Abstract
PRMT5 is a type II protein arginine methyltranferase that catalyzes monomethylation and symmetric dimethylation of arginine residues. PRMT5 is functionally involved in a variety of biological processes including embryo development and circadian clock regulation. However, the role of PRMT5 in oligodendrocyte differentiation and central nervous system myelination is unknown. Here we show that PRMT5 expression gradually increases throughout postnatal brain development, coinciding with the period of active myelination. PRMT5 expression was observed in neurons, astrocytes, and oligodendrocytes. siRNA-mediated depletion of PRMT5 in mouse primary oligodendrocyte progenitor cells abrogated oligodendrocyte differentiation. In addition, the PRMT5-depleted oligodendrocyte progenitor and C6 glioma cells expressed high levels of the inhibitors of differentiation/DNA binding, Id2 and Id4, known repressors of glial cell differentiation. We observed that CpG-rich islands within the Id2 and Id4 genes were bound by PRMT5 and were hypomethylated in PRMT5-deficient cells, suggesting that PRMT5 plays a role in gene silencing during glial cell differentiation. Our findings define a role of PRMT5 in glial cell differentiation and link PRMT5 to epigenetic changes during oligodendrocyte differentiation.
Keywords: Cell Differentiation, DNA Methylation, Gene Expression, Myelin, Oligodendrocytes
Introduction
Protein arginine methyltransferases (PRMTs)3 transfer methyl groups from S-adenosylmethionine to the nitrogen atoms of arginine, a positively charged amino acid that mediates hydrogen bonding and amino aromatic interactions (1). Arginine can be monomethylated or dimethylated (DMA). If two methyl groups are both placed on one of the terminal nitrogen atoms of the guanidino group, the derivative is an asymmetric DMA. Symmetric DMA is where one methyl group is located on each of the two nitrogens. There are currently nine mammalian PRMTs that are grouped as type I and type II enzymes (1, 2). Type I enzymes form monomethylated arginine and asymmetric DMA and include PRMT1, PRMT3, CARM1, PRMT6, and PRMT8. Type II enzymes, including PRMT5, PRMT7, and PRMT9, catalyze the production of monomethylated arginine and symmetric DMA. The enzyme activity of PRMT2 is not yet identified.
Symmetrical arginine dimethylation occurs less frequently than asymmetrical dimethylation, and among the type II enzymes, PRMT5 has been the most characterized. It is functionally involved in a broad spectrum of biological processes, but most importantly in early embryo development and later cellular differentiation as well as the circadian clock (3–6). Accumulating evidence reveals that PRMT5 recurrently exerts its biological functions through transcriptional repression of a large group of genes involved in these processes. PRMT5 associates with the cyclin E1 promoter and represses cyclin E1 transcription, leading to inhibition of cell proliferation (7). PRMT5 associates with BRG1-based SWItch/Sucrose NonFermentable (hSWI/SNF) chromatin remodeling complexes and promotes transcriptional repression of the Myc target gene cad (8). BRG1- and hBRM-associated PRMT5 also represses expression of ST7 and NM23 tumor suppressor genes and regulates cell growth and proliferation (9). PRMT5 is a key component of the SNAIL-silencing complex through binding to AJUBA and functions to repress the SNAIL target gene, E-cadherin (10, 11). PRMT5 suppresses the transcription of the RB family of tumor suppressors in leukemia and lymphoma cells (12). In addition, microarray studies demonstrate that many genes are up-regulated, but only a few genes are down-regulated when PRMT5 levels are reduced (9).
Histone 4 arginine 3 (H4R3) and histone 3 arginine 8 (H3R8) are two important PRMT5 substrates (12–15). As histone marks, symmetric dimethylation of H4R3 (H4R3sme2) and H3R8 (H3R8sme2) directly contributes to PRMT5 transcriptional repression functions. Indeed, global analysis of histone methylation ChIP sequencing data revealed recently that H4R3me2 globally represses gene expression (16). Along with 20 histone methylation events analyzed using two different methods, H4R3me2 was found to be frequently associated with global repressive marks (16). Moreover, methylation of non-histone proteins also interferes with gene expression. For example, PRMT5 methylation of transcription elongation factor SPT5 promotes its dissociation from the transcriptional complex (17).
Oligodendrocytes are the core myelin-producing cells in the CNS. Oligodendrocyte differentiation and myelin production are highly controlled by a complex network of signal transduction pathways that regulates the expression of an array of transcription factors inductive or inhibitory to the processes. Transcription factors that promote oligodendrocyte differentiation include the basic helix-loop-helix family members Olig1 and Olig2; Sox family members Sox8, Sox9, Sox10, and Sox17; and the homeobox containing (Hox) transcription factors Nkx2.2 and YY1, whereas differentiation inhibitors include members of the inhibitors of differentiation/DNA-binding (Id) family Id2 and Id4, as well as transcription factor 4 (Tcf4) (18–20).
Oligodendrocyte differentiation is also regulated at the epigenetic level, such as chromatin remodeling, histone modification, and DNA methylation (21). Inhibition of histone deacetylase activity in postnatal rats causes a delay in oligodendrocyte differentiation and myelination (22). Conditional deletion of histone deacetylase 1 and 2 in the oligodendrocyte lineage causes a loss of oligodendrocytes and development of severe hypomyelination (23). Recently, it has been reported that both symmetric and asymmetric dimethylation of H4R3 are dynamically distributed in murine developing oligodendrocyte precursors (24). However, so far, it remains unclear how protein arginine methylation is involved in the regulation of oligodendrocyte differentiation and myelin production. In this study, we uncovered that PRMT5 expression level in brain gradually increases during active myelination, and knockdown of PRMT5 expression leads to impaired oligodendrocyte differentiation. We further demonstrate that PRMT5 associates with Id2 and Id4 CpG islands and is required for maintaining their methylation status. These findings suggest that PRMT5 plays a role in gene silencing during oligodendrocyte differentiation.
