Abstract
Gonadotropin-regulated testicular RNA helicase (GRTH/DDX25), a testis-specific member of the DEAD-box family, is an essential post-transcriptional regulator of spermatogenesis. Failure of expression of Transition protein 2 (TP2) and Protamine 2 (Prm2) proteins (chromatin remodelers, essential for spermatid elongation and completion of spermatogenesis) with preservation of their mRNA expression was observed in GRTH-null mice (azoospermic due to failure of spermatids to elongate). These were identified as target genes for the testis-specific miR-469, which is increased in the GRTH-null mice. Further analysis demonstrated that miR-469 repressed TP2 and Prm2 protein expression at the translation level with minor effect on mRNA degradation, through binding to the coding regions of TP2 and Prm2 mRNAs. The corresponding primary-microRNAs and the expression levels of Drosha and DGCR8 (both mRNA and protein) were increased significantly in the GRTH-null mice. miR-469 silencing of TP2 and Prm2 mRNA in pachytene spermatocytes and round spermatids is essential for their timely translation at later times of spermiogenesis, which is critical to attain mature sperm. Collectively, these studies indicate that GRTH, a multifunctional RNA helicase, acts as a negative regulator of miRNA-469 biogenesis and consequently their function during spermatogenesis.
Keywords: MicroRNA; mRNA; RNA Helicase; Spermatogenesis; Translation Regulation; Drosha/DGCR8; Gonadotropin-regulated Testicular RNA Helicase, GRTH; Male Germ Cell Development; Testis-specific MicroRNA-469; MicroRNA Biogenesis
Introduction
Mammalian spermatogenesis is a complex process in which primary germ cells undergo mitotic and meiotic divisions to generate haploid round spermatids, and proceed to the differentiation process of spermiogenesis that produces elongating spermatids and mature sperm. This process is regulated at the transcriptional and post-transcriptional levels by the integrated expression of an array of testicular genes in a precise temporal sequence (1, 2). Chromatin compactation that occurs in elongated spermatids during spermiogenesis is essential for nuclear condensation to generate mature spermatozoa. This repackaging event is achieved by replacing histones with transition proteins (TP1 and TP2), which in turn are replaced by protamines (Prm1 and Prm2). The initial active transcription phase with translational repression is followed by cessation of transcription associated with chromatin modifications. mRNA of genes that are essential for later stages of spermiogenesis are generated well prior their translation. Several mRNAs that associate with messenger ribonuclear proteins are repressed translationally at cytoplasmic sites, presumably in the chromatoid body of round spermatids.
Gonadotropin-regulated testicular RNA helicase (GRTH2/DDX25), a testis-specific member of the DEAD (Asp-Glu-Gly-Asp)-box family present in Leydig and germ cells (meiotic spermatocytes and round spermatids) is regulated developmentally by androgen at the transcriptional level (3–6). GRTH is a multifunctional protein, and as component of messenger ribonuclear protein, it transports target mRNAs from the nucleus to cytoplasmic sites (chromatoid bodies, a perinuclear organelle of nuage structure in spermatids for storage/processing) and to polyribosomes for translation (7). GRTH is essential to govern the structure of the chromatoid body (CB) and to maintain systems that may participate in mRNA storage and their processing during spermatogenesis (8). GRTH knock-out male mice are sterile, with azoospermia caused by a complete arrest of spermiogenesis at step 8/9 of round spermatids and failure to elongate. The CB was condensed and greatly reduced in size in the round spermatids of GRTH-null mice (9). Accumulated evidence indicates that members of the small interfering RNA (siRNA) and microRNA (miRNA) pathway, which include Dicer to process si/miRNA precursors to mature small RNAs, the effector complex RNA-induced silencing complex, and mouse vasa homolog, another germ cell helicase, are present in the CB (10, 11). Preservation of the expression of relevant mRNAs with failure of protein expression such as angiotensin-converting enzyme, transition protein 1 (TP1), and transition protein 2 (TP2) in the GRTH-null mice indicate that GRTH is also involved in post-transcriptional events.
