Background: HCN2 and HCN4 respond to cAMP, whereas HCN1 does not.
Results: The C-linker plus CNBD of HCN2 and HCN4 show cAMP-induced tetramerization, whereas that of HCN1 contains prebound cAMP and is tetrameric.
Conclusion: HCN1 does not respond to the addition of cAMP because its CNBD contains cAMP already.
Significance: Tetramerization of the C terminus controls ligand gating in HCN channels.
Keywords: Crystallography; Cyclic Nucleotides; Gating; Ion Channels; Ligand-binding Protein; Potassium Channels; HCN Channels; Pacemaker; HCN1, cAMP Gating, CNBD Oligomerization, Crystal Structure
Abstract
Hyperpolarization-activated cyclic nucleotide-gated (HCN) channels are dually activated by hyperpolarization and binding of cAMP to their cyclic nucleotide binding domain (CNBD). HCN isoforms respond differently to cAMP; binding of cAMP shifts activation of HCN2 and HCN4 by 17 mV but shifts that of HCN1 by only 2–4 mV. To explain the peculiarity of HCN1, we solved the crystal structures and performed a biochemical-biophysical characterization of the C-terminal domain (C-linker plus CNBD) of the three isoforms. Our main finding is that tetramerization of the C-terminal domain of HCN1 occurs at basal cAMP concentrations, whereas those of HCN2 and HCN4 require cAMP saturating levels. Therefore, HCN1 responds less markedly than HCN2 and HCN4 to cAMP increase because its CNBD is already partly tetrameric. This is confirmed by voltage clamp experiments showing that the right-shifted position of V½ in HCN1 is correlated with its propensity to tetramerize in vitro. These data underscore that ligand-induced CNBD tetramerization removes tonic inhibition from the pore of HCN channels.
Introduction
Hyperpolarization-activated cyclic nucleotide-gated (HCN)2 channels underlie the If/Ih cation currents that control pacemaker activity in the heart and brain (1–3). HCN channels belong to the superfamily of the six-transmembrane domain segment voltage-gated K+ channels and are dually activated by membrane hyperpolarization and binding of cAMP to their cyclic nucleotide binding domain (CNBD) (4–6). Increase in the cytosolic cAMP concentration shifts the channel voltage dependence to more positive potentials, thereby increasing the open probability and current at a given voltage. In this respect, the three most studied isoforms, HCN1, HCN2, and HCN4, behave differently. HCN2 and HCN4 respond to saturating cAMP levels by shifting the activation curve of +17 mV, whereas HCN1 only shifts it by +4 mV (7). Interestingly, the behavior of HCN1 seems not to be due to a lower affinity of this isoform for cAMP because dose-response curves obtained in patch clamp recordings in inside-out configuration indicate similar nanomolar affinity for HCN1 and HCN2 activation (7, 8).
The current understanding of ligand-induced gating in HCN channels is that cAMP binding releases the tonic inhibition exerted by the cytoplasmic CNBD on the channel pore. This mechanism probably involves cAMP-induced tetramerization of the CNBD and is supported by two lines of evidence. On one side, removal of the cytosolic domain, by enzymatic digestion in the native If channels (9) or by gene manipulation in HCN clones (10), mimics the action of cAMP and shifts the activation curve of the channels to more depolarized potentials. On the other side, the addition of saturating cAMP concentrations promotes the tetramerization of the isolated cytosolic fragment (consisting of the CNBD and the C-linker) of HCN2 and HCN4 in solution (11, 12). Furthermore, it has been shown in HCN2 that promotion of tetramerization induced by a tripeptide mutation in the C-linker facilitated channel opening in the absence of cAMP (13).
With this background, we have focused our attention on HCN1 in order to explain its peculiar properties that include, besides a weak response to cAMP, also a right-shifted activation curve, when compared with the other isoforms (7, 8, 10), indicating that this isoform might partially lack the inhibition of the CNBD. To this end, we have solved the crystal structures of the cytosolic fragment (C-linker plus CNBD) of HCN1, HCN2, and HCN4 and compared them. Furthermore, we have systematically analyzed the ligand binding and ligand-induced tetramerization properties of the three proteins in solution. Our results show that the C-terminal fragment of HCN1 differs from those of HCN2 and HCN4 because it forms tetramers already at basal cAMP concentration, whereas tetramerization of the C-terminal domain of HCN2 and HCN4 requires saturating cAMP levels. This finding explains the modest response of HCN1 to the addition of cAMP in vivo and, in general, offers an experimental proof to the leading hypothesis that cAMP-induced tetramerization of the C-terminal domain of HCN channels removes the inhibition exerted by this domain on the pore gating.
EXPERIMENTAL PROCEDURES
Protein Preparation
The cDNA fragments comprised the C-linker and CNBD (CB) and encoded the following residues: 470–672 (human HCN2CB), 521–723 (human HCN4CB), 390–592 (mouse HCN1CB), and 441–592 (ΔC-linker HCN1CNBD).
The fragments were cloned into a modified pET-24b downstream of a double His6-maltose-binding protein (MBP) tag and transformed into Escherichia coli Rosetta strain. Cells were grown at 37 °C in Luria broth to 0.6 A600 and induced with 0.4 mm isopropyl 1-thio-β-d-galactopyranoside overnight at 20 °C. The cells were collected by centrifugation and resuspended in ice-cold lysis buffer (30 mm Hepes, pH 7.4, 500 mm NaCl, 10% glycerol, 1 mm β-mercaptoethanol) with the addition of 10 μg/ml DNase, 0.25 mg/ml lysozyme, 100 μm phenylmethylsulfonyl fluoride, 5 μm leupeptin, and 1 μm pepstatin. The cells were sonicated on ice 12 times for 20 s each, and the lysate was cleared by centrifugation. The proteins were purified by affinity chromatography on Ni2+-NTA and eluted in lysis buffer plus 300 mm imidazole. The His6-MBP was removed by HRV3C cleavage overnight at 4 °C. The cleavage reaction was loaded onto an amylose resin (New England Biolabs), and the flow-through was collected and loaded onto a HiLoad 16/60 Superdex 200 prep grade size exclusion column (GE Healthcare) that was equilibrated with lysis buffer. Analysis presented in Fig. 3 and Fig. 5 was performed with Superdex 200 5/150 GL (GE Healthcare). Protein concentration was 1.5 mg/ml. Buffer contained 100 mm NaCl, 20 mm Hepes, pH 7.0, 10% glycerol with or without cAMP.
FIGURE 3.
