Background: PhoP is global regulator of Mycobacterium tuberculosis physiology. However, the role of phosphorylation of PhoP remains unknown.
Results: PhoP activates complex lipid biosynthesis only upon phosphorylation.
Conclusion: PhoP regulates lipid biosynthesis by a phosphorylation-dependent mechanism to contribute to morphology of the bacilli.
Significance: This study sheds light on the unexplored role of phosphorylation of PhoP in regulating biosynthesis of lipids unique to M. tuberculosis.
Keywords: DNA-binding Protein, Gene Regulation, Lipid Synthesis, Phosphorylation, Transcription Factors
Abstract
Mycobacterium tuberculosis PhoP is essential for virulence and intracellular growth of the tubercle bacilli. Genetic evidence suggests that PhoP regulates complex lipid biosynthesis, and absence of some of these lipid molecules in a phoP mutant partly accounts for its attenuated growth in macrophages and/or mice. To investigate the mechanism of regulation, here we demonstrate the essentiality of phosphorylation of PhoP in the regulation of complex lipid biosynthesis. We show that phosphorylated PhoP activates transcription of pks2 and msl3, gene(s) encoding polyketide β-ketoacyl synthases through direct DNA binding at the upstream regulatory region(s) of the target genes. Our results identify the genetic determinants recognized by PhoP and show that activation of target genes requires interaction(s) of the phosphorylated regulator at the cognate binding sites. The fact that these sites within the regulatory region of respective genes do not bind in vitro with either unphosphorylated or phosphorylation-deficient PhoP protein is consistent with phosphorylation-dependent assembly of the transcription initiation complex leading to in vivo transcriptional activation. Together, these results reveal so far unknown molecular mechanisms of how PhoP contributes to M. tuberculosis cell wall composition by regulating complex lipid biosynthesis.
Introduction
Despite the apparent importance of two-component signaling (TCS)5 systems at various stages of the Mycobacterium tuberculosis life cycle (1–3), the functioning of TCS proteins is still poorly characterized. The PhoP-PhoR system has been implicated in the M. tuberculosis growth and survival in animal and cellular models because a phoP knock-out mutant of M. tuberculosis showed significant growth attenuation in both macrophages and in mice (4–7). Growing evidence indicates that PhoP regulates biosynthesis of sulfolipids (SL), diacyltrehaloses (DATs), and polyacyltrehaloses (PATs), and the absence of these complex lipid molecules in the phoP mutant is a major reason for the attenuated growth of the bacilli in a mouse model (5, 6, 8, 9). More recent studies suggest that PhoP, in addition to complex lipid biosynthesis, impacts numerous aspects of M. tuberculosis physiology, including early and enduring hypoxic responses (10), regulation of ESAT-6 secretion, specific T-cell recognition during virulence regulation of the bacilli (10–12), and pH sensing during intracellular adaptation (13). However, biochemical evidence showing the role of PhoP in transcription activation has been lacking.
PhoP typically consists of two domains: although the N-terminal receiver domain exhibits (α/β)5 topology with a conserved phosphorylation site and geometry (14), the C-terminal effector domain comprises a winged helix-turn-helix DNA-binding motif (14, 15) and is involved in transcriptional regulation of genes necessary to respond to environmental stimuli. Although M. tuberculosis PhoP regulates ∼114 genes acting both as a transcriptional activator and repressor (5), the role of phosphorylation in transcription regulation by PhoP remains unknown. In agreement with what was evident from the structural data of M. tuberculosis PrrA (16), the closest homologue of M. tuberculosis PhoP, we recently reported inter-domain interactions in PhoP leading to phosphorylation-dependent high affinity DNA binding of the regulator (17). However, molecular mechanism of how PhoP functions as a transcriptional regulator of its target genes remains undefined.
M. tuberculosis PhoP has been shown to be a regulator of the synthesis of three classes of polyketide-derived acyltrehaloses known as SL, DAT, and PAT (5, 6). Recent studies establish that a single nucleotide polymorphism within the DNA binding effector domain of PhoP in the avirulent M. tuberculosis H37Ra accounts for part of its attenuation (18) and absence of polyketide-derived acyltrehaloses compared with the virulent parent strain M. tuberculosis H37Rv (19). Together, these studies suggest that PhoP likely regulates expression of acyltransferase, polyketide synthase (pks), or pks-associated genes involved in the synthesis or transfer of methyl branched fatty acyl substituents found in SL, DAT, and PAT (5, 6). Furthermore, genetic studies on complementation of 1237ΔphoPR::hyg (a 774-bp fragment of M. tuberculosis genome encompassing part of the phoP and phoR coding sequences was replaced with a hygromycin resistance cassette; see Ref. 5) with phoP alone could restore lipid biosynthesis (5) suggesting that phosphorylation may not be essential for regulation of target genes as has been shown by PhoQ-independent activation of target genes of Salmonella enterica PhoP (20). However, there is evidence that response regulators often get phosphorylated by noncognate kinases belonging to different TCSs (for a review see Ref. 21). In contrast, Walters et al. (5) had clearly shown that the intracellular growth phenotype of phoP mutant in macrophages was complemented only in the presence of both phoP and phoR genes, and the phoP gene alone was unable to complement intracellular growth. Although recent studies strongly suggest a link between altered lipid compositions of the phoP mutant strain to its interaction with macrophages (22), the mechanism of how gene(s) involved in lipid biosynthesis are regulated by the important DNA binding transcription factor remains unknown.
Based on these studies, we predicted that PhoP functions as a regulator of genes involved in complex lipid biosynthesis. In this report, we demonstrate that only phosphorylated PhoP directly stimulates transcription of pks2 and msl3 by high affinity DNA binding to the upstream regulatory region of the target genes. Results reported here identify the genetic determinants recognized by PhoP in the effector regions of pks2 and msl3, and evidence is provided that the regulator-promoter interaction(s) at the newly identified PhoP-binding sites lead to activation of the genes. The results have implications for the mechanism of PhoP-mediated regulation of lipid biosynthesis, which is of critical importance to cell wall composition and morphology of the tubercle bacilli.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Media, and Growth Conditions
Growth of Mycobacterium smegmatis, M. tuberculosis H37Ra, and M. tuberculosis H37Rv and transformation by electroporation of test plasmids were as described previously (23). All M. tuberculosis-related work was carried out in a Biosafety Level 3 (BSL3) laboratory. Growth of Escherichia coli strains, cloning, and propagation of plasmids and overexpression of recombinant forms of PhoP proteins were as described (17). For mycobacterial strains, the antibiotics kanamycin and hygromycin were used at concentrations of 25 and 50 μg/ml, respectively; streptomycin was used at 20 μg/ml. For E. coli, kanamycin, hygromycin, and streptomycin were used at 50, 250, and 100 μg/ml, respectively.
General DNA Techniques
Cloning, restriction digestion using endonucleases, and DNA manipulations were carried out by standard techniques (24). Plasmid pJEM-phoP and pJEM-phoPD71N were used to complement H37Ra with PhoP and phosphorylation-deficient PhoP-D71N proteins of M. tuberculosis H37Rv, respectively. In these plasmids, the phoP genes were expressed under the control of its own promoter. To this effect, FPphoPup and RPphoP (supplemental Table S1) amplified the entire regulatory region of phoP and the complete phoP coding sequence of H37Rv (from −350, relative to the translation start site of phoP to the end of the phoP open reading frame). The PCR-amplified region was cloned between BamHI and KpnI sites of the promoter-less pJEM15 (25) to construct pJEM-phoP (Table 1).
TABLE 1.
Bacterial strains and plasmids used in this work
| Description | Source or Ref. | |
|---|---|---|
| Strains | ||
| E. coli DH5α | Novagen | |
| E. coli BL21(DE3) | 56 | |
| M. tuberculosis H37Ra | ATCC25177 | |
| M. tuberculosis H37Rv | ATCC27294 | |
| M. smegmatis mc2155 | 57 | |
| Plasmids | ||
| pET-phoP | PhoP residues 1–247 cloned in pET15ba | 34 |
| pET-phoPD71N | Asp71 codon mutated to Asn in pET-phoP | 17 |
| pME1mL1 | Mycobacterial protein expression vectorb | 27 |
| pME1mL1-phoP | PhoP residues 1–247 cloned in pME1mL1 | This work |
| pME1mL1-phoPD71N | Asp71 mutated to Asn in pME1mL1-PhoP | This work |
| pSM128 | Integrative promoter probe vector for mycobacteriac | 26 |
| pSM-pks2up1 | pks2up1-lacZ fusion in pSM128 | This work |
| pSM-pks2up1sDR2 | Mutant pks2up1-lacZ fusion carrying changes in DR2 site | This work |
| pSM-msl3up1 | msl3up1-lacZ fusion in pSM128 | This work |
| pSM-msl3up1sDR2 | Mutant msl3up1-lacZ fusion carrying changes in DR2 site | This work |
| pJEM15 | Promoter-less E. coli-mycobacteria shuttle vectord | 25 |
| pJEM-phoP | Entire regulatory region along with phoP encoding gene (residues 1–247) cloned in pJEM15 | This work |
| pJEM-phoPD71N | Asp71 codon of phoP mutated to Asn in pJEM-phoP | This work |
a Ampr indicates ampicillin resistance.
b Hygr indicates hygromycin resistance.
c Strr indicates streptomycin resistance.
d Kanr indicates kanamycin resistance.
