Background: Deubiquitinating enzymes (DUBs) remove polyubiquitin chains from proteins.
Results: Protein-attached monoubiquitin is rapidly removed by many DUBs, but polyubiquitin chains linked through lysine 48 are resistant to cleavage.
Conclusion: Intramolecular chain interactions limit DUB activity toward polyubiquitin chains.
Significance: High activity toward monoubiquitin allows many DUBs to minimize inappropriate ubiquitin signaling.
Keywords: Cell Cycle, Deubiquitination, Ubiquitin, Ubiquitin-dependent Protease, Ubiquitination
Abstract
The attachment of lysine 48 (Lys48)-linked polyubiquitin chains to proteins is a universal signal for degradation by the proteasome. Here, we report that long Lys48-linked chains are resistant to many deubiquitinating enzymes (DUBs). Representative enzymes from this group, Ubp15 from yeast and its human ortholog USP7, rapidly remove mono- and diubiquitin from substrates but are slow to remove longer Lys48-linked chains. This resistance is lost if the structure of Lys48-linked chains is disrupted by mutation of ubiquitin or if chains are linked through Lys63. In contrast to Ubp15 and USP7, Ubp12 readily cleaves the ends of long chains, regardless of chain structure. We propose that the resistance to many DUBs of long, substrate-attached Lys48-linked chains helps ensure that proteins are maintained free from ubiquitin until a threshold of ubiquitin ligase activity enables degradation.
Introduction
The reversible attachment of ubiquitin to proteins is central to the control of protein stability, protein sorting, and other cellular processes (1, 2). Ubiquitin is usually attached to proteins through an isopeptide bond that links the carboxyl terminus of ubiquitin to the ϵ-amino group of a lysine residue. This ligation is catalyzed by an enzymatic cascade involving a ubiquitin-activating enzyme (E1), conjugating enzyme (E2), and protein ligase (E3). Attachment of ubiquitin to a substrate lysine (monoubiquitination) is often the first step in formation of a polyubiquitin chain wherein additional ubiquitins are attached to one of seven lysines in ubiquitin itself.
The most common ubiquitin chain linkages are through Lys11, Lys48, or Lys63 (3). Lys11- and Lys48-linked chains of four or more ubiquitins generally result in protein degradation by the proteasome, whereas Lys63-linked chains are associated with signal transduction (3). Ubiquitin chains form distinct structures based on their linkage. Lys63-linked chains adopt an extended conformation with little interaction between adjacent ubiquitins (4). In contrast, Lys11- and Lys48-linked chains form compact, globular structures with significant ubiquitin-ubiquitin contact (5, 6).
Ubiquitination is reversed by deubiquitinating enzymes (DUBs),2 proteases that specifically hydrolyze the isopeptide bond to the C terminus of ubiquitin (7). Ubiquitin-specific proteases (USPs) comprise the largest fraction of DUBs, including 56 of the 79 predicted DUBs in humans and 16 of 20 yeast DUBs (7). USPs are generally large proteins that show little discrimination among chains of different linkage (4, 5, 7). Ovarian tumor proteases are the next largest DUB class and often show specificity for distinct chain linkages (7).
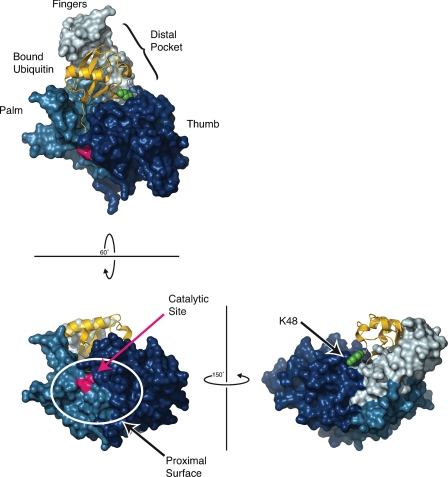
The structures of DUBs provide important clues about the molecular basis for their activity, regulation, and substrate specificity. Studies of USP7, also known as herpesvirus-associated USP, established a USP catalytic domain architecture that resembles a cupped right hand with separate thumb and fingers subdomains extending from the palm (Fig. 1) (8). In these DUBs, the active site resides in a cleft between the thumb and palm subdomains. In complexes with ubiquitin derivatives, residues in this cleft make contacts along the C-terminal tail of ubiquitin (7, 8). This distal or leaving ubiquitin occupies an extensive distal binding pocket established by the palm and fingers subdomains and abutting the thumb. On the opposite side of the catalytic cleft, a less well defined binding surface presumably interacts with the proximal ubiquitin (or other protein) that is conjugated through a lysine to the C terminus of the distal ubiquitin. This surface has several highly conserved residues that assist in catalysis but is otherwise quite variable (9).
FIGURE 1.
Structure of USP7 with bound ubiquitin. USP7/herpesvirus-associated USP (Protein Data Bank code 1NBF (8)) is shown as a surface rendering with thumb, palm, and fingers subdomains colored dark, medium, and light blue, respectively, with the catalytic Cys223/His464 residues colored red. Ubiquitin is shown in yellow bound to the distal binding pocket (S1 site) created between the tip of the fingers and base of the thumb subdomains. The side chain of Lys48 of this bound ubiquitin is shown in green. The proximal binding surface of USP7 (lower left panel; S1′ site) must accommodate the protein linked to the bound (distal) ubiquitin. Note the limited solvent accessibility of Lys48 of the bound ubiquitin (lower right panel). This illustration was made with MacPyMOL (29).
Our understanding of DUB activity and specificity has come primarily from studies using unanchored (unattached to substrate) ubiquitin chains. Little is known about the biochemical activities of DUBs toward ubiquitin chains on physiologically relevant E3 substrates. In this work, we set out to identify DUBs that oppose the budding yeast anaphase-promoting complex (APC/C), an E3 that controls progression through mitosis by building Lys48-linked chains on substrates. We screened yeast DUBs in vitro for activity toward mono- and polyubiquitinated APC/C substrates. Surprisingly, we found that many DUBs showed a preference for monoubiquitinated substrate and removed long chains very slowly. We propose that in the cell, efficient removal of monoubiquitin, coupled with the resistance to DUBs of long Lys48-linked chains, promotes the all-or-none generation of proteasomal signals on specific E3 substrates.
EXPERIMENTAL PROCEDURES
Immunopurification of DUB Complexes
Approximately 0.5 mg of lysate from yeast strains bearing tandem affinity purification-tagged DUBs was incubated with IgG-conjugated DynaBeads (Invitrogen), washed in lysis buffer (50 mm HEPES, pH 8.0, 150 mm NaCl, 0.2% Nonidet P-40, 10% glycerol, and 10 mm 2-mercaptoethanol), and then washed in 1× DUB buffer (see below) before use in DUB assays. Immunoblotting used the peroxidase anti-peroxidase antibody (Sigma-Aldrich) to detect the tandem affinity purification tag.
