Background: 4-Phenylbutyrate corrects trafficking of ΔF508-CFTR, most likely by modulation of chaperone expression.
Results: 4-Phenylbutyrate stimulates Elp2 expression and activation of STAT-3.
Conclusion: The transient stimulation of Hsp70 expression in CF epithelial cells with 4-phenylbutyrate is dependent on the Elp2 component of elongator.
Significance: Signaling through Elp2 and STAT-3 is a novel pathway by which 4-phenylbutyrate regulates Hsp70 expression.
Keywords: Cystic Fibrosis, Epithelial Cell, Heat Shock Protein, Molecular Chaperone, STAT3, 4-Phenylbutyrate, Elongator
Abstract
Sodium 4-phenylbutyrate (4PBA) corrects trafficking of ΔF508-CFTR in Cystic Fibrosis (CF) epithelia, which is hypothesized to, at least in part, result from increased expression of Hsp70 (stress-induced 70 kDa heat shock protein). To identify other 4PBA-regulated proteins that may promote correction of ΔF508 trafficking, we performed differential display RT-PCR on mRNA from IB3-1 CF bronchiolar epithelial cells treated for 0–24 h with 1 mm 4PBA. In this screen, a STAT-3 (signal transducer and activator of transcription-3)-interacting protein, StIP-1 that regulates STAT-3 activation had transiently increased expression. StIP-1 is identical to Elongator protein 2 (Elp2), a component of the Elongator complex that regulates RNA polymerase II. Previous studies have suggested that Elongator regulates Hsp70 mRNA transcription, and that the Hsp70 promoter contains functional STAT-3-binding sites. We therefore tested the hypothesis that 4PBA increases Hsp70 expression by an Elongator- and STAT-3-dependent mechanism. 4PBA treatment of IB3-1 CF bronchiolar epithelial cells caused transiently increased expression of Hsp70 protein, as well as Elp2 protein and mRNA. Elp2 depletion by transfection of small interfering RNAs, reduced both Elp2 and Hsp70 protein expression. 4PBA also caused transient activation of STAT-3, and increased abundance of nuclear proteins that bind to the STAT-3-responsive element of the Hsp70 promoter. Luciferase reporter assays demonstrated that both Elp2 overexpression and 4PBA increase Hsp70 promoter activity, while Elp2 depletion blocked the ability of 4PBA to stimulate Hsp70 promoter activity. Together, these data suggest that Elp2 and STAT-3 mediate, at least in part, the stimulation of Hsp70 expression by 4PBA.
Introduction
Cystic Fibrosis (CF)2 is the most common lethal autosomal recessive disease among Caucasians and results from a paucity of functional Cystic Fibrosis Transmembrane Conductance Regulator (CFTR). CFTR is a cAMP-activated chloride channel that is localized in the apical plasma membrane of epithelial cells where it has an integral role in regulating the transport of electrolytes and water. The most common mutation of CFTR, ΔF508-CFTR (deletion of a phenylalanine at position 508), is a temperature-sensitive trafficking mutant (1). ΔF508-CFTR is retained in the endoplasmic reticulum (ER), where it has increased associations with several cytosolic chaperones belonging to the Heat Shock Protein (Hsp) family (2–5) and with the ER-resident chaperones calnexin (6–9) and ERp29 (10). ΔF508-CFTR is targeted for rapid intracellular degradation (11) at least in part by the ubiquitin proteasome system (12, 13), and so mostly fails to reach its appropriate subcellular location at the apical membrane (14, 15). Targeted manipulation of molecular chaperone expression is a potential therapeutic strategy to facilitate improvement of ΔF508-CFTR trafficking.
Sodium 4-phenylbutyrate (4PBA) improves ΔF508-CFTR intracellular trafficking in CF epithelial cells such as the IB3-1 CF human bronchiolar epithelial cell line (genotype ΔF508/W1282X) as early as 4 h after exposure, and restores CFTR function at the plasma membrane without altering CFTR mRNA expression (16). Because CFTR mRNA expression was not altered by 4PBA, and because 4PBA and other butyrates are considered transcriptional regulators, we previously investigated whether 4PBA alters expression of molecular chaperones implicated in ΔF508-CFTR trafficking. We initially focused on Hsc70 (70 kDa heat shock cognate protein), a cytosolic chaperone involved in targeting a number of cellular proteins for ubiquitination and proteasome degradation (17). After 48 h of 4PBA treatment, both the expression of Hsc70 and complex formation between Hsc70 and ΔF508-CFTR decreased in IB3-1 cells. This Hsc70/ΔF508-CFTR complex may target ΔF508-CFTR for rapid intracellular degradation by the ubiquitin/proteasome pathway (5). While others suggested that 4PBA also caused increased expression of the Hsp70, the stress-induced 70 kDa heat shock protein, which could promote ΔF508-CFTR trafficking, (3), our data suggested unaltered Hsp70 expression after 48 h of 4PBA. We subsequently observed that 4PBA decreases the steady-state expression of Hsc70 by increasing the rate of Hsc70 mRNA turnover, and that this effect requires new mRNA synthesis (18). These data suggested that IB3-1 cells undergo a complex response to 4PBA, a notion that has since been confirmed in genomic and proteomic profiling experiments (19, 20); these profiling data also suggested that 4PBA's stimulation of Hsp70 expression was transient. Moreover these considerations suggested that multiple elements of this 4PBA response (i.e. 4PBA-regulated genes or targets) might influence ΔF508-CFTR trafficking.
To identify candidate 4PBA targets that might regulate or promote ΔF508 trafficking, we performed a limited differential display RT-PCR on 4PBA-treated IB3-1 CF bronchiolar epithelial cells. Among the differentially expressed mRNA, we isolated a cDNA clone for a human homologue of StIP-1 (STAT-3-interacting protein-1) that had transiently increased abundance at 4 h and decreased abundance by 24 h of treatment with 4PBA. StIP-1 is identical to Elp2, a subunit of the Elongator complex that regulates RNA polymerase II (RNA Pol-II). In humans, the elongator complex is composed by IKAP (Elp1), Elp2, Elp3, Elp4, and two additional proteins, and contains histone acetyltransferase activity (21). Interestingly, 4PBA has also been reported to have activity as a histone deacetylase inhibitor (22, 23). Previous data suggest that Elongator facilitates Hsp70 mRNA transcription by RNA polymerase II in yeast (24), and that the Hsp70 promoter contains functional STAT-3-binding sites that overlap with the binding site for the heat shock transcription factor (25, 26). We therefore tested the hypothesis that 4PBA increases Hsp70 expression by a mechanism that depends upon Elongator and activation of STAT-3.
EXPERIMENTAL PROCEDURES
Cell Culture
IB3-1 cells (27) were cultured as previously described (5, 18). Human colonic adenocarcinoma T84 cells (American Type Culture Collection (ATCC) cell line CCL248; Rockville, MD) were cultured in a 1:1 mixture of Dulbecco's modified Eagle's medium and Ham's F-12 medium (Invitrogen) supplemented with 100 units/ml penicillin, 0.1 mg/ml streptomycin, and 5% fetal bovine serum at 37 °C in 5% CO2. IB3-1 and T84 cells were studied as these endogenously express ΔF508 or wt CFTR, respectively.
Antibodies
Rabbit anti-Elp2 was from Millipore. Mouse STAT-3, clone F-2, was from Santa Cruz Biotechnology. Rabbit anti-STAT-3, anti-phospho-STAT-3 (Tyr-705), and anti-phospho-STAT-3 (Ser-727) were from Millipore Goat anti-c-Myc-fluorescein conjugate was from Bethyl Laboratories. Rhodamine Red™-X-conjugated mouse anti-rabbit IgG was from Jackson Immunoresearch. Fluorescein-conjugated anti-rabbit IgG was from Amersham Biosciences. Rabbit anti-Bip/GRP78 was from Sigma-Aldrich. Rabbit anti-Hsc70 and rabbit anti-Hsp70 were from Stressgen. Mouse anti-GAPDH was from Millipore, and mouse ant-V5 was from Abnova.