Transcriptional fusions to lacZ were obtained by cloning PCR-amplified fragments of the M. tuberculosis pks2 (pks2up1) and msl3 (msl3up1) regulatory regions into the ScaI site of pSM128 (a promoter-less integrative lacZ reporter vector (26) with a streptomycin resistance gene) using primer pairs FPpks2up1/RPpks2up1 and FPmsl3up1/RPmsl3up1, respectively (supplemental Table S1). pks2up1 and msl3up1 DNA fragments comprised upstream of the coding region included first 40 and 60 coding bases of the pks2 and msl3 genes, respectively. pSM128 was a kind gift by Dr. Issar Smith, Public Health Research Institute, University of Medicine and Dentistry of New Jersey. To examine the importance of PhoP-binding direct-repeat motif(s) within the pks2 and msl3 promoters, sequence variants that were altered in the downstream repeat unit were generated using the two-stage overlap extension method by interchanging all the As with Cs and all the Gs with Ts, and vice versa, keeping the other repeat unit unaltered (see supplemental Table S1 for mutagenic primers). All of the lacZ transcriptional fusion constructs were verified by restriction analysis and DNA sequencing.
To express M. tuberculosis PhoP in M. smegmatis under the control of TetR, lacZ of pME1mL1 (27) was replaced with phoP open reading frame. To this effect, the phoP gene was amplified from the genomic DNA using primers that introduced an NdeI site (phoPstart) at the start codon and a PstI site (MphoPstop) 3′ of the stop codon (supplemental Table S1). Prior to ligation of the phoP gene in pMElmL1, the 920-bp tetR gene with flanking NotI sites was excised out of the plasmid pME1mL1 by NotI restriction digestion as it included an NdeI site. Subsequently, the NdeI/PstI double-digested backbone fragment of pME1mL1 without the tetR gene was gel-purified and ligated to M. tuberculosis phoP using NdeI and PstI sites. Finally, tetR open reading frame was inserted back using NotI restriction sites to construct pME1mL1-phoP.
Promoter fragments of varying size were PCR-amplified from the full-length promoter templates as indicated under “Results” (see Fig. 1), purified by gel extraction (Qiagen), and end-labeled by T4 polynucleotide kinase (New England Biolabs) and [γ-32P]ATP (BRIT, India). Unincorporated free nucleotides were separated from labeled DNA fragments using a Sephadex G-50 quick spin column (GE Healthcare). When appropriate, PCR-amplified DNA probes were end-labeled as indicated above and digested with BamHI or KpnI to remove the radiolabel at one end. Finally, the end-labeled fragments were extracted with phenol/chloroform/isoamyl alcohol (25:24:1 (v/v/v)) and ethanol-precipitated.
FIGURE 1.
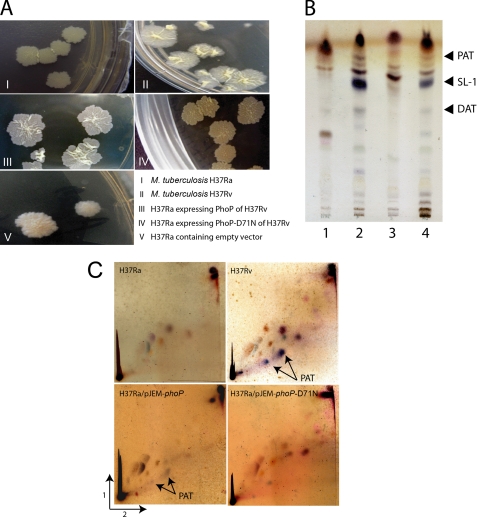
Role of M. tuberculosis PhoP in cell wall composition and complex lipid biosynthesis. A, complementation of M. tuberculosis H37Ra with the phoP gene of H37Rv reveals colony morphology similar to H37Rv. However, the phoPD71N gene of H37Rv fails to complement H37Ra. As a control, H37Ra carrying the empty vector (pJEM15) displays H37Ra-like colony morphology. B, TLC of indicated polar lipids of M. tuberculosis H37Ra (lane 1), H37Ra expressing PhoP or PhoP-D71N from H37Rv (lanes 2 and 3, respectively), and the M. tuberculosis H37Rv (lane 4). C, two-dimensional TLC of polar lipids of M. tuberculosis H37Ra, H37Ra expressing PhoP from H37Rv, or PhoP-D71N from H37Rv and the M. tuberculosis H37Rv. Note that glycolipids of M. tuberculosis strains displayed purple color on α-naphthol staining. Cell growth, lipid extraction, and analyses of lipid moieties were as described under “Experimental Procedures.”
Analysis of Complex Lipids of M. tuberculosis Strains
Extraction of lipids from M. tuberculosis strains, fractionation, and analysis were performed as described (28). The different organic phases were combined, washed extensively, and dried. The crude lipid extracts were subsequently analyzed by thin layer chromatography using chloroform/methanol/water (60:16:2, v/v/v) as described (5). Glycolipids were detected by spraying α-naphthol (in sulfuric acid), followed by baking. The identity of lipid moieties was confirmed by comparing previously reported relative migration of lipids under identical solvent conditions (5, 19). To detect and probe PATs in M. tuberculosis H37Ra, H37Rv, and phoP-complemented strains of H37Ra, total lipids were analyzed by two-dimensional TLC as described (19).
RNA Preparation and Primer Extension
Total RNA from M. tuberculosis grown in 7H9 was isolated and purified with some modifications of a previously reported protocol (29) as described elsewhere (30). Briefly, 25 ml of culture of each bacterial strain was grown in 7H9 media to mid-log phase (an optical density at 600 nm of ≈0.6). Cells were transferred to 30-ml conical tubes, pelleted down by centrifugation, and lysed by resuspending the cells in acetate/EDTA buffer containing glass beads and water-saturated phenol (Ambion). Following incubation at 65 °C for 30 min with severe vortexing after each 10 min, total RNA was chloroform-extracted and ethanol-precipitated. Resulting RNA was incubated with RNase-free DNase I (Promega) for 2 h at 37 °C to remove contaminating genomic DNA and purified by RNeasy column (Qiagen). The integrity of the RNA samples was checked by formaldehyde-agarose gel electrophoresis.
For primer extension, complementary strand oligonucleotides pks2PEx and msl3PEx (supplemental Table S1) that anneal 60 nucleotides (nt) downstream of pks2 and 120 nucleotides downstream of msl3, respectively, with respect to the corresponding translational start sites were end-labeled with [γ-32P]ATP as described above. The labeled primers were annealed to 1 μg of RNA and then extended with 1 unit of Moloney murine leukemia virus reverse transcriptase (Ambion) at 50 °C for 1 h in accordance with the manufacturer's recommendation. After heat inactivation at 92 °C for 10 min, primer extension products were extracted with phenol/chloroform/isoamyl alcohol (25:24:1 (v/v/v)). The nucleic acid was precipitated with 95% chilled ethanol, and the dried pellet was resuspended in formamide loading buffer. To determine the size of the extended product, both pks2up1 and msl3up1 DNA templates were sequenced using the respective primers (as used in the reverse transcription reactions, see supplemental Table S1) to generate a sequencing ladder by the dideoxy chain termination method using SequiTherm Excell II DNA sequencing kit (Epicenter). Primer extension products were loaded onto an 8% polyacrylamide sequencing gel along with the sequencing ladder and run for 2–3 h at 1800 V. After electrophoresis, the gel was dried and exposed to a phosphorimager screen overnight.