microRNAs, a class of ∼22-nt noncoding RNAs, participate in diverse biological functions by promoting degradation and inhibition of translation of target mRNAs. With the exception of miRNAs generated within introns of protein-coding genes, most miRNAs are derived from primary miRNA transcripts (pri-miRNAs) transcribed by RNA polymerase II, which are 5′-capped and polyadenylated (12, 13). pri-miRNAs are subsequently processed by the Drosha-DGCR8 complex in the nucleus to generate precursor miRNAs (pre-miRNAs), which are exported by Exportin-5 to the cytoplasm where mature miRNAs are generated via a Dicer-dependent or -independent route (14, 15). Recent findings of individual miRNAs expressed during spermatogenesis in a developmental stage-specific manner suggest the participation of miRNAs in male germ cell development through their contribution to cell type-specific profiles of protein expression during spermatogenesis (16, 17).
To elucidate regulatory actions of GRTH in miRNA processing during germ cell development, we first compared the differential expression profiles of miRNA in purified round spermatids from wild type and GRTH null mice. A panel of miRNAs and some of their corresponding pri-miRNAs was significantly increased in the GRTH-null mice. Among the various up-regulated miRNAs, the testis-specific miR-469 with cell expression similar to GRTH was selected to pursue further studies (16). miR-469 repressed the translation of TP2 and Prm2 through binding to the coding region of TP2 and Prm2 mRNAs. This is consistent with the preservation of TP2 and Prm2 mRNA with the failure of their protein expression found in the GRTH null mice (7). Also, GRTH appears to negatively regulate Drosha/DGCR8 gene expression to control microRNA maturation. This study has provided insights into a novel molecular control mechanism of GRTH through the microRNA pathway in the regulation of spermatogenesis.
EXPERIMENTAL PROCEDURES
Animals
Adult wild type (C57BL/6-SV129J) (Charles River Laboratories Inc.) and GRTH knock-out male mice (9) were housed in pathogen-free and temperature- and light-controlled conditions (20 °C; alternating light-dark cycle with 14 h of light and 10 h of darkness). All of the animal experiments were approved by the Animal Care and Use Committee of the NICHD. Mice were sacrificed by asphyxiation with CO2 and decapitated. The testes were removed and decapsulated for the purification of germ cells.
Purification of Round Spermatids
Testicular germ cells were prepared by collagenase/trypsin dispersion and purified by centrifugal elutriation (18). After collagenase dispersion, seminiferous tubules were minced and incubated in medium 199 containing 0.1% bovine serum albumin, 0.1% trypsin (Sigma) for 15 min in a rotary water bath (100 rpm, 35 °C). After the addition of 0.02% trypsin inhibitor (Sigma), the sample was filtered through a 300-, 90-, or 40-μm mesh screen and glass wool, and the cells were pelleted and resuspended in elutriation buffer containing 2 μg/ml DNase. The round spermatids were subsequently separated and purified by centrifugal elutriation using Beckman Avanti 21B centrifuge with elutriator rotor model J 5.0 as described previously (7). The first three fractions (fractions 1–3) were collected with flow rates of 13.5, 31.5, and 41.4 ml/min at 3000 rpm, and two additional fractions (fractions 4 and 5) were obtained with flow rates of 23.2 and 40 ml/min at 2000 rpm. Fraction 3 containing round spermatids at a purity of 86% was used for protein and RNA analyses.