Differential Display RT-PCR Screen
Differential display RT-PCR was performed using the RNAimage™ kit (Genhunter) on total RNA isolated from IB3-1 cells treated with 1 mm 4PBA for 0, 4, 8, or 24 h as previously described in detail (10). Differentially expressed cDNAs were extracted, re-amplified by PCR, and cloned into pCRII (Invitrogen). The cloned inserts were sequenced by automated analysis (Nucleic Acid Research Core, Children's Hospital of Philadelphia) and identified by Blast search.
Amplification of Full-length Elp2 and Cloning
Full-length Elp2 cDNA was isolated by 5′-RACE-PCR from IB3-1 cDNAs using the 5/3′-RACE kit (Roche). An Elp2-specific primer (5′-TTTTTTATTAAGTCCATTACAGTGC-3′) was used for reverse transcription. After addition of a 3′-poly(A) tail, the full-length cDNA was amplified by PCR using oligo dT-anchored and Elp2-specific primers (5′-CGGCGGATCCCCATTACAGTGCACATTT-3′, BamH I site underlined), cloned into pCRII, and sequenced. Elp2 was then subcloned into the HindIII/XhoI resctriction sites of mammalian expression vector pCDNA-4/TO/myc-his (Invitrogen).
RNase Protection
RNase protection was performed as described previously (5, 18) using the Direct Protect assay kit (Ambion). The Elp2-specific probe template comprised a 567 bp EcoR I fragment from pCRII-Elp2 subcloned into pSK(-) (Stratagene) and linearized by NotI. Probe was synthesized using a Maxiscript T7 kit (Ambion). Hybridization to 18 S rRNA served as an internal control.
Immunoblot
Cell lysates were prepared in RIPA buffer (NaCl 150 mm, Tris-HCl 50 mm pH 8, 1% Triton-X100, 1% sodium deoxycholate, 0.1% SDS) containing protease inhibitor mixture (Sigma). Lysates were passaged through a 22-gauge needle and cleared by centrifugation (15,000 × g, 15 min, 4 °C), and protein content determined using Bio-Rad DC reagents with bovine γ-globulin as a standard. Equal amounts of protein were resolved by SDS-PAGE and transferred to nitrocellulose as previously described (16). Nonspecific binding was blocked by incubation with 5% nonfat dry milk. Primary and horse radish peroxidase-conjugated secondary antibodies were applied in Tris-buffered saline (TBS 1×) (50 mm Tris-HCl, pH 7.4, and 150 mm NaCl.) 0.05% Tween-20, 1% bovine serum albumin. Immunoreactivity was detected by chemiluminescence (ECL, Amersham Biosciences) and fluorography, and was quantitated by densitometry (5, 18).
Immunoprecipitation
Cell lysates were prepared as described above except that SDS was omitted from the RIPA buffer. Mouse anti-STAT-3 (10 μl) or rabbit anti-Elp2 was incubated with the lysates (500 μg of protein in 0.5 ml) at 4 °C overnight. As a control, immunoprecipitations were also performed with mouse anti-V5 epitope as an unrelated antibody control. Immune complexes were captured with protein G-Sepharose (Amersham Biosciences) for STAT-3 and protein A-agarose (Invitrogen) for Elp2, released by boiling in SDS-PAGE sample buffer and resolved by SDS-PAGE.
Elp2 GST Pull-downs
A cDNA-encoding Elp2 amino acids 402–826 was amplified by PCR using sense (5′-CGGCGAGTCGACATTTATTATCACTGTT-3′, SalI site underlined) and antisense (5′-ATTAGCGGCCGCTTACAGTGCACA-3′, NotI site underlined) primers. A glutathione S-transferase fusion protein, (GST)-Elp2, was generated by cloning this cDNA into pGEX4T2 (Amersham Biosciences). GST or GST-Elp2 were expressed in Escherichia coli BL21, immobilized on glutathione-Sepharose 4B beads (Amersham Biosciences), and incubated overnight with IB3-1 cell lysates. Bound proteins were eluted by boiling in SDS-PAGE sample buffer and resolved by SDS-PAGE.
Immunofluorescence
IB3-1 cells grown on glass chamber slides (Falcon) were treated with 4PBA, fixed with 1% formalin in PBS (10 min) and permeabilized with 0.1% Nonidet P-40 in PBS (5 min). Nonspecific binding was blocked with 10% goat serum (Invitrogen) in PBS (1 h) at room temperature. STAT-3 was detected with mouse anti-STAT-3 (1/200) in TBS, 3% BSA, 0.2% Tween 20, and revealed with Alexa Fluor® 488 goat anti-mouse secondary antibody (1/200, Invitrogen). Nuclei were counterstained with DAPI (4′-6-diamidino-2-phenylindole). Fluorescence was viewed at room temperature at 510–560 nm on a Olympus IX81 inverted microscope, and images captured with a Hamamatsu digital camera using SlideBook software,
Electrophoretic Mobility Shift Assays (EMSAs)
Nuclear extracts were prepared from IB3-1 cells treated for 0, 2, 4, 8, and 24 h with 1 mm 4PBA as previously described (28). Protein concentrations were determined by the Bradford assay (Bio-Rad) according to the manufacturer's recommendations. Electrophoretic mobility shift assays (EMSAs) were performed as previously described (28) using a 32P-labeled oligonucleotide.
5′-GATCCAGCTTGAAAGTTCCAGAACGA-3′ encompassing the heat shock responsive element that overlaps with the STAT-3 binding site within the human Hsp70 promoter. DNA-protein complexes were resolved by PAGE in 1× TAE buffer (Tris 6.7 mm, sodium acetate 3.3 mm, EDTA 1 mm pH 7.9) at 4 °C. The specificity of the retarded band was tested by a supershift assay in which the nuclear extracts were preincubated with the STAT-3 antibody.
Depletion of Elp2 by siRNA
Elp2 expression was depleted using a commercial pool of 4 different Elp2 siRNA (Dharmacon/Thermo Fisher Scientific). Elp2 siRNA was delivered to IB3-1 cells by transfection with Lipofectamine RNAimax reagent (Invitrogen, Carlsbad, Ca), or to T-84 cells by electroporation according to a commercially available optimized protocol and reagents (amaxa, Lonza Cologne, Germany). Control siRNA (Dharmacon/Thermo Fisher Scientific) was delivered under identical conditions as the Elp2 siRNA. Cells were either lysed for immunoblot (described above) or used for luciferase reporter assays (described below) 48 h after transfection or electroporation.
Luciferase Reporter Assays
The human Hsp70 promoter (−2650/+150) was cloned into the pGL3 luciferase vector (Promega). Cells were transfected using the Lipofectamine plus reagent® (Invitrogen) using 1 μg of reporter plasmid and 500 ng of pCDNA4/TO/myc-his vector expressing Elp2, or 500 ng of “empty vector” pCDNA-4/TO/myc-his. Luciferase assays were performed with the Luciferase assay system (Promega) according the manufacturer's instructions. To account for possible differences in transfection efficiencies a cytomegalovirus-driven β-galactosidase expression plasmid (500 ng) was transfected simultaneously, and β-galactosidase expression was determined enzymatically.
Digital Images
Digital images were processed using Adobe Photoshop®(version 6), and included in figures using Adobe Illustrator® (version 9).
