Background: NDST1 and 2 are isoenzymes taking part in heparan sulfate and heparin N-sulfation during biosynthesis.
Results: NDST1+/− and NDST1−/− mast cells synthesize heparin with increased degree of sulfation.
Conclusion: Tentatively, the Golgi enzyme complex, “the GAGosome,” responsible for heparin biosynthesis, is more efficient when containing only NDST2.
Significance: Learning how biosynthesis is regulated is crucial to understanding the biological functions of heparin/heparan sulfate.
Keywords: Golgi, Heparan Sulfate, Heparin, Mast Cell, Protease, GAGosome, Heparin/Heparan Sulfate Biosynthesis, Mast Cell Protease
Abstract
Deficiency of the heparan sulfate biosynthesis enzyme N-deacetylase/N-sulfotransferase 1 (NDST1) in mice causes severely disturbed heparan sulfate biosynthesis in all organs, whereas lack of NDST2 only affects heparin biosynthesis in mast cells (MCs). To investigate the individual and combined roles of NDST1 and NDST2 during MC development, in vitro differentiated MCs derived from mouse embryos and embryonic stem cells, respectively, have been studied. Whereas MC development will not occur in the absence of both NDST1 and NDST2, lack of NDST2 alone results in the generation of defective MCs. Surprisingly, the relative amount of heparin produced in NDST1+/− and NDST1−/− MCs is higher (≈30%) than in control MCs where ≈95% of the 35S-labeled glycosaminoglycans produced is chondroitin sulfate. Lowered expression of NDST1 also results in a higher sulfate content of the heparin synthesized and is accompanied by increased levels of stored MC proteases. A model of the GAGosome, a hypothetical Golgi enzyme complex, is used to explain the results.
Introduction
Mast cells (MCs)4 can be divided into mucosal type and connective tissue type MCs. The two types of MC differ in their content of several components including MC proteases (1). In rodents, the two types of MC can also be distinguished based on glycosaminoglycan content; whereas mucosal MCs synthesize the proteoglycan serglycin with chondroitin sulfate chains, serglycin in connective tissue type MCs contains heparin (2). Also, during connective tissue type MC differentiation, glycosaminoglycan content differs between immature cells containing chondroitin sulfate and more highly differentiated cells that synthesize heparin (2).
Heparin is a highly sulfated variant of heparan sulfate (HS). Whereas heparin is found exclusively in the MC granules, HS is a ubiquitous component of cell surfaces and is also present in the extracellular matrix, predominantly in basement membranes. HS/heparin biosynthesis is a complex process with many different Golgi enzymes involved (3). The HS-polymerases EXT1 and EXT2 synthesize a polysaccharide backbone consisting of repeating units of glucuronic acid and N-acetylglucosamine. The first modifying event is the N-deacetylase/N-sulfotransferase (NDST) reaction, where N-acetyl groups of selected N-acetylglucosamine residues are removed and replaced by sulfate groups (4). After N-sulfation, the C5-epimerase converts glucuronic acid residues to iduronic acid. Sulfation at the 2-O position of iduronic acid residues and some glucuronic acid is then carried out by a 2-O-sulfotransferase, followed by glucosamine 6-O-sulfation and, more rarely, 3-O-sulfation. Little is known about the organization of the biosynthesis enzymes in the Golgi stacks. It has been suggested that the enzymes together with other yet unidentified components form GAGosomes, molecular machines responsible for elongation as well as modification of the glycosaminoglycan chains (5). Different compositions of the GAGosome may result in different modification patterns of the HS chain. In support of the GAGosome concept, several interactions between pairs of HS/heparin biosynthesis enzymes have been demonstrated. For example, the polymerases EXT1 and EXT2 are known to form a functional complex (6, 7). Interactions have also been demonstrated between the C5-epimerase and the 2-O-sulfotransferase (8) and between the xylosyltransferase and galactosyltransferase-I (9). In addition, we recently demonstrated an interaction between NDST1 and EXT2 that greatly influenced the structure of the polysaccharide formed (10).
During recent years it has become evident that HS has a key role in embryonic development (11). As first demonstrated for FGFs, the proteoglycans act as co-receptors for signaling molecules (12). In addition, HS has been shown to create and maintain gradients of morphogens and cytokines (13). Also, extracellular matrix proteins, proteases, protease inhibitors, lipases, lipoproteins, and microbial proteins are known to show affinity for HS (5). Knock-out mice have been particularly useful to study the role of HS in development. Targeted disruption of the EXT1 and EXT2 genes, respectively, leads to a complete lack of HS and an early embryonic lethality (14, 15). Four different NDSTs have been recognized (4). Both NDST1 and NDST2 have broad expression patterns and are found in most cell types and tissues during embryonic development and adult life, whereas NDST3 and NDST4 have a much more restricted expression pattern (16).
Mouse strains deficient in NDST1 (17–19) have been characterized extensively (17–31). Complete lack of NDST1 results in perinatal lethality, forebrain defects, skeletal malformation, and lung hypoplasia. Vascular development, endothelial cell function, lipid metabolism, lacrimal gland induction, lens development, and neural tube fusion have also been shown to be impaired in NDST1-deficient animals. In contrast, NDST2-deficient mice are healthy and fertile, but their connective tissue type MCs lack sulfated heparin and contain reduced levels of histamine and the MC-specific proteases, (32, 33). Also, mice with a targeted deletion of NDST3 develop normally with only subtle symptoms, such as lowered cholesterol and HDL levels (34). No knock-out strain carrying a targeted deletion of NDST4 has yet been described.
Crosses between NDST2−/− mice and NDST1+/− and analyses of their offspring demonstrated that lack of both isoforms results in early embryonic lethality (35). To be able to determine the cause of the early embryonic lethality, we recently established embryonic stem (ES) cell lines deficient for both NDST1 and NDST2 and showed that the HS produced lacks N-sulfation but contains low levels of 6-O-sulfate groups (35). In the present paper, we have studied the role of NDST1 and NDST2 in MC development in vitro. Although no MCs were formed when ES cells deficient in both NDST1 and NDST2 were differentiated in vitro, deficiency in either NDST1 or NDST2 was compatible with MC differentiation. In vitro differentiated NDST2−/− MCs had an altered morphology compared with control cells and lacked or showed reduced expression of MC proteases. These cells synthesized HS/heparin with a lower sulfation degree than polysaccharide isolated from control cells. In contrast, in vitro differentiated NDST1−/− as well as NDST1+/− MCs contained increased levels of connective tissue type MC proteases compared with control cells and synthesized heparin with a higher degree of sulfation.