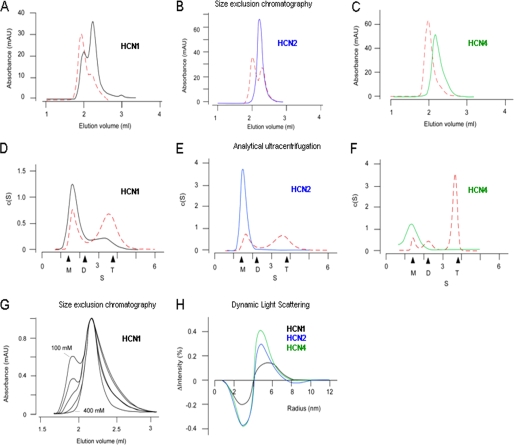
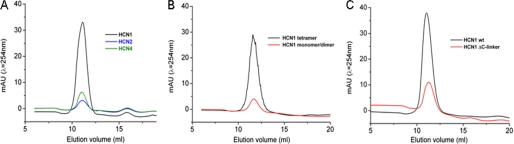
HCN1CB tetramerizes in the absence of added cAMP. A–C, size exclusion chromatography profiles of HCN1CB (black), HCN2CB (blue), and HCN4CB (green) proteins in the absence (solid line) and in the presence (dashed red line) of 1 mm cAMP; D–F, distribution of the sedimentation coefficient (c(s)), as calculated from sedimentation velocity experiments for HCN1CB, HCN2CB, and HCN4CB in the absence (solid line) and in the presence (dashed red line) of 0.3 mm cAMP; black arrowheads indicate from left to right the theoretical values for monomer, dimer, and tetramer (supplemental Table III); G, size exclusion chromatography profiles of HCN1CB obtained in the absence of cAMP at increasing NaCl concentration: 100, 150, 200, 300, and 400 mm. The absorbance values are normalized to the peak at 2.2 ml of elution volume. H, differential curves showing the cAMP-induced change in protein radius estimated by dynamic light scattering. Curves were obtained by fitting individual dynamic light scattering spectra in the presence and absence of 2.5 mm cAMP with single Gaussians. After normalizing the fitted curves to the same ordinate, the difference spectra were obtained by subtracting the curves in the presence from those in the absence of cAMP. Each pair includes ≥4 data sets.
FIGURE 5.
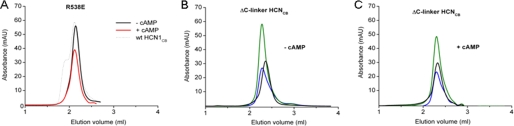
Size exclusion chromatography profile of HCNCB mutants. A, elution profile of HCN1CB R538E mutant, in the presence (red lines) and in the absence (black lines) of 1 mm cAMP. The profile of HCN1CB WT is shown for comparison as a dotted line. Shown are elution profiles of ΔC-linker HCNCB proteins (HCN1 (black), HCN2 (blue), and HCN4 (green)) in the absence (B) and in the presence (C) of 1 mm cAMP.
Fluorescence Polarization
The direct fluorescence polarization (FP) assay was performed following in principle the procedure from Moll et al. (14). 8-Fluo-cAMP and 8-Fluo-cGMP were purchased from the Biolog Life Science Institute (Bremen, Germany). Fine chemicals (research grade) were purchased from Sigma. The measurements were performed in 150 mm NaCl, 20 mm MOPS, 0.005% (v/v) CHAPS, pH 7.0, using the FusionTM α-FP microtiter plate reader at room temperature in a 384-well microtiter plate (PerkinElmer, Optiplate, black). The protein concentration was varied (from 50 μm to 12 pm), and the concentration of 8-Fluo-cAMP/8-Fluo-cGMP was fixed at 1 nm. The FP signal was detected for 2 s at excitation of 485 nm and emission FP filter of 535 nm with a photomultiplier tube voltage of 1,100. Data were analyzed with GraphPad Prism 5.03 (GraphPad Software, San Diego, CA) by plotting the FP signal in millipolarization units (mPol) against the logarithm of the HCN concentration.
Isothermal Titration Calorimetry
Measurements were carried out at 25 °C using an iTC200 microcalorimeter (MicroCal, GE Healthcare). The volume of sample cell was 0.2 ml; the reference cell contained water. The proteins were extensively dialyzed against PBS plus 10% glycerol, and the same buffer was used to dissolve cAMP. The proteins (11–75 μm) were titrated with cAMP (250–900 μm) using injection volumes of 1–2 μl. Calorimetric data were analyzed with Origin software (version 7, MicroCal), using equations described for the single-site binding model (15).
Surface Plasmon Resonance Measurements
Protein/nucleotide interactions were monitored as solution competition experiments and were performed using a Biacore 3000 instrument (Biacore GE Healthcare) at 20 °C.
A carboxymethylated sensor chip surface (CM5, research grade, Biacore GE Healthcare) was activated for covalent coupling of 8-AHA-cAMP as described previously (16). Running buffer was PBS, pH 7.4, plus 0.005% (v/v) surfactant P20. Proteins (100 nm) were preincubated with varying concentrations of free cAMP and injected over the 8-AHA-cAMP surface. Surface plasmon resonance (SPR) signals indicate binding of the respective proteins to the cAMP analog 8-AHA-cAMP covalently linked to the sensor chip. The association was monitored for 3 min, and the binding signal was collected at the end of the association phase. The resulting binding signals were plotted against the logarithm of the free cAMP concentration, and the EC50 values were calculated form the dose-response curve.
Crystallization
Crystallization trials were set up in 96-well sitting drop plates (Greiner) using the Orxy 8.0 crystallization robot (Douglas Instruments) and stored at 4 °C. Crystals of the mHCN1-cAMP complex (protein concentration 10 mg/ml, 5 mm cAMP) were grown by vapor diffusion, using as a precipitant solution 20–22% PEG 3350, 400 mm sodium acetate buffer, pH 5.0. Crystals usually grew in 2 weeks and were cryoprotected with the same crystallization well solution supplemented with 30% glycerol prior to cryocooling in liquid nitrogen. A full data set was collected to 2.9 Å resolution using synchrotron radiation (ID29 beamline, ESRF, Grenoble, France). Crystals of the human HCN2-cAMP complex (protein concentration 10–13 mg/ml, 5 mm cAMP) were grown by vapor diffusion, using as a precipitant solution 20% PEG 8000, 500 mm NaCl, 100 mm citrate buffer, pH 4.6, 10% glycerol. A full data set was collected to 2.3 Å resolution (ID23-2 beamline, ESRF, Grenoble, France). Crystals of the human HCN4-cAMP complex (protein concentration 10 mg/ml, 5 mm cAMP) were grown by vapor diffusion using as a precipitant solution 25% PEG 3350, 400 mm sodium acetate buffer, pH 5.0 (with/without 500 mm dibasic ammonium phosphate). A full data set was collected to 2.5 Å resolution (ID29 beamline, ESRF, Grenoble, France). Raw data were processed with Mosflm (17) and Scala (18), and the structures were solved by molecular replacement using the program MolRep (19). The crystal structure of mouse HCN2 in complex with cAMP (11) (Protein Data Bank entry 1Q43) was used as search model. To avoid model bias, the bound cAMP was removed from the search model, and the side chains were truncated to Ala in cases of mismatch between the amino acid sequences. Several cycles of manual rebuilding, using the program COOT (20), and refinement, using the program REFMAC (21) (rigid body and restrained refinement), were carried out to improve the electron density map, and the side chains omitted in the search model were rebuilt into the electron density.