EXPERIMENTAL PROCEDURES
Antibodies and Reagents
Anti-A2B5 antibody was obtained from Dr. T. E. Kennedy (McGill University). Anti-galactocerebroside (GalC) antibody was obtained from Dr. V. W. Yong (University of Calgary). Anti-glial fibrillary acidic protein mouse antibody was purchased from DAKO Diagnostics, and anti-neurofilament H monoclonal antibody was purchased from Santa Cruz Biotechnology Inc. Anti-MBP monoclonal antibody was purchased from Sternberger Monoclonals. Anti-Myc rabbit polyclonal antibody was purchased from Abcam. Anti-QKI-5 anti-QKI-6, anti-PRMT1, anti-PRMT5, and anti-MBP rabbit polyclonal antibodies were obtained from Millipore Inc. Anti-CARM1 antibody was from Active Motif Inc. The secondary antibodies used were goat anti-mouse Cy3 (Jackson Immunoresearch), goat anti-rabbit Cy3 (Jackson Immunoresearch), and goat anti-rabbit Alexa 488 (Molecular Probes). Nuclear Hoechst dye (bis-benzamide) was purchased from Molecular Probes.
Cell Culture and siRNA Transfection
Primary oligodendrocyte progenitor cells (OPCs) were prepared from newborn Sprague-Dawley rat brains as described previously (25). The cells were plated on poly-d-lysine-coated dishes and grown in serum-free DMEM-F12 mixture (1:1) medium supplemented with 10 mm HEPES, 0.1% bovine serum albumin, 25 μg/ml of human transferrin, 30 nm triiodothyronine, 20 nm hydrocortisone, 20 nm progesterone, 10 nm biotin, 5 μg/ml of insulin, 16 μg/ml of putrescine, 30 nm selenium, and 2.5 ng/ml of each of PDGFAA and basic FGF. The progenitors are proliferative under these conditions, whereas removal of the mitogens initiates their differentiation. Culture medium is replaced every 2 days under all experimental conditions.
A pool of three siRNAs designed to target PRMT5 (5′-GAG CAC AGC ACU UCC UGA AAG AUG A-3′, 5′-AGA CGU GGU UGU GGU GGC AUA ACU U-3′, and 5′-CCA UCC CAA CCG AGA UCC UAU GAU U-3′) or to target QKI (5′-GGA CUU ACA GCC AAA CAA C-3′) and luciferase specific control siRNA (5′-CGU ACG CGG AAU ACU UGA-3′) with or without 5′-tagged AlexaFluor488 fluorophore were purchased from Invitrogen. Primary OPCs were transfected using the siIMPORTER reagent (Upstate Biotechnology, Lake Placid, NY) and 120 nm siRNAs first for 48 h prior to growth factor removal and for 48 h after growth factor removal according to the manufacturer's instructions. The cells were harvested 48 h postdifferentiation unless otherwise specified.
C6 rat glioma cells were cultured in DMEM (Hyclone) supplemented with 10% bovine calf serum (Hyclone). The cells were transfected with 120 nm of indicated siRNAs with Lipofectamine RNAiMAX (Invitrogen), using the reverse method according to the manufacturer's instructions. After 24 h transfection, the cell medium was replaced with serum free DMEM for 3 days.
Immunoblotting
Samples were lysed with lysis buffer (50 mm HEPES, 150 mm NaCl, 1% Triton, and protease inhibitors) and sonicated. Fifty micrograms of protein were separated by 10% SDS-PAGE, transferred onto a nitrocellulose membrane (Bio-Rad), and immunoblotted as described (26).
Tissue Processing and Immunohistochemistry
The mice were handled and sacrificed in accordance with a protocol approved by the Animal Care Committee at McGill University. Anesthetized mice were perfused with ice-cold PBS followed by 4% paraformaldehyde. The extracted brains were cryoprotected in 4% paraformaldehyde overnight at 4 °C and then in 30% sucrose at 4 °C until dehydrated. Tissue blocks were embedded in OCT compound (Tissue-Tek) over dry ice in acetone. Tissues were cryostat sectioned at 10 μm and collected on +/+ slides (Fisher). Tissue sections were blocked in 10% goat serum in TBS containing 0.5% Triton X-100 for 1 h followed by incubation with primary antibodies in TBS overnight at room temperature. The slides were incubated with Alexa-fluor 488 or 546 IgG in TBS at a dilution of 1:400 for 4 h.
Immunofluorescence
Live cell staining was performed for antibodies against surface antigens A2B5 (1:30), O4 (1:50), and GalC (1:30), respectively, in culture medium at 37 °C for 15 min. The cells were then fixed with 4% paraformaldehyde. For cytoplasmic antigens, the cells were permeablized with 0.1% Triton X-100 in PBS or cold acetone, and then blocked in 10% goat serum in PBS. Fixed cells were stained with antibodies against intracellular antigens PRMT5 (1:200) and QKI-5/QKI-6 (1:200) at room temperature for 1 h. Appropriate secondary antibodies were applied at 4 °C for 30 min. The cell nuclei were identified using Hoechst dye (1:1,000). The cells were mounted onto glass slides and visualized with a Carl Zeiss M1 microscope.
Quantitative Real Time PCR
Total RNA was extracted using the RNeasy Plus Minikit (Qiagen), treated with DNase (Thermo Scientific), and reverse transcribed with Super Script II reverse transcriptase (Invitrogen) according to the manufacturer's protocols. The qPCR primers designed for GAPDH, PRMT5, PRMT7, and p27Kip1 were purchased from Qiagen, and the primers for Id2 (5′-GGA CAG AAC CAA ACG TCC AG-3′ and 5′-TAA GCT CAG AAG GGA ATT CAG AC-3′), Id4 (5′-GGG CGA CAG CAT TCT CTG-3′ and 5′-CTC TGG CCC TCC CTT TCT-3′), Tcf4 (5′-CAC TCC TCG GCA GAC ATC AA-3′ and 5′-TTT GTG CCA CTT CGA TGG AA-3′), Sox10 (5′-ATG TCA GAT GGG AAC CCA GA-3′ and 5′-GTC TTT GGG GTG GTT GGA G-3′), Nkx2.2 (5′-GCC TGC CCC TTA AGA GTC CTT-3′ and 5′-GGC CAG CCA GCG AGT GTA-3′), and Olig2 (5′-CCT CCT GTC TCT CCT GTC GAT T-3′ and 5′-CAT GTG GTC TGA AAA GGA ACA TTC-3′) were used for qPCR using Quantitect SYBR Green qPCR kit (Qiagen) on Applied Biosystem 7500 real time PCR system. The relative expression values for each gene of interest normalized to GAPDH were analyzed by ABI SDS software using the comparative CT method.