EXPERIMENTAL PROCEDURES
Mice
For the generation of MCs from mouse embryos the following mouse strains were used: C57BL/6, NDST1+/− (N10 on C57BL/6) (17), and NDST1+/−2−/− (N10 on C57BL/6) (32). NDST1+/−2−/− mice were obtained by crossing NDST1+/− with NDST2−/− mice. All animal experiments were conducted with the approval of the local animal ethical committee in Uppsala.
Mast Cell Differentiation from Mouse Embryos
Embryo-derived MCs were obtained by culturing of day 10.5 to day 12.5 embryos in defined medium in a humidified cell culture chamber in 37 °C and 5% CO2 essentially as described by Vial et al. (36). The embryos were collected and transferred into a 24-well plate containing 0.5 ml of Complete medium (DMEM supplemented with 10% heat-inactivated FBS, 4 mm l-glutamine, 0.5 × 10−6 m β-mercaptoethanol, 10% NCTC 109 medium (Invitrogen), 0.1 mm nonessential amino acids, 1 mm sodium pyruvate, 50 μg/ml G418, 10 ng/ml recombinant mouse (rm) IL-3 and 25 ng/ml rmSCF). Embryo tissues were left to adhere to the plastic for 2 days and then trypsinized to disrupt embryonic structures and to get a dispersed cell suspension. Trypsinization was stopped by addition of 1.5 ml (10× the volume of trypsin) of complete medium. The dispersed cells quickly adhered to the tissue culture dish, and after ∼4 days to 1 week nonadherent cells appearing in the cultures were transferred into 6-well plates and expanded for 2–4 weeks. Aliquots of cells were removed from the developing MC culture corresponding to each embryo. Cells were put on cytospin glasses and stained with May-Grünwald/Giemsa for morphological examination of MC differentiation. In addition, DNA was purified and analyzed by PCR, as described previously (35), to define the genotype of the cultures.
Mast Cell Differentiation from Embryonic Stem Cells
NDST1−/−2−/− ES cells were cultivated as described previously (35). NDST1+/−, NDST2+/−, and WT (R1) (37) ES cells were used as controls. Before differentiation, ES cells were cultured in DMEM with GlutaMAX-1, sodium pyruvate, 4,500 mg/liter glucose, and pyridoxine (Invitrogen) supplemented with 1× nonessential amino acids (Invitrogen), 20% FCS (ES cell qualified; Invitrogen), 10−4 m β-mercaptoethanol (Sigma), and 1,000 units/ml leukemia inhibitory factor (mouse recombinant, Chemicon). To induce MC development, a slight modification of the protocol described previously by Tsai et al. was used (38). In different experiments, 2,000 or 20,000 ES cells were plated in bacterial dishes in 100-μl droplets of Iscove's modified Dulbecco's medium (Sigma) containing 15% FCS, 2 mm l-glutamine (Sigma), 5 ng/ml rmIL-11 (R&D Systems), 50 ng/ml rmSCF (Peprotech), 50 μg/ml G418 (Invitrogen), and 450 μm monothioglycerol (Sigma) to enable formation of embryoid bodies (EBs). The plating of cells on nonadherent bacterial Petri dishes is referred to as day 0. Two to 3 days after the initiation of EB formation, more medium was added to achieve a floating EB culture. Six days after initiation, the EB cultures were boosted with 1 volume of Iscove's modified Dulbecco's medium containing 15% FCS, 2 mm l-glutamine, 30 ng/ml rmIL-6 (Peprotech), 30 ng/ml rmIL-3 (Peprotech), 50 ng/ml rmSCF (Peprotech), 50 μg/ml G418, and 450 μm monothioglycerol (Sigma). At day 12, the EBs were transferred to tissue culture plates for adherent growth and differentiation in a defined MC medium (DMEM containing 10% FCS, 2 mm l-glutamine, 50 μg/ml G418, and 450 μm monothioglycerol (Sigma), 50 ng/ml rmSCF (Peprotech), and 30% WEHI-3 cell conditioned medium as an IL-3 supplement. Once a week, half of the medium containing the newly developed MCs was removed from the dishes, and new medium was added. Transferred nonadherent cells were grown continuously in MC medium. Starting from 2 weeks, an aliquot was checked weekly by May-Grünwald/Giemsa for MC morphology. The derived MCs were used for different experiments from week 5 to 9 after the initiation of MC differentiation.
Quantitative PCR Analysis
RNA purification from MCs, ES cells, and mouse 11-day embryo total RNA (Clontech) was performed with the E.Z.N.A. total RNA kit (Omega). The amount of RNA obtained was estimated based on absorbance at 260 nm. One μg of RNA was used as template for cDNA synthesis using SuperscriptTM II (Invitrogen,) reverse transcriptase with random hexamers. The Bio-Rad MiniOpticon system together with the Bio-Rad enzyme mix was used for quantitative PCR analysis. cDNA was amplified using the following primers: NDST1 (226 bp) forward, 5′-CCA CAA CTA TCA CAA AGG CAT CG-3′ and reverse, 5′-GAA AGG TTG ACT TTA GGG CCA C-3′; NDST2 (240 bp) forward, 5′-GTG TGG CAG AAT CCC TGT G-3′ and reverse, 5′-GTG CAG GCT CAG GAA GAA GT-3′; NDST3 (143 bp) forward, 5′-GGA GCT CTT CTT CAC TGT GGT T-3′ and reverse, 5′-TCT GAA GAC GCA GGT TGG T-3′; NDST4 (170 bp) forward, 5′-GGA GAA AAC CTG TGA CCA TTT AC-3′ and reverse, 5′-CCT TGT GAT AGT TGT TGC CAT TA-3′. The PCR was performed by denaturating the DNA at 95 °C for 10 min before amplification for 40 cycles, each cycle consisting of 95 °C, 30 s; 60 °C, 30 s; 72 °C, 30 s.