microRNA Microarray Analysis
Total RNA enriched for small RNAs was isolated from purified round spermatids either from wild type or GRTH knock-out mice by using the mirVanaTM miRNA isolation kit (Ambion). The quality of RNA was assessed by identification of 28 S rRNA, 18 S rRNA, and small RNA peaks on Agilent 2100 Bioanalyzer with an RNA 6000 nano kit (Agilent Technologies). RNA samples with an RNA integrity number between 8 and 10 were further used for analysis. Rodent TaqMan® Low Density miRNA Arrays A (TLDA) version 2.0 was used to detect and quantify the mature miRNAs expression level in accordance with the manufacturer's instructions (19). All reagents were obtained from Applied Biosystems. Briefly, 500 ng of total RNA were reverse-transcribed using the miRNA reverse transcription kit with the Megaplex pool containing about 380 stem-looped reverse transcription (RT) primers. No prior miRNA pre-amplification step was needed. For each cDNA sample, 384 small RNAs were profiled using a gene maximization PCR plate (384-well plate). The Rodent Arrays A plate contains 335 individual mouse miRNAs in each well, 4 wells of U6 small nucleolar RNA as internal controls, and other small nucleolar RNAs or rat miRNAs. 6 μl of RT product and 450 μl of 2× TaqMan PCR Master Mix, No AmpErase UNG were mixed with 444 μl of nuclease-free water. 100 μl was loaded into each port of the appropriate 384-well TLDA array, centrifuged 1 for min at 300× g to distribute samples to the multiple wells, and then sealed with a micro-fluidic card staker. The arrays were run on an ABI Prism 7900 HT sequence detection system (Applied Biosystems). miRNAs were excluded from the analysis if cycle threshold (Ct) values were above 35 or high variation was found. Relative quantification (RQ) values of miRNA expression levels were calculated by the comparative 2−ΔΔCt method with U6 small nucleolar RNA as endogenous controls. The experiment was performed in duplicate from two independent samples.
microRNA Quantitative Real Time RT-PCR
To validate the accuracy of microarray data, single qPCR experiments for representative miRNAs were performed. Total RNA enriched for small RNAs was extracted from round spermatids of wild type or GRTH knock-out mice using miRNeasy mini kit (Qiagen). The RNA quality was checked by Agilent 2100 Bioanalyzer (Agilent Technologies). 100 ng of total RNA was reverse-transcribed (RT) with 50 nm miRNA-specific stem-loop primers containing common sequences described previously (5′-GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACTGGATACGAC) (20) or U6 small nuclear RNA random primers using TaqMan® microRNA reverse transcription kit (Applied Biosystems) according to the manufacturer's instructions. microRNA levels were measured in triplicate using SYBR Green PCR Master Mix (Applied Biosystems) with miRNA-specific forward primers designed based on miRBase 14 release and a universal reverse primer (5′-AGTGCAGGGTCCGAGG) on an ABI 7500 real time PCR system (Applied Biosystems) with the following conditions: 2 min at 50 °C and then 10 min at 95 °C followed by 40 cycles of 95 °C for 15 s and 60 °C for 60 s. PCR specificity was checked by melting curves and agarose gel electrophoresis. miRNA levels were normalized to U6 small nuclear RNA, and fold change was determined by the comparative threshold method (2−ΔΔCt). All miRNA-specific and U6 random primers are listed in supplemental Table 1.
Assessment of Primary microRNA and mRNA by Quantitative Real Time RT-PCR
Total RNA was extracted from round spermatids or culture cell pellets using RNeasy micro kit (Qiagen), and the samples were treated with the RNase-free DNase set (Qiagen) as per the manufacturer's instructions. cDNA synthesis was performed with 1 μg of total RNA using SuperScriptTM III first-strand synthesis system and oligo(dT) primers (Invitrogen) according to the manufacturer's instructions. The amounts of primary microRNA or mRNA were measured using SYBR Green PCR Master Mix (Applied Biosystems) with β-actin as an internal control on the ABI 7500 real time PCR system (Applied Biosystems). The primary microRNA-specific primers were designed upstream of the precursor microRNA hairpin based on the Ensembl ortholog data base. The primer sequences were used for primary microRNA, and mRNA qRT-PCRs are available in supplemental Table 2.