The final R-factor/R-free for HCN1-cAMP, HCN2-cAMP, and HCN4-cAMP are 20.5%/27.5%, 20.2%/27.1%, and 19.3%/27.7%, respectively (supplemental Table I). Residue 586 of HCN4-cAMP has been modeled and refined as S-hydroxycysteine (probably induced by x-ray oxidation). The program Procheck (22) was used to assess protein stereochemical quality. The program PISA (23) was used to identify and analyze the quaternary assemblies. Atomic coordinates and structure factors have been deposited with the Protein Data Bank, with entry codes 3U0Z, 3U10, and 3U11 for HCN1-cAMP, HCN2-cAMP, and HCN4-cAMP, respectively.
Analytical Ultracentrifugation
Sedimentation Velocity experiments were performed at 20 °C using a Optima XL-I Ultracentrifuge (Beckman Coulter) with an An50 Ti rotor at a rotor speed of 50,000 rpm. Data were acquired by monitoring absorbance at 280 nm through sapphire cell windows, in Epon sectors. Proteins (2.5 mg/ml) were run at 100 μm, and the related gel filtration buffer was used as a blank (20 mm Hepes, pH 7.0, 150 mm NaCl, 10% glycerol). cAMP at a concentration of 0.3 mm was added directly to the protein and reference solutions. Data were analyzed with the software SEDFIT (24) to determine the sedimentation coefficients of the proteins in solution. Solvent density (1.028 g/ml), partial specific volume (0.7354 ml/g), and viscosity (0.013284 g/s/cm) were calculated using the SEDNTERP program (available on the World Wide Web). The oligomeric states of the proteins were determined by comparing the sedimentation coefficients from SEDFIT with those calculated using the crystallographic coordinates and the program HYDROPRO (25).
Dynamic Light Scattering
Experiments were conducted using a Protein Solutions DynaPro 99 instrument with a DynaProMSTC200 microsampler (Protein Solutions, Charlottesville, VA). Protein concentration was 1 mg/ml. Buffer contained 20 mm Hepes, pH 7.0, 150 mm NaCl, 10% glycerol with or without 2.5 mm cAMP. Acquisition was performed at 10 °C with Dynamics 5, 30–50 scans, 30 s/scan. Data analysis was performed with Dynamics 6 software.
Determination of cAMP Content
cAMP was released by boiling the protein sample (∼1 mg) in 100 μl of lysis buffer for 2 min. The boiled sample was centrifuged at maximum speed for 10 min at room temperature, and the supernatant was collected and added with 900 μl of 5 mm ammonium bicarbonate. The sample was loaded onto an anion exchange chromatography column (HiTrapQ (1 ml), GE Healthcare), previously equilibrated with 5 mm ammonium bicarbonate. Unbound molecules were washed out with 7 ml of 5 mm ammonium bicarbonate. The cAMP was eluted with a linear gradient of ammonium bicarbonate (5–1000 mm) in 20 column volumes.
Electrophysiology
Site-directed mutagenesis on pGHE::mHCN1 was performed using the QuikChange mutagenesis kit (Stratagene). All of the constructs were linearized and transcribed into cRNA using T7 polymerase (T7 Riboprobe® system, Promega). Oocyte preparation and cRNA transcription were performed according to standard procedures as reported previously (26). Oocytes were injected with 50 nl of cRNA solution each, at a concentration of 0.5 μg/μl. Two-electrode voltage clamp recordings were performed 2 days after cRNA injection using a GeneClamp 500 amplifier (Axon Instruments) and digitized at 50 kHz with a Digidata 1200 (Axon Instruments). Data acquisition and analysis were done using the pCLAMP8 software package (Axon Instruments). Microelectrodes filled with 3 m KCl had resistances of 0.5–2.0 megaohms. Oocytes were bathed in extracellular solution containing 98 mm KCl, 2 mm NaCl, 5 mm HEPES, pH 7.5, 1.8 mm CaCl2, and 1 mm MgCl2. Three-second-long voltage steps were applied in 10-mV increments from a holding potential of −30 mV. Peak tail current amplitudes were measured at −40 mV, and tail current I-V curves were fitted using the Boltzmann equation, I(V) = A1 + A2/(1 + exp((V − V½)/s)).
RESULTS
cAMP Binding Affinity Measurements
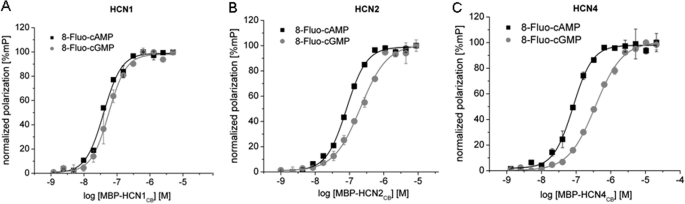
We have prepared three constructs, hereafter termed HCN1CB, HCN2CB, and HCN4CB, comprising the C-linker and the CNBD domains of mouse HCN1, human HCN2, and human HCN4, respectively. The three constructs differ by 26 substitutions in total, 14 of which occur in HCN1CB (supplemental Fig. 1). The purified proteins were tested for cyclic nucleotide binding with three different techniques: FP, SPR, and isothermal titration calorimetry (ITC). The three proteins showed similar binding affinities when tested with the same technique (Table 1), but different techniques gave quite different absolute values (12). In FP measurements, all three proteins bind the 8-Fluo-cAMP analog in the nanomolar range (Table 1). Notably, the highest affinities were measured with the maltose-binding protein fused at the N terminus of the HCN proteins (MBP-HCNCB). Fig. 1 shows exemplary curves in which KD values of 39.7, 82.0, and 39.2 nm were calculated for MBP-HCN1, MBP-HCN2, and MBP-HCN4 constructs, respectively. These results are remarkably similar to the K½ values obtained in patch clamp experiments: 60 and 100 nm for HCN1 and HCN2, respectively (8).
TABLE 1.