Western Blot Analysis
Approximately 1 × 106 MCs were solubilized in 100 μl of 1 × SDS-PAGE sample buffer containing 5% β-mercaptoethanol. Equal volumes of the cell extracts were subjected to SDS-PAGE on 12% gels. The separated proteins were then blotted onto nitrocellulose membranes and blocked with 5% milk powder in TBS/0.1% Tween 20 for 1 h at room temperature. Following blocking, the membranes were incubated with antisera against carboxypeptidase A3 (CPA3), the chymase MCPT5 (also designated mMCP-5), and the tryptase MCPT6 (also designated mMCP-6), diluted 1:2,000 in TBS/2% BSA/0.1% Tween 20, at 40 °C overnight (the antisera were kindly provided by Dr. Lars Hellman, Uppsala University). Membranes were then washed extensively with TBS/0.1% Tween 20, and the secondary anti-rabbit antibody conjugated with horseradish peroxidase was left to bind for 1 h. After extensive washing, the blots were developed using a Bio-Rad detection system.
Glycosaminoglycan Isolation and Analysis
35S-Labeled Glycosaminoglycans
5 × 106 MCs of relevant genotypes, cultured in the MC-inducing medium described above, were metabolically labeled overnight with 200 μCi of carrier-free [35S]sulfate (Amersham Biosciences). After incubation, cells were pelleted by centrifugation for 10 min at 300 × g, washed with cold PBS, incubated in 2 ml of solubilization buffer (50 mm Tris-HCl, pH 7.5, 1% Triton X-100, 0.10 m NaCl), and centrifuged at 800 × g for 15 min. The supernatant containing radiolabeled macromolecules was recovered, and 35S-labeled glycosaminoglycans were isolated from the solubilized cell lysate on a 0.3-ml column of DEAE-Sephacel (Amersham Biosciences), equilibrated with 50 mm Tris-HCl, pH 7.4, 0.1% Triton X-100, 0.10 m NaCl. After washing the column with equilibration buffer followed by a second washing step with 50 mm acetate buffer, pH 4.0, containing 0.1% Triton X-100 and 0.10 m NaCl, the 35S-labeled glycosaminoglycans were eluted with 50 mm acetate buffer, pH 4.0, containing 0.1% Triton X-100 and 2 m NaCl. A portion of the eluted 35S-labeled glycosaminoglycans was treated with alkali (0.5 m NaOH) as described previously (39). After desalting in water on PD10 columns (Amersham Biosciences), followed by lyophilization, the 35S-labeled glycosaminoglycan chains were subjected to digestion with 0.1 unit of chondroitinase ABC (Seikagaku) as described previously (40) or treated with nitrous acid at pH 1.5 (41). Untreated and treated 35S-labeled glycosaminoglycans were then analyzed by gel chromatography on Sephadex G50 eluted with 0.2 m NH4HCO3.
Unlabeled Glycosaminoglycans
Cells (10 × 106) were dissolved in 0.5 ml of Pronase buffer (1% Triton X-100, 50 mm Tris-HCl, pH 8.0, 1 mm CaCl2, 0.8 mg/ml Pronase) and incubated end-over-end for 19 h at 55 °C. After heat inactivation of the enzyme for 5 min at 96 °C, MgCl2 was added to a final concentration of 2 mm. After addition of Benzonase (12 milliunits), the sample was incubated for 2 h at 37 °C followed by heat inactivation for 5 min at 96 °C. The NaCl concentration was then adjusted to 0.1 m, and the sample was centrifuged at 13,000 × g for 10 min. The supernatant was diluted with 0.5 ml of 50 mm Tris-HCl, pH 8.0, 0.1 m NaCl, and applied to a Sep-Pak® C18 cartridge (Waters) that had been primed first with methanol, then with water, and finally with 50 mm Tris-HCl, pH 8.0, 0.1 m NaCl. The cartridge was washed with 2 ml of the Tris-HCl buffer, and the washing fraction was combined with the nonbinding fraction and used for purification of glycosaminoglycans. The sample was applied to a 0.2-ml DEAE-Sephacel column equilibrated in loading buffer (50 mm Tris-HCl, pH 8, 0.1 m NaCl, 0.1% Triton X-100). After washing with 6 column volumes of loading buffer, 6 volumes of low pH buffer (50 mm sodium acetate, pH 4.0, 0.1 m NaCl, 0.1% Triton X-100), and 6 volumes of loading buffer without Triton X-100, the glycosaminoglycans were eluted with 0.6 ml of elution buffer (50 mm Tris-HCl, pH 8.0, 1.5 m NaCl). After desalting on an NAP-10 column (Amersham Biosciences) equilibrated in water, the glycosaminoglycans were dried by SpeedVac centrifugation. The glycosaminoglycan pool was then digested with 50 milliunits of chondroitinase ABC in 100 μl of 40 mm Tris acetate buffer, pH 8.0. The digestion was allowed to proceed for 4 h at 37 °C, and the sample was then boiled to stop the reaction. After removal of 10 μl for chondroitin sulfate analysis by RPIP-HPLC, as described previously (20), heparin/HS was recovered after a second round of DEAE-Sephacel chromatography, as described for total glycosaminoglycan isolation. Purified heparin/HS was dissolved in 200 μl of heparinase buffer (5 mm Hepes buffer, pH 7.0, 50 mm NaCl, 1 mm CaCl2) and divided into two equal aliquots. One of the aliquots was treated with 0.4 milliunit each of heparinases I, II, and III and incubated for 16 h at 37 °C. The other aliquot (control sample) was incubated under the same conditions without enzymes. After heat inactivation for 5 min at 96 °C, the samples were analyzed by RPIP-HPLC (20).
Statistical Analyses
Statistical analyses were performed using Prism 5.0 (GraphPad Software, San Diego, CA). Significant differences were determined using an unpaired t test. Results are presented as mean ± S.E. and calculated p values indicated as not significant (ns) p > 0.05 or significant *, p < 0.05.