DNA Constructs
Mouse transition protein 1 (TP1, NM_009407, nt 40–389), transition protein 2 (TP2, NM_013694, nt 58–535), protamine 1 (Prm1, NM_013637, nt 97–383), protamine 2 (Prm2, NM_008933, nt 103–594), phosphor-glycerate kinase 2 (PGK2, NM_031190, nt 951–1568), and H1 histone family, member N, testis-specific (H1fnt, NM_027304, nt 441–1313) cDNA, including coding region and 3′-untranslated region (UTR) were amplified by PCR using cDNA template prepared from round spermatids. To construct luciferase reporter plasmids, various fragments of the potential target gene cDNA or a series of deletion and annealed oligomers were cloned into psiCHECK-2 (Promega) at restriction sites XhoI and NotI, directly 3′ downstream of Renilla luciferase. The wild type of Prm2 or TP2 coding region (CDs) were cloned into pCMV-tag2 vector (Invitrogen) at restriction sites EcoRI and XhoI. To generate each CD mutant construct, PCR-based site-directed mutagenesis used the pCMV-FLAG-Prm2 or pCMV-FLAG-TP2 as templates with a pair of primers containing the mutant MRE sequence using QuikChange II XL site-directed mutagenesis kit (Stratagene). The PCR products of mutant CDs amplified from the mutant construct were subsequently cloned into psiCHECK-2 (Promega) at restriction sites XhoI and NotI, directly 3′ downstream of Renilla luciferase. In some cases, annealed oligonucleotides containing wild type or mutated sequence were directly subcloned downstream of Renilla luciferase. The nucleotide sequences of the constructs were confirmed by DNA sequencing analyses. The primer sequences used for construct are shown in supplemental Table 3.
Cell Culture and Transfection
NIH3T3 cells (American Type Culture Collection) were maintained in Dulbecco's modified Eagle's medium plus 10% heat-inactivated fetal bovine serum and penicillin/streptomycin at 37 °C with 5% CO2. All cell culture reagents and culture plasticware were from Invitrogen and Corning Inc., respectively, unless otherwise specified. For the qRT-PCR and Western blot, the cells were seeded into 6-well plates 24 h before transfection at a density of 1 × 105 cells/well in an antibiotic-free medium. Pre-miRNAs or the scrambled RNA oligomer (Ambion) was cotransfected at a final concentration of 50–100 nm with 0.1–0.5 μg of pCMV-tag-Prm2 or pCMV-tag-TP2 construct using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The microRNA expression was confirmed by the stem-loop qPCR.
Dual-Luciferase Assay
NIH3T3 cells were seeded 24 h before transfection at a density of 6 × 103 cells/well in 96-well plates. pre-miRNAs (5 pmol) or scrambled RNA oligomer (5 pmol) or miRCURY LNATM miRNA-469 inhibitor (5 pmol, Exiqon) or control miRCURY LNA miRNA inhibitor (5 pmol, Exiqon) was cotransfected at a final concentration of 33 nm with 2 ng of the psiCHECK-2/GENE construct. In all cases, a constitutively expressed firefly luciferase gene in psiCHECK-2 was used as a normalization control for transfection efficiency. Forty eight hours after transfection, cells were washed with PBS and lysed in Passive Lysis Buffer (Promega). Firefly and Renilla luciferase activities were measured consecutively with the Dual-Luciferase reporter system (Promega) using a luminometer (Mithras LB940; Berthold Technologies). All luciferase assays were repeated a minimum of three times with four replicates each.