Protein Purification
Recombinant proteins carrying GST and/or His6 tags (see supplemental “Experimental Procedures”) were expressed in Rosetta 2 Escherichia coli (EMD Chemicals, Gibbstown, NJ). Frozen cell pellets were lysed with sonication and 50 units/ml DNase I in GST lysis buffer (50 mm HEPES, pH 8.0, 500 mm NaCl, 10% glycerol, 1 mm MgCl2, 10 mm 2-mercaptoethanol) or His6 lysis buffer (same as GST lysis buffer, plus 0.1% Nonidet P-40 and 7.5 mm imidazole) with protease inhibitors (1 mm PMSF, 1 μm aprotinin, 0.63 mg/ml benzamidine, and 1 μg/ml each of pepstatin A and leupeptin). Lysates were cleared by centrifugation, bound to resin (GST (GE Healthcare); His6, Ni-NTA-agarose (Qiagen, Valencia CA)), washed extensively with lysis buffer (for His6 wash, imidazole was increased to 20 mm), and eluted with 20 mm reduced glutathione or 300 mm imidazole (His6). Affinity tags were removed by incubation with His6-tobacco etch virus protease or GST-PreScission (GE Healthcare) proteases and dialyzed into HBS (20 mm HEPES, pH 8.0, 150 mm NaCl, 10% glycerol, 10 mm 2-mercaptoethanol).
Following GST purification, DUBs were flowed over glutathione resin to remove GST and PreScission protease and then bound to Ni-NTA resin and purified with their C-terminal His6 tags as above. Ubiquitin preparations were flowed over glutathione resin to remove GST, flowed over Ni-NTA resin to remove tobacco etch virus protease, heated at 65 °C for 20 min to denature any contaminants, held on ice for 10–30 min, and centrifuged (100,000 × g, 30 min, 4 °C), essentially as described (10). Pds1–110 and Pds1-Lys33 were treated like ubiquitin (omitting flow through glutathione resin), followed by purification over a MonoS column (GE Healthcare) and size-exclusion chromatography. Cyclin B (CycB) purification has been described elsewhere (11). CycB-Lys60 was purified as in the DUB purifications, followed by heating as in the ubiquitin purification. All proteins were dialyzed to HBS, concentrated with Centriprep spin concentrators (Millipore, Billerica, MA) and stored at −80 °C.
Human Ube1 was expressed in E. coli and purified with ubiquitin-agarose (Boston Biochem, Cambridge, MA) as described (12). Human USP7 preparations were purchased from Boston Biochem.
Radiolabeling Substrates
32P-labeled diubiquitin and CycB-Lys60 were phosphorylated as described (13). 25 μg of diubiquitin (bearing a PKA site on the distal ubiquitin; see below) or PKA-CycB-Lys60 were incubated with 10 μCi of [γ-32P]ATP, 1.7 nmol of ATP, and 2 μl of PKA (New England Biolabs) in 50 μl, using the provided reaction buffer. Reactions were terminated by heating as for ubiquitin purification. ATP was removed by gel filtration through G25 Sephadex. 125I-radiolabeled substrates were prepared as described (11).
Generation of Ubiquitinated Substrates
APC/C reaction components (E1, Ubc1, Ubc4, APC/C, Cdh1) were purified as described (11), except that the final calmodulin-binding step was omitted from the APC/C preparation. Standard ubiquitination reactions contained E1 (1 μm Uba1), E2 (25 μm Ubc4 and 5 μm Ubc1), 1 mg/ml ubiquitin (Boston Biochem), 1 mg/ml ATP, APC/C, Cdh1, and radiolabeled substrate. 25–90% methyl ubiquitin (Boston Biochem) was substituted for wild-type ubiquitin to limit the length of wild-type chains. Reactions were performed in QAH8 (25 mm HEPES, pH 7.8, 100 mm NaCl, 1 mm MgCl2). E2 enzymes were charged at room temperature for 15 min before starting APC/C reactions.
APC/C reactions were terminated to eliminate competition with DUBs. Removing ATP with G25 Sephadex spin columns (GE Healthcare) was sufficient to stop APC/C activity but yielded ubiquitin ladders with poorly resolved bands (data not shown). Using the protocol of Rodrigo-Brenni et al. (10) for purification of phosphorylated ubiquitin stopped APC/C activity and retained sharp ubiquitin ladder banding. Reactions were thus terminated by heating at 65 °C for 20 min, chilling on ice for 10 min, and centrifugation for 10 min at 20,000 × g. Supernatants containing ubiquitinated substrates were divided into aliquots and stored at −80 °C. Heat treatment of ubiquitinated substrates did not affect the length selectivity of Ubp15 (data not shown).
The substrates used in Fig. 6, A–H, were made with the APC/C in two steps. First, Ubc4 was used with the APC/C to ligate G76A or wild-type (i.e. Gly76) monoubiquitin to CycB-Lys60. Reactions were terminated with heating as described above. To make diubiquitinated CycB-Lys60, Ubc1 was used with the APC/C to ligate a second ubiquitin to the first. Reactions were terminated with heating as described above. Ubiquitin bearing K48R was used for monoubiquitin-only substrates and as the terminal ubiquitin in diubiquitin chains. Ubc1 and Ubc4 were charged with ubiquitin bearing G76A at 37 °C for 30 min prior to APC/C reactions.
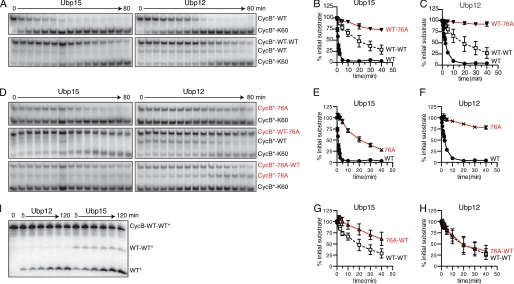
FIGURE 6.
Ubp15 and Ubp12 show distinct base- and tip-directed activities. A, Ubp15 and Ubp12 deubiquitination of mono- (top panels) or diubiquitinated CycB (bottom panels). Time course of 125I-labeled CycB-Lys60 deubiquitination by 0.4 nm Ubp15 (left) or 0.5 nm Ubp12 (right). K48R ubiquitin was used for monoubiquitination and as the distal ubiquitin of diubiquitin chains. APC/C reactions with Ubc4 attached the first ubiquitin to CycB; Ubc1 was substituted for Ubc4 to attach a second ubiquitin. APC/C reactions were terminated with heating. B and C, quantitation of data from A and D. Levels of each ubiquitin conjugate are normalized to their initial values (in arbitrary units) at time zero. Error bars denote S.E. from two to three replicates. D, G76A ubiquitin conjugates reveal base versus tip selectivity. Shown is the time course as described in A, substituting ubiquitin bearing G76A where indicated. E–H, quantitation of data from A and D, as described in B. I, time course of CycB diubiquitin hydrolysis in which the distal ubiquitin, rather than CycB, is radiolabeled. K48R ubiquitin bearing a PKA phosphorylation site was ligated to wild-type ubiquitin with Cdc34 and purified by ion exchange chromatography. After phosphorylation, this diubiquitin was ligated to unlabeled CycB-Lys60 in an APC/C reaction with Ubc4. After gel purification, 50 nm CycB diubiquitin was incubated with 200 nm Ubp12 (left) or Ubp15 (right). See supplemental Table S1 for initial rates.