Densitometric Analysis
Fluorographic images were digitized using an Alphaimager 2200 digital analysis system (AlphaInnotech, San Leandro, CA). Densitometric analysis of these images was performed using Alphaimager analysis software (version 5.5, AlphaInnotech) with two-dimensional integration of the selected band. For comparison within an immunoblot experiment, the density of the 0 h time point or undtreated control was arbitrarily set to 1.0, with the remaining densities expressed relative to this reference density. For comparison within a ribonuclease protection experiment, the density of the Elp2 was initially normalized by the density of the 18 S hybridization within the same sample. The relative Elp2/18S density of the 0 h time point was then arbitrarily set to 1.0, and the remaining densities were again expressed relative to this reference density. In the figures, these data are expressed as mean ± S.E. Student's t test or a one-way ANOVA was used to determined statistical significance of changes in density of fluorographic bands as appropriate (SigmaStat software, version 2.03).
RESULTS
Hsp70 Expression Is Transiently Increased by 4PBA
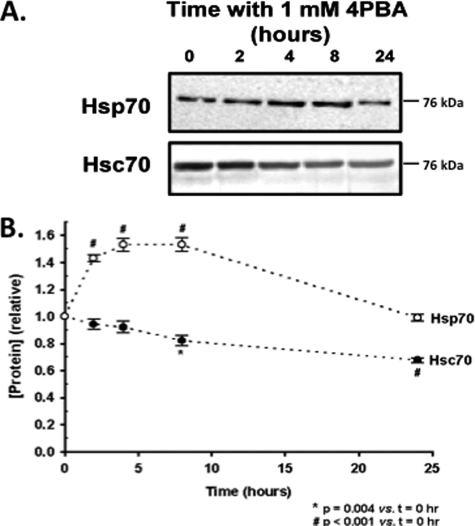
Others have suggested that 4PBA may promote ΔF508-CFTR by increasing Hsp70 expression (3), while our previous data suggested that Hsp70 expression was unaltered in IB3-1 cells after 48 h of treatment with 1 mm 4PBA (5). Gene profiling experiments suggested that 4PBA induced a transient increase in Hsp70 mRNA expression that returned to baseline by 24 h (19, 20). We therefore examined Hsp70 protein expression in IB3-1 cells over a 24-h exposure to 1 mm 4PBA (Fig. 1). Hsp70 expression increased transiently by ∼50%; peak expression occurred after 4–8 h of exposure. Hsp70 expression returned to baseline by 24 h of exposure. In contrast, Hsc70 expression tonically decreased over this time period, with an ∼30% reduction after 24 h; this reduction is consistent with our previous report that Hsc70 expression decreases ∼40% after 48 h of exposure to 4PBA (5).
FIGURE 1.
Expression of Hsp70 and Hsc70 in response to 4PBA. IB3-1 cells were incubated for the indicated times with 1 mm 4PBA. Whole cell lysates were prepared, and Hsp70 and Hsc70 were quantitated by immunoblot as described under “Experimental Procedures.” A, representative immunoblot. B, densitometry of the indicated number of independent time course experiments (mean ± S.E.). Statistical significance was determined by 1-way ANOVA in comparison to t = 0.
Elp2 mRNA and Protein Expression Is Transiently Increased by 4PBA
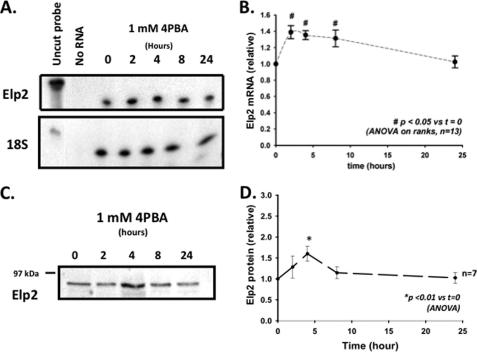
In IB3-1 cells, 4PBA decreases steady-state Hsc70 mRNA expression by increasing the rate of Hsc70 mRNA turnover; this effect requires new mRNA synthesis, suggesting that 4PBA causes a more global cellular adaptation (18). To characterize the elements of this hypothetical cellular adaptation, we performed differential display RT-PCR on RNA isolated from IB3-1 cells treated with 1 mm 4PBA for 0, 4, 8, and 24 h. Among the differentially expressed mRNA species, we isolated a cDNA clone for a hypothetical human STAT-3 (signal transducer and activator of transcription-3) interacting protein (StIP-1). StIP-1 is identical to Elongator protein 2 (Elp2); Elongator is a protein complex that facilitates mRNA transcription by RNA polymerase II, and may also have roles in regulation of polarized vesicular transport (30) and modification of tRNAs (31, 32). In the differential display experiments, this clone's mRNA had increased abundance at 8 h and decreased abundance at 24 h (data not shown). This 4PBA-induced transient increase in Elp2 mRNA abundance was confirmed by ribonuclease protection, which suggested an ∼30–40% (n = 13, p < 0.05 versus t = 0, Fig. 2, A and B) increased Elp2 mRNA expression after 2, 4, and 8 h of treatment with 1 mm 4PBA. We also observed transiently increased expression of Elp2 at the protein level by immunoblot in IB3-1 cells after incubation with 1 mm 4PBA (Fig. 2, C and D), with a peak increase of ∼50% after 4 h treatment with 4PBA (n = 7, p < 0.01 versus t = 0).
FIGURE 2.
Elp2 mRNA and protein level are transiently increased in IB3-1 cells treated with 4PBA. A, Elp2 mRNA was quantitated by ribonuclease protection in IB3-1 cells treated for the indicated times with 1 mm 4PBA. The hybridization of an 18 S rRNA as an internal standard was constant under these conditions. A representative fluorogram is shown. B, relative amount of Elp2–1 mRNA compared with the zero time point was determined by densitometry of 13 total experiments using 5 independent sets of samples. For analysis, the density of the Elp2 was initially normalized by the density of the 18 S hybridization within the same sample. The relative Elp2/18S density of the 0 h time point was then arbitrarily set to 1.0, and the remaining densities were again expressed relative to this reference density. Means + S.E. are shown, with p values determined by one-way ANOVA in comparison with t = 0. C, representative fluorogram of equal amounts of whole cell lysates of IB3-1 cells treated with 1 mm 4PBA for 0, 2, 4, 8, and 24 h resolved by SDS-PAGE and quantitated by immunoblot using a specific Elp2 antiserum as described under “Experimental Procedures.” D, densitometric analysis (mean ± S.E.) of seven independent time course experiments. In this analysis the density of the 0 h time point was arbitrarily set to 1.0, with the remaining densities expressed relative to this reference density. Statistical significance was determined by a 1-way ANOVA in comparison with t = 0.
Expression of Phosphorylated/Activated STAT-3 Is Increased by 4PBA
The murine homologue of Elp2 regulates the activation of STAT-3 in response to IL-6 by hypothetically serving as a scaffold protein that promotes the interaction between Janus kinases and their STAT-3 substrate (33). Activation of STAT-3 is correlated with specific tyrosine phosphorylation (at residue Y705), and additional serine phosophorylation (at residue S727). Such phosphorylation appears to promote STAT-3 dimerization and subsequent nuclear translocation and regulation of transcription (34). We therefore examined whether 4PBA caused activation of STAT-3 in IB3-1 cells.
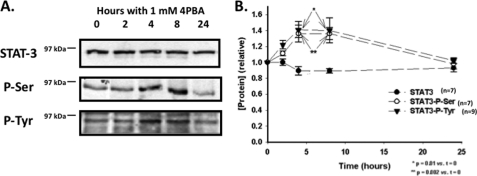
We performed immunoblots on whole cell lysates prepared from IB3-1 cells treated for 0–24 h with 1 mm 4PBA using antisera targeting STAT-3, phospho-S727-STAT-3, and phospho-Y705-STAT-3 (Fig. 3). Total STAT-3 expression did not change during these time course experiments. However, the abundance of phospho-S727-STAT-3, and phospho-Y705-STAT-3 STAT-3 increased ∼40% after 4–8 h of treatment with 4PBA (n = 7, p = 0.01 (phospho-S) and p = 0.002 (phospho-Y) versus t = 0). These data suggest that there is transiently increased phosphorylation and activation of STAT-3 in IB3-1 cells in response to 4PBA.