Protein Extraction and Western Blot Analysis
Cell pellets harvested from transfected NIH3T3 cells or purified round spermatids were washed in chilled PBS and incubated for 20 min at 4 °C with rotation in 1× RIPA buffer (500 nm Tris-HCl, pH 7.4; 150 nm NaCl; 0.25% deoxycholic acid; 1% Nonidet P-40; 1 mm EDTA) (Millipore) containing freshly added protease inhibitor mixture (Roche Applied Science). Lysates were cleared by centrifugation at 4 °C for 15 min at 16,000 × g, and protein concentrations were determined using Bradford dye (Bio-Rad). For Western blot analysis, 30 μg of total protein was size-fractionated by SDS-PAGE on 8–16% Tris-glycine gels in the running buffer (Invitrogen) and transferred to nitrocellulose membranes (Invitrogen) in Tris-glycine transfer buffer (Invitrogen) containing 10% methanol. The membrane was blocked for 1 h at room temperature in 5% milk and probed with specific primary antibodies obtained from commercial sources, including anti-mouse antisera, FLAG (1:1000, Sigma) and β-actin (1:5000, Sigma); anti-rabbit antisera, Drosha (1:1000, Cell Signaling) and DGCR8 (1:1000, ProteinTech). The blots were developed with goat anti-mouse or goat anti-rabbit IgG horseradish peroxidase-conjugated antibodies (1:2000–10,000, Santa Cruz Biotechnology), respectively. Reacted proteins were visualized by the electrochemiluminescence system (Pierce). The intensity of each protein band from Western blots from three independent experiments was scanned by densitometry (GS-800 calibrated densitometer, Bio-Rad) in the linear range of optical density and quantitated by a software package (Quantity One version 4.2.1). The values obtained from quantitation of optical densities of Western blot signals were normalized by endogenous β-actin. KO values were expressed relative to WT (100%).
Statistical Analysis
The significance of the differences on the expression of luciferase activity, mRNAs, and proteins between groups was determined by Dunnett's multiple-comparison test (one-way analysis of variance).
RESULTS
Differential Expression of miRNAs from Round Spermatids in Wild Type and GRTH Knock-out Mice
miRNA expression profiles for round spermatids of wild type and GRTH knock-out mice were determined by Rodent TaqMan® Low Density miRNA Arrays A version 2.0 (TLDA, Applied Biosystem). miRNAs were considered to have significant differential expression if they were up- or down-regulated at least 4-fold (accession number GSE 33969). A total of 119 miRNAs was found to be up-regulated in the GRTH knock-out mice compared with the wild type; however, no down-regulated miRNAs were observed in this study (supplemental Table 4).
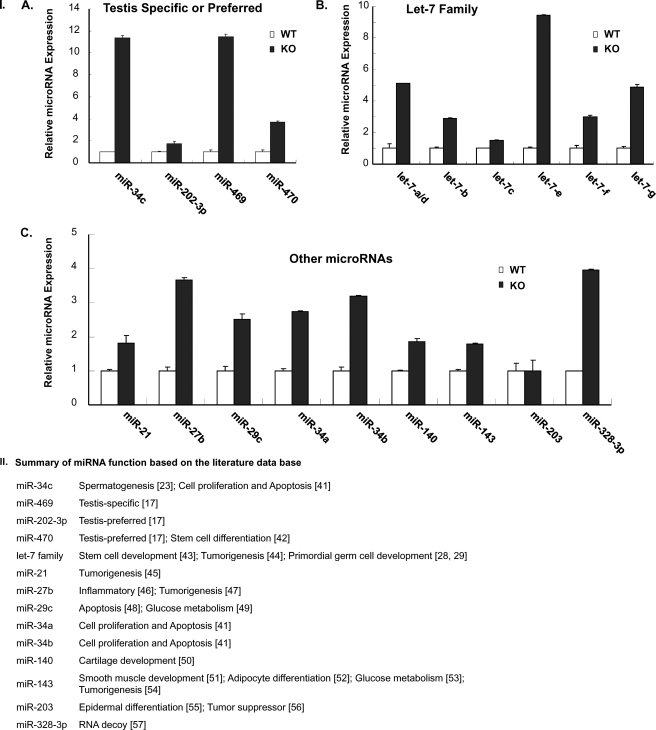
To verify the results obtained by microarray analysis, quantitative real time RT-PCR analysis was carried out to measure the differential expression level of miRNAs known to be testis-specific or others in round spermatids of WT versus KO mice. Some of these with postulated functions reported in the literature are shown in Fig. 1II. The qRT-PCR validation of selected differentially expressed miRNAs (Fig. 1I) showed that testis-specific (miR-469) or preferred (miR-34C and miR-470) (Fig. 1I, panel A) (16) members of the let-7 family with a 1–2-nucleotide difference (Fig. 1I, panel B) and others, including miR-21, -27b, -29C, -34a, -34b, -140, -143, and -328-3p (Fig. 1I, panel C), were significantly up-regulated in GRTH KO mice. In contrast, miRNA-202–3p and let-7c showed a minor increase, and no change in miRNA-203 level was observed. These findings were consistent with those derived from microRNA array expression profiles.