Real Time Quantitative PCR
Specific primer sets for target genes were synthesized by Sigma and are listed in supplemental Table S1. Both cDNA synthesis and PCRs were performed by using 0.2 μg of DNase-treated RNA in a reaction buffer containing 2 units of Superscript III Platinum-SYBR Green one-step quantitative RT-PCR kit (Invitrogen) with gene-specific primer pairs (final concentration of 0.2 μm) according to the manufacturer's recommendation. Control reaction mixtures containing identical reactions with 2 units of Platinum TaqDNA polymerase (Invitrogen) instead of reverse transcriptase were also carried out to confirm the absence of genomic DNA in all of the samples. All PCRs were performed using the iCycler real time PCR detection system (Bio-Rad), and melting curve analysis was carried out to confirm amplification of a single product. To compensate for variations in transcript numbers, pks2 and msl3 expression level between strains were normalized to that of sigA (31), expression of which is known to be constitutive in M. tuberculosis (32).
Promoter Regulation by M. tuberculosis PhoP in M. smegmatis
To express PhoP in M. smegmatis, electrocompetent cells were transformed with pME1mL1-phoP expressing wild type PhoP with the Pmyc1tetO promoter under the control of TetR repressor. Cultures of M. smegmatis strains harboring different lacZ fusions and transformed with PhoP expression construct (or no expression plasmid as control) were grown to an absorbance at 600 nm ≈1.5 as described earlier (27). Cultures were inoculated in fresh medium (1:100) either in the absence or in presence of 50 ng/ml of anhydrotetracycline (ATc) and allowed to grow. Aliquots were withdrawn from this culture at indicated time points and centrifuged, and cell pellets were washed with phosphate-buffered saline. Cells were resuspended in 0.5 ml of phosphate buffer (pH 7.2) containing 75 mm NaCl, and cell suspensions were sonicated. β-Galactosidase activity of the cell extracts were determined by using chromogenic substrate 2-nitrophenyl-β-d-galactoside at a final concentration of 1 mg/ml. The reaction mixtures were incubated at 37 °C for 10 min, and the reactions were terminated by adding 0.2 m Na2CO3. The absorbance at 420 nm (A420) values of the supernatant was determined, and β-galactosidase activity was calculated in Miller units as described (33).
To examine TetR-controlled PhoP expression, crude lysates from M. smegmatis (≈5 μg of protein) expressing PhoP were resolved by 12% SDS-PAGE and transferred to PVDF membrane for Western blot analysis. The blots were probed with anti-PhoP primary and HRP-conjugated anti-rabbit IgG secondary antibodies (Abexome Biosciences, India) and developed with Luminata Forte Chemiluminescence reagent (Millipore).
Gel Mobility Shift and DNase I Protection Assays
DNA binding by PhoP proteins, expressed and purified as described earlier (17, 34), were evaluated by gel shift assays as described (17, 34). For DNase I footprint mapping, binding reaction mixtures contained ∼50 fmol of 32P-labeled PCR-amplified DNA fragment and the indicated amounts of purified PhoP in 50 μl of reaction buffer as used in gel shift assays. Reaction mixtures were incubated for 20 min at 15 °C. Following incubation, 50 μl of a solution containing 5 mm CaCl2 and 10 mm MgCl2 was added to the reaction mixtures at 15 °C, and DNase I digestion was carried out with 0.75 units of DNase I for 1 min. The digestion reactions were terminated by addition of 100 μl of stop solution (200 mm NaCl, 30 mm EDTA, 1% SDS) and extracted with phenol/chloroform/isoamyl alcohol (25:24:1 (v/v/v)). The nucleic acids were ethanol-precipitated with 95% chilled ethanol, and the dried pellets were resuspended in 5 μl of formamide loading buffer (1% bromphenol blue, 1% xylene cyanol, 10 mm EDTA in 98% formamide). After electrophoresis on an 8% denaturing polyacrylamide sequencing gel, the gel was dried, and the footprinting reactions were analyzed by autoradiography. To identify the protected sequence on each strand, footprinting reactions were analyzed alongside a DNA sequencing ladder generated by SequiTherm Excell II DNA sequencing kit (Epicenter) on both 32P-labeled DNA fragments. Primers used to generate DNA fragments are described in supplemental Table S1.
RESULTS
Phosphorylation of PhoP Regulates Cell Wall Composition by Controlling Lipid Biosynthesis in M. tuberculosis
Studies aimed at investigating phenotypic differences between M. tuberculosis H37Ra and the pathogenic H37Rv showed striking morphological differences displaying smaller colony size and lesser wrinkling on the colony surface for H37Ra compared with H37Rv. Later, these observations were extended to show that a similar and distinct difference exists between H37Rv and a phoP knock-out mutant of H37Rv when cultured on 7H10 Middlebrook agar plates (4, 18, 35). More interestingly, two independent studies show that much of the difference was significantly reduced when PhoP from H37Rv was expressed in H37Ra (18, 19). We extended these results to gain insight into the mechanism of how PhoP contributes to M. tuberculosis morphology. In agreement with previous studies, morphological differences in H37Ra and H37Rv disappeared significantly when H37Ra was transformed with pJEM15 (25) expressing PhoP of H37Rv (pJEM-phoP) from its endogenous promoter (see under “Experimental Procedures” for details) (Fig. 1A). In striking contrast, H37Ra-expressing phosphorylation-deficient PhoP-D71N of H37Rv failed to show morphological properties like H37Rv. It should be noted that the M. tuberculosis PhoP and PhoR proteins form a functional TCS, which in their simplest form utilize a histidine-aspartate phosphorelay between two modular proteins (for reviews, see Refs 36, 37), and the primary site of covalent phosphorylation of PhoP has been mapped to Asp71 (38, 39). Substitution of Asp71 to Asn71 alters the side chain functional group from carboxylic acid to carboxyamide moiety, thus rendering PhoP phosphorylation-deficient (39). As expected, H37Ra transformed with the empty vector pJEM15 displayed H37Ra-like morphology. Together, these observations clearly suggest that expression of PhoP and its phosphorylation is necessary and sufficient to restore H37Rv-like morphology of H37Ra.
The fact that a single nucleotide polymorphism of PhoP in the avirulent H37Ra explains the absence of polyketide-derived acyltrehaloses compared with the parent strain H37Rv (19) prompted us to investigate and compare glycolipid moieties of H37Ra and H37Rv. As expected, H37Rv showed the presence of SL1, DAT, and PAT (as indicated on the figure), which were completely absent in H37Ra (compare lane 1 with lane 4, Fig. 1B). However, in agreement with a previous report (19), H37Ra-expressing PhoP of H37Rv showed the presence of SL1, DAT, and PAT in the thin layer chromatography experiments (lane 2, Fig. 1B). Strikingly, expression of PhoP-D71N was unable to restore SL1, DAT, and PAT biosynthesis in H37Ra (Fig. 1B, compare lane 2 and lane 3). The presence of PATs in H37Rv PhoP-complemented strain of H37Ra, but not that of PhoP-D71N, was further confirmed by analysis of total lipids using two-dimensional thin layer chromatography (Fig. 1C). Thus, we conclude that PhoP regulates complex lipid biosynthesis of M. tuberculosis, and phosphorylation of PhoP appears to be essential for its regulatory role in lipid biosynthesis.