Bisulfite Sequencing
DNA was extracted from cells 48 h post-siRNA transfection. Purified DNA was bisulfite-treated using a CpGenome fast DNA modification kit (Millipore) following the manufacturer's protocol. The following nonmethylation specific primers were designed using the Methyl Primer Express program. The resulting PCR products were cloned into pBluescript SK (+), and at least 20 clones were sequenced (Genome Quebec). The significance between different siRNA-transfected cultures was calculated by Fisher's exact test. The primers used were: for Id2-1 (−316 to −134), 5′-GGG TTT TGT TAG TTT TGG AAA TTA G-3′ and 5′-CCT CCC ATT AAT AAA TAA AAA CTC TC-3′; for Id2-2 (−1950 to −1772), 5′-TTT TGG TAT GAT AGT TAG AGA AAG AAA-3′ and 5′-TTC CAA CAA AAA AAC CCT AAA CTA A-3′; for Id2-8 (+592 to +898), 5′-ATG AAT TTG TTT AAT GGG ATT TG-3′ and 5′-CCC TTC ATA CCT CTC TAA AAA C-3′; and for Id 4–8 (+1211 to +1505), 5′-GGT TTG ATA TTT ATA AAG AGG GAT G-3′ and 5′-TTC TAC TCT AAC CCT CCC TTT C-3′.
Chromatin Immunoprecipitation
C6 rat glioma cells were grown to 40% confluence in DMEM supplemented with 10% bovine calf serum, where upon the medium was replaced with serum-free culture medium to allow the cells to differentiate. After 3 days of differentiation, the cells were trypsinized and incubated with 1% formaldehyde in DMEM for 10 min at room temperature. Cross-linking was stopped with the addition of glycine, and the cells were washed with cold 1× PBS. The cells were resuspended in swelling buffer containing 50 mm Tris, pH 8.0, 85 mm KCl, and 0.5% Nonidet P-40 supplemented with protease inhibitors (Roche Applied Science) and passed through an insulin syringe multiple times. The nuclei were suspended in nuclei lysis solution (50 mm Tris, pH 8.0, 10 mm EDTA, 1% SDS, and protease inhibitors) and sonicated four times for 10 s at 50% duty to generate chromatin 200–500 bp in length. The samples were then incubated with 2 μg of anti-PRMT5 (Millipore) or control IgGs (Vector Labs). DNA was isolated through column purification (Qiagen) and subjected to qPCR. The following primers were used: Id2-2 (5′-CAG GAA TGG TCT TGG GAG AA-3′ and 5′-GCC AGC CTC ATT ATT CCA AC-3′), Id2-8 (5′-TGC TTT TTA TCC TCT TTC TCT CCA-3′ and 5′-CCG CAA ACA CTC ACC ATT TA-3′), Id4-8 (5′-TTT TGG GTT GTT GAT CAT GC-3′ and 5′-CAC CTG TGG AAG AGG AAT GAA-3′), p27 (5′-GCC GTT TGG CTA GTT TGT TT-3′ and 5′-GAG GTG TAC GAC TGC CAA CA-3′), and Tcf4 (5′-GAT CTT GGC TGT GTG TCT GC-3′ and 5′-CAC CAC CAC CTC CTC CAA C-3′).
RESULTS
Elevated PRMT5 Expression in Brain Coincides with CNS Myelination
To identify a role for protein arginine methylation during CNS myelination, we examined the expression of the major PRMTs in brain lysates in postnatal mice. Whole brain lysates from postnatal day 1 (P1), P4, P11, P14, and P21 and adult mice were analyzed for PRMT1, CARM1, and PRMT5 expression by immunoblotting (Fig. 1). The onset of CNS myelination occurs ∼2 weeks after birth in mice, as monitored by the expression of the myelin component MBP (Fig. 1). The expression of PRMT1 and CARM1, type I enzymes that generate asymmetric DMA, gradually decreased after birth in whole brain lysates (Fig. 1). In contrast, the brain expression of PRMT5, a type II enzyme that generates symmetric DMA, gradually increased after P11 and peaked in adult mice (Fig. 1). These findings show that elevated PRMT5 expression coincides with CNS myelination, and we focused our studies on PRMT5.
FIGURE 1.
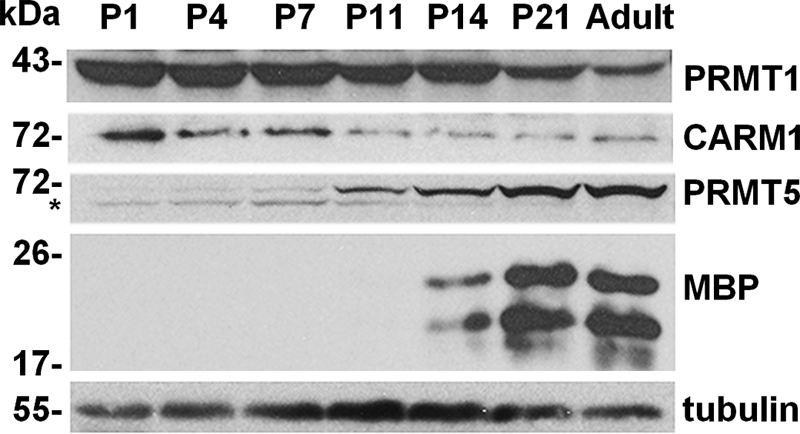
PRMT1, CARM1, and PRMT5 expression in the developing mouse brain. Total brain lysates were obtained from P1, P4, P7, P11, P14, P21, and adult mice. The expression of PRMT1, CARM1, and PRMT5 was analyzed by immunoblotting. MBP isoforms, observed as the two classical major isoforms, were used as markers of myelination, and β-tubulin was used as a loading control. The asterisk denotes a nonspecific band. The molecular mass markers are shown in kDa on the left.