FIGURE 3.
Phosphorylation of STAT-3 is increased by 4PBA. A, equal amounts of whole cell lysates of IB3-1 cells treated with 1 mm 4PBA for 0, 2, 4, 8, and 24 h were resolved by SDS-PAGE, and the abundance of STAT-3, phosphoserine STAT-3 and phosphotyrosine STAT-3 were determined by immunoblot using specific antisera as described under “Experimental Procedures.” Representative immunoblots are shown. B, densitometric analysis (mean ± S.E.) of 7 independent immunoblot experiments. The density of the 0 h time point was arbitrarily set to 1.0, with the remaining densities expressed relative to this reference density. Statistical significance was determined by a 1-way ANOVA in comparison with t = 0.
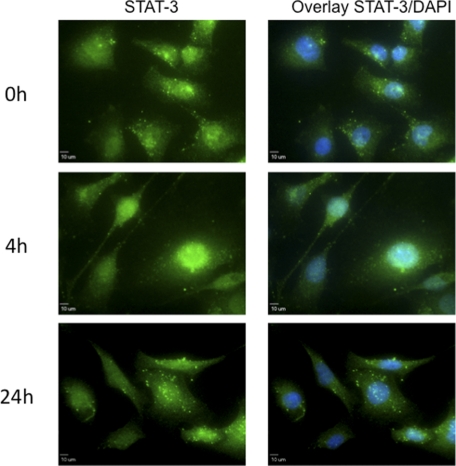
Activation of STAT-3 is also associated with translocation of STAT-3 from the cytoplasm to the nucleus. We therefore assessed the influence of 4PBA on the subcellular localization of STAT-3 in IB3-1 cells treated with 1 mm 4PBA for 0, 4, or 24 h by immnunofluorescence (Fig. 4). In untreated (0 h) cells, STAT-3 immunofluorescence was predominately contained within the cytoplasm, and did not overlap with the nuclear counter stain. In presence of 1 mm 4PBA, STAT-3 is increasingly detected in the nucleus, with 4 h of treatment with 4PBA. At 24 h, STAT-3 was again predominately localized to the cytoplasm. These data are also consistent with 4PBA causing transient activation of STAT-3 in IB3-1 cells.
FIGURE 4.
Transient nuclear localization of STAT-3 with 4PBA treatment. Immunofluorescent detection of STAT-3 in IB3-1 cells treated with 1 mm 4PBA for the indicated time period was performed as described under “Experimental Procedures.” Binding of the primary STAT-3 antibody was visualized with an Alexa Fluor® 488-conjugated secondary antibody. Nuclei were counterstained with DAPI. These data are representative of n = 5 independent experiments.
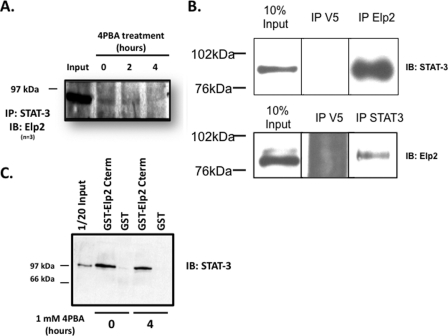
Interaction of STAT-3 and Elp2 in IB3-1 Cells
Association of murine Elp2 and STAT-3 is suggested to be required for STAT-3 activation (33). We therefore assessed whether Elp2 and STAT-3 interact in IB3-1 cells, as well as the effect of 4PBA on this interaction. As shown in Fig. 5A, Elp2 was recovered when STAT-3 was immunoprecipitated from lysates of untreated IB3-1 cells (“0” lane). Less Elp2 was recovered with STAT-3 by co-immunoprecipitation from lysates of IB3-1 cell after 2 h treatment with 1 mm 4PBA, and no Elp2 was detected after STAT-3 immunoprecipitation after 4 h of exposure to 4PBA, the time at which STAT-3 activation appears maximal. In control experiments, Elp2 was not detected on immunoblots when an unrelated antibody (anti-V5) was used for immunoprecipitation (Fig. 5B, top panel). Similarly STAT-3 was detected on immunoblots when immunoprecipitations were performed with anti-Elp2, but not with anti-V5 (Fig. 5B, bottom panel). These suggest are consistent with Elp2 interacting with inactive STAT-3, and with activation of STAT-3 by 4PBA promoting STAT-3 dissociation from Elp2.
FIGURE 5.
Association of STAT-3 and Elp2 in IB3-1 cells treated with 4PBA. A, IB3-1 cells were incubated with 1 mm 4PBA for the indicated times and whole cell lysates were prepared as described in the Immunoprecipitation section under “Experimental Procedures.” STAT-3 and its interacting proteins were then immunoprecipitated from equal amounts of whole cell lysate protein with a specific STAT-3 mouse monoclonal antibody and resolved by SDS-PAGE, again as described under “Experimental Procedures.” Immunodetection of Elp2 was performed with a rabbit polyclonal antibody. These data are representative of n = 5 independent experiments. B, top panel: equal amounts of IB3-1 cell lysate were subject to immunprecipitation with either anti-STAT-3 or anti-V5. Elp2 was readily detected by immunoblot in whole cell lysate (10% input) and in the proteins precipitated by anti-STAT-3, but not in the proteins precipitated by anti-V5. The panel presents identically exposed non-contiguous lanes of the same immunblot. Bottom panel: equal amounts of IB3-1 cell lysate were subject to immunprecipitation with either anti-Elp2 or anti-V5. STAT-3 was readily detected by immunoblot in whole cell lysate (10% input) and in the proteins precipitated by anti-Elp2, but not in the proteins precipitated by anti-V5. The panel presents identically exposed non-contiguous lanes of the same immunblot. C, glutathione S-transferase (GST)-Elp2-(aa 402–826) fusion protein was constructed, expressed in E. coli and purified on glutathione-Sepharose beads as described under “Experimental Procedures.” Native GST was similarly expressed in E. coli and purified. GST or GST-Elp2-(aa 402–826) immobilized on glutathione-Sepharose beads was incubated with equal amounts of whole cell lysates of IB3-1 cells that had been treated with 1 mm 4PBA for the indicated times. Precipitated proteins were resolved by SDS-PAGE, and STAT-3 detected by immunoblot as described under “Experimental Procedures.” These data are representative of n = 3 experiments.
Elp2 contains 12 WD40 repeats. Proteins that contain WD40 repeats may serve as scaffolds for coordinating multi-protein complex assemblies, and the WD40 repeat units are implicated in mediating these protein-protein interactions (35, 36). Collum et al. have suggested that WD40 repeats 10 to 12 of murine Elp2 interact with STAT-3 (33). To test whether these domains also interact with STAT-3 in IB3-1 cells, and to assess the potential influence of 4PBA on this interaction, we created a GST-Elp2 (aa 402–826) fusion protein construct that contains these WD40 repeats. We then expressed this protein in E. coli, and performed GST pull-down experiments with lysates of IB3-1 cells treated for 0 or 4 h with 1 mm 4PBA. As shown in Fig. 5C, STAT-3 was precipitated from lysates of IB3-1 cells untreated with 4PBA by the GST-Elp2-(aa 402–826) fusion protein, but not GST alone. This interaction appeared slightly decreased in lysates of IB3-1 cells treated for 4 h with 4PBA, but this decrease did not achieve statistical significance in densitometric analysis (not shown). These data are consistent with previous data (33) suggesting that WD40 domains 10 to 12 of Elp2 are sufficient for Elp2 interaction with STAT-3.
