 283, 10026–10036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lee D., Oh E. S., Woods A., Couchman J. R., Lee W. (1998) J. Biol. Chem. 273, 13022–13029 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.