PRMT5 Is Expressed in Myelinating Oligodendrocytes
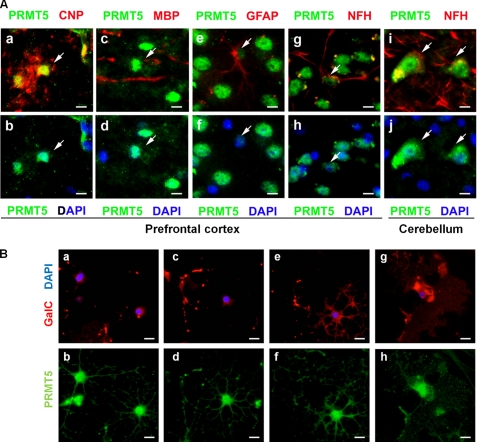
We next examined whether PRMT5 was expressed within glial or neuronal cells by immunohistochemistry. Prefrontal cortex sections immunolabeled with 2′,3′-cyclic nucleotide 3′ phosphodiesterase, a marker of oligodendrocytes, showed elevated nuclear expression of PRMT5 with expression in the cell body and the processes of oligodendrocytes (Fig. 2A, panels a and b). The actively myelinating oligodendrocytes in proximity to MBP-positive axon fibers also had a similar expression pattern (Fig. 2A, panels c and d). Astrocytes identified by glial fibrillary acidic protein also expressed PRMT5 but at much lower levels compared with oligodendrocytes (Fig. 2A, panels e and f). Neurofilament H-positive neurons in the prefrontal cortex were also intensely stained by the PRMT5 antibody (Fig. 2A, panels g and h). Furthermore, the glutamatergic neurons in the cerebellum also expressed significant levels of PRMT5 (Fig. 2A, panels i and j). In summary, PRMT5 is expressed in mature, myelinating oligodendrocytes, as well as in neurons and astrocytes.
FIGURE 2.
PRMT5 is expressed in oligodendrocytes, as well as in neurons and astrocytes. A, prefrontal cortex (panels a–h) and cerebellar sections (panels i and j) of wild-type P30 mice were immunostained with anti-PRMT5 (green), anti-2′,3′-cyclic nucleotide 3′ phosphodiesterase (CNP, red), MBP (red), anti-glial fibrillary acidic protein (GFAP, red), and anti-neurofilament H (NFH, red) antibodies and DAPI (blue), and visualized by fluorescence microscopy. Scale bar, 10 μm. B, oligodendrocyte cultures differentiated for 2 days were immunostained with GalC (red), PRMT5 (green), and DAPI (blue). Typical images of GalC-negative OPC with few primary branches (panels a and b), GalC-negative OPC with primary and secondary branching (panels c and d), GalC-positive immature oligodendrocytes with complex branch formations (panels e and f) and GalC-positive mature oligodendrocytes with sheet-like membrane structures (panels g and h) are shown. The sections were visualized by fluorescence microscopy. Scale bar, 10 μm.
Because oligodendrocytes are the myelinating cells of the CNS, we next examined the intracellular localization of PRMT5 within cultured oligodendrocytes. Early progenitors, characterized by the absence of GalC expression and the presence of only a few primary branches, expressed PRMT5 within both the processes and the cell body with a high nuclear concentration (Fig. 2B, panels a and b). A similar staining pattern was observed in the GalC-negative late progenitors (Fig. 2B, panels c and d). In the GalC-positive immature oligodendrocytes, a significant proportion of PRMT5 was concentrated within the processes, including the distal process tips (Fig. 2B, panels e and f). In myelinating oligodendrocytes, PRMT5 also accumulated within the sheet-like myelin structures (Fig. 2B, panels g and h). These results indicate that PRMT5 is highly concentrated within the nucleus and the distal processes of oligodendrocytes.
PRMT5 Deficiency Induces the Expression of Id2 and Id4 in C6 Glioma Cells
The increase in PRMT5 expression observed during the period of active myelination led us to evaluate whether PRMT5 plays a role in the process of glial cell maturation. C6, a rat glioma cell line, retains the characteristics of undifferentiated glioma and express astrocyte and oligodendrocyte markers (27–30). Moreover, these cells can be induced to commit to the oligodendrocyte lineage with elevated myelin gene expression, and they are easily transfectable. Furthermore, stimulating the lipid homeostasis pathways of C6 cell line induces the expression of MBP proteins (31). Thus C6 glioma makes an ideal proof-of-concept system to study the participation of PRMT5 in glial cell differentiation.
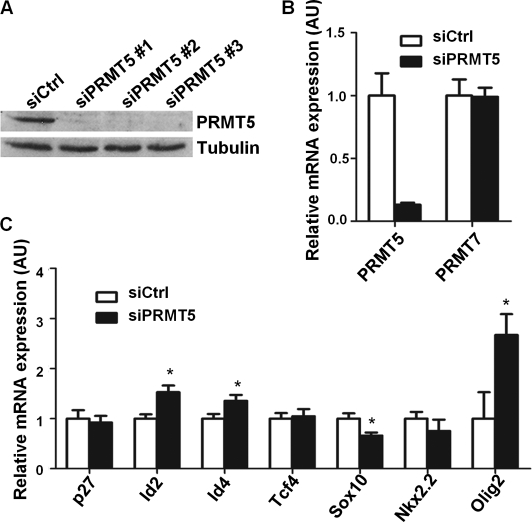
We first transfected C6 cells with three separate PRMT5-specific siRNAs (siPRMT5) and luciferase-specific siRNA (siCtrl) as a control. We then cultured the cells in high density under serum-free conditions to promote the expression of myelin genes. Each individual siPRMT5 was able to lower the expression of PRMT5 protein by >90% (Fig. 3A). Moreover, using quantitative PCR, we detected ∼80% reduction in PRMT5 mRNA levels, whereas the transcriptional level of another type II protein arginine methyltransferase, PRMT7, remained unaffected (Fig. 3B). We then examined the expression of transcriptional regulators involved in oligodendrocyte differentiation such as inhibitors of differentiation/DNA binding (Id) family Id2 and Id4, as well as the transcription factor 4 (Tcf4), Sox family member Sox10, the homeobox-containing (Hox) transcription factor Nkx2.2, and the basic helix-loop-helix family member Olig2 (18–20). The expression of Id2 and Id4 was significantly elevated in siPRMT5-transfected C6 cells, whereas the expression of Tcf4 remained constant (Fig. 3C). On the other hand, the levels of the differentiation promoters Sox10 and Nkx2.2 were decreased in the siPRMT5-transfected cells (Fig. 3C). The level of Olig2, a promoter of oligodendrocyte differentiation was significantly increased (Fig. 3C), which possibly reflects compensational mechanisms for the loss of differentiation potential. Although the level of cell cycle inhibitor p27Kip1 was not altered in PRMT5 knocked down cells (Fig. 3C), PRMT5 promotes cell cycle arrest in a p53-dependent manner (32), and PRMT5 also negatively regulates cyclin E1 promoter activity (7). Taken together, PRMT5 deficient C6 glioma cells harbored gene expressions of immature, undifferentiated, proliferating glial cells.