PhoP Regulates Expression of pks2 and msl3 in Vivo in a Phosphorylation-dependent Manner
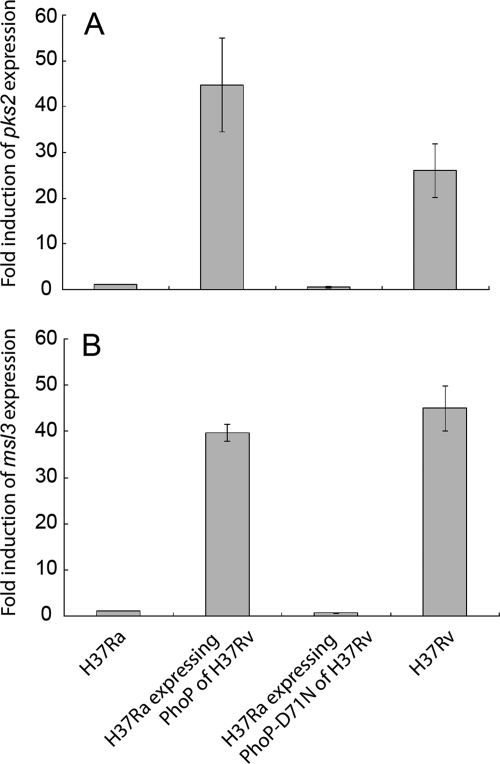
pks2 and msl3 gene(s) of M. tuberculosis encode enzymes that have been implicated in the synthesis of multiple methyl branched fatty acids essential for biosynthesis of cell wall lipids, like SL (40), DAT, and PAT (41). Furthermore, genetic data coupled with biochemical evidence show essential regulatory role of PhoP in the biosynthesis of SL, DAT, and PAT (5, 6). To this end, we examined relative expression of pks2 and msl3 in vivo by real time reverse transcription-PCR from the M. tuberculosis H37Rv and H37Ra. Interestingly, H37Rv showed a significant up-regulation of pks2 and msl3 expression of 25.9 ± 5.9- and 45 ± 4.9-fold, respectively, relative to H37Ra cultures (Fig. 2). To study if PhoP from H37Rv contributes to the variation of expression, pks2 and msl3 expression was quantified in the H37Ra complemented with wild type H37Rv phoP allele (pJEM-phoP). Strikingly, both pks2 and msl3 expressions could be restored to wild-type levels when H37Ra expressed phoP from H37Rv. In fact, consistent with enhanced production of PATs, DATs, and SLs (Fig. 1B), H37Ra expressing PhoP from H37Rv displayed a higher level of pks2 expression (≈1.7-fold) and comparable msl3 expression (≈0.9-fold) with respect to wild-type H37Rv. Together, these in vivo results are in agreement with previous microarray data suggesting effect of PhoP on regulation of pks2 and msl3 expression (5). In sharp contrast, H37Ra transformed with pJEM-phoPD71N plasmid was completely ineffective in complementing pks2 and msl3 expression. From these results we conclude that phosphorylation of PhoP is essential for regulation of expression of both pks2 and msl3 in vivo.
FIGURE 2.
PhoP regulates pks2 (A) and msl3 (B) expression in vivo in M. tuberculosis by a phosphorylation-dependent mechanism. H37Ra, H37Ra-expressing wild type phoP allele from H37Rv, H37Ra carrying the mutant phoP-D71N allele, and wild-type H37Rv were grown to mid-log phase. Following cell growth, total RNA was extracted from bacterial cells for quantitative real time reverse transcription PCR (see “Experimental Procedures” for details). The values shown here represent the indicated fold induction of expression of pks2 and msl3 with respect to their expression levels in H37Ra and are derived from at least three independent experiments using at least two different RNA preparations.
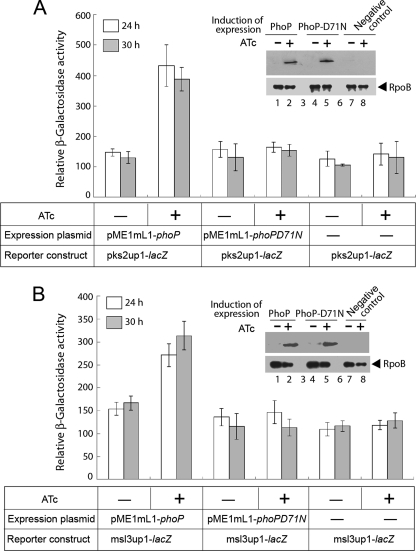
To investigate if PhoP functions as a direct regulator of M. tuberculosis pks2 and msl3 genes in vivo, we next constructed transcriptional fusions to lacZ by cloning PCR-amplified fragments of the pks2 and msl3 regulatory regions at the ScaI site of pSM128, an integrative promoter probe vector for mycobacteria (26). To this end, PCR-amplified DNA fragments, pks2up1 and msl3up1, comprising −206 to +40 and −350 to +60, respectively, with respect to their corresponding translational start sites were considered as regulatory region(s) of pks2 and msl3 genes. To express PhoP in M. smegmatis, strains harboring the transcriptional fusions were transformed with pME1mL1-phoP, an inducible expression system (27) expressing wild-type PhoP from the Pmyc1tetO promoter under the control of TetR repressor. Transformed cells were then grown in 7H9 medium containing appropriate antibiotics in the absence or presence of 50 ng/ml ATc as inducer of PhoP expression. Strikingly, the pks2up1-lacZ fusion was significantly activated with induction of PhoP at indicated time points, as the β-galactosidase levels obtained in the presence of ATc was ≈3-fold higher than that obtained in absence of ATc (Fig. 3A). Similarly, cells carrying msl3up1-lacZ fusion showed an ≈2-fold enhanced β-galactosidase activity when PhoP expression was induced compared with the uninduced sample (Fig. 3B). However, both reporter constructs, under identical experimental conditions, failed to show promoter activation when expression of phosphorylation-deficient PhoP-D71N was induced (1.04 ± 0.2- and 0.98 ± 0.2-fold change in β-galactosidase activity, Fig. 3, A and B, respectively). Insets in Fig. 3, A and B, show comparable expression of M. tuberculosis PhoP and its mutant in M. smegmatis in the presence of ATc, thus ruling out the possibility that altered expression of the mutant could account for the failure to activate transcription. From these results we conclude that phosphorylation of PhoP alone is essential to activate expression of pks2 and msl3 in vivo.
FIGURE 3.
PhoP regulates expression of pks2 (A) and msl3 (B) in vivo in M. smegmatis. M. smegmatis strains harboring indicated constructs along with wild-type and mutant PhoP expression plasmids were grown in the absence or presence of ATc, and β-galactosidase activity from the transcription fusion was measured at indicated times. The values shown here are averages of at least three independent experiments. Insets compare expression of PhoP in crude extracts (24-h time point) containing equal amounts of total protein (as determined by Bradford assay) by Western blot using anti-PhoP antibody (Abexome Biosciences). As a loading control, the same crude extracts were probed with antibody against the β-subunit of RNA polymerase (Abcam).
PhoP Recognizes Regulatory Regions of pks2 and msl3
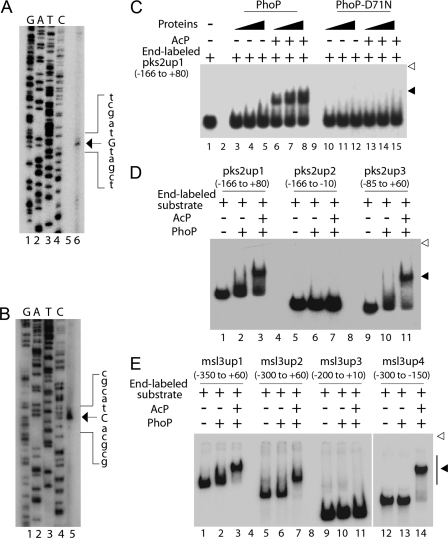
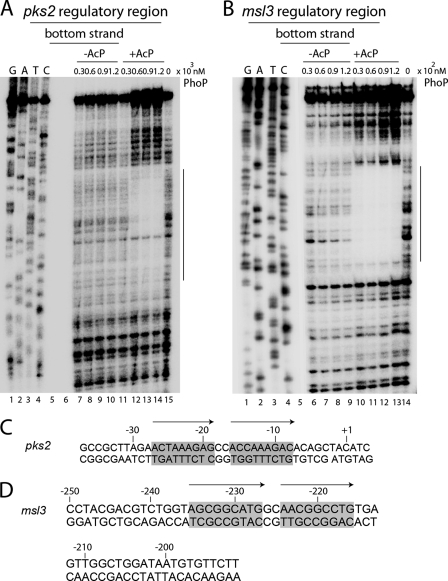
To precisely locate and identify regulatory regions of pks2 and msl3, the origin(s) of transcription of pks2 and msl3 were determined by primer extension using total RNA from M. tuberculosis H37Rv. A 100-bp extension product was obtained, which localized the pks2 transcription start site to a guanine 40 nucleotides upstream of the ATG start site (Fig. 4A). However, a primer extension experiment generated a 127-bp extension product that localized the msl3 transcription start site to the first guanine of the GTG start site (Fig. 4B).
FIGURE 4.