Comparison of binding constants for cyclic nucleotide binding to the C termini of HCN proteins (HCNCB) derived from FP, SPR, and ITC measurements
|
KD (from FP) ± S.E.a |
EC50 (from SPR) ± S.D. for cAMP | KD (from ITC) ± S.D. for cAMP | ||
|---|---|---|---|---|
| 8-Fluo-cAMP | 8-Fluo-cGMP | |||
| nm | μm | μm | ||
| MBP-HCN1CB | 116 ± 26 (n = 4) | 208 ± 51 (n = 5) | NDb | Not measurable |
| MBP-HCN2CB | 115 ± 18 (n = 4) | 286 ± 34c (n = 5) | ND | 3.06 ± 0.7 (n = 9) |
| MBP-HCN4CB | 111 ± 36 (n = 4) | 414 ± 56d (n = 3) | ND | 0.97 ± 0.7 (n = 4) |
| HCN1CB | 382 ± 87 (n = 4) | 386 ± 76 (n = 3) | 5 ± 1e (n = 5) | Not measurable |
| HCN2CB | 606 ± 219 (n = 3) | 752 ± 236 (n = 3) | 10 ± 4 (n = 3) | 3.6 ± 1.3 (n = 3) |
| HCN4CB | 167 ± 36 (n = 4) | 694 ± 121d (n = 4) | 11 ± 2 (n = 3) | ND |
| HCN1CB ΔC-linker | ND | ND | 5 ± 2e (n = 3) | ND |
| HCN1CB G510S/S515G/S516N | ND | ND | 7.3 ± 1.6e (n = 3) | ND |
a FP experiments were in duplicate or triplicate.
b ND, not determined.
c Mean p < 0.05; values are significantly different from that of 8-Fluo-cAMP (analyzed by paired t test, p value two-tailed, confidence interval 99%, GraphPad Prism version 5.03).
d Mean p < 0.01; values are significantly different from that of 8-Fluo-cAMP (analyzed by paired t test, p value two-tailed, confidence interval 99%, GraphPad Prism version 5.03).
e Protein concentration was 500 nm instead of 100 nm.
FIGURE 1.
Determination of the dissociation constant KD for 8-Fluo-cAMP (black square) and 8-Fluo-cGMP (gray circle) to MBP-tagged HCN1CB (A), HCN2CB (B), and HCN4CB (C) using fluorescence polarization. KD values for 8-Fluo-cAMP and 8-Fluo-cGMP are as follows: for MBP-HCN1CB, KD = 40 and 60 nm; for MBP-HCN2CB, KD = 82 and 209 nm; for MBP-HCN4CB, KD = 40 nm and 135 nm.
The measured affinity of the HCN proteins for 8-Fluo-cGMP was significantly lower than that for 8-Fluo-cAMP, at least for HCN2 and HCN4, but still in the nanomolar range. Notably, patch experiments had previously reported micromolar K½ for HCN2 (11, 27). This observation suggests that the higher K½ values for cGMP might be related to a lower efficacy of this ligand in promoting gating.
When we measured binding with other techniques, we obtained 10 times lower affinities. Binding of cAMP by ITC showed micromolar affinity with and without the MBP constructs: for MBP-HCN2CB, KD = 3.06 ± 0.7 μm; for MBP-HCN4CB, KD = 0.97 ± 0.7 μm; and for HCN2CB, KD = 3.6 ± 1.3 μm (Table 1 and supplemental Fig. 2). For unclear reasons, we were not able to record binding with the HCN1CB constructs. Micromolar binding was also measured by SPR with immobilized 8-AHA-cAMP in a solution competition experiment. In this case, the signal obtained with HCN1CB was low but measurable. The calculated EC50 values where roughly the same for the three proteins: 5 ± 1, 10 ± 4, and 11 ± 2 μm for HCN1CB, HCN2CB, and HCN4CB, respectively.
Altogether, the binding data show no dramatic difference in the binding affinities of the three proteins. Notably, different techniques yielded different results; the most plausible explanation is that different cAMP and cGMP analogs indeed have different affinities for the proteins, as reported for HCN2 (28). But these results are not necessarily due to technical reasons. They may highlight a high and a low affinity state of the same protein. The low to absent signal of HCN1 observed in two cases (SPR and ITC measurements) highlights a difference from HCN2 and HCN4, probably in the fraction of protein available for binding. We did not observe a systematic difference in binding due to the presence of the MBP (e.g. see the ITC results for HCN2 in Table 1).
CNBD Structural Comparison
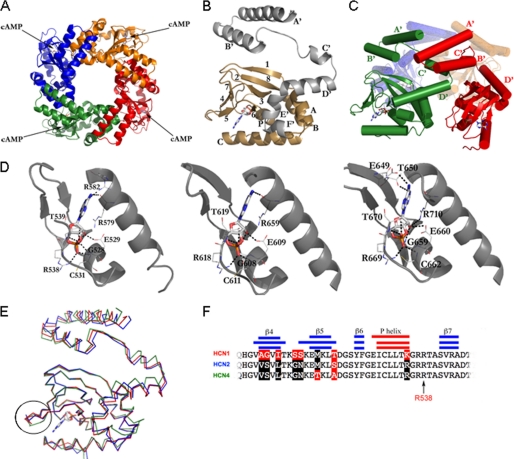
We next determined the crystal structures of HCN1CB, HCN2CB, and HCN4CB (at 2.9, 2.3, and 2.5 Å resolutions, respectively) (supplemental Table I). Fig. 2A shows a top view of the three-dimensional structure of mouse HCN1CB. It crystallized as a tetramer in the presence of 5 mm cAMP, as shown previously for mouse HCN2 (11) and, more recently, for human HCN4 (12). Fig. 2B shows the tertiary structure of the HCN1CB protomer, with the seven α-helices of the C-linker (A′–F′) in gray and the CNBD, composed of four α-helices (A, P, B, and C) and eight β-sheets (numbered 1–8) in gold. Fig. 2C shows a side view of the tetramer, highlighting the contacts provided by the C-linker, where the A′ and B′ helices contact the C′ and D′ of the neighboring subunit. The cAMP binding mode (with the cAMP molecule adopting the anti-conformation) and the most relevant protein-ligand interactions are conserved in HCN1CB, HCN2CB, and HCN4CB (Fig. 2D and supplemental Table II). The tertiary structure of HCN1CB superimposes well with those of HCN2CB (average root mean square deviation of 0.85 Å over 198 Cα atoms) and of HCN4CB (average root mean square deviation of 1.05 Å over 197 Cα atoms) (Fig. 2E). The only deviations of the Cα backbones emerge for two short regions located at the C-linker and at the loop between strands β4 and β5 (facing the cAMP binding pocket, circled in Fig. 2E). Such discrepancies are not surprising because most of the residue substitutions of HCN1CB cluster in these regions (supplemental Fig. 1). Analysis of the subunit interfaces reveals that all HCN structures share similar buried surface areas between adjacent subunits (average of 1085 Å2) and mostly conserved polar interactions.
FIGURE 2.