FIGURE 3.
PRMT5-deficient C6 glioma cells lead to elevated Id2 and Id4 expression. A, C6 glioma cells were transfected with three separate siPRMT5 and siCtrl, respectively. The cells were then cultured in serum-free conditions for 48 h and immunoblotted for PRMT5 expression. The expression of β-tubulin was used as a loading control. B and C, total RNA was extracted from C6 cells transfected with pooled siPRMT5 and siCtrl, respectively. Normalized to GAPDH, the relative expression level of each indicated transcript was assessed by quantitative RT-PCR. (The asterisk denotes a significant difference from siCtrl-treated cells; p < 0.05, Student's t test.)
Knockdown of PRMT5 Inhibits Oligodendrocyte Differentiation
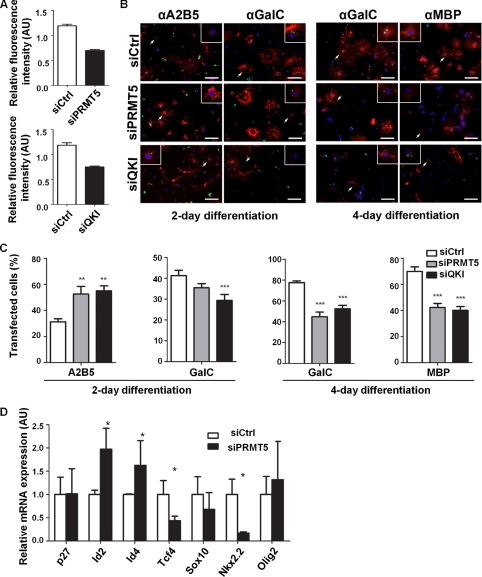
We next examined whether PRMT5 is required for oligodendrocyte differentiation. Primary OPCs were transfected with siPRMT5, luciferase-specific siRNA, and an siRNA against the QKI RNA binding proteins. The QKI RNA binding proteins are known to be required for oligodendrocyte differentiation (33) and served as a positive control. Two days later, another round of siRNA transfection was performed, and growth factor withdrawal-mediated differentiation was initiated. Because of the low transfection efficiency of primary OPCs, Alexa Fluor-488 fluorescent dye-tagged siRNAs were used to identify transfected cells. By quantifying the fluorescence intensity, a polyclonal antibody against PRMT5 detected ∼40% reduction of PRMT5 in siPRMT5-transfected oligodendrocytes in comparison to the siCtrl-transfected cells (Fig. 4A). An ∼30% knockdown efficiency was detected in siQKI transfected OPCs, using a QKI-specific polyclonal antibody (Fig. 4A). We monitored oligodendrocyte differentiation by using immunocytochemistry with known markers. As expected, at day 2 postdifferentiation, knockdown of QKI prevented oligodendrocyte differentiation, as indicated by an increase (p < 0.0001) of A2B5-positive cells, an early oligodendrocyte progenitor marker (Fig. 4, B and C). Similarly, a reduction of GalC-positive immature oligodendrocytes (p = 0.0068) was observed in the siQKI-transfected cells (Fig. 4, B and C). These effects were further augmented at day 4 postdifferentiation, where the QKI knockdown diminished populations of both GalC-positive immature oligodendrocytes (p < 0.0001) and MBP-positive mature oligodendrocytes (p < 0.0001) (Fig. 4, B and C). The knockdown of PRMT5 also prevented primary oligodendrocyte maturation. At day 2 postdifferentiation, PRMT5 knockdown significantly increased the proportion of the early A2B5-positive progenitors (p = 0.004) and slightly reduced the proportion of GalC-positive immature oligodendrocytes (p = 0.0923; Fig. 4, B and C). By day 4 the knockdown of PRMT5 significantly decreased the immature GalC-positive oligodendrocytes and the mature MBP-positive oligodendrocytes (p < 0.0001 for both; Fig. 4, B and C). Together, these data suggest that PRMT5 is indispensible for oligodendrocyte differentiation.
FIGURE 4.
PRMT5 is required for oligodendrocyte differentiation. Primary OPCs isolated from P1 rat brains were transfected twice with Alexa Fluor 488-conjugated siPRMT5, siCtrl, and siQKI. Forty-eight hours after the first transfection, the cells were retransfected with a second round and then induced for OPC differentiation by growth factor withdrawal. The cultures were differentiated for 2 or 4 days. A, To quantify the knockdown efficacy, at day 2 after induction of differentiation, the OPCs were fixed, permeablized, and immunostained with anti-QKI and PRMT5 antibodies (red). Quantified by Zeiss AxioVision program, the intensity of each QKI or PRMT5 transfected cell was averaged and normalized to that of the nontransfected cells. B and C, to demonstrate OPC maturation levels, the transfected OPCs were immunostained (red) for A2B5 and GalC on day 2 and for GalC and MBP on day 4 of differentiation. A representative image for each is shown. The cells that retained the siRNA are indicated by the green staining and counted as transfected cells. The percentages of A2B5, GalC, and MBP-positive cells in the transfected cell populations was calculated. Approximately 500 transfected cells were counted from nine separate fields on three coverslips in total for each staining. A typical experiment is shown in B, and the average percentage with ± S.E. is plotted in C. *, significantly different from siCtrl treated cells, p < 0.05; **, p < 0.01; ***, p < 0.001, Student's t test. Scale bar, 50 μm. D, total RNA was extracted from OPCs transfected with siPRMT5 and siCtrl. Normalized to GAPDH, the relative expression level of each indicated transcript was assessed by quantitative PCR. (Asterisk denotes significantly different from siCtrl treated cells; p < 0.05, Student's t test.)