Mapping transcription start sites (A and B) and core PhoP-binding sites (C–E) within the regulatory regions of pks2 and msl3. To determine transcription start sites, total RNA isolated from M. tuberculosis H37Rv was used in primer extension experiments using antisense oligonucleotide primers (see supplemental Table S1) located within pks2(A) and msl3 (B) genes as described under “Experimental Procedures.” Sequencing samples as markers were prepared with the same labeled primer as used for the reverse transcription and resolved in lanes 1–4, adjacent to the primer extension product (lane 6 for A and lane 5 for B). The unique band identified in each primer extension reaction is indicated on the figure. C, EMSA of radiolabeled pks2up1 for binding of increasing concentrations of PhoP (lanes 3–8) or phosphorylation-deficient PhoP-D71N (lanes 10–15) preincubated in phosphorylation mixture with or without AcP, respectively. Lane 1 shows the free probe. Note that binding mixtures in lanes 3–5, 6–8, 10–12, and 13–15 contained the indicated proteins at 0.1, 0.2 and 0.3 μm. To probe the core-binding site of PhoP within the regulatory region of pks2 (D) and msl3 (E), PhoP binding to indicated DNA fragments was carried out with 200 and 50 nm of PhoP, respectively, preincubated in phosphorylation mixture with or without AcP. The position of the radioactive material was determined by exposure to a phosphor storage screen, and bands were quantified in the phosphorimager (Fuji). Open and filled arrowheads indicate origins of the polyacrylamide gel and slower moving complexes with band shifts produced in presence of PhoP, respectively.
To study whether PhoP acts as a regulator by direct binding to the regulatory region(s) of target genes, we next examined the ability of PhoP to recognize pks2up1 by EMSA. Although purified PhoP was unable to generate a complex stable to gel electrophoresis, PhoP preincubated in a phosphorylation mixture containing acetyl phosphate (AcP) as the phospho-donor displayed efficient DNA binding with end-labeled pks2up1 by forming a complex of reduced electrophoretic mobility (compare lanes 3–5 with lanes 6–8, Fig. 4C). Previously, we provided a direct demonstration of phosphorylation of PhoP at Asp71 using AcP as the phospho-donor (39). A quantitative analysis suggests at least 20-fold stimulation of DNA binding by phospho-PhoP with the pks2up1 compared with the unphosphorylated protein (based on the limits of detection in this assay). As expected, the PhoP-D71N mutant (with impaired phosphorylation) preincubated in the phosphorylation mixture with or without AcP, failed to generate a PhoP-DNA complex with identical DNA substrate (lanes 10–15, Fig. 4C). Thus, in conjunction with previous results (17) and the results shown here, we conclude that specific interaction(s) between PhoP and the msl3 and/or pks2 regulatory region is dependent on PhoP phosphorylation.
Using a number of PCR-amplified overlapping fragments, we next addressed a more precise characterization of the pks2 and msl3 regulatory regions important for PhoP binding. Fig. 4D shows that in contrast to pks2up1 (−166 to +80 with respect to the transcription origin) as positive control, pks2up2 (−166 to −10) was not shifted upon incubation with phospho-PhoP (compare lanes 1–3 and lanes 5–7). However, pks2up3 (−85 to +60) generated a stable slower moving complex with phospho-PhoP (Fig. 4D, lanes 9–11), indicating that there was no high affinity PhoP binding sequence upstream of −85 nucleotide of the pks2 regulatory region.
However, whereas msl3up1 (−350 to +60 with respect to the transcription origin) and msl3up2 (−300 to +60) fragments were shifted by phospho-PhoP (lanes 1–3 and lanes 5–7, respectively; Fig. 4E), msl3up3 (−200 to +10) was not shifted upon incubation with phospho-PhoP (lanes 9–11). In contrast, incubation of phospho-PhoP with an ≈150-bp msl3up4 DNA fragment (−300 to −150) induced complete band shift of the DNA fragment (lanes 12–14). Thus, phosphorylated PhoP is apparently binding to the msl3 regulatory region spanning −300 to −150 relative to the transcription start site. Together, these EMSA experiments suggest that PhoP binding is likely to be sequence-specific because purified PhoP was able to recognize the specific regulatory region of pks2 and msl3 and not any sequence stretch within the regulatory regions.
PhoP Protects Nucleotide Sequences Containing a Direct Repeat Motif Upstream of pks2 and msl3 Genes
To determine whether a specific nucleotide sequence motif within ≈145-bp pks2up3 and ≈150-bp msl3up4 is recognized by PhoP, the precise area of DNA bound by the protein was determined using DNase I footprint mapping. The purified DNA substrates carrying radioactive label either at the top or the bottom strands were incubated with PhoP, and DNase I was added to partially digest the DNA. The resulting fragments were analyzed on a denaturing sequencing gel. Fig. 5A shows that phospho-PhoP, in a concentration-dependent manner, protected a stretch of ≈41-bp of the bottom strand of pks2up3, beginning 78 nucleotides upstream of the ATG start site. An almost identical pattern of protection was observed with phospho-PhoP when the top strand-labeled substrate was used during DNase I footprinting (supplemental Fig. S1A). Similarly, a stretch of ≈60 bp from both the bottom and top strands of msl3up4 was protected by PhoP, beginning 250 nucleotides upstream of the GTG start site (Fig. 5B and supplemental Fig. S1B, respectively). In agreement with the EMSA data (Fig. 4), clearly protection of the nucleotide sequence from both DNA substrates was significantly influenced by phosphorylation of PhoP. It is noteworthy that we consistently observed an ≈10-fold difference in protein concentration to achieve comparable protection to DNase I cleavage at msl3 and pks2 regulatory regions. This is in agreement with the EMSA experiments suggesting that PhoP shares ∼5–10-fold higher binding affinity with msl3 regulatory region.
FIGURE 5.
DNase I protection mapping of PhoP binding to pks2 and msl3 regulatory region(s). ≈50 fmol of pks2up3 (A) and msl3up4 (B) fragments each carrying the label at the bottom strands were incubated with increasing concentrations of PhoP preincubated in phosphorylation mixture in the absence (lanes 7–10 for pks2up3; lanes 6–9 for msl3up4) or presence of AcP (lanes 11–14 for pks2up3; lanes 10–13 for msl3up4) and in the absence of PhoP protein (lane 15 for pks2up3; lane 14 for msl3up4) prior to digestion with DNase I as described under “Experimental Procedures.” G, A, T, and C designate the DNA sequencing ladder generated for each strand. The protected regions on each of the bottom strands are indicated by vertical lines. Nucleotide sequences within the DNase I-protected region(s) of pks2up3 (C) and msl3up4 (D) (numbered with respect to the transcription origins) include a conserved 9-bp direct repeat motif shaded in gray and indicated with arrows.
When we examined each protected region more closely, strikingly a conserved 9-bp direct repeat motif was apparent within each PhoP-protected sequence. For example, the PhoP-protected pks2 promoter region contained a direct repeat motif with two 9-bp repeat units (comprising nucleotides −28 to −20 and −17 to −9 relative to the transcription start site) separated by an intervening spacer sequence of two nucleotides (Fig. 5C). Similarly a 9-bp direct repeat motif (comprising nucleotides −235 to −227 and −224 to −216 relative to the transcription start site) separated by a spacer of two nucleotides (Fig. 5D) was identified within the PhoP-protected sequence of the msl3 regulatory region. From these results, we surmise that PhoP directly binds to regulatory regions of pks2 and msl3 containing a 9-bp direct repeat motif. It should be noted that although the two repeat units consisting of a direct repeat motif, both within the pks2 and msl3 regulatory regions, share a significant similarity in nucleotide sequence (7 of 9 nucleotides are identical in both cases), there exists striking differences in the nucleotide sequence of the repeat units present within the regulatory regions of two genes (8 of 9 nucleotides are different).
Transcriptional Activation of pks2 and msl3 Involves Recognition of the Direct Repeat Motif by M. tuberculosis PhoP
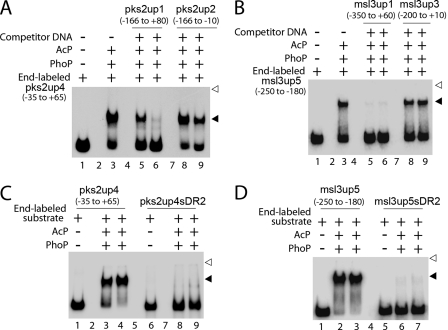
To investigate whether the 9-bp direct repeat motif upstream of both pks2 and msl3 alone was critical for DNA binding, PhoP binding to the 95-bp pks2up4 (−35 to +65 relative to the pks2 transcription start site) and the 70-bp msl3up5 (−250 to −180 relative to the msl3 transcription start site) DNA fragments containing the putative direct repeat motif(s) was investigated by EMSA in the absence (lane 3, Fig. 6A) or presence of either a 12.5- and 25-fold excess of specific (lanes 5 and 6) or nonspecific competitor (lanes 8 and 9). Although unlabeled pks2up1 at a 25-fold molar excess efficiently competed PhoP binding (only 5 ± 0.5% of binding; Fig. 6A, lane 6), identical fold excess of pks2up2 as nonspecific competitor DNA resulted in a minor variation of DNA binding efficiency (57 ± 0.8% binding; lane 9) compared with no competitor control (68 ± 1% binding; lane 3), suggesting that PhoP recruitment at pks2up4 is sequence-specific. Similarly, whereas unlabeled msl3up1 at a 12.5-fold molar excess efficiently competed PhoP binding to msl3up5 (<2% of binding; lane 5, Fig. 6B), even 25-fold excess of msl3up3 as nonspecific competitor DNA resulted in an insignificant variation of 12(±2)% DNA binding efficiency compared with no competitor control (compare lane 9 to lane 3, Fig. 6B), suggesting that PhoP recruitment at msl3up5 is sequence-specific.