Products bearing I44A ubiquitin chains were not heat stable, so these reactions were terminated by treatment with 1 mm N-ethylmaleimide and 5 mm hydroxylamine for 30 min, followed by 4 mm DTT to quench unreacted N-ethylmaleimide. CycB-Lys60 bearing Lys63-linked chains was generated using standard APC/C reactions, omitting Ubc1 and charging Ubc4 for 30 min at 37 °C with Lys63-linked chains (Enzo Life Sciences, Farmingdale, NY).
Synthesis of Phosphorylated Diubiquitin CycB
The substrate for Fig. 6I (diubiquitinated CycB-Lys60 in which only the distal ubiquitin was labeled) was made as described in supplemental “Experimental Procedures”. Briefly, Lys48-linked diubiquitin (a proximal wild-type ubiquitin ligated to a distal PKA-K48R ubiquitin) was synthesized in a Cdc34-dependent reaction essentially as described (14). This diubiquitin was phosphorylated as described above and ligated to unlabeled CycB-Lys60 using APC/C and Ubc4. The final product was purified using Tris-Tricine PAGE followed by gel extraction.
Deubiquitination Activity Assays
DUBs were activated by incubation at room temperature for 10 min in 1× DUB buffer (50 mm HEPES, pH 7.8, 100 mm NaCl, 10 mm DTT, 0.1 mg/ml BSA). Activated DUBs were mixed with ubiquitinated substrates at room temperature, and reactions were terminated with PAGE sample buffer, followed by heating (3 min, 95 °C) and 15% SDS-PAGE (see Figs. 3E and 4, A and F) or 10% Tris/Tricine PAGE. Reaction products were visualized with a Typhoon PhosphorImager, and data were quantified using ImageQuant software (GE Healthcare). Data were processed with Excel (Microsoft, Redmond, WA). Curve-fitting analyses and graphs were produced with Prism5 (GraphPad Software, La Jolla, CA).
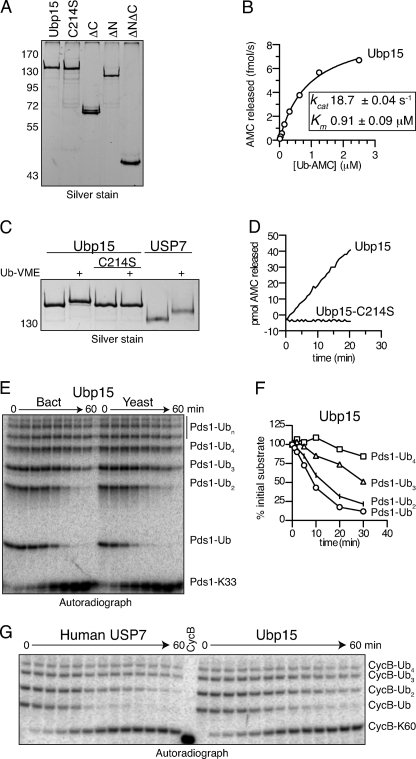
FIGURE 3.
Ubp15 and human USP7 show similar preferences for substrate-attached monoubiquitin and short chains. A, purity of recombinant Ubp15 isoforms. Shown is a silver-stained gel showing 50 ng of the indicated Ubp15 isoforms after affinity purification from E. coli lysates. C214S indicates a catalytically inactive isoform of Ubp15 wherein the catalytic Cys214 was mutated to Ser (9, 23). B, characterization of Ub-AMC hydrolysis by Ubp15. Recombinant Ubp15 (10 pm) was incubated with Ub-AMC (0.02–2.5 μm) for 30 min at room temperature and release of fluorescent AMC was monitored. Initial velocities were calculated and plotted against Ub-AMC concentration. Estimated values of Km and kcat were calculated using Prism. C, catalytic Cys214 is required for Ub-VME binding to Ubp15. Ubp15 or Ubp15-C214S (50 ng) was exposed to Ub-VME, an activity-based probe that irreversibly cross-links to the catalytic Cys residue in active USPs (25). Human USP7 serves as a positive control for Ub-VME reactivity. D, catalytic Cys214 is required for Ub-AMC hydrolysis by Ubp15. Ubp15 or Ubp15-C214S (33 pm) was exposed to Ub-AMC (2.5 μm) for 30 min at room temperature, and release of fluorescent AMC was monitored. E, comparison of recombinant and endogenous yeast Ubp15. Shown is the time course of 125I-labeled Pds1-Lys33 deubiquitination by 4 nm recombinant (Bact) or 10 nm immunoprecipitated (Yeast) Ubp15. Pds1-Lys33 ubiquitin conjugates were made as described in Fig. 2G. Deubiquitinated Pds1-Lys33 (bottom band) and bands corresponding to 1 or more ubiquitins on Pds1-Lys33 are indicated. F, quantitation of recombinant Ubp15 activity from E. Levels of each ubiquitin conjugate are normalized to their initial values (in arbitrary units) at time zero. G, comparison of Ubp15 with human USP7. Time course of ubiquitin removal from 125I-labeled CycB-Lys60 by 130 nm recombinant Ubp15 or 6 nm USP7 as described in E. Wild-type CycB is shown as molecular weight reference. See supplemental Table S1 for initial rates. All results are representative of two to three experiments.
FIGURE 4.
Ubp15 and USP7 flanking domains are not required for substrate selectivity, and Ubp15 shows complementary activity to the 26 S proteasome. A, time course of 125I-labeled Pds1-Lys33 deubiquitination as described in Fig. 3E by 160 nm full-length Ubp15 or truncated isoforms: ΔN (residues 199–1230) or ΔC (residues 1–562). B and C, quantitation of A. Levels of mono- (B) or tetraubiquitinated (C) Pds1-Lys33 are normalized to their initial values (in arbitrary units) at time zero. D, time course of 0.5 μm Ub-AMC hydrolysis by 50 pm full-length Ubp15 or truncated isoforms: ΔN, ΔC, or isolated catalytic domain (ΔNΔC, residues 199–562). E, time course of 125I-labeled CycB-Lys60 deubiquitination by 6 nm full-length USP7 or 20 nm Ubp15 as compared with the isolated catalytic domain of USP7 (ΔNΔC; residues 213–549) at 20 μm, as indicated. F, Ubp15 preferentially targets ubiquitin conjugates that are not degraded by the 26 S proteasome. Time course of 125I-labeled Pds1-Lys33 deubiquitination by 160 nm recombinant Ubp15 as shown in A or 1 μl of immunoprecipitated yeast 26 S proteasome. G and H, quantitation of F. Levels of each ubiquitin conjugate are normalized to their initial values (in arbitrary units) at time zero for Ubp15 (G) and proteasome (H). See supplemental Table S1 for initial rates. All results are representative of two to three experiments.