Profiling some key transcriptional regulators, we observed that PRMT5-deficient OPCs also displayed up-regulation of Id2 and Id4 and a reduction of Tcf4, Sox10, and Nkx2.2 (Fig. 4D). We also observed an increase in Olig2 transcripts albeit to a lower level than in C6 glioma cells (Fig. 4D). The level of the OPC cell cycle regulator p27Kip1 remained constant in siPRMT5-transfected OPCs (Fig. 4D). These data suggest that the knockdown of PRMT5 in primary OPCs results in an immature transcription factor expression profile consistent with a role for PRMT5 in gene regulation during oligodendrocyte differentiation.
PRMT5 Regulates DNA Methylation in the CpG Islands within Id2 and Id4 Genes
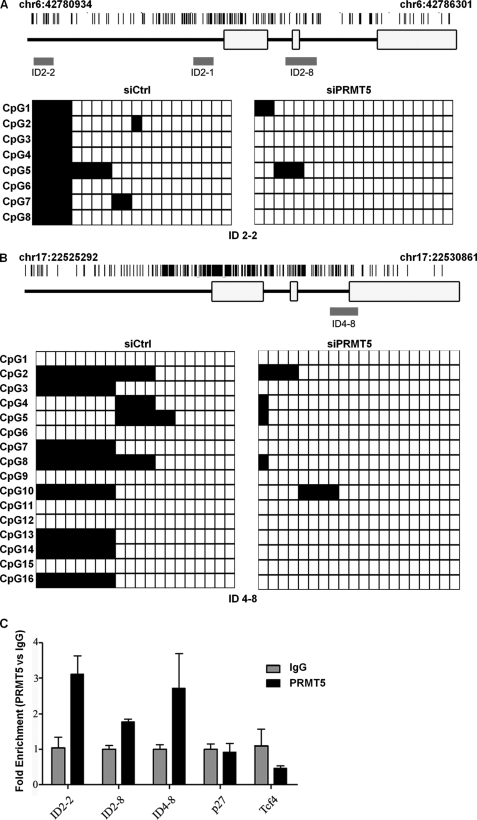
We next evaluated the underlying mechanism by which PRMT5 promotes oligodendrocyte differentiation. PRMT5-mediated H4R3 methylation directly recruits DNA methyltransferase DNMT3A, linking PRMT5 function to epigenetic gene silencing (34). To determine whether PRMT5 uses a similar mechanism in the regulation of oligodendrocyte differentiation, we examined whether PRMT5-deficient glial cells harbored hypomethylated CpG islands within Id2 and Id4 genes using bisulfite sequencing. We identified a CpG region 1.8 kb upstream of the transcriptional start site of Id2 gene (Id2-2; Fig. 5A) and one at the intron 2/exon 3 junction of Id4 gene (Id4-8; Fig. 5B) to be significantly methylated in C6 glioma cells, whereas two other CpG regions of the Id2 gene (Id2-1 and Id2-8; Fig. 5A) were unmethylated in C6 glioma cells (not shown). Interestingly, PRMT5 deficiency led to hypomethylation of both Id2-2 and Id4-8 regions within the Id2 and Id4 genes, respectively (Fig. 5, A and B, p < 0.001). We next examined whether PRMT5 was associated with the CpG islands within Id2 and Id4 genes. Indeed we identified PRMT5 association with the Id2-2 and Id4-8 genomic regions by ChIP using C6 glioma cells (Fig. 5C). As a negative control, PRMT5 did not associate with the p27Kip1 genomic region (Fig. 5C), consistent with the fact that p27Kip1 levels were not regulated by PRMT5 during OPC differentiation. We also showed that the specific Id genomic region, Id4-8, harbored increased DNA methylation in serum-free differentiating conditions (supplemental Fig. S1), consistent with our findings that Id CpG methylation is associated with glial cell differentiation. These findings show that PRMT5 inhibits the expression of Id2 an Id4 by regulating region-specific DNA methylation.
FIGURE 5.
PRMT5-deficient C6 glioma cells harbor hypomethylated DNA in the CpG islands of Id2 and Id4 genes. C6 glioma cells were transfected with siPRMT5 and siCtrl, respectively. 48 h after transfection, genomic DNA was extracted and subjected to bisulfite-sequencing as described under “Experimental Procedures.” A and B, the location of each sequenced region within the gene is denoted in red. Each row shows the methylation status (black, methylated; white, nonmethylated) of individual CpG dinucleotides derived from sequence analysis of at least 20 individual cloned PCR products of the Id2 (A) or Id4 (B) genes following bisulfite modification. The differences between the siCtrl and siPRMT5 panels are highly significant (p < 0.001, Fisher exact test). C, PRMT5 associates within chromatin areas of Id2 and Id4. Cross-linked chromatin prepared from C6 rat glioma cells were subjected to ChIP analysis using anti-PRMT5 or control IgGs. Immunoprecipitated DNA was analyzed in triplicate by qPCR with primers specific for genes Id2, Id4, p27, and Tcf4. The mean values are expressed as fold enrichment.
DISCUSSION
In this study, we demonstrate that PRMT5 is required for oligodendrocyte differentiation, supported by several observations. First, PRMT5 expression level is elevated in the central nervous system during the period of active myelination. Second, PRMT5 deficiency leads to impaired primary mouse OPC differentiation. Finally, knockdown of PRMT5 in C6 rat glioma cells or OPCs results in significant changes of the genes typically involved in the oligodendrocyte differentiation toward an expression profile of a more immature phenotype, such as increased expression of Id2 and Id4. Moreover, studies of detailed molecular mechanisms reveal that PRMT5 positively regulates DNA methylation within the CpG islands of Id2 and Id4 genes and contributes to the inhibitory regulation of Id2 and Id4 expression during oligodendrocyte differentiation.
PRMT5 has been shown to function as a transcriptional repressor. When associated with the SWI-SNF chromatin remodeling complex members such as BRG and BRM, it methylates H3R8 and H4R3, which repress various genes (9). The symmetrically dimethylated H4R3 (H4R3sme2) serves to recruit the DNA methyltransferase DNMT3A, which methylates neighboring CpG islands leading to gene silencing (34). These findings couple PRMT5 function to DNA methylation. Here, we demonstrate that Id2 and Id4 gene methylation is regulated by PRMT5 in glial cells, resulting in silencing of Id2 and Id4 expression.