FIGURE 6.
EMSA experiments examined sequence-specific binding of phospho-PhoP to pks2up4 (A) and msl3up5 (B) in the absence (lane 3) or presence of 12.5- and 25-fold excess of specific (lanes 5 and 6) and nonspecific (lanes 8 and 9) competitors at 100 and 25 nm of PhoP, for A and B, respectively. Lane 1 shows the free probe. Additional EMSA experiments examined indicated end-labeled probes pks2up4 and pks2up4sDR2 (C) and msl3up5 and msl3up5sDR2 (D) for binding of phospho-PhoP at 0.1 and 0.2 μm PhoP (lanes 3 and 4, and lanes 8 and 9 (C)) and 50 and 100 nm of PhoP (lanes 2 and 3, and lanes 6 and 7 (D)), respectively. Lanes 1 and 6 of C and lanes 1 and 5 of D show indicated free probes used. Open and filled arrowheads indicate origins of the polyacrylamide gel and slower moving complexes with band shifts produced in the presence of PhoP, respectively. Note that nucleotides outside the direct repeats, in both cases, included identical extensions of natural sequence at both the 5′- and 3′-ends. The gels are representative of at least three independent experiments.
We next examined PhoP binding to variants of pks2up4 and msl3up5, which were altered in the downstream repeat sequence of the direct repeat motif (pks2up4sDR2 and msl3up5sDR2, respectively) as described under “Experimental Procedures.” Although pks2up4 was shifted completely by phospho-PhoP, pks2up4sDR2 probe was completely ineffective for PhoP binding (<2 and <5% of DNA binding at 0.1 and 0.2 μm PhoP, respectively; compare lanes 3 and 4 to lanes 8 and 9, Fig. 6C). Similarly, compared with msl3up5, msl3up5sDR2 probe was significantly ineffective for PhoP binding (undetectable binding at both 0.05 and 0.1 μm PhoP (based on the limits of detection in this assay); compare lanes 2 and 3 to lanes 6 and 7, Fig. 6D). Thus, specific interaction(s) of PhoP at the regulatory region(s) of pks2 and msl3 is exclusively dependent on the presence of the direct repeat motif(s).
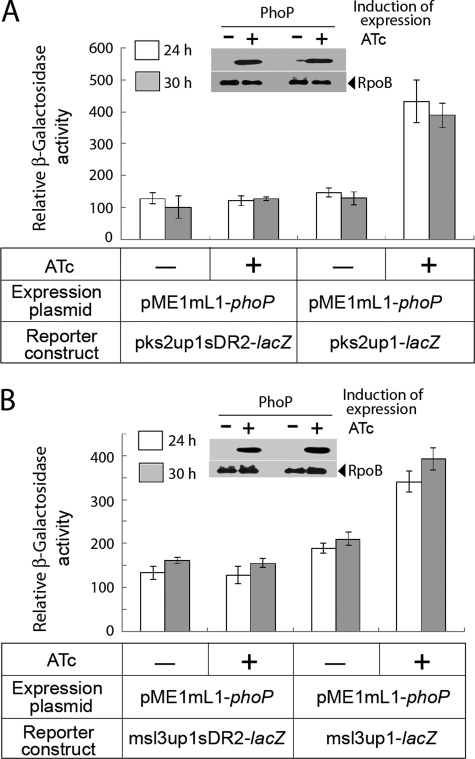
To investigate the role of the newly identified PhoP-binding site in PhoP-coupled transcription activation of pks2 and msl3, changes were introduced in the nucleotide sequence of the downstream repeat unit of pks2up1 and msl3up1 template DNA as described under “Experimental Procedures.” The DNA fragments carrying changes in the PhoP-binding site (pks2up1sDR2 and msl3up1sDR2, respectively) were subsequently cloned in pSM128 and used as transcription fusions to examine the effect of PhoP on the regulation of expression (Fig. 7). Interestingly, with induction of PhoP expression by ATc, there was no significant change in the level of β-galactosidase activity from the promoters carrying changes at PhoP-binding sites. Multiple replicates of experiments confirmed that the difference of β-galactosidase activity in the presence and absence of induction of PhoP expression was 0.94(±0.05)-fold (compare empty columns) and 1.07(±0.03)-fold (compare filled columns) from the pks2up1sDR2-lacZ at 24 and 30 h, respectively (Fig. 7A). Likewise, we obtained 0.95(±0.01)-fold (compare empty columns) and 0.96(±0.03)-fold (compare filled columns) difference of β-galactosidase activity at 24 and 30 h, respectively, from the msl3up1sDR2-lacZ construct in presence and absence of induction of PhoP expression (Fig. 7B). These results are in striking contrast with PhoP-dependent 2.9(±0.2)- and 1.9(±0.3)-fold activation of wild-type pks2up1 and msl3up1 expression, respectively, under identical conditions examined (Fig. 7, A and B, respectively). From these results, we conclude that phosphorylated PhoP binding at the newly identified repeat motif within the regulatory regions of pks2 and msl3 is necessary and sufficient for transcription activation of these genes. The fact that PhoP-binding site(s) within the regulatory regions of pks2 and msl3 are located at distinctly different distances from the corresponding transcription start sites possibly indicates mechanistic variation of PhoP-mediated activation in the presence of the RNA polymerase.
FIGURE 7.
PhoP regulates expression of pks2 and msl3 by specific recognition of direct repeat motif. To examine the importance of predicted PhoP-binding sites in PhoP-dependent transcription regulation of pks2 and msl3, M. smegmatis strains harboring pks2up1sDR2 (A) and msl3up1sDR2 (B) (see “Results” for details), as well as the corresponding wild-type regulatory regions, were grown in the absence or presence of inducing PhoP expression as indicated on the figure and described in the legends to Fig. 3. β-Galactosidase activity from the transcription fusion was measured at indicated times. Insets compare expression of PhoP in crude extracts (24-h time point) containing equal amounts of total protein by Western blot using anti-PhoP antibody (Abexome Biosciences). As loading control, crude extracts were probed with antibody against β-subunit of RNA polymerase (Abcam).
Notably, within the msl3 regulatory region, the PhoP-binding site is located very far from the promoter (Fig. 5D), suggesting that any regulatory role of PhoP on transcription would require constraints on DNA topology.
DISCUSSION
Interesting similarities in growth attenuation and morphological and cytochemical properties between H37Ra and phoP mutant of M. tuberculosis H37Rv have been proposed to be a direct consequence of the absence of three kinds of complex lipids, SL, DAT, and PAT (5, 6, 18). It is noteworthy that these lipids are relatively restricted to the virulent strains of the M. tuberculosis complex, suggesting their role in pathogenicity of the tubercle bacilli (42, 43). These reports coupled with recent studies show that among the reasons for the attenuation of M. tuberculosis the H37Ra strain is a single nucleotide polymorphism within the phoP gene that (a) is incapable of restoring polyketide-derived acyltrehalose synthesis in a phoP-phoR knock-out mutant of H37Rv (19) and (b) inhibits secretion of proteins that are important for virulence (18). Although these results establish a strong link between PhoP with lipid biosynthesis and virulence regulation, studies using a pks2–3/4 knock-out mutant of M. tuberculosis H37Rv clearly show that not polyketide-derived acyltrehaloses but rather phthiocerol dimycocerosates, which are not regulated by PhoP, are the major contributor of virulence of the tubercle bacillus (19, 43, 44). However, it should be noted that proteins encoded by genes of the pks2 cluster (Rv3825c-3824c-3823c) and msl3 cluster (Rv1180-Rv1182-Rv1183) (supplemental Fig. S2) are shown to be regulated by PhoP (5), and some of these proteins critically contribute to regulation of M. tuberculosis virulence (45, 46). In agreement with these studies, more recently acyltrehaloses have been shown to influence phagosome maturation in macrophages (47).