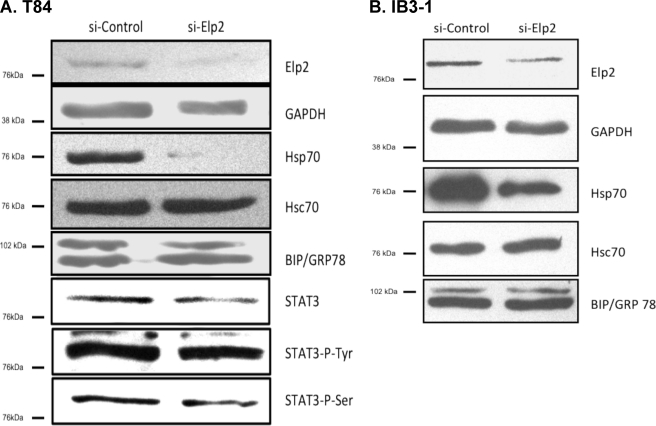
Depletion of Elp2 Decreases Hsp70 Expression
4PBA improves the aberrant trafficking of ΔF508-CFTR in CF epithelial cells (16) at least part of this effect is hypothesized by some to result from increased expression of Hsp70 (3). Elp2 is also necessary for activation of STAT-3 in response to IL-6, and the Hsp70 promoter contains a functional STAT-responsive element. We therefore investigated the regulation of Hsp70 by Elp2 using a small interfering RNA (siRNA) approach. As shown in Fig. 6A, delivery of Elp2-directed siRNA to T84 colonic adenocarcinoma cells (where efficiency of siRNA delivery by electroporation approaches 100%) decreased both Elp2 and and Hsp70 whole cell expression relative to cells transfected with control siRNA, but had no effect on the expression of Hsc70 (representative of n = 3 experiments). Similarly, in IB3-1 cells (where siRNA transfection efficiency is at best 50%, data not shown), Elp2-directed siRNA also resulted in decreased Elp2 expression and Hsp70 expression, again without effect on Hsc70 expression (Fig. 6B, representative of n = 3 experiments). The expression of BiP/grp78 was not altered in either T84 or IB3-1 cells by delivery of Elp2 siRNA (Fig. 6), suggesting that the siRNA-mediated depletion of Elp2 did not cause a global stress or unfolded protein response.
FIGURE 6.
Elp2 depletion with siRNA. Elp2-specific or control siRNA was delivered to T84 colonic adenocarcinoma or IB3-1 CF bronchiolar epithelial cells as described under “Experimental Procedures.” After 48 h cells were lysed, and equal amounts of whole-cell lysate protein were resolved by SDS-PAGE. Elp2, BiP/grp78, Hsc70, Hsp70, STAT3, phosphotyrosine STAT3, phosphoserine STAT3 and, as a loading control, GAPDH were detected by immunoblot in lysates of T84 cells (panel A) and IB3-1 cells (panel B). Data representative of three independent experiments are shown.
Because 4PBA transiently stimulates Hsp70 expression and there is a STAT-3-responsive element into Hsp70 promoter, we also assessed the influence of Elp2 depletion on the expression and activation of STAT-3. As shown in Fig. 6A, the expression of STAT-3, p-Tyr-STAT3, and p-Ser-STAT-3 all appeared slightly decreased, respectively, compared with cells transfected with control siRNA (representative of n = 3 experiments), but while this trend was a consistent observation, these decreases were not statistical significant in quantitative densitometric analyses (data not shown). These data are therefore consistent with the decrease in Hsp70 expression after Elp2 depletion in T84 cells being mediated by a decrease in the expression of Elp2, but without a significant decrease in STAT-3 expression.
Regulation of the Hsp70 Promoter by STAT-3 and Elp2
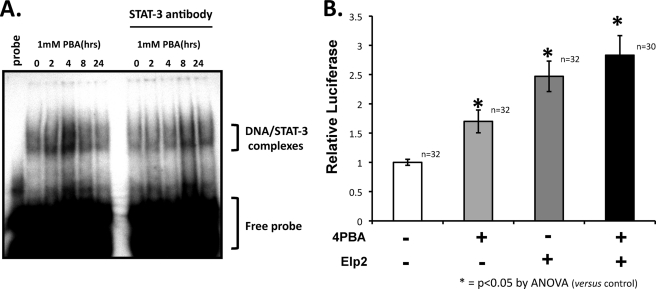
Previous data suggest that STAT-3 may bind to the Hsp70 promoter between −206 and −187 bp (37). We tested whether the binding of STAT3 to Hsp70 promoter is affected by 4PBA. We performed an electrophoretic mobility shift assays (EMSA) using a 32P-labeled oligonucleotide encompassing the STAT-3 response elements in the Hsp70 promoter and nuclear extracts prepared from IB3-1 cells treated for 0, 2, 4, 8, and 24 h with 1 mm 4PBA. As shown in Fig. 7A, nuclear protein binding to the Hsp70 promoter was transiently increased by 4PBA; again, this increase was greatest in nuclear extracts of cells that had been treated with 4PBA for 4 h. Preincubation with anti-STAT-3 decreased this 4PBA-stimulated nuclear protein binding to the Hsp70 promoter (Fig. 7A, right panel) suggesting that STAT-3 is at least a component of these proteins that bind to the Hsp70 promoter with 4PBA treatment.
FIGURE 7.
Regulation of the Hsp70 promoter by STAT-3 and Elp2. A, nuclear extracts were prepared from IB3-1 cells treated for the indicated times with 1 mm 4PBA. Electrophoretic mobility shift assays (EMSAs) were performed using 32P-labeled oligonucleotides encompassing the STAT-3 response elements within the human Hsp70 promoter. The specificity of the retarded band was tested in a supershift assay by preincuation with a STAT-3-specific antibody, which decreased the formation of this band. B, human Hsp70 promoter (nucleotides −2650/+150) was cloned into the pGL3 luciferase vector (Promega). IB3-1 cells were transfected with 1 mg of this reporter plasmid, 500 ng of a cytomegalovirus-driven β-galactosidase expression plasmid, and 500 ng of either pCDNA4/TO/myc-his vector expressing Elp2 or empty pCDNA-4/TO/myc-his vector; protein expression from this pCDNA4 vector is constitutive in the absence of the tet repressor. Luciferase assays were performed with the Luciferase assay system (Promega), and were normalized for transfection efficiency by assay of β-galactosidase activity.
We next tested the effect of Elp2 onto Hsp70- promoter function using a luciferase reporter assay. The human Hsp70 promoter (−2650/+150) was cloned into a pGL3 luciferase reporter vector. Both 4PBA treatment and Elp2 overexpression increased Hsp70 promoter activity (Fig. 7B), with overexpression of Elp2 having a greater effect. The combination of Elp2 overexpression and treatment with 4PBA did not appear to have additive effects on Hsp70 promoter activity (Fig. 7B, p = ns for comparison of cells transfected with Elp2 −/+ 4PBA by ANOVA).
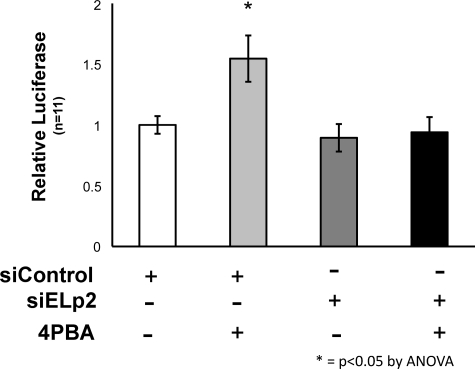
Elp2 Depletion Blocks Stimulation of the Hsp70 Promoter by 4PBA
Because 4PBA treatment and overexpression of Elp2 did not have an additive effect on Hsp70 promoter activity, we tested whether Elp2 was required for 4PBA to stimulate the Hsp70 promoter, or whether such regulation might also occur through an alternate pathway. We performed luciferase reporter assays in IB3-1 cells that were transfected with Elp2-directed or non-target control siRNA 24 h prior the transfection with the Hsp70-promoter-luciferase reporter (and β-galactosidase control) plasmids. As shown in Fig. 8 siRNA-mediated depletion of Elp2 in IB3-1 cells prevented 4PBA from stimulating Hsp70 promoter activity. These data suggest that Elp2 is required for 4PBA to stimulate Hsp70 expression, and that alternate pathways for stimulation of the Hsp70 promoter by 4PBA are not present in IB3-1 cells.