The Id family of proteins consists of four members: Id1, Id2, Id3, and Id4. Because Id2 and Id4 are inhibitors of differentiation, down-regulation of their expression plays an important role in oligodendrocyte differentiation (18, 35, 36). Overexpression of Id2 or Id4 significantly blocks oligodentrocyte differentiation, whereas the absence of Id2 or Id4 induces premature oligodentrocyte differentiation (18, 35). However, the molecular mechanism by which Id2 and Id4 expression is down-regulated during oligodendrocyte differentiation has remained unknown. Interestingly, accumulating evidence suggests that Id4 expression is repressed by DNA methylation at neighboring CpG islands (37, 38). Hypermethylation at the Id4 gene is associated with a loss of Id4 expression in a few types of cancers (37, 39, 40). The methylation of the Id2 gene is less understood. However, its promoter region is enriched with GC-rich elements, such as bone morphogenetic protein (BMP) response element (GGCGCC) (41). The same BMP response element was also identified in other Id family members (42). Id family proteins are direct target genes of BMP, the members of TGF-β family of growth factors. Their expression is induced by BMP stimulation (41, 43). We show that the CpG islands within Id2 gene are methylated. As a result, this methylation can repress Id2 expression by forming compact inactive chromatin as a general mechanism by which DNA methylation represses gene expression. Alternatively, DNA methylation may interfere with the BMP response through methylation of the GC-rich BMP response elements.
It is known that induction of Id2 and Id4 mediates the inhibitory effect of BMP signaling on oligodendrocyte differentiation and the stimulatory effect on the generation of astrocytes (44, 45). Interestingly, we found that the expression level of PRMT5 is much lower in astrocytes than in oligodendrocytes (Fig. 2A). Because PRMT5 is required for the methylation of Id2 and Id4 genes (Fig. 5), it is possible that Id2 and Id4 genes are hypomethylated in astrocytes where PRMT5 is less expressed. Therefore, the epigenetic regulation of Id family proteins by DNA methylation may antagonize the inhibitory effect of BMP signaling on oligodendrocyte differentiation, whereas the BMP signal is required for generation of astrocytes. This mechanism can contribute to the precise regulation of lineage selection between oligodendrocyte and astrocyte during the differentiation of neural precursors in the developing brain.
PRMT5 has been also shown to positively regulate gene expression, although less is known about the mechanism of the action. PRMT5 is required for the activation of myogenin expression and MyoD-mediated skeletal muscle differentiation (46). PRMT5 dimethylates H3R8 at the myogenin gene promoter and facilitates BRG1-dependent chromatin remodeling for transcriptional activation of the target genes (46). Our studies reveal that PRMT5 deficiency leads to reduced expression of the differentiation promoters Sox10 and Nkx2.2, suggesting that PRMT5 positively regulates Sox10 and Nkx2.2 expression. Therefore, PRMT5 might also regulate oligodendrocyte differentiation through modulating Sox10 and Nkx2.2 expression, although the molecular mechanism remains to be studied. Finally, we found that the expression level of the differentiation promoter Olig2 is increased in both PRMT5 knocked down C6 cells and OPCs. It is not clear how the altered level of Oligo2 expression is related to PRMT5 function. However, it remains possible that the increased expression reflects a compensational mechanism for the loss of differentiation potential. In conclusion, our studies define a role for PRMT5 during oligodendrocyte differentiation. We show that PRMT5 is required for maintaining the methylation status of CpG islands of Id2 and Id4 leading to gene silencing during glial cell differentiation.
Supplementary Material
Acknowledgment
We thank Christina Gavino for editorial assistance.
This work was supported by Grant MOP-67070 from the Canadian Institutes of Health Research (to S. R.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. S1.
- PRMT
- protein arginine methyltransferase
- OPC
- oligodendrocyte progenitor cell(s)
- DMA
- dimethylated arginine
- H4R3
- histone 4 arginine 3
- H3R8
- histone 3 arginine 8
- GalC
- galactocerebrosidase
- MBP
- myelin basic protein
- qPCR
- quantitative PCR
- Pn
- postnatal day(s)
- BMP
- bone morphogenetic protein.
REFERENCES
- 1. Bedford M. T., Richard S. (2005) Mol. Cell 18, 263–272 [DOI] [PubMed] [Google Scholar]
- 2. Bedford M. T., Clarke S. G. (2009) Mol. Cell 33, 1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Tee W. W., Pardo M., Theunissen T. W., Yu L., Choudhary J. S., Hajkova P., Surani M. A. (2010) Genes Dev. 24, 2772–2777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huddleston J. E. (2011) Nat. Rev. Mol. Cell Biol. 12, 76. [DOI] [PubMed] [Google Scholar]
- 5. Sanchez S. E., Petrillo E., Beckwith E. J., Zhang X., Rugnone M. L., Hernando C. E., Cuevas J. C., Godoy Herz M. A., Depetris-Chauvin A., Simpson C. G., Brown J. W., Cerdán P. D., Borevitz J. O., Mas P., Ceriani M. F., Kornblihtt A. R., Yanovsky M. J. (2010) Nature 468, 112–116 [DOI] [PubMed] [Google Scholar]