For Ub-AMC hydrolysis assays, DUBs were activated as above and mixed with Ub-AMC in 1× DUB buffer. Reaction progress was monitored as an increase in fluorescence (excitation, 380 nm; emission, 460 nm) by a Synergy4 plate reader (BioTek). Data were analyzed as described above.
For Ub-vinyl methyl ester (Ub-VME; Boston Biochem) binding assays, 10 mm TCEP was used in place of DTT in the assay buffer to maintain Ub-VME reactivity. In a volume of 10 μl, 50 ng of each DUB and 90 ng of Ub-VME (∼30-fold molar excess) were allowed to react for 20 min at room temperature.
Proteasome Assays
26 S proteasome was purified from SDL145 using IgG-Sepharose as described (15).
RESULTS
Multiple DUBs Can Deubiquitinate Securin and Cyclin B
Our initial goal was to identify yeast DUBs that deubiquitinate two major APC/C targets, securin (Pds1 in yeast), and cyclin. We immunopurified the 20 yeast DUBs, using an equal amount of lysate in each purification. Most DUBs were readily purified, yielding discrete bands near or greater than the expected molecular weight (Fig. 2, A and B). Many DUBs co-purified with other proteins, in agreement with earlier studies (16, 17). DUB protein levels in these preparations varied widely, reflecting a range of expression levels (18). In general, high enzyme levels did not predict high DUB activity, but very low levels often resulted in little apparent activity (see below).
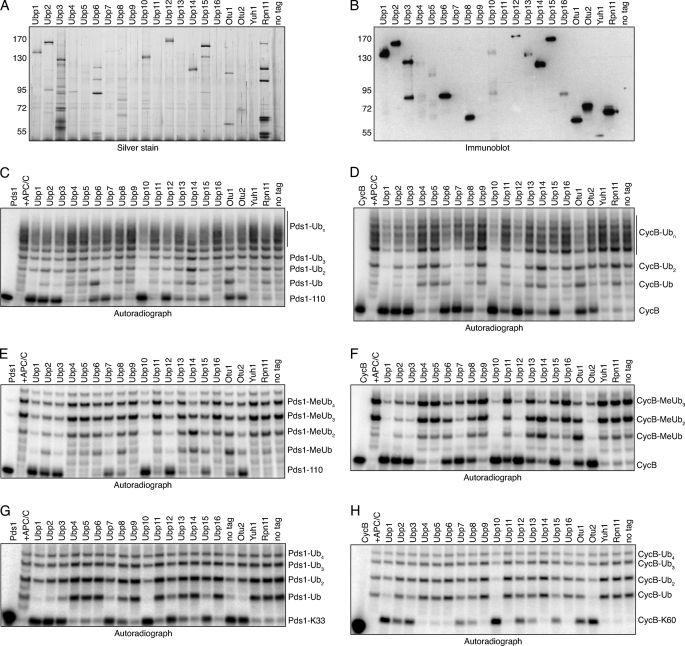
FIGURE 2.
Yeast DUBs show distinct patterns of ubiquitin removal from securin and cyclin. A, silver-stained gel showing DUB complexes, each immunoprecipitated from ∼0.5 μg of yeast lysate. B, immunoblot showing relative expression levels of tandem affinity purification-tagged DUBs purified as described in A. C, removal of polyubiquitin chains from securin. The indicated DUBs were immunopurified from yeast as described in A and incubated for 1 h at room temperature with 125I-labeled Pds1–110 that had been ubiquitinated in APC/CCdh1-dependent reactions (+APC/C). APC/C reactions were terminated with heating. Pds1–110 is an N-terminal fragment of yeast Pds1 (residues 1–110). Control reactions were performed with mock-purified lysate from a strain lacking a tandem affinity purification-tagged DUB (no tag). Fully deubiquitinated Pds1–110 (lowest band) and bands corresponding to 1 or more ubiquitins on Pds1–110 are indicated. D, removal of polyubiquitin chains from cyclin. The DUB assay was performed as described in C, substituting 125I-labeled CycB. CycB is an N-terminal fragment of sea urchin cyclin B (residues 10–113). E, removal of proximal ubiquitins from Pds1–110. Deubiquitination assay was performed as described in C, using 100% methyl ubiquitin (MeUb) to prevent chain formation. F, removal of proximal ubiquitins from CycB. Deubiquitination assay was performed as described in D, using MeUb as described in E. G, deubiquitination of single ubiquitin chains on Pds1–110. Deubiquitination assays were performed as described in C, substituting 125I-labeled Pds1-Lys33, a Pds1–110 isoform bearing just one ubiquitin-accepting lysine (see text). H, deubiquitination of single ubiquitin chains on CycB. Deubiquitination assays were performed as described in D, substituting 125I-labeled CycB-Lys60, a CycB isoform bearing just one ubiquitin-accepting lysine (see text). All autoradiographs show representative results from two to three experiments.
In pilot experiments, we found it difficult to quantify DUB activities using polyubiquitinated full-length securin, which migrates by SDS-PAGE as a highly heterogeneous smear (19). To facilitate a detailed analysis of DUB activity, we prepared an amino-terminal fragment of securin encompassing residues 1–110 (Pds1–110), which includes D- and KEN-box elements important for ubiquitination by the APC/C (20). We radiolabeled Pds1–110 with 125I and built Lys48-linked polyubiquitin chains on it using the APC/C and its two partner E2 enzymes, Ubc1 and Ubc4 (13).
The accumulation of deubiquitinated Pds1–110 (lowest band on the gel) showed that most of the purified DUBs could deubiquitinate Pds1–110, with some enzymes (e.g. Ubp1, -2, -3, -10, and -12) displaying high activity and others (e.g. Ubp4, -5, -11, and -16) showing little or no activity (Fig. 2C). We also measured deubiquitination of an N-terminal fragment (residues 13–110) of sea urchin CycB, a well characterized model APC/C substrate. The enzymes that deubiquitinated Pds1–110 also targeted CycB with similar activity (Fig. 2D).
Most DUBs showing activity generated fully deubiquitinated Pds1–110 and CycB, indicating that they removed proximal ubiquitins (i.e. directly attached to substrate) along with any polyubiquitin chains. Several enzymes (e.g. Ubp6 and Ubp14) seemed to accumulate mono-, di-, or triubiquitinated products and might therefore be less efficient at removing proximal ubiquitins. To directly test this, we substituted methyl ubiquitin in the APC/C reaction to prevent chain extension. This confirmed the restricted activity seen with Ubp6 and Ubp14 and showed that the ability of a DUB to remove monoubiquitin from a substrate generally correlated with its activity toward mixed chain lengths (Fig. 2, E and F).