RESULTS
Embryo-derived Mast Cells from NDST1+/− and NDST1+/−/2−/− Intercrosses
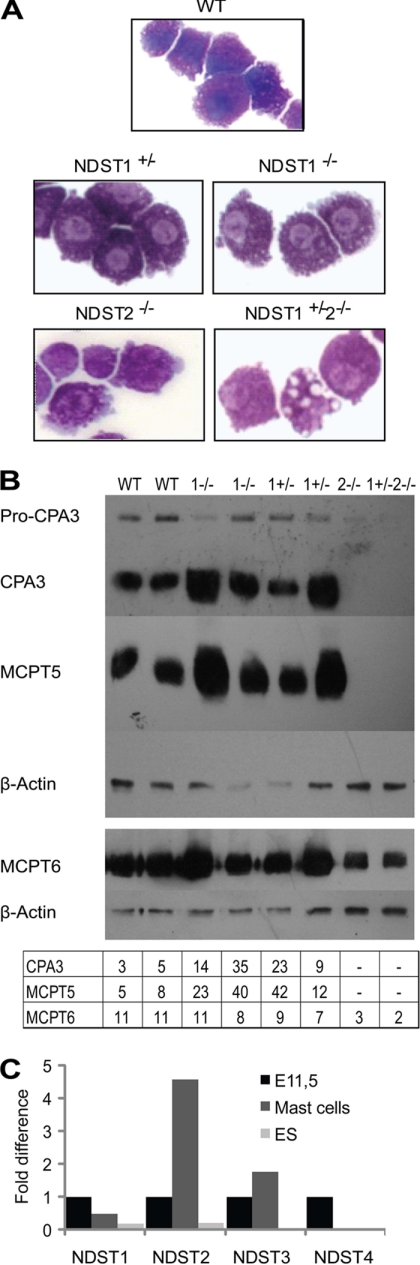
Using an in vitro cultivation protocol (36), embryos obtained from NDST1+/− and NDST1+/−2−/− intercrosses, isolated at embryonic (E) stages E10.5 to E12.5, were used as a starting point for differentiation of embryo-derived MCs. Because NDST1−/−2−/− embryos do not survive beyond E5.5, this genotype was not represented among the cultures. The embryos were placed into 24-well plates, and after 2 days of in vitro cultivation the embryo tissues were dispersed by trypsinization, and the cells were left to adhere again. Approximately 4 days to 1 week after readherence, MC differentiation was observed in all genotypes tested. No major morphological differences could be seen between cells of the different genotypes except for the presence of some “empty” vacuoles in the cells lacking NDST2 (Fig. 1A).
FIGURE 1.
Embryo-derived MCs. A, May Grünwald/Giemsa staining of WT, NDST1+/−, NDST1−/−, NDST2−/−, and NDST1+/−2−/− embryo-derived MCs. B, expression of MC proteases analyzed by Western blotting as described under “Experimental Procedures.” Two separate runs are shown. β-Actin expression served as a loading control. In the table below the blot, the level of expression of CPA3, MCPT5, and MCPT6 compared with β-actin is shown. C, quantitative PCR of NDST expression. Data are displayed as the -fold difference between expression of the respective NDST isoform in total embryo at day E11.5 (set at 1.0) and WT embryo-derived MCs and undifferentiated ES cells, respectively. C(t) values for the different samples were: NDST1, E11.5: 8.29, MCs: 9.32, ES cells: 10.73; NDST2, E11.5: 5.87, MCs: 3.68, ES cells: 8.10; NDST3, E11.5: 8.17, MCs: 7.35, ES cells: 13.26. NDST4, E11.5: 13.59. The experiment has been repeated with similar results.
Analysis of the MC proteases at the protein level demonstrated that the WT embryo-derived MCs resemble true connective tissue type MCs and express CPA3 as well as high levels of the chymase MCPT5 and the tryptase MCPT6 (Fig. 1B). In agreement with previous work (32, 33), cells lacking NDST2, the NDST2−/−, and NDST1+/−2−/− embryo-derived MCs were devoid of activated CPA3 and MCPT5 and contained reduced levels of MCPT6 (Fig. 1B). Remarkably, NDST1−/− as well as NDST1+/− MCs seemed to store higher amounts of CPA3 and MCPT5 than WT MCs (Fig. 1B).
Increased Heparin Synthesis in NDST1−/− and NDST1+/− Embryo-derived MCs
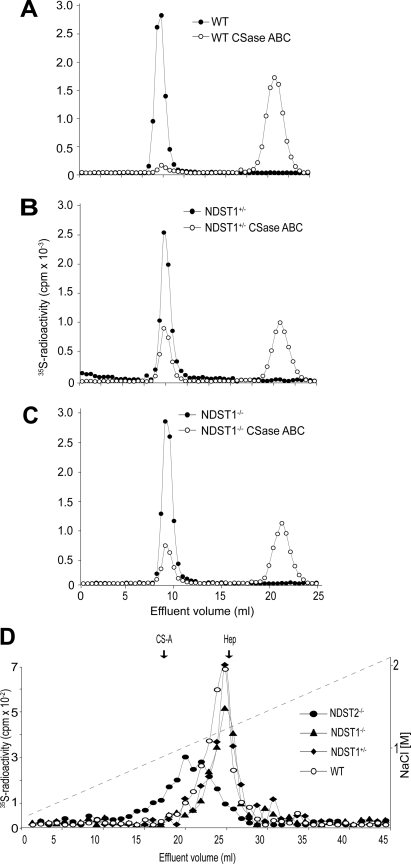
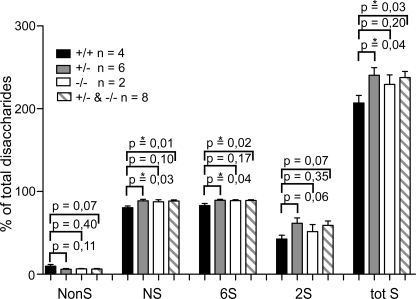
Characterization of the glycosaminoglycans produced by the MCs in the presence of [35S]sulfate also gave interesting results. Compared with WT MCs where only 4% of the total 35S-labeled glycosaminoglycans resisted degradation with chondroitinase ABC and thus was identified as heparin/HS, ∼30% of the glycosaminoglycans produced by NDST1−/− and NDST1+/− cells was heparin/HS (Fig. 2, A–C). Analysis with ion exchange chromatography of the chondroitinase ABC-resistant 35S-glycosaminoglycans (representing HS and/or heparin) demonstrated that the NDST2-deficient cells produced HS with a lower charge density (Fig. 2D). In contrast, 35S-labeled heparin/HS produced by NDST1−/− and NDST1+/− MCs were eluted at higher ionic strength and were more homogeneous than WT heparin/HS, which in addition contained some material of lower charge density. To characterize in more detail the structure of the heparin/HS produced by the embryo-derived MCs, glycosaminoglycans from the cells were analyzed by RPIP-HPLC (20). Glycosaminoglycans from four WT, six NDST1+/−, and two NDST1−/− MC cultures were separately analyzed (Fig. 3 and supplemental Fig. 1). Confirming the results obtained with ion exchange chromatography (Fig. 2D), the total sulfation of heparin/HS isolated from NDST1+/− and NDST1−/− cells is higher than that of heparin/HS from WT cells. Importantly, N-sulfation increased with ≈10%. Because the 2- and 6-O-sulfotransferases are largely dependent on N-sulfation for substrate recognition, an increase in O-sulfation would be expected. This was also the case, resulting in an overall increase in total sulfation with ≈15% (Fig. 3). Chondroitin sulfate sulfation was, as expected, largely unaffected by NDST1 ablation, even though a smaller increase in 4-O-sulfation was noted (supplemental Fig. 1).