FIGURE 8.
Stimulation of Hsp70 promoter activity by 4PBA is blocked by Elp2 depletion. 24 h prior the transfection with pHsp70-pGL3 luciferase reporter and β-galactosidase plasmids (as described in the legend to Fig. 7), IB3-1 cells were transfected with non-target control or Elp2-directed siRNA as described under “Experimental Procedures.” Luciferase assays were performed and were normalized by β-galactosidase expression, also as described under “Experimental Procedures.”
DISCUSSION
ΔF508 ability to transport chloride in the ER (38) led to the hypothesis that “correction” of ΔF508's aberrant trafficking could improve CFTR function in CF epithelia. In fact, ΔF508 trafficking can be improved by a number of physical or pharmacologic manipulations (see (39) for review). Sodium 4-phenylbutyrate (4PBA), a FDA-approved pharmaceutical for patients with urea cycle disorders, corrects ΔF508 trafficking and restores CFTR function to the plasma membrane of CF epithelial cells (16). Two clinical trials have also demonstrated that 4PBA causes small but significant improvements in nasal epithelial chloride transport in ΔF508-homozygous CF patients (40, 41). However, the mechanism of 4PBA action remains elusive.
In IB3-1 CF bronchiolar epithelial cells, improvements in ΔF508 trafficking are seen as early as 4 h after exposure to 4PBA, but occur without alteration of CFTR mRNA expression (16). This was an unexpected finding, as 4PBA, like other butyrates, is hypothesized to regulate gene transcription, perhaps by inhibiting histone deacetylase and activating transcription of chromatin-silenced genes (23). Instead, 4PBA treatment decreased both the expression of Hsc70 (70 kDa heat shock cognate protein) and complex formation between Hsc70 and ΔF508-CFTR. This complex may target ΔF508-CFTR for degradation by the ubiquitin/proteasome pathway (5), as Hsc70 is a necessary co-factor for targeting a number of cellular proteins for ubiquitination and proteasome degradation.
While we initially focused on the decreasing expression of Hsc70 with 4PBA as a mechanism by which ΔF508-CFTR trafficking improves (5, 18), others Choo-Kang, 2001 286/id} suggested that 4PBA might improve ΔF508-CFTR trafficking by increasing expression of Hsp70. Here, we confirm that 4PBA causes a decrease in Hsc70 expression and a transient increase in Hsp70 expression; these changes are also consistent with the genomic and proteomic profiling data of others (19, 20). We have also shown that such alterations in Hsc70 and Hsp70 expression can modulate the intracellular trafficking of the epithelial sodium channel, ENaC (68). Together, these observations suggest that 4PBA causes a more global cellular adaptation or response, one result of which is improved ΔF508 trafficking in CF epithelia. However, the details of this adaptation, as well as the mechanism by which 4PBA signals the cell to adapt, are not known. This is likely a fundamental mechanistic issue, as 4PBA also promotes trafficking and/or secretion of ENaC (42), π-Z-α1-antitrypsin (43), mutant surfactant protein C (44), the ENaC-homologous acid sensitive ion channel 2 (45), and mutant ABCA3 transporters (46).
Using differential display RT-PCR, we sought to identify additional 4PBA-regulated species in IB3-1 CF epithelial cells that could contribute to improved ΔF508-CFTR trafficking. We recently demonstrated that one 4PBA-regulated species identified in this screen, ERp29 (endoplasmic reticular protein of 29 kDa), is a novel chaperone of the endoplasmic reticulum that promotes the trafficking of wt and ΔF508 CFTR, the first such luminal chaperone to have this role (10). We also, as described here, identified Elp2 as a potential 4PBA-stimulated regulator of Hsp70.
Elp2, which is also known as StIP-1, was first identified in a mouse myelomonocyte cDNA library as a scaffold protein that promotes the interactions between Janus kinases and STAT-3, and that is required for STAT-3 activation in response to IL-6 (33). In IB3-1 cells, exposure to 1 mm 4PBA led to a transient activation of STAT-3 that peaks at 4 h of treatment; this transient STAT-3 activation was coincident with a transient increase in Elp2 mRNA and protein expression. We also demonstrated that 4PBA leads to increased binding of STAT-3 to and activation of the Hsp70 promoter. Furthermore, depletion of Elp2 using a pool of specific siRNAs blocked both the transient activation of STAT-3 and the increased in Hsp70 promoter activity in response to 4PBA, as well as decreased Hsp70 protein expression.
The siRNA-mediated depletion of Elp2 and Hsp70 expression was less robust in IB3-1 cells than in T84 cells. We feel this likely reflects experimental differences in transfection efficiencies; our methods for transfecting IB3-1 cells (lipofection) at best achieves transfection of ∼50% of cells, while delivery of siRNA to T84 cells approaches 100% efficiency using electroporation (data not shown). This issue of lesser transfection efficiency of IB3-1 cells does not influence the assays of Hsp70 promoter activity, as only luciferase-transfected cells (that presumably also took up siRNA) are apparent in the assay. Thus, these data suggest that Elp2 regulates Hsp70 expression, and furthermore is required for 4PBA's activation of STAT-3 and transient stimulation of Hsp70 protein expression in IB3-1 cells, as it is for the activation of STAT-3 in response to IL-6.
Elongator is a complex comprised of six proteins, Elp1–6. In Saccharomyces cerevisiae, Elongator has roles in regulation of transcriptional elongation via RNA polymerase II (Pol II), exocytosis, and tRNA modification (30, 31, 47–51). Homologs of Elongator subunits proteins are present in other eukaryotes, and a six-subunit Elongator complex is present in human (21, 53). Elongator, via its regulation of Pol II, also directly influences Hsp70 expression in yeast. Elp3-induced histone H3 acetylation increases Hsp70 gene transcription (24, 54). Similarly, decreasing human Elp3 and Elongator complex expression suppresses the expression of Hsp70 in human cells as a result of histone H3 hypoacetylation (24). As Hsp70 interacts with wt and ΔF508-CFTR and may promote their trafficking (3), Elongator function could also influence CFTR trafficking via regulation of Hsp70 expression. Recent data also suggest a direct interaction of Elongator with DNAJB11 (55), a member of the Hsp40 family of co-chaperones (56). Thus, in addition to regulating Hsp70 expression, Elongator may also regulate Hsp70 function.
In humans, tissue-specific splicing mutations of Elp1 cause Familial Dysautonomia (FD), an autosomal recessive disease characterized by defects in the development and maintenance of neurons of the autonomic and sensory systems (57–59). Although the molecular and cellular mechanisms underlying FD remain largely undefined, impaired cell mobility and migration may contribute to the FD phenotype, as human cells depleted for Elp1 often have defects in migration (60).
These considerations suggest that Elp2 may act in multiple cellular compartments, and may shuttle between the cytoplasm and nucleus. In the cytoplasm, Elp2 acts as a scaffold for ligand-dependent STAT-3 activation, while in the nucleus Elp2 may coordinate Elongator complex formation and/or Elongator/RNA Pol II interactions. In fact, 4PBA's regulation of Elp2 may also portend a more general regulation of transcription via increased histone acetylation and stimulation of RNA Pol II. As murine Elp2 interacts with several STAT proteins (33, 33) Elp2 may also serve as a general binding partner and/or chaperone for STAT proteins during the process of activation and nuclear translocation.