Our results indicate that several yeast DUBs possess activity toward APC/C substrates, raising the possibility of redundancy in DUB function in the cell. Indeed, none of the 16 UBP genes is required for viability (21), and we found that no single deletion mutant displays a clear cell cycle defect.3 We also were unable to detect genetic interactions between the APC/C and Ubp family members: for example, the growth defects of hypomorphic APC/C mutants (doc1–4A and cdc26Δ) were not suppressed by deletion of individual UBP genes. We therefore suspect that APC/C substrate modification in the cell is controlled by multiple DUBs.
Long Lys48-linked Ubiquitin Chains Attached to Substrates Are Resistant to Many DUBs
Pds1–110 and CycB form complex ubiquitination patterns because chains can be built on multiple lysines. To allow more rigorous analysis, we produced various Pds1–110 isoforms bearing only one lysine that can accept ubiquitin. We found that the KEN box lysine (Lys8) is not ubiquitinated but is required for APC/C ubiquitination at several other lysines (Lys33, Lys41, Lys50, Lys62, Lys74, and Lys109).4 The first five of these were rapidly deubiquitinated by Ubp15. We used Pds1–110 bearing only lysines 8 and 33 (Pds1-Lys33) for further work.
Ubiquitination of Pds1-Lys33 by the APC/C, together with the Lys48-specific E2, Ubc1, yielded a ladder of reaction products reflecting assembly of Lys48-linked chains on Lys33 of Pds1 (Fig. 2G) (13). Exposing these products to the panel of DUBs revealed that enzymes with activity toward Pds1–110 also targeted Pds1-Lys33. We also constructed a CycB isoform (CycB-Lys60) retaining just one lysine, Lys60, a target the APC/C.5 CycB-Lys60 ubiquitin conjugates were targeted by the DUB panel in a similar manner to Pds1-Lys33 conjugates (Fig. 2H).
Interestingly, several DUBs with significant activity toward single-lysine substrates appeared to be more efficient at cleaving short ubiquitin chains than long ones. This was most clearly seen in reactions with Ubp1, Ubp15, and Otu2, in which most monoubiquitin was removed, whereas tri- and tetraubiquitin chains were more stable (Fig. 2, G and H). Thus, long Lys48-linked chains seemed resistant to cleavage by several DUBs. Ubp12 appeared to be an exception, showing activity toward both short and long chains.
Ubp15 and USP7 Preferentially Remove Monoubiquitin and Short Chains from Substrates
To more rigorously understand chain length preference, we carried out detailed studies of Ubp15, which displayed a preference for short substrates and was also of interest because it is known to associate with the APC/C activator Cdh1, suggesting a possible regulatory interaction with the APC/C (22, 23). We expressed Ubp15 in bacteria, and two affinity purification steps yielded active enzyme that was 95% pure (Fig. 3A). Using the model substrate Ub-AMC, we measured a kcat/Km value of 2.1 × 107 m−1 s−1 (Fig. 3B), similar to that seen in recent studies of Ubp15 and 10-fold higher than its human ortholog, USP7 (23, 24). Ubp15 reacted with Ub-VME and hydrolyzed Ub-AMC in a manner dependent on its catalytic Cys214 residue, consistent with earlier studies (Fig. 3C, D) (23, 25). Recombinant Ubp15 showed a pattern of Pds1-Lys33 deubiquitination similar to that of Ubp15 immunoprecipitated from yeast, indicating that its activity and specificity do not depend on modification or binding partners from yeast cells (Fig. 3E, supplemental Table S1).
Recombinant Ubp15 removed monoubiquitin faster than di- or triubiquitin, and chains of tetraubiquitin or longer were removed very slowly (Fig. 3F). Estimation of initial rates indicated that monoubiquitin levels fell 7-fold faster than tetraubiquitin (supplemental Table S1). We also found that human USP7 showed similar selectivity, removing monoubiquitin from CycB-Lys60 30-fold faster than tetraubiquitin (Fig. 3G, supplemental Table S1).
For Ubp15, this chain length selectivity did not depend on the N- or C-terminal domains flanking the catalytic core, although these domains were required for full activity, as seen with Ub-AMC (Fig. 4, A–D) (23). Similarly, the isolated catalytic core of USP7 retained selectivity, despite an ∼1000-fold loss of overall activity (Fig. 4E). We propose that kinetic chain length discrimination may explain why other DUBs in this class (Ubp1, Ubp10, etc.) preferentially target monoubiquitin and short chains.
The 26 S proteasome is known to recognize Lys48-linked chains of four or more ubiquitins (26). Thus, the low activity of Ubp15 toward longer chains suggests that it will not prevent proteasomal destruction of substrates carrying long chains. Consistent with this idea, we observed that the ubiquitinated substrates most resistant to Ubp15 are the preferred substrates of the proteasome in vitro (Fig. 4, F–H).
Ubp12 Efficiently Hydrolyzes Both Short and Long Lys48-linked Chains on Substrates
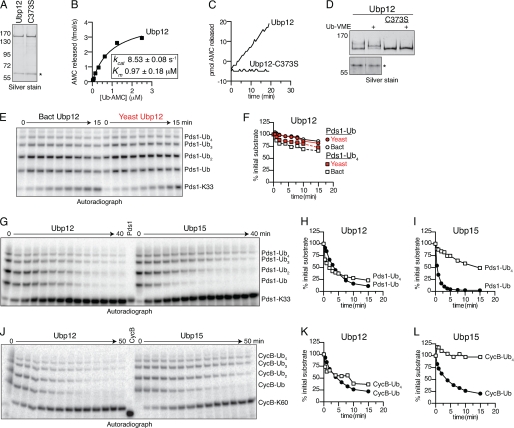
Our initial studies of single-lysine substrates suggested that Ubp12 displayed little chain length selectivity (Fig. 2, G and H). To explore this issue in detail, we purified recombinant Ubp12, which displayed ∼2-fold lower activity toward Ub-AMC than did Ubp15 (kcat/Km = 8.8 × 106 m−1 s−1) (Fig. 5, A and B). Ubp12 hydrolyzed Ub-AMC and reacted with Ub-VME in a manner dependent on its predicted catalytic Cys373 residue (Fig. 5, C and D) (9, 25). Recombinant Ubp12 activity was comparable with that of Ubp12 purified directly from yeast, indicating that activity does not depend on modification or binding partners from yeast cells (Fig. 5, E and F).
FIGURE 5.