- 6. Hong S., Song H. R., Lutz K., Kerstetter R. A., Michael T. P., McClung C. R. (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 21211–21216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fabbrizio E., El Messaoudi S., Polanowska J., Paul C., Cook J. R., Lee J. H., Negre V., Rousset M., Pestka S., Le Cam A., Sardet C. (2002) EMBO Rep. 3, 641–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pal S., Yun R., Datta A., Lacomis L., Erdjument-Bromage H., Kumar J., Tempst P., Sif S. (2003) Mol. Cell Biol. 23, 7475–7487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pal S., Vishwanath S. N., Erdjument-Bromage H., Tempst P., Sif S. (2004) Mol. Cell Biol. 24, 9630–9645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hou Z., Peng H., Ayyanathan K., Yan K. P., Langer E. M., Longmore G. D., Rauscher F. J., 3rd (2008) Mol. Cell Biol. 28, 3198–3207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hou Z., Peng H., White D. E., Negorev D. G., Maul G. G., Feng Y., Longmore G. D., Waxman S., Zelent A., Rauscher F. J., 3rd (2010) Proc. Natl. Acad. Sci. U.S.A. 107, 2938–2943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang L., Pal S., Sif S. (2008) Mol. Cell Biol. 28, 6262–6277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Le Guezennec X., Vermeulen M., Brinkman A. B., Hoeijmakers W. A., Cohen A., Lasonder E., Stunnenberg H. G. (2006) Mol. Cell Biol. 26, 843–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ancelin K., Lange U. C., Hajkova P., Schneider R., Bannister A. J., Kouzarides T., Surani M. A. (2006) Nat. Cell Biol. 8, 623–630 [DOI] [PubMed] [Google Scholar]
- 15. Tae S., Karkhanis V., Velasco K., Yaneva M., Erdjument-Bromage H., Tempst P., Sif S. (2011) Nucleic Acids Res. 39, 5424–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Xu X., Hoang S., Mayo M. W., Bekiranov S. (2010) BMC Bioinformatics 11, 396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kwak Y. T., Guo J., Prajapati S., Park K. J., Surabhi R. M., Miller B., Gehrig P., Gaynor R. B. (2003) Mol. Cell 11, 1055–1066 [DOI] [PubMed] [Google Scholar]
- 18. Kondo T., Raff M. (2000) EMBO J. 19, 1998–2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wegner M., Stolt C. C. (2005) Trends Neurosci. 28, 583–588 [DOI] [PubMed] [Google Scholar]
- 20. Wegner M. (2008) J. Mol. Neurosci. 35, 3–12 [DOI] [PubMed] [Google Scholar]
- 21. Yu Y., Casaccia P., Lu Q. R. (2010) Epigenetics 5, 124–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Shen S., Li J., Casaccia-Bonnefil P. (2005) J. Cell Biol. 169, 577–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ye F., Chen Y., Hoang T., Montgomery R. L., Zhao X. H., Bu H., Hu T., Taketo M. M., van Es J. H., Clevers H., Hsieh J., Bassel-Duby R., Olson E. N., Lu Q. R. (2009) Nat. Neurosci. 12, 829–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chittka A. (2010) PLoS One 5, e13807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Almazan G., Afar D. E., Bell J. C. (1993) J. Neurosci. Res. 36, 163–172 [DOI] [PubMed] [Google Scholar]
- 26. Yu Z., Chen T., Hébert J., Li E., Richard S. (2009) Mol. Cell Biol. 29, 2982–2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Nave K. A., Lemke G. (1991) J. Neurosci. 11, 3060–3069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Coyle D. E. (1995) J. Neurosci. Res 41, 374–385 [DOI] [PubMed] [Google Scholar]
- 29. McMorris F. A. (1977) Proc. Natl. Acad. Sci. U.S.A. 74, 4501–4504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Milner R. J., Lai C., Nave K. A., Lenoir D., Ogata J., Sutcliffe J. G. (1985) Cell 42, 931–939 [DOI] [PubMed] [Google Scholar]
- 31. Leisewitz A. V., Urrutia C. R., Martinez G. R., Loyola G., Bronfman M. (2008) J. Cell Physiol. 217, 367–376 [DOI] [PubMed] [Google Scholar]
- 32. Jansson M., Durant S. T., Cho E. C., Sheahan S., Edelmann M., Kessler B., La Thangue N. B. (2008) Nat. Cell Biol. 10, 1431–1439 [DOI] [PubMed] [Google Scholar]
- 33. Larocque D., Galarneau A., Liu H. N., Scott M., Almazan G., Richard S. (2005) Nat. Neurosci. 8, 27–33 [DOI] [PubMed] [Google Scholar]
- 34. Zhao Q., Rank G., Tan Y. T., Li H., Moritz R. L., Simpson R. J., Cerruti L., Curtis D. J., Patel D. J., Allis C. D., Cunningham J. M., Jane S. M. (2009) Nat. Struct. Mol. Biol. 16, 304–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang S., Sdrulla A., Johnson J. E., Yokota Y., Barres B. A. (2001) Neuron 29, 603–614 [DOI] [PubMed] [Google Scholar]
- 36. Swiss V. A., Nguyen T., Dugas J., Ibrahim A., Barres B., Androulakis I. P., Casaccia P. (2011) PLoS One 6, e18088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chan A. S., Tsui W. Y., Chen X., Chu K. M., Chan T. L., Chan A. S., Li R., So S., Yuen S. T., Leung S. Y. (2003) Oncogene 22, 6946–6953 [DOI] [PubMed] [Google Scholar]
- 38. Yu L., Liu C., Vandeusen J., Becknell B., Dai Z., Wu Y. Z., Raval A., Liu T. H., Ding W., Mao C., Liu S., Smith L. T., Lee S., Rassenti L., Marcucci G., Byrd J., Caligiuri M. A., Plass C. (2005) Nat. Genet. 37, 265–274 [DOI] [PubMed] [Google Scholar]
- 39. Noetzel E., Veeck J., Niederacher D., Galm O., Horn F., Hartmann A., Knüchel R., Dahl E. (2008) BMC Cancer 8, 154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Carey J. P., Asirvatham A. J., Galm O., Ghogomu T. A., Chaudhary J. (2009) BMC Cancer 9, 173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakahiro T., Kurooka H., Mori K., Sano K., Yokota Y. (2010) Biochem. Biophys. Res. Commun. 399, 416–421 [DOI] [PubMed] [Google Scholar]
- 42. Korchynskyi O., ten Dijke P. (2002) J. Biol. Chem. 277, 4883–4891 [DOI] [PubMed] [Google Scholar]
- 43. Hollnagel A., Oehlmann V., Heymer J., Rüther U., Nordheim A. (1999) J. Biol. Chem. 274, 19838–19845 [DOI] [PubMed] [Google Scholar]
- 44. Nakashima K., Takizawa T., Ochiai W., Yanagisawa M., Hisatsune T., Nakafuku M., Miyazono K., Kishimoto T., Kageyama R., Taga T. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 5868–5873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Samanta J., Kessler J. A. (2004) Development 131, 4131–4142 [DOI] [PubMed] [Google Scholar]
- 46. Dacwag C. S., Ohkawa Y., Pal S., Sif S., Imbalzano A. N. (2007) Mol. Cell Biol. 27, 384–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.