Our data are consistent with Elp2 and STAT-3 most robustly associating when STAT-3 is unphosphorylated, and that phosphorylation and activation of STAT-3 may destabilize this assocation. Our data do not address where this dissociation might occur. Dissociation might occur prior to nuclear translocation, as STAT-3 has a nuclear localization signal that appears to become functional upon STAT-3 activation by phosphorylation, and yeast Elp1 also has several putative nuclear localization signals (61). In contrast, Elp2 harbors only a nuclear export signal as predicted using the NetNES 1.1 Server.
Alternatively, dissociation could occur after the STAT-3/elongator complex translocates to the nucleus. In this latter hypothetical case, the activated STAT-3 would then bind to its target promoter sequence(s) and simultaneously deliver Elp-2 (and Elongator) near the promoter of the STAT-responsive gene. This STAT-3-targeted delivery of Elongator would then locally increase histone acetylation and RNA Pol II-mediated transcription.
The STATs may also regulate expression of molecular chaperones such as Hsc70, Hsp70, and Hsp90 (25, 26, 62–64). Our group and others have suggested that interactions between CFTR (wild type or mutant) and these chaperones regulate CFTR intracellular trafficking (4, 5, 18, 29, 52, 65–67). Modulations of these interactions by altering chaperone abundance may allow repair of ΔF508 intracellular trafficking (3).
In summary, our data confirm that 4PBA transiently increases Hsp70 expression in CF epithelial cells, which had been previously suggested by genomic and proteomic expression profiling experiments. This transiently increased Hsp70 expression likely results from transient activation and nuclear translocation of STAT-3 where it can bind to and activate the Hsp70 promoter. This process is dependent on the Elp2 component of elongator, which itself has transiently increased expression in response to 4PBA. Taken together, these data support the hypothesis that 4PBA improves ΔF508-CFTR intracellular trafficking in CF epithelial cells by regulation of Elp2, transient activation of STAT-3, and a resulting alteration in molecular chaperone expression.
This work was supported, in whole or in part, by Grants R01 DK058046 and R01 DK073185 (to R. C. R.) from the NIDDK, National Institutes of Health, from the Pennsylvania-Delaware Chapter of the American Heart Association (to L. S.), and the Cystic Fibrosis Foundation (to R. C. R.).
- CF
- cystic fibrosis
- CFTR
- cystic fibrosis transmembrane conductance regulator
- STAT-3
- signal transducer and activator of transcription-3
- StIP-1
- STAT-3-interacting protein-1
- Elp2
- elongator protein 2
- Hsc70
- heat shock cognate 70-kDa protein
- ER
- endoplasmic reticulum
- RT-PCR
- reverse transcriptase polymerase chain reaction
- 4PBA
- sodium 4-phenylbutyrate
- ns
- not significant
- aa
- amino acids.
REFERENCES
- 1. Denning G. M., Anderson M. P., Amara J. F., Marshall J., Smith A. E., Welsh M. J. (1992) Nature 358, 761–764 [DOI] [PubMed] [Google Scholar]
- 2. Yang Y., Janich S., Cohn J. A., Wilson J. M. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 9480–9484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Choo-Kang L. R., Zeitlin P. L. (2001) Am. J. Physiol. Lung. Cell Mol. Physiol. 281, L58-L68 [DOI] [PubMed] [Google Scholar]
- 4. Loo M. A., Jensen T. J., Cui L., Hou Y., Chang X. B., Riordan J. R. (1998) EMBO J. 17, 6879–6887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Rubenstein R. C., Zeitlin P. L. (2000) Am. J. Physiol. Cell Physiol. 278, C259–C267 [DOI] [PubMed] [Google Scholar]
- 6. Okiyoneda T., Wada I., Jono H., Shuto T., Yoshitake K., Nakano N., Nagayama S., Harada K., Isohama Y., Miyata T., Kai H. (2002) FEBS Lett. 526, 87–92 [DOI] [PubMed] [Google Scholar]
- 7. Okiyoneda T., Harada K., Takeya M., Yamahira K., Wada I., Shuto T., Suico M. A., Hashimoto Y., Kai H. (2004) Mol. Biol. Cell 15, 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pind S., Riordan J. R., Williams D. B. (1994) J. Biol. Chem. 269, 12784–12788 [PubMed] [Google Scholar]
- 9. Rosser M. F., Grove D. E., Chen L., Cyr D. M. (2008) Mol. Biol. Cell 19, 4570–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Suaud L., Miller K., Alvey L., Yan W., Robay A., Kebler C., Kreindler J. L., Guttentag S., Hubbard M. J., Rubenstein R. C. (2011) J. Biol. Chem. 286, 21239–21253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ward C. L., Kopito R. R. (1994) J. Biol. Chem. 269, 25710–25718 [PubMed] [Google Scholar]
- 12. Jensen T. J., Loo M. A., Pind S., Williams D. B., Goldberg A. L., Riordan J. R. (1995) Cell 83, 129–135 [DOI] [PubMed] [Google Scholar]
- 13. Ward C. L., Omura S., Kopito R. R. (1995) Cell 83, 121–127 [DOI] [PubMed] [Google Scholar]
- 14. Cheng S. H., Gregory R. J., Marshall J., Paul S., Souza D. W., White G. A., O'Riordan C. R., Smith A. E. (1990) Cell 63, 827–834 [DOI] [PubMed] [Google Scholar]
- 15. Kartner N., Augustinas O., Jensen T. J., Naismith A. L., Riordan J. R. (1992) Nat. Genet. 1, 321–327 [DOI] [PubMed] [Google Scholar]
- 16. Rubenstein R. C., Egan M. E., Zeitlin P. L. (1997) J. Clin. Invest. 100, 2457–2465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bercovich B., Stancovski I., Mayer A., Blumenfeld N., Laszlo A., Schwartz A. L., Ciechanover A. (1997) J. Biol. Chem. 272, 9002–9010 [DOI] [PubMed] [Google Scholar]
- 18. Rubenstein R. C., Lyons B. M. (2001) Am. J. Physiol. Lung. Cell Mol. Physiol. 281, L43–L51 [DOI] [PubMed] [Google Scholar]
- 19. Singh O. V., Vij N., Mogayzel P. J., Jr., Jozwik C., Pollard H. B., Zeitlin P. L. (2006) J. Proteome. Res. 5, 562–571 [DOI] [PubMed] [Google Scholar]
- 20. Wright J. M., Zeitlin P. L., Cebotaru L., Guggino S. E., Guggino W. B. (2004) Physiol. Genomics 16, 204–211 [DOI] [PubMed] [Google Scholar]
- 21. Hawkes N. A., Otero G., Winkler G. S., Marshall N., Dahmus M. E., Krappmann D., Scheidereit C., Thomas C. L., Schiavo G., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2002) J. Biol. Chem. 277, 3047–3052 [DOI] [PubMed] [Google Scholar]
- 22. Rahmani M., Dai Y., Grant S. (2002) Exp. Cell Res. 277, 31–47 [DOI] [PubMed] [Google Scholar]
- 23. Chen W. Y., Bailey E. C., McCune S. L., Dong J. Y., Townes T. M. (1997) Proc. Natl. Acad. Sci. U.S.A. 94, 5798–5803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Han Q., Hou X., Su D., Pan L., Duan J., Cui L., Huang B., Lu J. (2007) Acta Biochim. Biophys. Sin. 39, 453–461 [DOI] [PubMed] [Google Scholar]
- 25. Stephanou A., Isenberg D. A., Nakajima K., Latchman D. S. (1999) J. Biol. Chem. 274, 1723–1728 [DOI] [PubMed] [Google Scholar]