Crystal structure of the soluble portion CB (C-linker plus CNBD) of HCN1 bound to cAMP. A, HCN1CB tetramer viewed parallel to the 4-fold axis. Each subunit is shown in a different color. B, the protomer of HCN1CB with cAMP. The C-linker, helices A′–F′, is in gray; the CNBD, β-sheets 1–8 and helices A, B, P, and C, is in gold. C, subunit-subunit interactions mediated in the tetramer by the C-linker; helices A′ and B′ form a helix-turn-helix motif that interacts with the helix-turn-helix motif formed by the C′ and D′ helices of the neighboring subunit. The C-linker of one monomer (red) contacts two other subunits in the tetramer (green and orange). Cylinders and arrows represent helices and strands, respectively. D, from left to right, cAMP binding site of HCN1CB, HCN2CB, and HCN4CB, showing hydrogen bonds and salt bridges that stabilize the cAMP molecule. The binding amino acids are conserved among the three HCN isoforms. In particular, six polar contacts are strictly preserved between the same atoms (see supplemental Table II for the list of interactions). In addition, the residue Cys-531 (HCN1) makes a salt bridge (not present in the other two isoforms) between its nitrogen and the O1P of the cAMP, stabilizing the cyclic nucleotide inside the binding pocket. Another stabilizing element is the polar contact between residue Arg-582 (Arg-659 in HCN2 and Arg-632 in HCN4) and the atom N1 of the cAMP. The same atom contacts residue Arg-579, fundamental for the affinity for the cNMP (27). Also visible in our HCN2 structure are two more polar contacts (not reported in Ref. 11) between the atoms O2* and O2P (cAMP) and the nitrogen atoms of the residues Cys-611 and Gly-608, completing the range of strictly preserved amino acids mentioned above. E, differences in the structures highlighted by the superimposition of HCN1CB (red), HCN2CB (blue), and HCN4CB (green). The β4-β5 loop at the entrance of the cAMP binding site in the CNBD is circled. F, alignment of the amino acid sequences of the three proteins in the circled region. Secondary structures are indicated by the blue/red bars. Residues identical in HCN1 and HCN2 (black) but different in HCN1 (red) are highlighted. The conserved arginine, Arg-538 in HCN1, important for cAMP binding, is indicated by an arrow.
In conclusion, the comparative structural analysis of cAMP-bound HCN proteins (i.e. the end products of cAMP binding and tetramerization processes) did not highlight systematic structural differences that would group HCN1CB separately from HCN2CB and HCN4CB. This prompted us to look for possible differences between HCN1CB and the other isoforms in the apo-form, a state in which HCN1CB diverges substantially from the other two, as for cAMP-induced gating in vivo.
CNBD Oligomerization Propensity
To test this hypothesis, a solution experimental approach was undertaken on the apo-forms of the three constructs. Fig. 3 summarizes three independent lines of evidence highlighting that HCN1CB has different oligomerization properties from the others: HCN1CB is present in a tetrameric form already in the absence of added cAMP, whereas HCN2CB and HCN4CB form tetramers only in its presence. In size exclusion chromatography, HCN2CB and HCN4CB elute as a single peak (Fig. 3, B and C, solid line), whereas HCN1CB elution shows an additional peak at higher molecular weight (Fig. 3A, solid line). The higher molecular weight peak appears in HCN2CB and HCN4CB only upon the addition of cAMP (Fig. 3, A–C, dashed red lines). Sedimentation velocity analytical ultracentrifugation studies assign a proper size to the two forms (Fig. 3, D–F, and supplemental Table III). In the absence of added cAMP, HCN2CB and HCN4CB were mainly identified as monomers, with only a small amount of dimeric component (Fig. 3, E and F, solid line). In contrast, HCN1CB exists as a mixture of monomers, dimers, and tetramers (Fig. 3D, solid line). These oligomeric states are observed for HCN2CB and HCN4CB only after adding saturating amounts of cAMP (Fig. 3, D–F, dashed red line). Interestingly, the equilibrium between monomer/dimer and tetramer observed in the HCN1CB protein depends on the ionic strength of the solution. Fig. 3G shows the effect of increasing salt concentration, from 100 to 400 mm, on the elution profile of HCN1CB. The disappearance of the tetrameric form of the protein implies an electrostatic nature for the stabilizing interactions. This is strengthened by the observation that, in the tetramer of HCN1CB, all 14 substitutions that distinguish this isoform from the other two are exposed to the solvent, and five of them (Ile-432, Asn-437, Asp-444, Ala-496, and Val-497) are at the interface between monomers (not shown).
Finally, dynamic light scattering analysis indicates that, in the absence of added cAMP, HCN2CB and HCN4CB are mainly dimers, whereas HCN1CB is a mixture of dimers and tetramers. The addition of cAMP promotes the full tetramerization of HCN2CB and HCN4CB as well as the residual tetramerization of HCN1CB (supplemental Table IV). The differential signal in the dynamic light scattering peaks, which is due to the addition of cAMP, is illustrated in Fig. 3H, indicating the presence of tetrameric HCN1CB protein in the absence of added cAMP.
cAMP Retention after Purification
On the basis of the above reported evidence, a clear and systematic difference emerges between HCN1CB and the other two proteins; whereas HCN2CB and HCN4CB form tetramers only after the addition of saturating concentrations of cAMP, HCN1CB is found as a tetramer already without the addition of exogenous cAMP. This raises the question of whether HCN1CB has a high propensity to tetramerize in the apo-state or if this behavior is due to some tightly prebound cAMP. We then analyzed the endogenous cAMP content associated with the protein after purification. As a negative control, we prepared the mutant protein R538E HCN1CB. Mutation of this invariant amino acid (Arg-591 in HCN2 and Arg-669 in HCN4) that interacts with the exocyclic oxygen of the cAMP phosphate (29) reduces the affinity of HCN CNBD for the ligand by more than 3 orders of magnitude (7, 27, 30). Fig. 4A shows that HCN1CB contains a significantly higher amount of endogenous cAMP than HCN2CB and HCN4CB. The measured molar ratio (protein/cAMP) in HCN1CB is 6. In contrast, this ratio is 24 in HCN4CB and 50 in HCN2CB. The value found in the negative control HCN1CB R538E is 40 (Table 2). Furthermore, separate analysis of the two forms of HCN1CB, monomer/dimer and tetramer, showed that almost all cAMP is in the tetrameric form, where we found a protein/cAMP ratio of 2 (Fig. 4B and Table 2). From these experiments, we must conclude that the tetrameric, but not the monomeric/dimeric, form of HCN1CB contains endogenous cAMP and binds it with high affinity because the ligand is not released during the purification procedure. Altogether our data confirm that cAMP binding to CNBD promotes tetramerization of the C-terminal fragment (11, 12) and suggest that cAMP binds with high affinity to the HCN1CB tetramer, resulting in ligand trapping. We further tackled this point by checking 1) the tetramer formation of an HCN1 mutant with a reduced affinity for cAMP and 2) the cAMP content in a protein with altered tetramerization properties.