Ubp12 shows little chain length selectivity. A, purity of recombinant Ubp12. Shown is a silver-stained gel showing 50 ng of the indicated Ubp12 isoforms after affinity purification from E. coli lysates. C373S indicates a catalytically inactive isoform of Ubp12 wherein the predicted catalytic Cys373 was mutated to Ser (9). The asterisk indicates a co-purifying protein that does not have DUB activity (see D). B, kinetic characterization of Ub-AMC hydrolysis by Ubp12. Recombinant Ubp12 (10 pm) was incubated with Ub-AMC (0.02–2.5 μm) for 30 min at room temperature, and the release of fluorescent AMC was monitored. Initial velocities were calculated and plotted against Ub-AMC concentration. Estimated values of Km and kcat were calculated using Prism. C, catalytic Cys373 is required for Ub-AMC hydrolysis by Ubp12. Ubp12, or Ubp12-C373S (33 pm) was exposed to Ub-AMC (2.5 μm) for 30 min at room temperature, and the release of fluorescent AMC was monitored. D, catalytic Cys373 is required for Ub-VME binding to Ubp12. Ubp12 or Ubp12-C373S (50 ng) was exposed to Ub-VME as shown for Ubp15 (Fig. 3C). The lower panel shows that the co-purifying 60 kDa protein does not react with Ub-VME. E, comparison of recombinant and endogenous yeast Ubp12. Time course of 125I-labeled Pds1-Lys33 deubiquitination by 30 nm recombinant (Bact) or 10 nm immunoprecipitated (Yeast) Ubp12. Pds1-Lys33 ubiquitin conjugates were made as in Fig. 2G. Deubiquitinated Pds1-Lys33 (bottom band) and bands corresponding to one or more ubiquitins on Pds1-Lys33 are indicated. F, quantitation of E. Levels of each ubiquitin conjugate are normalized to their initial values (in arbitrary units) at time zero. G–L, comparison of recombinant Ubp12 and Ubp15. Shown are time courses of ubiquitin removal from 125I-labeled Pds1-Lys33 (G) or 125I-labeled CycB-Lys60 (J) by 40 nm Ubp12 or 90 nm Ubp15 as described for E. H, I, K, and L are quantitation of data from G and J as described for F. See supplemental Table S1 for initial rates. All results are representative of two to three experiments.
Direct comparisons of Ubp12 and Ubp15 revealed distinct chain length specificities. Levels of monoubiquitinated Pds1 fell just 2-fold faster than those of tetraubiquitin with Ubp12, whereas the difference with Ubp15 was 20-fold (Fig. 5, G–I, and supplemental Table S1). The same patterns held for CycB conjugates (Fig. 5, J–L, supplemental Table S1).
One explanation for these results is that Ubp12 removes monoubiquitin more slowly than does Ubp15. Alternatively, monoubiquitinated substrate could accumulate in Ubp12 reactions as the product of longer chain degradation. If monoubiquitin is hydrolyzed faster than longer chains (as was observed with Ubp15), then the effect of any accumulation might be minimal. However, because Ubp12 degraded long chains efficiently, its actual rate of monoubiquitin cleavage could be higher than that observed here. Thus, the principal difference between Ubp15 and Ubp12 might lie in their activities toward long chains.
Ubp15 and Ubp12 Show Distinct Base- and Tip-directed Activities
To assess how chain length affects activity, we built homotypic substrates of either mono- or diubiquitin attached to CycB-Lys60 and exposed them to Ubp15 or Ubp12. Both enzymes removed monoubiquitin much faster than diubiquitin (Fig. 6. A–C, supplemental Table S1). The high rate of monoubiquitin cleavage seen here contrasts with the observed rates in complex mixtures, where Ubp15 removed monoubiquitin just 3-fold faster than diubiquitin (versus 10-fold in this experiment) and Ubp12 showed no difference (versus 9-fold in this experiment). We suspect, as described above, that tip-directed activity in complex substrate mixtures reduces the apparent rate of short chain hydrolysis by producing intermediate length isoforms.
To explore further the ability of these DUBs to cleave at the tip or base of chains, we used ubiquitin in which the terminal glycine was mutated to alanine (G76A), which is known to reduce DUB activity (25). G76A impeded the cleavage of monoubiquitin from CycB by both Ubp15 and Ubp12 (9- and 32-fold, respectively) (Fig. 6, D–F, supplemental Table S1). By incorporating G76A ubiquitin at the base or tip of a diubiquitin chain, we then tested the effects of inhibiting cleavage at the proximal or distal scissile bond. Diubiquitin hydrolysis by Ubp15 was significantly slowed when G76A was used as the distal ubiquitin, indicating that Ubp15 has tip-directed activity (Fig. 6B). However, placing G76A at the base of the chain also slowed diubiquitin cleavage, suggesting base-directed activity (Fig. 6G). Because neither placement displayed wild-type cleavage rates, it appears that the wild-type rate is due to both tip- and base-directed activity. Recall, however, that Ubp15 has very little activity with longer Lys48-linked chains, indicating that chain length itself affects Ubp15 activity.
Ubp12 had negligible activity with substrates in which G76A was placed at the tip of a diubiquitin chain (Fig. 6C), whereas placement of G76A at the base of the chain allowed wild-type rates of cleavage (Fig. 6H). Interestingly, G76A monoubiquitin accumulated in the latter reactions as Ubp12 removed the distal wild-type ubiquitin (Fig. 6D, lower right panel). Thus, Ubp12 activity is directed toward the tip of substrate-attached Lys48-linked diubiquitin chains, suggesting that its high activity toward longer chains could also be tip-directed.
We also assessed tip- and base-directed activities using a diubiquitinated CycB-Lys60 substrate in which only the terminal ubiquitin was radiolabeled. Consistent with the results above, treatment of this substrate with Ubp15 resulted in the generation of both mono- and diubiquitin, whereas treatment with Ubp12 generated primarily monoubiquitin (Fig. 6I).
Chain Structure and Linkage Affect Ubp15 and Ubp12 Activities
Substrate-attached Lys48-linked chains are resistant to Ubp15. In these chains, the hydrophobic patches of adjacent ubiquitins interact to form a dimer, which can then interact with an adjacent dimer to form a globular tetramer (6). We hypothesized that this closed structure restricts Ubp15 access, inhibiting cleavage of longer chains.
Mutations in the ubiquitin hydrophobic patch (Leu8, Ile44, and Val70) disrupt the closed structure of Lys48-linked chains (27). We used an I44A ubiquitin mutant to build short Lys48-linked chains on CycB-Lys60. Upon exposure to Ubp15, levels of I44A monoubiquitinated CycB fell just 2-fold faster than triubiquitin levels (Fig. 7, A and B). This contrasts with the kinetics of wild-type chain hydrolysis by Ubp15, where monoubiquitin levels declined 7–11-fold faster than triubiquitin (supplemental Table S1). Thus, the structure of wild-type Lys48-linked chains (rather than the linkage itself) impedes Ubp15 activity.
FIGURE 7.