FIGURE 2.
Altered 35S-labeled heparin/HS production in NDST1-deficient MCs. A–C, analytical gel chromatography on Sephadex G50 of 35S-labeled glycosaminoglycans from WT (A), NDST1+/− (B), and NDST1−/− MCs (C) before (filled circles) and after treatment with chondroitinase ABC (CSase ABC; open circles). The column was eluted in 0.2 m NH4HCO3, and fractions of 0.5 ml were collected and analyzed for [35S] radioactivity. D, ion exchange chromatography on Mono Q of 35S-labeled heparin/HS isolated from WT (filled circles), NDST2−/− (filled diamonds), NDST1−/− (filled triangles), and NDST1+/− (open circles) as described under “Experimental Procedures.”. The samples were applied to a Mono Q column (Amersham Biosciences) equilibrated in 50 mm Tris-HCl, pH 8.0, 0.4 m NaCl and eluted in the same buffer with a gradient ranging from 0.4 to 2.1 m NaCl. The vertical arrows indicate the elution position of chondroitin sulfate (CS-A) and heparin (Hep) standards.
FIGURE 3.
Increased sulfation of heparin/HS in NDST1+/− and NDST1−/− MCs. Heparin/HS disaccharide composition was determined by RPIP-HPLC as described under “Experimental Procedures,” and the percentage of heparin/HS disaccharides with different modifications was calculated from the data (see detailed disaccharide composition in supplemental Fig. 1). Mean values ± S.E. (error bars) were calculated from separate analyses of glycosaminoglycans from four WT (black columns), six NDST1+/− (gray columns), and two NDST1−/− MC cultures (white columns). The striped columns represent the mean values obtained for the six NDST1+/− and the two NDST1−/− analyses added together. NonS, percentage of disaccharides that do not contain any sulfate group; NS, percentage of disaccharides that are N-sulfated; 6S, percentage of disaccharides that contain a 6-O-sulfate group; 2S, percentage of disaccharides that contain a 2-O-sulfate group; tot S, NS + 6S + 2S; *, significant difference (p < 0.05).
Expression of NDST Transcripts in Embryo-derived MCs
To investigate whether the increased sulfation of heparin in the NDST1-deficient MCs was due to increased levels of any of the other NDST isoforms, their expression was quantified by real-time PCR. As shown in Fig. 1C for embryo-derived MCs, the expression of NDST1 transcript in WT was similar to the expression of this isoform in whole embryo. This was also the case for NDST3 expression, whereas ≈5 times more NDST2 transcript was expressed in the MCs compared with the whole embryo (Fig. 1C). No, or very low levels of NDST4 were detected in the embryo-derived MCs. As a comparison, WT embryonic stem cells produced lower levels of both NDST1 and NDST2 transcripts than whole embryo, and NDST3 and NDST4 expression was below detection (Fig. 1C). In NDST1−/− and NDST1+/− MCs, the level of NDST3 was slightly increased whereas the level of NDST2 transcript was unaltered, and the level of NDST4 expression too low to be quantified (Table 1). Altered expression of other NDST isoforms thus did not seem to explain the increased heparin sulfation in NDST1−/− MCs.
TABLE 1.
Expression of NDST2 and NDST3 in embryo-derived mast cells
Transcript levels were quantitated with real-time PCR as described under “Experimental Procedures.” Mean values of two determinations are shown. The experiment has been repeated once with similar results, which have also been confirmed by RT-PCR.
| Genotype | NDST2 |
NDST3 |
||
|---|---|---|---|---|
| ΔCT (CT NDST2 − CT actin) | -Fold difference/average WT = 1 | ΔCT (CT NDST3 − CT actin) | -Fold difference/average WT = 1 | |
| WT | 4.8 ± 0.4 | 1.0 ± 0.3 | 9.3 ± 0.1 | 1.0 ± 0.04 |
| NDST1−/− | 4.8 ± 0.1 | 1.0 ± 0.1 | 8.2 ± 0.2 | 2.2 ± 0.3 |
| NDST1+/− | 4.1 ± 0.5 | 1.7 ± 0.5 | 8.5 ± 0.2 | 1.7 ± 0.2 |
MC Differentiation Capacity of NDST1/2-deficient ES Cells
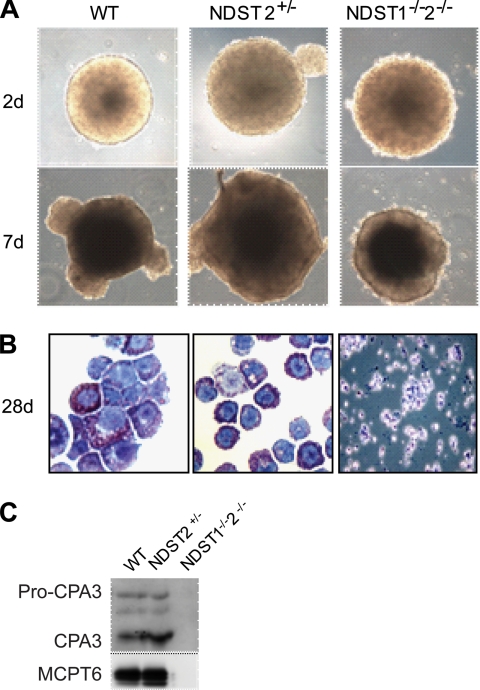
Next we investigated how lack of both NDST1 and 2 affects MC differentiation. Because NDST1/2-deficient embryos die around day E6, we used in vitro differentiation of established NDST1/2-deficient ES cells, taking advantage of a protocol described by Tsai et al. (38). To initiate differentiation, ES cells of different genotypes were first aggregated in nonadherent bacterial Petri dishes. The ES cells tested included NDST1−/−2−/− (three separately isolated clones), NDST1+/−, NDST2+/−, and control WT ES cells (the ES cell clones R1 and GSI (Genome Systems), as well as in-house established WT ES cells from C57BL/6 mice blastocysts). Forming embryoid bodies were left to maturate for 12–14 days in defined medium. ES cells of all the different genotypes had equal aggregation properties, as seen in Fig. 4A. After replating the EBs for adherent growth, we observed the rapid differentiation of fibroblast-like cells growing out from the EB center in all except in the NDST1−/−2−/− cultures. On these fibroblast-like cells, small rounded cells were normally seen after 1 week. The small adherent cells continued to differentiate, and with increased size they detached from the feeder layer. The differentiating cultures were screened weekly for MC development by morphological examination. After transfer to a new plate the cells expanded rapidly. After 2–3 weeks of culturing in defined medium, >95% of the cells showed MC morphology as they stained positively with May-Grünwald/Giemsa. However, in contrast, the EBs deficient in both NDST1 and NDST2 lost their defined outer border and displayed a rugged surface phenotype (Fig. 4A). They also showed poor adhesion when plated into cell culture dishes, and no MCs were observed in the NDST1−/−2−/− cytospun slide stainings. Instead, in the defined medium the NDST1−/−2−/− EBs disintegrated, resulting in cultures filled with small poorly differentiated and fragmented apoptotic cells (Fig. 4B). The WT controls and EBs heterozygous for either NDST1 or NDST2 jumped into MC differentiation within the first week after replating (Fig. 4B). To evaluate the degree of MC development further we checked for the expression of the MC proteases MCPT6 and CPA3. All control cultures expressed MCPT6 and CPA3, whereas no expression of these proteases was seen in the NDST1/2-deficient MC cultures (Fig. 4C).