FIGURE 4.
Quantification of the cAMP molecules released by the HCN proteins. Cyclic AMP content was estimated by the absorbance at λ = 254 nm (calibration curve shown in supplemental Fig. 3). A, absorbance profiles of cAMP released by the three HCNCB proteins after boiling (for experimental details, see “Experimental Procedures”). B, comparison between the cAMP content of the monomer/dimer and of the tetramer form of HCN1CB. C, comparison between the cAMP content of the WT HCN1CB and the HCN1 ΔC-linker protein (see Table 2 for molar ratios).
TABLE 2.
Cyclic AMP content in purified HCNCB proteins expressed as molar ratio (protein/cAMP)
| Molar ratio (protein/cAMP) | n | |
|---|---|---|
| HCN1CB | 6 ± 2 | 5 |
| HCN2CB | 49.5 ± 8.5 | 4 |
| HCN4CB | 24 ± 5 | 3 |
| HCN1CB monomer/dimer | 17 ± 2.7 | 3 |
| HCN1CB tetramer | 2.2 ± 0.36 | 3 |
| HCN1CB ΔC-linker | 25 ± 3 | 3 |
| HCN1CB R538E | 40 | 1 |
Point one was addressed by testing the elution profile in size exclusion chromatography of the R538E mutant that, as previously mentioned, has a decreased affinity for cAMP. As shown in Fig. 5A, R538E protein elutes as a monomer/dimer and does not show the tetramer peak found in the wild type. As expected, the mutant also does not tetramerize upon the addition of 1 mm cAMP.
In regard to point two, in order to prevent tetramerization, we modified the C-linker, because it establishes extensive interactions that mediate the assembly of the four subunits (Fig. 2C). We deleted the first three helices (A′, B′, and C′) and tested the resulting protein fragment, ΔC-linker HCN1CB, for tetramerization and cAMP binding. For control, we have also performed the same deletion in HCN2CB and HCN4CB. We found that ΔC-linker HCN1CB does not show the tetramer peak in size exclusion chromatography and that addition of cAMP does not induce its tetramerization (Fig. 5, B and C, and supplemental Table IV). The same behavior was found for ΔC-linker HCN2CB and ΔC-linker HCN4CB (data not shown). It is worth noting that ΔC-linker HCN1CB is still able to dimerize, as determined by dynamic light scattering (supplemental Table IV). The results of these experiments show that tetrameric assembly requires the presence of an intact C-linker region, in agreement with the proposed role of the C-linker in oligomerization (11, 13). We then tested if this construct could trap cAMP. Interestingly, ΔC-linker HCN1 has a very low content of cAMP (protein/cAMP ratio = 25) (Fig. 4C and Table 2), showing that an intact C-linker is needed to establish the high affinity state, which traps cAMP in the CNBD. It is worth noting that this construct shows micromolar affinity for cAMP by SPR (Table 1), indicating that this affinity value corresponds to that of the low affinity state. The question is why HCN1CB behaves differently from the other two isoforms. To answer this question, we have reconsidered the divergence in the β4-β5 loop found in the holo-structures because it was recently reported that this linker modulates binding in HCN4 (12). Three residues that are conserved in the β4-β5 region of HCN2 and HCN4 but not in HCN1 have been tested by performing the following mutations in HCN1: the single mutation G510S, the double mutation S515G/S516N, and the triple mutation G510S/S515G/S516N. We tested their impact on cAMP binding to HCN1CB and on the electrophysiological properties of the full-length channel expressed in Xenopus oocytes. None of the mutations changed the binding properties of HCN1CB or the tetramerization behavior in gel filtration and the cAMP content of the protein (data not shown). Thus, we have to conclude that the β4-β5 loop does not determine the peculiar biochemical properties of HCN1CB. Similar results were obtained by measuring, in intact oocytes, the activation curves of wild type and mutant channels. Table 3 reports the calculated values for half-activation voltage of WT HCN1 and the mutants. None of the mutations affected this parameter. We further extended our analysis to the β5 strand and tested the mutation M519T, previously reported to affect K½ in HCN2 without affecting the binding (12). Again, we observed no effect either on V½ (Table 3) or on binding (not shown). As controls for the voltage clamp experiments, we used the wild type HCN2 and the low affinity mutant HCN1 R538E. Both channels showed an equally left-shifted V½ (−76 mV) when compared with HCN1, as reported previously (7). From these experiments, we conclude that the right-shifted position of the V½ of HCN1 is correlated with its property of eluting as a tetramer in vitro. This is in line with the finding that the two controls, HCN2 and HCN1 R538E, that do not form tetramers, are left-shifted in vivo.
TABLE 3.
Effect of mutations in the β4-β5 linker and in β-sheet 5 on the half-activation voltage of HCN1 (V1/2), measured by two-electrode voltage clamp in intact oocytes
| Channel | V1/2 | n |
|---|---|---|
| HCN1 WT | −67.5 ± 0.13 | 11 |
| G510S | −66.5 ± 0.25 | 4 |
| S515G/S516N | −65.6 ± 0.23 | 5 |
| G510S/S515G/S516N | −67.9 ± 0.34 | 5 |
| M519T | −66.5 ± 0.65 | 3 |
| R538E | −75.9 ± 0.35 | 4 |
| HCN2 WT | −76.5 ± 0.04 | 7 |
DISCUSSION
In this paper, we have addressed the behavior of the pacemaker channel isoform HCN1, which, unlike HCN2 and HCN4, shows a right-shifted activation curve and responds weakly to saturating cAMP levels in vivo. According to the current view, this behavior is reminiscent of a lack of inhibition by the CNBD. To test this hypothesis, we have solved the crystal structure of the isolated C-terminal domain of HCN1 and compared it with those of HCN2 and HCN4; furthermore, we have tested their biophysical and biochemical properties in vitro and validated them by electrophysiology.
In the cAMP-bound form, all three HCN isoforms show similar tertiary and quaternary structures. The holoproteins are tetrameric, with the C-linkers mediating the “elbow on shoulder” assembly of the four subunits (11). A deviation of the Cα backbones emerges only in the β4-β5 loop, where the substitutions among the three proteins are clustered (Fig. 2).