Chain structure affects Ubp15 and Ubp12 activity. A, deubiquitination of disordered Lys48-linked chains by Ubp15. Shown is a time course of removal of ubiquitin bearing the hydrophobic patch mutation, I44A, from 125I-labeled CycB-Lys60 by 4 nm Ubp15. I44A ubiquitin was attached to CycB-Lys60 in an APC/C reaction with Ubc4 and Ubc1. The APC/C reaction was terminated with hydroxylamine and N-ethylmaleimide. B, quantitation of data from A. Levels of each ubiquitin conjugate are normalized to their initial values (in arbitrary units) at time zero. Note the short time scale relative to other graphs in this figure. C, deubiquitination of Lys63-linked chains. Shown is the time course of removal of monoubiquitin or Lys63-linked ubiquitin chains from 32P-labeled CycB-Lys60 by 0.4 nm Ubp15. Preformed chains of Lys63-linked ubiquitin were attached to CycB-Lys60 using Ubc4 in an APC/C-dependent reaction. The APC/C reaction was terminated with heat. D, quantitation of data in C as shown in B. E, deubiquitination of disordered Lys48-linked chains by Ubp12. Shown is the time course of I44A ubiquitin removal as described in A, using 20 nm Ubp12. F, quantitation of data in E as shown in B. G, deubiquitination of Lys63-linked chains. Shown is the time course of removal of monoubiquitin or Lys63-linked ubiquitin chains as shown in C, using 0.4 nm Ubp12. H, quantitation of data in G as shown in B. See supplemental Table S1 for initial rates. All results are representative of two to three experiments.
These results suggest that natural chain linkages with an open conformation should offer little resistance to Ubp15. To test this, we attached chains of Lys63-linked ubiquitin to CycB-Lys60. Ubp15 removed all chain lengths at similar rates (Fig. 7, C and D), suggesting that the structure of long ubiquitin chains, dictated by their linkage, determines resistance to Ubp15.
Levels of long and short I44A chains also declined at similar rates with Ubp12, although the amounts of di- and monoubiquitinated species rose before they began to fall (Fig. 7, E and F). The cleavage of Lys63-linked chains by Ubp12 also showed a rise in monoubiquitinated CycB as longer chain levels fell (Fig. 7, G and H). The accumulation of short products in these reactions is consistent with the tip-directed activity seen above for Ubp12. This contrasts with Ubp15 and suggests that for Ubp12, the tips of unstructured chains may be more rapidly hydrolyzed than monoubiquitinated substrate.
DISCUSSION
Understanding how DUBs target physiological substrates has been difficult because of their complex ubiquitination patterns. Using unanchored chains to characterize DUB activity has been fruitful, but unanchored chains are not the relevant substrates for most DUBs (7). By using simplified physiological substrates bearing single ubiquitin chains, we unexpectedly found that many DUBs, including Ubp12, Ubp15, and human USP7, preferentially remove monoubiquitin from protein substrates. This activity suggests an efficient and general means for DUBs to functionally oppose E3 ligases by preventing chain elongation.
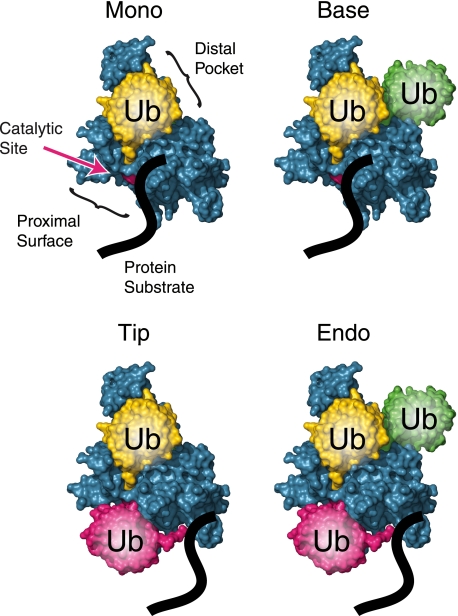
To help explain our results with longer chains, it is useful to consider that ubiquitinated substrates can interact with a DUB in four major ways (Fig. 8). Ubp15 and Ubp12 both displayed high monoubiquitin-directed activity, indicating that in both enzymes, the proximal binding surface accommodates the unstructured polypeptide chain of the securin or cyclin substrate. Ubp15 has base- and tip-directed activity, suggesting that its proximal surface can also bind ubiquitin and that its distal pocket allows binding of proximal as well as terminal ubiquitins. However, substrates are excluded from productive interaction with Ubp15 if they bear long Lys48-linked chains. This may indicate difficulty in capturing the proximal or terminal ubiquitin of chains that are folded into higher-order structures by interubiquitin contacts. Many USPs in this study showed a similar restriction, suggesting that reduced activity toward longer Lys48-linked chains on substrates could be a common feature of these enzymes.
FIGURE 8.
Models of substrate-attached chain deubiquitination. Deubiquitinating enzymes can use four different mechanisms to remove ubiquitin from substrates. The simplest reaction is monoubiquitin removal, in which the protein substrate (e.g. securin or cyclin; shown as a black line) interacts with the proximal binding surface (S1′ site) of the DUB, whereas the distal pocket (S1 site) binds ubiquitin. Base-directed activity is similar, but with additional ubiquitin(s) attached to the one in the distal pocket. For tip-directed activity, the distal pocket of the DUB binds the terminal ubiquitin of a chain, leaving the adjacent ubiquitin to interact with the proximal binding surface. Finally, for endo-deubiquitination, both sites are occupied with ubiquitin, with additional ubiquitin (shown in green) attached to the distal one, potentially interacting with an S2 site on the DUB (30). Shown are illustrations of Protein Data Bank code 1NBF with MacPyMOL (8, 29).
We found that Ubp12 can cut from the tips of long chains in addition to removing monoubiquitin but shows negligible base-directed activity. We infer that its proximal site is permissive for ubiquitin or substrate, but its distal pocket may not accommodate extensions, at least from Lys48, beyond the bound ubiquitin. This latter restriction would be sufficient to explain the lack of appreciable base-directed activity but not the ability to access the tips of long Lys48-linked chains. We speculate that the ability of Ubp12 to access the tips of long Lys48-linked chains might reflect a gain of function relative to other DUBs, as it has several structural features that are not found together in other DUBs in this study (9).
Many E3 ligases build chains on substrates in two steps, using distinct E2 enzymes (28). In the case of the yeast APC/C, Ubc4 is specialized for attaching monoubiquitin to substrates, priming them for Lys48-linked chain extension by Ubc1 (13). The APC/C binds with extremely high affinity to specific targets, allowing processive assembly of long polyubiquitin chains in a single substrate-binding event (19). In contrast, nonspecific, low affinity interactions between E3 ligases (or E2 enzymes) and other proteins are likely to generate monoubiquitin or short polyubiquitin chains on those proteins. In the cell, the ability of Ubp12, Ubp15, and numerous other DUBs to rapidly remove monoubiquitin might provide an effective mechanism for reversing the effects of these nonspecific ligation reactions, thereby preventing inappropriate protein destruction. Similarly, many DUBs are found in complexes with E3 enzymes, and monoubiquitin-directed DUB activity could create a high threshold for those E3 enzymes to build polyubiquitin chains and may also oppose E3 autoubiquitination (16, 17).
Our finding that long Lys48-linked chains on substrates are resistant to Ubp15, human USP7, and other yeast DUBs could suggest that the same chains that target substrates to the proteasome also resist removal by many DUBs. We propose that such functional opposition helps create an all-or-none mechanism in which substrates are either free from ubiquitin or degraded due to attachment of DUB-resistant long Lys48 chains.