FIGURE 4.
ES cell-derived MCs. A, EBs of different genotypes cultured in suspension photographed at day 2 and at day 7 after plating. B, May Grünwald/Giemsa staining of WT and NDST2+/− MCs, and NDST1−/−2−/− cells from EB outgrowths placed on cytospin glasses. C, MC protease expression analyzed by Western blotting as described under “Experimental Procedures.”
DISCUSSION
Work published more than 10 years ago demonstrated that the NDST2 enzyme is required for heparin synthesis in MCs (32, 33). The number of connective tissue type MCs in mice lacking this enzyme is reduced, and the MCs formed contain dramatically reduced numbers of granules, less histamine, and they lack MC proteases. We now show that deficiency of the isoenzyme NDST1 in embryo-derived MCs has the opposite effects. The heparin production in the embryo-derived NDST1−/− MCs increases from a few percent to ∼30% of total glycosaminoglycans (Fig. 2, A–C), and the sulfate content of the heparin synthesized is increased (Figs. 2D and 3). Most likely as a result of the altered heparin biosynthesis, the cells contain elevated levels of proteases characteristic for connective tissue type MCs (Fig. 1B). How is this possible?
NDST1 and NDST2 both have N-deacetylase as well as N-sulfotransferase activities and are co-expressed in most tissues and cell types (42). Therefore, the restricted phenotype of the NDST2−/− mice, where only connective tissue type MCs were affected, was surprising (32, 33). Detailed analyses of a number of different organs of NDST2−/− mice failed to identify any changes in HS structure (20). In contrast, NDST1 deficiency resulted in a systemic effect on HS biosynthesis (17, 21).
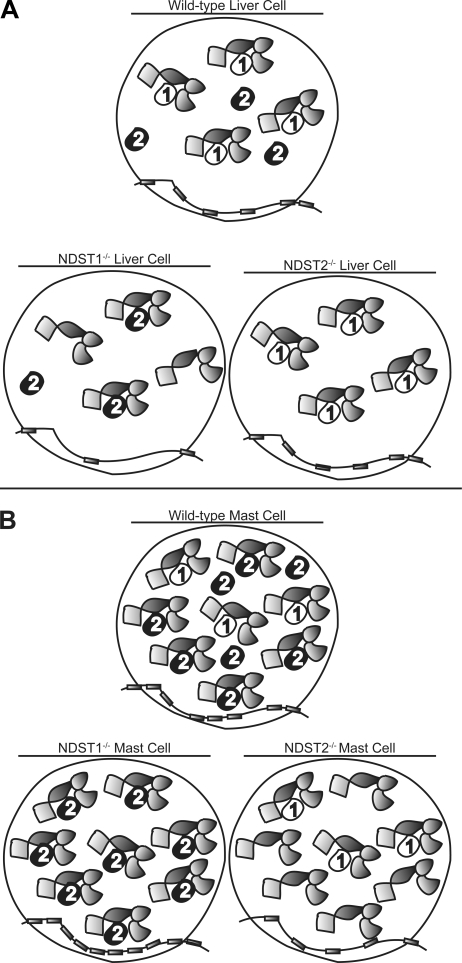
As discussed above, NDST1 appears to have a vital role for HS synthesis in most cells whereas NDST2 is crucial for heparin sulfation in MCs. We also know that the degree of N-sulfation depends both on isoform and the level of enzyme expressed; overexpression of NDST1 and NDST2 in 293 cells both result in increased HS N-sulfation, but the level of N-sulfation reached is higher in NDST2-overexpressing cells (40, 43). Investigating the roles of NDST1 and NDST2 in liver HS biosynthesis, we previously could conclude that in the absence of NDST1 (and NDST3 and 4, not expressed in the liver), NDST2 was responsible for HS N-sulfation (21). In NDST2−/− liver, only NDST1 was active. The structure of HS in control and NDST2−/− liver was the same, whereas liver HS from NDST1−/− mice was undersulfated. Similar to the MCs (Table 1), disruption of the gene for one NDST isoform did not influence expression of the other NDSTs. Based on these results, we hypothesized that NDST1 was preferred for incorporation into the GAGosome and that NDST2 only in the absence of NDST1 would be present in the enzyme complex (Fig. 5A). We argue that the same hypothesis can explain the results presented in this paper (Fig. 5B). In control MCs, much more NDST2 than NDST1 is expressed (Fig. 1C) (42), and NDST2 will therefore be the dominating isoform in the GAGosomes, despite its lower preference for GAGosome incorporation. Because the level of NDST enzymes is higher in MCs than in liver cells, the polysaccharide produced will have a high content of sulfate groups. In the NDST1−/− and NDST1+/− MCs, even more GAGosomes will house NDST2, and the heparin produced will be more heavily sulfated than in the control cells (Figs. 2D and 3). In contrast, low sulfated HS will be produced by the NDST2-deficient MCs, because only low levels of NDST1 are expressed by the cells (Fig. 2D).