This loop is in proximity to the ligand binding pocket and, therefore, could potentially control ligand affinity (12); our mutational analysis of this region has shown no relevant changes in the properties of the purified HCN1CB fragment and, notably, also of the channel expressed in oocytes. Therefore, we can conclude that, at least in HCN1, there is no obvious correlation between the β4-β5 loop and the processes of ligand binding and/or gating. Because the fully saturated cAMP-bound form of the HCN tetramer is similar in all isoforms, the structural differences able to justify the different biochemical behaviors of HCNs should reside in the apo-form of the proteins. Unfortunately, any attempt to crystallize HCNs in the unbound state failed. Only the HCN2 structure has been reported also in the cAMP-free state, but its structure is identical to the holo-form due to the presence of two bromide ions located in the cyclic nucleotide binding pocket, one of which occupied the position equivalent to the phosphate group of cAMP in the holo-state structure (31). Size exclusion chromatography, sedimentation velocity analytical ultracentrifugation, and dynamic light scattering were used to analyze the HCN isoforms in their unbound state in solution. Our results indicate that in the absence of added cAMP, HCN1CB has a different oligomerization behavior relative to HCN2CB and HCN4CB. HCN2CB and HCN4CB were mainly identified as monomers with only a small amount of dimeric component (Fig. 3, E and F, solid line). In contrast, HCN1CB exists as a mixture of monomers, dimers, and tetramers (Fig. 3D, solid line). These oligomeric states are observed for HCN2CB and HCN4CB only after adding saturating amounts of cAMP (Fig. 3, D–F, dashed red line). Thus, one of the main findings is that HCN1CB has a higher propensity to tetramerize than HCN2CB and HCN4CB in the same experimental conditions. Remarkably, this property clearly pools HCN1CB separately from HCN2CB and HCN4CB and closely mirrors the grouping established by the in vivo response to cAMP, with HCN2 and HCN4 linked by the strong response to cAMP and separated from HCN1.
The analysis of the different HCN species separated by size exclusion chromatography revealed that HCN1CB contains a significantly higher amount of endogenous cAMP than HCN2CB and HCN4CB (molar protein/cAMP ratios of 6, 24, and 50, respectively) (Table 2) and that almost all endogenous cAMP is bound to the HCN1CB tetramer, with a cAMP/tetramer molar ratio of 2. The correlation between tetramerization and cAMP binding is unraveled by analyzing the behavior of the HCN1CB R538E mutant and the ΔC-linker HCN1CB. The first mutant has an altered cAMP-binding site, resulting in a reduced affinity for cAMP. The HCN1CB R538E mutant elutes as a monomer/dimer (Fig. 5A), and tetramerization does not occur even upon the addition of 1 mm cAMP. The ΔC-linker HCN1CB mutant construct has an intact cAMP-binding site but cannot tetramerize because it is missing the C-linker region. ΔC-linker HCN1CB shows a strong reduction in cAMP content, comparable with those of the other HCN isoforms. We can conclude that cAMP binding promotes tetramerization and that in the tetrameric form, cAMP is bound tightly, because the complex survives extensive dialysis during purification. On the contrary, cAMP-trapping is not a property of the monomeric/dimeric HCN isoforms. Our data suggest that cAMP-induced tetramerization, which is mediated by the C-linker, forces HCN1CB into a high affinity state that in turn stabilizes the tetramer. This finding is in line with the reports of state-dependent affinity of HCN channels for their ligand (30, 32) and agrees with the cyclic allosteric model proposed for HCN channels that postulates the coexistence of the protein in resting and active states with different affinities for the ligand (30, 33, 34). Furthermore, the presence of a stable tetrameric structure agrees with the finding that cAMP traps channels in an open state that kinetically affects the depolarization-dependent deactivation (35).
As for the possibility of measuring in vitro these two affinity states, our data show that when probed with the same technique, the affinities of the three C-terminal fragments appear similar and do not highlight an obvious difference between HCN1 and the others. However, as to the absolute values, different techniques yielded very different results. Whereas the KD and EC50 values obtained by ITC and SPR are in the micromolar range, we obtained much higher affinities, in the nanomolar range, by FP. The possibility that the fluorescein-modified analog of cAMP (8-Fluo-cAMP) used in FP experiments has a higher affinity than cAMP because of its chemical modification is ruled out by the evidence that 8-Fluo-cAMP and cAMP produced similar results in ITC when tested on mHCN2 (12). We can speculate that different techniques might highlight a high and a low affinity state of the same protein. In this context, the observation that HCN1CB yielded very poor to absent signals in SPR and ITC but not in FP may be relevant.
The presence of different cAMP affinities, depending on the oligomerization state, can be a general property of HCNs, but HCN1CB differs from HCN2CB and HCN4CB because it tetramerizes already at basal cAMP concentration and with a cAMP/tetramer ratio of 2. Thus, in full-length HCN channels, the CNBD of HCN1, but not that of HCN2 and HCN4, should exist as tetramers already at basal cAMP concentrations. This property of HCN1 protein fully explains the right-shifted position of the activation curve and the weak response to an increase of cAMP level in vivo. In regard to the cAMP/tetramer ratio of 2 found in HCN1CB, half-saturation of the tetramer appears more likely than half of the tetramers being fully saturated because the second hypothesis would imply that tetramerization is not mandatorily cAMP-driven. Tetramer formation with only half of the binding sites being occupied by cAMP is, on the contrary, consistent with other reports (36) and would explain other experimental findings, such as the small additional shift (2–4 mV) induced by saturating cAMP on the already right-shifted activation curve of HCN1. In this view, full saturation of the tetramer by high cAMP promotes a residual conformational change that, in vivo, results in the additional shift in activation curve. The evidence that a tetramer can form even if it is not fully saturated by cAMP has a general relevance for HCN channels that extends beyond the peculiar properties of HCN1. This is in agreement with the report that in HCN2, increasing the number of available binding sites from 1 to 4 progressively increases the number of mV in the shift (36). More recently, Kusch et al. (32) have shown by patch clamp fluorimetry that only two of the four binding sites in the tetrameric HCN2 channel have to be occupied for maximal activation. Ulens and Siegelbaum (36) have suggested, on the basis of the stoichiometry of cAMP gating, that a 4-fold symmetric gating ring forms from a 2-fold symmetric dimer of dimers.
CONCLUSIONS
We showed that the C-terminal domain of HCN1 is different from those of HCN2 and HCN4 because it partly purifies as a tetramer, half-saturated with cAMP. Given the well known effect of cAMP-induced tetramerization in removing the inhibition exerted by this domain on the pore gating, we conclude that the property of the HCN1 CNBD to accumulate in the tetrameric state can explain the right-shifted position observed in HCN1 before the addition of saturating cAMP, whereas its half-saturation with cAMP can explain the weak response to saturating cAMP levels. Specifically, we propose the following model that recapitulates well known properties of HCN channels. The C-terminal region exerts a tonic inhibition on the pore when the C-linker plus CNBD domains are in a non-tetrameric form (probably dimers (this paper) (13, 36)). When the cAMP concentration increases, the C-linker/CNBD region tetramerizes and releases the inhibition from the pore gate. This event results in the positive shift of 16–20 mV in the activation curve of HCN2 and HCN4. In the case of HCN1, a large fraction of C-linker/CNBD is already tetrameric at basal cAMP concentrations. Therefore, it does not exert inhibition and retains the right shifted position of the activation curve. The addition of cAMP saturates the tetramerization process and affects the HCN1 activation curve only marginally, reflected in the modest shift of 2–4 mV.