Given the link that exists between M. tuberculosis PhoP and complex lipid biosynthesis on the one hand (5, 6) and between the presence of polyketide-derived lipids and maintenance of cell morphology on the other hand (18, 19), we sought to investigate the mechanism of how PhoP regulates expression of genes involved in lipid biosynthesis. To circumvent the problem of PhoP being phosphorylated in vivo by a noncognate sensor kinase or by some other mechanisms, we utilized the phosphorylation-deficient PhoP-D71N mutant in both in vivo and in vitro experiments. Our results show that PhoP in the phosphorylated form directly regulates expression of pks2 and msl3, products of which are essential components of M. tuberculosis cell wall (40, 41). What offers a new mechanistic insight is the finding that phosphorylation at Asp71 of PhoP is critically important for the control mechanism of complex lipid biosynthesis, which in turn regulates cell morphology. This is clearly shown by our results on cell morphology of M. tuberculosis H37Ra, H37Rv, and H37Ra complemented with H37Rv phoP along with analyses of lipid profile (Fig. 1) and concomitant restoration of expression of pks2 and msl3 (Fig. 2). Thus, PhoP contributes to the regulation of cell morphology of the tubercle bacilli most likely through its regulatory influence on complex lipid biosynthesis and facilitates an integrated view of our results. However, additional experiments are needed to examine and understand the signal sensed by the cognate kinase PhoR (38), which in turn regulates phosphorylation of PhoP. The fact that H37Ra carries an identical copy of phoR as H37Rv appears to suggest that PhoR, under the conditions examined, triggers phosphorylation of PhoP. However, PhoP-dependent up-regulation of pks2 and msl3 in M. smegmatis in the absence of PhoR is most likely suggestive of PhoP phosphorylation by cross-talk with other two-component systems (21). It is tempting to speculate that a relatively lower level of activation of pks2 and msl3 (Fig. 3) by PhoP is possibly due to the low level of phosphorylation of PhoP in the absence of cognate kinase.
Phosphorylation of response regulators plays an important role to activate or repress gene expression in vivo (48). Results reported here validate and expand upon our previous work on msl3 promoter recognition (17) to show that phospho-PhoP, through sequence-specific recognition of the regulatory regions, directly regulates transcription of pks2 and msl3, the two genes of related metabolic function involving complex lipid biosynthesis. In agreement with data on in vivo regulation (Figs. 1–3), phosphorylation of PhoP showed a striking impact on in vitro DNA binding, a result that was further confirmed by DNase I footprinting analyses. Interestingly, these results are consistent with the crystal structure of M. tuberculosis PrrA (Protein Data Bank code 1YS6 (16)), which shows that the recognition helix is involved in interacting with the regulatory domain and thereby on phosphorylation, significantly enhancing binding affinity of the protein to the regulatory region of the prrA-prrB operon (49). Although DNase I footprint mapping clearly identified genetic determinants recognized by phospho-PhoP (Fig. 5), additional EMSA experiments coupled with in vivo reporter assays using transcription fusion of wild type and mutant promoters (carrying mutations at the PhoP binding sites; Fig. 7) to lacZ clearly establishes the following: (i) the identified direct repeat motif alone is likely responsible for PhoP-DNA interaction(s) at the relevant regulatory regions, and (ii) recruitment of phospho-PhoP at these sites is essential for PhoP-dependent regulation of pks2 and msl3. The structural data derived from the co-crystals of protein-DNA complexes of E. coli PhoB-DNA (50) and Bacillus subtilis Spo0A-DNA (51) also exhibit tandem binding of the protein on adjacent repeat motifs where a single protomer recognizes each repeat sequence. Thus, recruitment of PhoP protomers on two adjacently arranged repeat sites consisting of a direct repeat motif provides an interesting example of how similar sequence modules with variations of nucleotide sequence enable functional diversification by the same family of response regulator.
Although expression of PhoP is known to be critical for M. tuberculosis virulence (4–8), the genetic determinant(s) in the downstream effector regions that are recognized by PhoP remain largely unknown. PhoP belongs to the OmpR/PhoB subfamily, members of which often recognize repeat DNA sequences. M. tuberculosis DosR, a regulator of genes involved in response to hypoxia and NO exposure had been shown to recognize a 20-bp palindromic sequence that is present upstream of almost all genes regulated by it (1, 52). In contrast, M. tuberculosis MprA recognizes a direct repeat motif comprising two tandemly arranged 8-bp repeat units, a sequence present upstream of mprA and pepD genes (53). Here, we show recruitment of phosphorylated PhoP to a 20-bp stretch of pks2 and msl3 regulatory region(s) comprising a 9-bp direct-repeat motif separated by a two-nucleotide spacer. Together, these results suggest that differences in nucleotide sequences, orientation of repeat units, and/or intervening spacer lengths at the recognition sites of different promoters most likely regulate transcription factors with largely conserved structures to control diverse biological responses. Interestingly, our results on newly identified PhoP-binding sites are in broad agreement with the presence of three 9-bp repeat units within the PhoP-protected region of the phoP promoter, including a 23-bp sequence containing a 9-bp direct repeat motif (38). However, the nucleotide sequences of the repeat sites identified here (within the regulatory regions of pks2 and msl3; Fig. 5, C and D), consistent with their variable affinity of interaction with PhoP (Fig. 5, A and B), are significantly different from each other and also from what we had identified previously within the phoP promoter itself. Thus, these results suggest that differences in nucleotide sequences, in addition to displaying differential affinity for the same transcription factor (PhoP), contribute to regulatory mechanisms of variable physiological functions.
In conclusion, our results show that direct interactions between the phosphorylated PhoP and the newly identified PhoP-binding site(s) are essential for activation of the genes, a result confirmed by reporter assays using transcriptional fusion of promoters to lacZ. More importantly, we demonstrate a critical role of phosphorylation of a single residue of PhoP contributing to regulation of cell morphology most likely by influencing lipid biosynthesis, a novel result of unusual significance with implications on the molecular mechanism of action of the key regulator. It should be noted that earlier studies link PhoP-PhoR of Streptomyces lividans and Streptomyces coelicolor to phosphate control of lipid biosynthesis (54, 55). Together, these results provide an interesting example of how similar mechanisms in actinomycetes exist and operate to regulate biosynthesis of polyketides or polyketide-derived products of diverse structure and function.
Supplementary Material
Acknowledgments
We thank Dr. Issar Smith (Public Health Research Institute, University of Medicine and Dentistry of New Jersey) for the kind gift of pSM128, Dr. Sabine Ehrt (Weill Medical College of Cornell University) for pME1mL1 expression vector, Renu Sharma and Mahendra K. Yadav for technical assistance, and Renu Sharma for help with the preparation of the manuscript.
This work was supported in part by Council of Scientific and Industrial Research Supra Institutional Project SIP-10 and by research grants from the Department of Biotechnology and Department of Science and Technology, Government of India (to D. S.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table S1 and Figs. S1 and S2.
- TCS
- two-component system
- ATc
- anhydrotetracycline
- DAT
- diacyltrehalose
- PAT
- polyacyltrehalose
- SL
- sulfolipid
- AcP
- acetyl phosphate.