FIGURE 5.
GAGosome model. A, in the WT liver cell both NDST1 and NDST2 are expressed. NDST1 is preferred as GAGosome component leaving NDST2 outside the functional enzyme complex. Because NDST2−/− cells and WT cells both have identical NDST1-containing GAGosomes, they synthesize HS with identical structure (21). A few of the GAGosomes in NDST1−/− cells contain NDST2 and synthesize a low sulfated HS. B, in the heparin/HS biosynthesis machinery of WT MCs, NDST2 is the dominating NDST isoform, but a few GAGosomes contain NDST1. In the NDST2−/− cells, only the remaining NDST1-containing GAGosomes can modify the heparin/HS chains, and a low sulfated product is synthesized. When NDST1 is lacking, all GAGosomes contain NDST2, and a polysaccharide with a sulfate content exceeding that of heparin produced by WT MCs is produced.
The increased sulfation of heparin in the NDST1−/− and NDST1+/− MCs was paralleled by an increased amount of CPA3 and MCPT5 (Fig. 1B). We hypothesize that the increased levels of proteases simply is due to the availability of a greater number of heparin binding sites for the enzymes. In the NDST2−/− MCs obtained after differentiation, the level of MC-specific proteases, as shown by Western blotting, was low compared with control cells (Fig. 1B). Similar results were obtained previously when freshly isolated peritoneal MCs were studied (32). In this paper it was also demonstrated that although the proteases were absent from the granules, the protease transcript levels were the same in control and NDST2-deficient cells. It was speculated that the high negative charge of the normally sulfated heparin was required for the assembly of the positively charged proteases. The lowered sulfation of the heparin/HS polysaccharide synthesized by the NDST2−/− MCs (Fig. 2D) could thus explain the decreased levels of proteases in these cells.
As shown in Fig. 1B, the MCs devoid of NDST2−/− contain low amounts of pro-CPA3 but lack the ability to process the zymogen into active enzyme. This is in line with previous observations using bone marrow-derived MCs (44, 45). Thus, NDST2-driven heparin synthesis seems to be essential for correct processing of CPA3. However, the exact mechanism of activation remains elusive, although cathepsin E has been suggested to play a major role (46).
Finally, the inability of ES cells lacking both NDST1 and NDST2 to form MCs is probably unrelated to GAGosome composition but may instead reflect a defect in mesodermal differentiation (47). Interestingly, we have recently shown that the ES cells lacking both NDST1 and NDST2 can form osteoblasts but not adipocytes,5 demonstrating that the defect is not general.
Supplementary Material
This work was supported by the Swedish Research Council (to L. K. and M. Å.), the Swedish Research Council Formas (to M. Å.), the Swedish Cancer Society (to L. K.), the Göran Gustafsson Foundation (to M. Å.), Gustaf V:s 80-årsfond (to L. K.), the Magnus Bergvall Foundation (to M. Å.), and Polysackaridforskning AB (to L. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
M. Forsberg, K. Holmborn, A. Dagälv, L. Kjellén, and K. Forsberg-Nilsson, unpublished data.
- MC
- mast cell
- CPA3
- carboxypeptidase A3
- E
- embryonic day
- EB
- embryoid body
- HS
- heparan sulfate
- MCPT
- mouse mast cell protease
- NDST
- N-deacetylase/N-sulfotransferase
- rm
- recombinant mouse
- SCF
- stem cell factor
- RPIP
- reversed-phase ion pair.
REFERENCES
- 1. Gurish M. F., Austen K. F. (2001) J. Exp. Med. 194, F1–5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Kolset S. O., Prydz K., Pejler G. (2004) Biochem. J. 379, 217–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Lindahl U., Kusche-Gullberg M., Kjellén L. (1998) J. Biol. Chem. 273, 24979–24982 [DOI] [PubMed] [Google Scholar]
- 4. Grobe K., Ledin J., Ringvall M., Holmborn K., Forsberg E., Esko J. D., Kjellén L. (2002) Biochim. Biophys. Acta 1573, 209–215 [DOI] [PubMed] [Google Scholar]
- 5. Esko J. D., Selleck S. B. (2002) Annu. Rev. Biochem. 71, 435–471 [DOI] [PubMed] [Google Scholar]
- 6. McCormick C., Duncan G., Goutsos K. T., Tufaro F. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 668–673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Senay C., Lind T., Muguruma K., Tone Y., Kitagawa H., Sugahara K., Lidholt K., Lindahl U., Kusche-Gullberg M. (2000) EMBO Rep. 1, 282–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Pinhal M. A., Smith B., Olson S., Aikawa J., Kimata K., Esko J. D. (2001) Proc. Natl. Acad. Sci. U.S.A. 98, 12984–12989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Schwartz N. B., Rodén L., Dorfman A. (1974) Biochem. Biophys. Res. Commun. 56, 717–724 [DOI] [PubMed] [Google Scholar]
- 10. Presto J., Thuveson M., Carlsson P., Busse M., Wilén M., Eriksson I., Kusche-Gullberg M., Kjellén L. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 4751–4756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lin X. (2004) Development 131, 6009–6021 [DOI] [PubMed] [Google Scholar]
- 12. Pellegrini L. (2001) Curr. Opin. Struct. Biol. 11, 629–634 [DOI] [PubMed] [Google Scholar]
- 13. Lander A. D., Nie Q., Wan F. Y. (2002) Dev. Cell 2, 785–796 [DOI] [PubMed] [Google Scholar]