Supplementary Material
Acknowledgments
We thank Dan Minor (University of California, San Francisco) and Bina Santoro (Columbia University, New York) for helpful discussion and the generous gift of MBP and mHCN1 clones, Michaela Hansch and Maike Vetter for technical assistance, and Xention (Cambridge, UK) for human HCN2 and HCN4 clones.
This work was supported in part by European Union Grant EDICT (European Drug Initiative on Channels and Transporters) and Progetto di Cooperazione Scientifica e Tecnologica Regione Lombardia (to A. M.) and European Union grant Affinity Proteome and Affinomics (to F. W. H.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables I–IV and Figs. 1–3.
The atomic coordinates and structure factors (codes 3U0Z, 3U10, 3U11) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University,NewBrunswick, NJ (http://www.rcsb.org/).
- HCN
- hyperpolarization-activated cyclic nucleotide-gated
- CNBD
- cyclic nucleotide binding domain
- MBP
- maltose-binding protein
- FP
- fluorescence polarization
- SPR
- surface plasmon resonance
- ITC
- isothermal titration calorimetry
- 8-AHA-cAMP
- 8-(6-aminohexylamino)adenosine-3′,5′-cyclic monophosphate.
REFERENCES
- 1. DiFrancesco D. (1993) Annu. Rev. Physiol. 55, 455–472 [DOI] [PubMed] [Google Scholar]
- 2. Robinson R. B., Siegelbaum S. A. (2003) Annu. Rev. Physiol. 65, 453–480 [DOI] [PubMed] [Google Scholar]
- 3. Santoro B., Baram T. Z. (2003) Trends Neurosci. 26, 550–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Santoro B., Liu D. T., Yao H., Bartsch D., Kandel E. R., Siegelbaum S. A., Tibbs G. R. (1998) Cell 93, 717–729 [DOI] [PubMed] [Google Scholar]
- 5. Gauss R., Seifert R., Kaupp U. B. (1998) Nature 393, 583–587 [DOI] [PubMed] [Google Scholar]
- 6. Ludwig A., Zong X., Jeglitsch M., Hofmann F., Biel M. (1998) Nature 393, 587–591 [DOI] [PubMed] [Google Scholar]
- 7. Chen S., Wang J., Siegelbaum S. A. (2001) J. Gen. Physiol. 117, 491–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang J., Chen S., Siegelbaum S. A. (2001) J. Gen. Physiol. 118, 237–250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Barbuti A., Baruscotti M., Altomare C., Moroni A., DiFrancesco D. (1999) J. Physiol. 520, 737–744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wainger B. J., DeGennaro M., Santoro B., Siegelbaum S. A., Tibbs G. R. (2001) Nature 411, 805–810 [DOI] [PubMed] [Google Scholar]
- 11. Zagotta W. N., Olivier N. B., Black K. D., Young E. C., Olson R., Gouaux E. (2003) Nature 425, 200–205 [DOI] [PubMed] [Google Scholar]
- 12. Xu X., Vysotskaya Z. V., Liu Q., Zhou L. (2010) J. Biol. Chem. 285, 37082–37091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou L., Olivier N. B., Yao H., Young E. C., Siegelbaum S. A. (2004) Neuron 44, 823–834 [DOI] [PubMed] [Google Scholar]
- 14. Moll D., Prinz A., Gesellchen F., Drewianka S., Zimmermann B., Herberg F. W. (2006) J. Neural Transm. 113, 1015–1032 [DOI] [PubMed] [Google Scholar]
- 15. Wiseman T., Williston S., Brandts J. F., Lin L. N. (1989) Anal. Biochem. 179, 131–137 [DOI] [PubMed] [Google Scholar]
- 16. Moll D., Schweinsberg S., Hammann C., Herberg F. W. (2007) Biol. Chem. 388, 163–172 [DOI] [PubMed] [Google Scholar]
- 17. Leslie A. G. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 18. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 19. Vagin A., Teplyakov A. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 22–25 [DOI] [PubMed] [Google Scholar]
- 20. Emsley P., Cowtan K. (2004) Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 21. Murshudov G. N., Vagin A. A., Dodson E. J. (1997) Acta Crystallogr. D Biol. Crystallogr. 53, 240–255 [DOI] [PubMed] [Google Scholar]
- 22. Laskowski R. A., MacArthur M. W., Moss D. S., Thornton J. M. (1993) J. Appl. Cryst. 26, 283–291 [Google Scholar]
- 23. Krissinel E., Henrick K. (2005) in CompLife LNBI 3695 (Berthold M. R., Glen R., Diederichs K., Kohlbacher O., eds) pp. 163–174, Springer, Berlin [Google Scholar]
- 24. Schuck P., Perugini M. A., Gonzales N. R., Howlett G. J., Schubert D. (2002) Biophys. J. 82, 1096–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. García De La Torre J., Huertas M. L., Carrasco B. (2000) Biophys. J. 78, 719–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kang M., Moroni A., Gazzarrini S., DiFrancesco D., Thiel G., Severino M., Van Etten J. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 5318–5324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Zhou L., Siegelbaum S. A. (2007) Structure 15, 655–670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Scott S. P., Shea P. W., Dryer S. E. (2007) Biochemistry 46, 9417–9431 [DOI] [PubMed] [Google Scholar]
- 29. Weber I. T., Steitz T. A. (1987) J. Mol. Biol. 198, 311–326 [DOI] [PubMed] [Google Scholar]
- 30. Wu S., Vysotskaya Z. V., Xu X., Xie C., Liu Q., Zhou L. (2011) Biophys. J. 100, 1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taraska J. W., Puljung M. C., Olivier N. B., Flynn G. E., Zagotta W. N. (2009) Nat. Methods 6, 532–537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kusch J., Biskup C., Thon S., Schulz E., Nache V., Zimmer T., Schwede F., Benndorf K. (2010) Neuron 67, 75–85 [DOI] [PubMed] [Google Scholar]
- 33. Wang J., Chen S., Nolan M. F., Siegelbaum S. A. (2002) Neuron 36, 451–461 [DOI] [PubMed] [Google Scholar]
- 34. Altomare C., Bucchi A., Camatini E., Baruscotti M., Viscomi C., Moroni A., DiFrancesco D. (2001) J. Gen. Physiol. 117, 519–532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wicks N. L., Wong T., Sun J., Madden Z., Young E. C. (2011) Proc. Natl. Acad. Sci. U.S.A. 108, 609–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ulens C., Siegelbaum S. A. (2003) Neuron 40, 959–970 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.