Supplementary Material
Acknowledgments
We thank M. Lopez, M. Rodrigo-Brenni, M. Matyskiela, D. Toczyski, J. Tenthorey, S. Foster, and G. Yaakov for assistance, discussions, and comments on the manuscript and V. Van Voorhis, M. Rodrigo-Brenni, and M. Lopez for reagents and sharing of unpublished data.
This work was supported by funding from the National Institute of General Medical Sciences (R01-GM053270) and the National Cancer Institute (T32-CA09043).

The on-line version of this article (available at http://www.jbc.org) contains supplemental “Experimental Procedures,” Table S1, and additional references.
J. B. Schaefer, unpublished results.
V. Van Voorhis, unpublished results.
M. Rodrigo-Brenni, unpublished results.
- DUB
- deubiquitinating enzyme
- APC/C
- anaphase promoting complex/cyclosome
- CycB
- cyclin B
- PKA
- cAMP-dependent protein kinase A
- Ub-AMC
- ubiquitin-7-amido-4-methylcoumarin
- Ub-VME
- ubiquitin vinyl methyl ester
- USP
- ubiquitin-specific protease
- Ni-NTA
- nickel-nitrilotriacetic acid.
REFERENCES
- 1. Deshaies R. J., Joazeiro C. A. (2009) Annu. Rev. Biochem. 78, 399–434 [DOI] [PubMed] [Google Scholar]
- 2. Pickart C. M., Cohen R. E. (2004) Nat. Rev. Mol. Cell Biol. 5, 177–187 [DOI] [PubMed] [Google Scholar]
- 3. Xu P., Duong D. M., Seyfried N. T., Cheng D., Xie Y., Robert J., Rush J., Hochstrasser M., Finley D., Peng J. (2009) Cell 137, 133–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Komander D., Reyes-Turcu F., Licchesi J. D., Odenwaelder P., Wilkinson K. D., Barford D. (2009) EMBO Rep. 10, 466–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bremm A., Freund S. M., Komander D. (2010) Nat. Struct. Mol. Biol. 17, 939–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Eddins M. J., Varadan R., Fushman D., Pickart C. M., Wolberger C. (2007) J. Mol. Biol. 367, 204–211 [DOI] [PubMed] [Google Scholar]
- 7. Komander D., Clague M. J., Urbé S. (2009) Nat. Rev. Mol. Cell Biol. 10, 550–563 [DOI] [PubMed] [Google Scholar]
- 8. Hu M., Li P., Li M., Li W., Yao T., Wu J. W., Gu W., Cohen R. E., Shi Y. (2002) Cell 111, 1041–1054 [DOI] [PubMed] [Google Scholar]
- 9. Ye Y., Scheel H., Hofmann K., Komander D. (2009) Mol. Biosyst. 5, 1797–1808 [DOI] [PubMed] [Google Scholar]
- 10. Rodrigo-Brenni M. C., Foster S. A., Morgan D. O. (2010) Mol. Cell 39, 548–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Carroll C. W., Morgan D. O. (2005) Methods Enzymol. 398, 219–230 [DOI] [PubMed] [Google Scholar]
- 12. Ciechanover A., Elias S., Heller H., Hershko A. (1982) J. Biol. Chem. 257, 2537–2542 [PubMed] [Google Scholar]
- 13. Rodrigo-Brenni M. C., Morgan D. O. (2007) Cell 130, 127–139 [DOI] [PubMed] [Google Scholar]
- 14. Komander D., Lord C. J., Scheel H., Swift S., Hofmann K., Ashworth A., Barford D. (2008) Mol. Cell 29, 451–464 [DOI] [PubMed] [Google Scholar]
- 15. Leggett D. S., Hanna J., Borodovsky A., Crosas B., Schmidt M., Baker R. T., Walz T., Ploegh H., Finley D. (2002) Mol. Cell 10, 495–507 [DOI] [PubMed] [Google Scholar]
- 16. Sowa M. E., Bennett E. J., Gygi S. P., Harper J. W. (2009) Cell 138, 389–403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Ventii K. H., Wilkinson K. D. (2008) Biochem. J. 414, 161–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Ghaemmaghami S., Huh W. K., Bower K., Howson R. W., Belle A., Dephoure N., O'Shea E. K., Weissman J. S. (2003) Nature 425, 737–741 [DOI] [PubMed] [Google Scholar]
- 19. Matyskiela M. E., Morgan D. O. (2009) Mol. Cell 34, 68–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hornig N. C., Knowles P. P., McDonald N. Q., Uhlmann F. (2002) Curr. Biol. 12, 973–982 [DOI] [PubMed] [Google Scholar]
- 21. Amerik A. Y., Li S. J., Hochstrasser M. (2000) Biol. Chem. 381, 981–992 [DOI] [PubMed] [Google Scholar]
- 22. Ho Y., Gruhler A., Heilbut A., Bader G. D., Moore L., Adams S. L., Millar A., Taylor P., Bennett K., Boutilier K., Yang L., Wolting C., Donaldson I., Schandorff S., Shewnarane J., Vo M., Taggart J., Goudreault M., Muskat B., Alfarano C., Dewar D., Lin Z., Michalickova K., Willems A. R., Sassi H., Nielsen P. A., Rasmussen K. J., Andersen J. R., Johansen L. E., Hansen L. H., Jespersen H., Podtelejnikov A., Nielsen E., Crawford J., Poulsen V., Sørensen B. D., Matthiesen J., Hendrickson R. C., Gleeson F., Pawson T., Moran M. F., Durocher D., Mann M., Hogue C. W., Figeys D., Tyers M. (2002) Nature 415, 180–183 [DOI] [PubMed] [Google Scholar]
- 23. Bozza W. P., Zhuang Z. (2011) Biochemistry 50, 6423–6432 [DOI] [PubMed] [Google Scholar]
- 24. Ma J., Martin J. D., Xue Y., Lor L. A., Kennedy-Wilson K. M., Sinnamon R. H., Ho T. F., Zhang G., Schwartz B., Tummino P. J., Lai Z. (2010) Arch. Biochem. Biophys. 503, 207–212 [DOI] [PubMed] [Google Scholar]
- 25. Love K. R., Catic A., Schlieker C., Ploegh H. L. (2007) Nat. Chem. Biol. 3, 697–705 [DOI] [PubMed] [Google Scholar]
- 26. Thrower J. S., Hoffman L., Rechsteiner M., Pickart C. M. (2000) EMBO J. 19, 94–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Beal R., Deveraux Q., Xia G., Rechsteiner M., Pickart C. (1996) Proc. Natl. Acad. Sci. U.S.A. 93, 861–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ye Y., Rape M. (2009) Nat. Rev. Mol. Cell Biol. 10, 755–764 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. DeLano W. L. (2010) The PyMOL Molecular Graphics System, version 1.3r1, Schrödinger, LLC, New York [Google Scholar]
- 30. Ye Y., Akutsu M., Reyes-Turcu F., Enchev R. I., Wilkinson K. D., Komander D. (2011) EMBO Rep. 12, 350–357 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.