- 14. Lin X., Wei G., Shi Z., Dryer L., Esko J. D., Wells D. E., Matzuk M. M. (2000) Dev. Biol. 224, 299–311 [DOI] [PubMed] [Google Scholar]
- 15. Stickens D., Zak B. M., Rougier N., Esko J. D., Werb Z. (2005) Development 132, 5055–5068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Aikawa J., Grobe K., Tsujimoto M., Esko J. D. (2001) J. Biol. Chem. 276, 5876–5882 [DOI] [PubMed] [Google Scholar]
- 17. Ringvall M., Ledin J., Holmborn K., van Kuppevelt T., Ellin F., Eriksson I., Olofsson A. M., Kjellen L., Forsberg E. (2000) J. Biol. Chem. 275, 25926–25930 [DOI] [PubMed] [Google Scholar]
- 18. Fan G., Xiao L., Cheng L., Wang X., Sun B., Hu G. (2000) FEBS Lett. 467, 7–11 [DOI] [PubMed] [Google Scholar]
- 19. Grobe K., Inatani M., Pallerla S. R., Castagnola J., Yamaguchi Y., Esko J. D. (2005) Development 132, 3777–3786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ledin J., Staatz W., Li J. P., Götte M., Selleck S., Kjellén L., Spillmann D. (2004) J. Biol. Chem. 279, 42732–42741 [DOI] [PubMed] [Google Scholar]
- 21. Ledin J., Ringvall M., Thuveson M., Eriksson I., Wilén M., Kusche-Gullberg M., Forsberg E., Kjellén L. (2006) J. Biol. Chem. 281, 35727–35734 [DOI] [PubMed] [Google Scholar]
- 22. Wang L., Fuster M., Sriramarao P., Esko J. D. (2005) Nat. Immunol. 6, 902–910 [DOI] [PubMed] [Google Scholar]
- 23. Pan Y., Woodbury A., Esko J. D., Grobe K., Zhang X. (2006) Development 133, 4933–4944 [DOI] [PubMed] [Google Scholar]
- 24. Pallerla S. R., Pan Y., Zhang X., Esko J. D., Grobe K. (2007) Dev. Dyn. 236, 556–563 [DOI] [PubMed] [Google Scholar]
- 25. Fuster M. M., Wang L., Castagnola J., Sikora L., Reddi K., Lee P. H., Radek K. A., Schuksz M., Bishop J. R., Gallo R. L., Sriramarao P., Esko J. D. (2007) J. Cell Biol. 177, 539–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. MacArthur J. M., Bishop J. R., Stanford K. I., Wang L., Bensadoun A., Witztum J. L., Esko J. D. (2007) J. Clin. Invest. 117, 153–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Hu Z., Yu M., Hu G. (2007) Bone 40, 1462–1474 [DOI] [PubMed] [Google Scholar]
- 28. Abramsson A., Kurup S., Busse M., Yamada S., Lindblom P., Schallmeiner E., Stenzel D., Sauvaget D., Ledin J., Ringvall M., Landegren U., Kjellén L., Bondjers G., Li J. P., Lindahl U., Spillmann D., Betsholtz C., Gerhardt H. (2007) Genes Dev. 21, 316–331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pan Y., Carbe C., Powers A., Zhang E. E., Esko J. D., Grobe K., Feng G. S., Zhang X. (2008) Development 135, 301–310 [DOI] [PubMed] [Google Scholar]
- 30. Hu Z., Wang C., Xiao Y., Sheng N., Chen Y., Xu Y., Zhang L., Mo W., Jing N., Hu G. (2009) J. Cell Sci. 122, 1145–1154 [DOI] [PubMed] [Google Scholar]
- 31. Ringvall M., Kjellén L. (2010) Prog. Mol. Biol. Transl. Sci. 93, 35–58 [DOI] [PubMed] [Google Scholar]
- 32. Forsberg E., Pejler G., Ringvall M., Lunderius C., Tomasini-Johansson B., Kusche-Gullberg M., Eriksson I., Ledin J., Hellman L., Kjellén L. (1999) Nature 400, 773–776 [DOI] [PubMed] [Google Scholar]
- 33. Humphries D. E., Wong G. W., Friend D. S., Gurish M. F., Qiu W. T., Huang C., Sharpe A. H., Stevens R. L. (1999) Nature 400, 769–772 [DOI] [PubMed] [Google Scholar]
- 34. Pallerla S. R., Lawrence R., Lewejohann L., Pan Y., Fischer T., Schlomann U., Zhang X., Esko J. D., Grobe K. (2008) J. Biol. Chem. 283, 16885–16894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Holmborn K., Ledin J., Smeds E., Eriksson I., Kusche-Gullberg M., Kjellén L. (2004) J. Biol. Chem. 279, 42355–42358 [DOI] [PubMed] [Google Scholar]
- 36. Vial D., Oliver C., Jamur M. C., Pastor M. V., da Silva Trindade E., Berenstein E., Zhang J., Siraganian R. P. (2003) J. Immunol. 171, 6178–6186 [DOI] [PubMed] [Google Scholar]
- 37. Nagy A., Rossant J., Nagy R., Abramow-Newerly W., Roder J. C. (1993) Proc. Natl. Acad. Sci. U.S.A. 90, 8424–8428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Tsai M., Wedemeyer J., Ganiatsas S., Tam S. Y., Zon L. I., Galli S. J. (2000) Proc. Natl. Acad. Sci. U.S.A. 97, 9186–9190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Bengtsson J., Eriksson I., Kjellén L. (2003) Biochemistry 42, 2110–2115 [DOI] [PubMed] [Google Scholar]
- 40. Cheung W. F., Eriksson I., Kusche-Gullberg M., Lindhal U., Kjellén L. (1996) Biochemistry 35, 5250–5256 [DOI] [PubMed] [Google Scholar]
- 41. Shively J. E., Conrad H. E. (1976) Biochemistry 15, 3943–3950 [DOI] [PubMed] [Google Scholar]
- 42. Kusche-Gullberg M., Eriksson I., Pikas D. S., Kjellén L. (1998) J. Biol. Chem. 273, 11902–11907 [DOI] [PubMed] [Google Scholar]
- 43. Pikas D. S., Eriksson I., Kjellén L. (2000) Biochemistry 39, 4552–4558 [DOI] [PubMed] [Google Scholar]
- 44. Henningsson F., Ledin J., Lunderius C., Wilén M., Hellman L., Pejler G. (2002) Biol. Chem. 383, 793–801 [DOI] [PubMed] [Google Scholar]
- 45. Henningsson F., Hergeth S., Cortelius R., Abrink M., Pejler G. (2006) FEBS J. 273, 4901–4912 [DOI] [PubMed] [Google Scholar]
- 46. Henningsson F., Yamamoto K., Saftig P., Reinheckel T., Peters C., Knight S. D., Pejler G. (2005) J. Cell Sci. 118, 2035–2042 [DOI] [PubMed] [Google Scholar]
- 47. Jakobsson L., Kreuger J., Holmborn K., Lundin L., Eriksson I., Kjellén L., Claesson-Welsh L. (2006) Dev. Cell 10, 625–634 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.