- 26. Stephanou A., Latchman D. S. (1999) Gene. Expr. 7, 311–319 [PMC free article] [PubMed] [Google Scholar]
- 27. Zeitlin P. L., Lu L., Rhim J., Cutting G., Stetten G., Kieffer K. A., Craig R., Guggino W. B. (1991) Am. J. Respir. Cell Mol. Biol. 4, 313–319 [DOI] [PubMed] [Google Scholar]
- 28. Bossu J. P., Chartier F. L., Fruchart J. C., Auwerx J., Staels B., Laine B. (1996) Biochem. J. 318, 547–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meacham G. C., Patterson C., Zhang W., Younger J. M., Cyr D. M. (2001) Nat. Cell Biol. 3, 100–105 [DOI] [PubMed] [Google Scholar]
- 30. Rahl P. B., Chen C. Z., Collins R. N. (2005) Mol. Cell 17, 841–853 [DOI] [PubMed] [Google Scholar]
- 31. Huang B., Johansson M. J., Byström A. S. (2005) RNA 11, 424–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Esberg A., Huang B., Johansson M. J., Byström A. S. (2006) Mol. Cell 24, 139–148 [DOI] [PubMed] [Google Scholar]
- 33. Collum R. G., Brutsaert S., Lee G., Schindler C. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 10120–10125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wen Z., Zhong Z., Darnell J. E., Jr. (1995) Cell 82, 241–250 [DOI] [PubMed] [Google Scholar]
- 35. Li D., Roberts R. (2001) Cell Mol. Life Sci. 58, 2085–2097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Smith T. F., Gaitatzes C., Saxena K., Neer E. J. (1999) Trends Biochem. Sci. 24, 181–185 [DOI] [PubMed] [Google Scholar]
- 37. Yamagishi N., Fujii H., Saito Y., Hatayama T. (2009) FEBS J. 276, 5870–5880 [DOI] [PubMed] [Google Scholar]
- 38. Pasyk E. A., Foskett J. K. (1995) J. Biol. Chem. 270, 12347–12350 [DOI] [PubMed] [Google Scholar]
- 39. Rubenstein R. C. (2006) Mol. Diagn. Ther. 10, 293–301 [DOI] [PubMed] [Google Scholar]
- 40. Rubenstein R. C., Zeitlin P. L. (1998) Am. J. Respir. Crit. Care Med. 157, 484–490 [DOI] [PubMed] [Google Scholar]
- 41. Zeitlin P. L., Diener-West M., Rubenstein R. C., Boyle M. P., Lee C. K., Brass-Ernst L. (2002) Mol. Ther. 6, 119–126 [DOI] [PubMed] [Google Scholar]
- 42. Prulière-Escabasse V., Planès C., Escudier E., Fanen P., Coste A., Clerici C. (2007) J. Biol. Chem. 282, 34048–34057 [DOI] [PubMed] [Google Scholar]
- 43. Burrows J. A., Willis L. K., Perlmutter D. H. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 1796–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Wang W. J., Mulugeta S., Russo S. J., Beers M. F. (2003) J. Cell Sci. 116, 683–692 [DOI] [PubMed] [Google Scholar]
- 45. Vila-Carriles W. H., Kovacs G. G., Jovov B., Zhou Z. H., Pahwa A. K., Colby G., Esimai O., Gillespie G. Y., Mapstone T. B., Markert J. M., Fuller C. M., Bubien J. K., Benos D. J. (2006) J. Biol. Chem. 281, 19220–19232 [DOI] [PubMed] [Google Scholar]
- 46. Cheong N., Madesh M., Gonzales L. W., Zhao M., Yu K., Ballard P. L., Shuman H. (2006) J. Biol. Chem. 281, 9791–9800 [DOI] [PubMed] [Google Scholar]
- 47. Krogan N. J., Greenblatt J. F. (2001) Mol. Cell Biol. 21, 8203–8212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Otero G., Fellows J., Li Y., de Bizemont T., Dirac A. M., Gustafsson C. M., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (1999) Mol. Cell 3, 109–118 [DOI] [PubMed] [Google Scholar]
- 49. Winkler G. S., Petrakis T. G., Ethelberg S., Tokunaga M., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2001) J. Biol. Chem. 276, 32743–32749 [DOI] [PubMed] [Google Scholar]
- 50. Winkler G. S., Kristjuhan A., Erdjument-Bromage H., Tempst P., Svejstrup J. Q. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 3517–3522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wittschieben B. O., Otero G., de Bizemont T., Fellows J., Erdjument-Bromage H., Ohba R., Li Y., Allis C. D., Tempst P., Svejstrup J. Q. (1999) Mol. Cell 4, 123–128 [DOI] [PubMed] [Google Scholar]
- 52. Farinha C. M., Nogueira P., Mendes F., Penque D., Amaral M. D. (2002) Biochem. J. 366, 797–806 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Kim J. H., Lane W. S., Reinberg D. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 1241–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Han Q., Lu J., Duan J., Su D., Hou X., Li F., Wang X., Huang B. (2008) Biochem. J. 409, 779–788 [DOI] [PubMed] [Google Scholar]
- 55. Johansen L. D., Naumanen T., Knudsen A., Westerlund N., Gromova I., Junttila M., Nielsen C., Bøttzauw T., Tolkovsky A., Westermarck J., Coffey E. T., Jäättelä M., Kallunki T. (2008) J. Cell Sci. 121, 854–864 [DOI] [PubMed] [Google Scholar]
- 56. Cyr D. M., Lu X., Douglas M. G. (1992) J. Biol. Chem. 267, 20927–20931 [PubMed] [Google Scholar]
- 57. Slaugenhaupt S. A., Gusella J. F. (2002) Curr. Opin. Genet. Dev. 12, 307–311 [DOI] [PubMed] [Google Scholar]
- 58. Anderson S. L., Coli R., Daly I. W., Kichula E. A., Rork M. J., Volpi S. A., Ekstein J., Rubin B. Y. (2001) Am. J. Hum. Genet. 68, 753–758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Slaugenhaupt S. A., Blumenfeld A., Gill S. P., Leyne M., Mull J., Cuajungco M. P., Liebert C. B., Chadwick B., Idelson M., Reznik L., Robbins C., Makalowska I., Brownstein M., Krappmann D., Scheidereit C., Maayan C., Axelrod F. B., Gusella J. F. (2001) Am. J. Hum. Genet. 68, 598–605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Close P., Hawkes N., Cornez I., Creppe C., Lambert C. A., Rogister B., Siebenlist U., Merville M. P., Slaugenhaupt S. A., Bours V., Svejstrup J. Q., Chariot A. (2006) Mol. Cell 22, 521–531 [DOI] [PubMed] [Google Scholar]
- 61. Fichtner L., Jablonowski D., Schierhorn A., Kitamoto H. K., Stark M. J., Schaffrath R. (2003) Mol. Microbiol. 49, 1297–1307 [DOI] [PubMed] [Google Scholar]
- 62. Madamanchi N. R., Li S., Patterson C., Runge M. S. (2001) J. Biol. Chem. 276, 18915–18924 [DOI] [PubMed] [Google Scholar]
- 63. Ripley B. J., Stephanou A., Isenberg D. A., Latchman D. S. (1999) Immunology 97, 226–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Stephanou A., Isenberg D. A., Akira S., Kishimoto T., Latchman D. S. (1998) Biochem. J. 330, 189–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Strickland E., Qu B. H., Millen L., Thomas P. J. (1997) J. Biol. Chem. 272, 25421–25424 [DOI] [PubMed] [Google Scholar]
- 66. Jiang C., Fang S. L., Xiao Y. F., O'Connor S. P., Nadler S. G., Lee D. W., Jefferson D. M., Kaplan J. M., Smith A. E., Cheng S. H. (1998) Am. J. Physiol. 275, C171–C178 [DOI] [PubMed] [Google Scholar]
- 67. Brown C. R., Hong-Brown L. Q., Biwersi J., Verkman A. S., Welch W. J. (1996) Cell Stress. Chaperones. 1, 117–125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Goldfarb S. B., Kashlan O. B., Watkins J. N., Suaud L., Yan W., Kleyman T. R., Rubenstein R. C. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 5817–5822 [DOI] [PMC free article] [PubMed] [Google Scholar]