REFERENCES
- 1. Sherman D. R., Voskuil M., Schnappinger D., Liao R., Harrell M. I., Schoolnik G. K. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 7534–7539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zahrt T. C., Deretic V. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12706–12711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Betts J. C., Lukey P. T., Robb L. C., McAdam R. A., Duncan K. (2002) Mol. Microbiol. 43, 717–731 [DOI] [PubMed] [Google Scholar]
- 4. Pérez E., Samper S., Bordas Y., Guilhot C., Gicquel B., Martín C. (2001) Mol. Microbiol. 41, 179–187 [DOI] [PubMed] [Google Scholar]
- 5. Walters S. B., Dubnau E., Kolesnikova I., Laval F., Daffe M., Smith I. (2006) Mol. Microbiol. 60, 312–330 [DOI] [PubMed] [Google Scholar]
- 6. Gonzalo Asensio J., Maia C., Ferrer N. L., Barilone N., Laval F., Soto C. Y., Winter N., Daffé M., Gicquel B., Martín C., Jackson M. (2006) J. Biol. Chem. 281, 1313–1316 [DOI] [PubMed] [Google Scholar]
- 7. Ludwiczak P., Gilleron M., Bordat Y., Martin C., Gicquel B., Puzo G. (2002) Microbiology 148, 3029–3037 [DOI] [PubMed] [Google Scholar]
- 8. Martin C., Williams A., Hernandez-Pando R., Cardona P. J., Gormley E., Bordat Y., Soto C. Y., Clark S. O., Hatch G. J., Aguilar D., Ausina V., Gicquel B. (2006) Vaccine 24, 3408–3419 [DOI] [PubMed] [Google Scholar]
- 9. Ryndak M., Wang S., Smith I. (2008) Trends Microbiol. 16, 528–534 [DOI] [PubMed] [Google Scholar]
- 10. Gonzalo-Asensio J., Mostowy S., Harders-Westerveen J., Huygen K., Hernández-Pando R., Thole J., Behr M., Gicquel B., Martín C. (2008) PLoS ONE 3, e3496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Frigui W., Bottai D., Majlessi L., Monot M., Josselin E., Brodin P., Garnier T., Gicquel B., Martin C., Leclerc C., Cole S. T., Brosch R. (2008) PLoS Pathog. 4, e33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li A. H., Waddell S. J., Hinds J., Malloff C. A., Bains M., Hancock R. E., Lam W. L., Butcher P. D., Stokes R. W. (2010) PLoS ONE 5, e11066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Abramovitch R. B., Rohde K. H., Hsu F. F., Russell D. G. (2011) Mol. Microbiol. 80, 678–694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Menon S., Wang S. (2011) Biochemistry 50, 5948–5957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Wang S., Engohang-Ndong J., Smith I. (2007) Biochemistry 46, 14751–14761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nowak E., Panjikar S., Konarev P., Svergun D. I., Tucker P. A. (2006) J. Biol. Chem. 281, 9659–9666 [DOI] [PubMed] [Google Scholar]
- 17. Pathak A., Goyal R., Sinha A., Sarkar D. (2010) J. Biol. Chem. 285, 34309–34318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee J. S., Krause R., Schreiber J., Mollenkopf H. J., Kowall J., Stein R., Jeon B. Y., Kwak J. Y., Song M. K., Patron J. P., Jorg S., Roh K., Cho S. N., Kaufmann S. H. (2008) Cell Host Microbe 3, 97–103 [DOI] [PubMed] [Google Scholar]
- 19. Chesne-Seck M. L., Barilone N., Boudou F., Gonzalo Asensio J., Kolattukudy P. E., Martín C., Cole S. T., Gicquel B., Gopaul D. N., Jackson M. (2008) J. Bacteriol. 190, 1329–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lejona S., Castelli M. E., Cabeza M. L., Kenney L. J., García Véscovi E., Soncini F. C. (2004) J. Bacteriol. 186, 2476–2480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bijlsma J. J., Groisman E. A. (2003) Trends Microbiol. 11, 359–366 [DOI] [PubMed] [Google Scholar]
- 22. Ferrer N. L., Gomez A. B., Neyrolles O., Gicquel B., Martin C. (2010) PLoS ONE 5, e12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Goude R., Parish T. (2008) in Mycobacteria Protocols (Parish T., Brown A. C., eds) 2nd Ed., pp. 203–215, Humana Press, London, UK [Google Scholar]
- 24. Sambrook J., Fritsch E., Maniatis T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 25. Timm J., Lim E. M., Gicquel B. (1994) J. Bacteriol. 176, 6749–6753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Dussurget O., Timm J., Gomez M., Gold B., Yu S., Sabol S. Z., Holmes R. K., Jacobs W. R., Jr., Smith I. (1999) J. Bacteriol. 181, 3402–3408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ehrt S., Guo X. V., Hickey C. M., Ryou M., Monteleone M., Riley L. W., Schnappinger D. (2005) Nucleic Acids Res. 33, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Besra G. S. (1998) Methods Mol. Biol. 101, 91–107 [DOI] [PubMed] [Google Scholar]
- 29. Bowtell D., Sambrook J. (2003) DNA Microarrays: A Molecular Cloning Manual, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Mannan M. A., Sharma S., Ganesan K. (2009) Anal. Biochem. 389, 77–79 [DOI] [PubMed] [Google Scholar]
- 31. Manganelli R., Dubnau E., Tyagi S., Kramer F. R., Smith I. (1999) Mol. Microbiol. 31, 715–724 [DOI] [PubMed] [Google Scholar]
- 32. Hu Y., Coates A. R. (1999) J. Bacteriol. 181, 469–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Miller J. H. (1972) Experiments in Molecular Genetics, Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 34. Gupta S., Pathak A., Sinha A., Sarkar D. (2009) J. Bacteriol. 191, 7466–7476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Glickman M. S., Cox J. S., Jacobs W. R., Jr. (2000) Mol. Cell 5, 717–727 [DOI] [PubMed] [Google Scholar]
- 36. Gao R., Mack T. R., Stock A. M. (2007) Trends Biochem. Sci. 32, 225–234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Gao R., Stock A. M. (2010) Curr. Opin. Microbiol. 13, 160–167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Gupta S., Sinha A., Sarkar D. (2006) FEBS Lett. 580, 5328–5338 [DOI] [PubMed] [Google Scholar]
- 39. Sinha A., Gupta S., Bhutani S., Pathak A., Sarkar D. (2008) J. Bacteriol. 190, 1317–1328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Sirakova T. D., Thirumala A. K., Dubey V. S., Sprecher H., Kolattukudy P. E. (2001) J. Biol. Chem. 276, 16833–16839 [DOI] [PubMed] [Google Scholar]
- 41. Dubey V. S., Sirakova T. D., Kolattukudy P. E. (2002) Mol. Microbiol. 45, 1451–1459 [DOI] [PubMed] [Google Scholar]
- 42. Graham J. E., Clark-Curtiss J. E. (1999) Proc. Natl. Acad. Sci. U.S.A. 96, 11554–11559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rousseau C., Neyrolles O., Bordat Y., Giroux S., Sirakova T. D., Prevost M. C., Kolattukudy P. E., Gicquel B., Jackson M. (2003) Cell. Microbiol. 5, 405–415 [DOI] [PubMed] [Google Scholar]
- 44. Rousseau C., Turner O. C., Rush E., Bordat Y., Sirakova T. D., Kolattukudy P. E., Ritter S., Orme I. M., Gicquel B., Jackson M. (2003) Infect. Immun. 71, 4684–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Converse S. E., Mougous J. D., Leavell M. D., Leary J. A., Bertozzi C. R., Cox J. S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6121–6126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Domenech P., Reed M. B., Dowd C. S., Manca C., Kaplan G., Barry C. E., 3rd (2004) J. Biol. Chem. 279, 21257–21265 [DOI] [PubMed] [Google Scholar]
- 47. Brodin P., Poquet Y., Levillain F., Peguillet I., Larrouy-Maumus G., Gilleron M., Ewann F., Christophe T., Fenistein D., Jang J., Jang M. S., Park S. J., Rauzier J., Carralot J. P., Shrimpton R., Genovesio A., Gonzalo-Asensio J. A., Puzo G., Martin C., Brosch R., Stewart G. R., Gicquel B., Neyrolles O. (2010) PLoS Pathog. 6, e1001100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Av-Gay Y., Deretic V. (2005) in Tuberculosis and the Tubercle Bacillus (Cole S. T., ed) pp. 359–367, American Society for Microbiology, Washington, D. C [Google Scholar]
- 49. Ewann F., Locht C., Supply P. (2004) Microbiology 150, 241–246 [DOI] [PubMed] [Google Scholar]
- 50. Blanco A. G., Sola M., Gomis-Rüth F. X., Coll M. (2002) Structure 10, 701–713 [DOI] [PubMed] [Google Scholar]
- 51. Zhao H., Msadek T., Zapf J., Madhusudan Hoch J. A., Varughese K. I. (2002) Structure 10, 1041–1050 [DOI] [PubMed] [Google Scholar]
- 52. Voskuil M. I., Schnappinger D., Visconti K. C., Harrell M. I., Dolganov G. M., Sherman D. R., Schoolnik G. K., (2003) J. Exp. Med. 198, 705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. He H., Zahrt T. C. (2005) J. Bacteriol. 187, 202–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sola-Landa A., Moura R. S., Martín J. F. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 6133–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Martín J. F. (2004) J. Bacteriol. 186, 5197–5201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Studier F. W., Moffatt B. A. (1986) J. Mol. Biol. 189, 113–130 [DOI] [PubMed] [Google Scholar]
- 57. Snapper S. B., Melton R. E., Mustafa S., Kieser T., Jacobs W. R., Jr. (1990) Mol. Microbiol. 4, 1911–1919 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.