FIGURE 1.
I, RT-quantitative PCR of individual miRNAs from round spermatids of GRTH-null (KO) and wild type (WT) mice. Panel A, miRNAs known to be testis-specific (miR-469) or preferred (miR-34c, miR-202–3p, and miR470). Panel B, members of the let-7 family. Panel C, other microRNAs. Results show a representative experiment in triplicate from three independent experiments. Data represent means ± S.E. II, summary of known functions corresponding to miRNAs differentially expressed in round spermatids of wild type versus GRTH KO mice derived from miRNA microarray analysis (17, 28, 29, 41–57).
Differential Expression of pri-miRNAs and Drosha-DGCR8 in Round Spermatids between Wild Type and GRTH Knock-out Mice
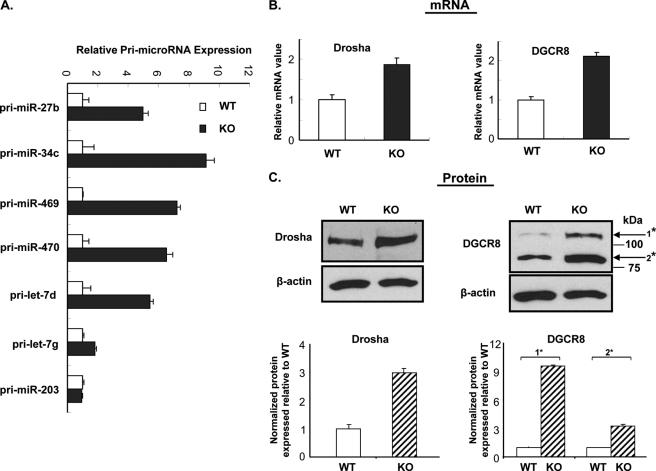
To elucidate the processing mechanism leading to the up-regulation of miRNAs, presented in Fig. 1, pri-miRNAs corresponding to selected up-regulated and unchanged (control) miRNAs from the different groups were analyzed in round spermatids of GRTH-KO and WT mice by the qRT-PCR with specific gene primers located upstream to the hairpin of each precursor (Fig. 2). Primary precursors for the testis-specific (miR-469) or preferred (miR-34c and -470) members of let-7 family (pri-let-7d and -7g) and pri-miR-27b showed increased levels consistent with those observed for the up-regulated mature miRNA observed in the round spermatids of GRTH KO (p < 0.05). No change in the pri-miR-203, as was the case for its mature miR-203 counterpart, was observed (Fig. 1I). Other primary precursors for the miRNAs shown in Fig. 1 were not analyzed in this study. The expression of Drosha and DGCR8 proteins of the microprocessor complex, required for the generation of precursor pre-miRNA, were significantly increased at both mRNA (Fig. 2B, ∼2-fold) and protein levels (Fig. 2C, ∼3–9-fold) in the absence of GRTH (GRTH KO versus WT). In contrast, no changes in Dicer or Ago protein was observed (8). These results indicated that GRTH negatively regulates miRNA biogenesis at levels of the synthesis of primary miRNA (pri-miRNA) and of enzymes of the microprocessor complex processing complex (Drosha and DGCR8) for generation of mature miRNA.
FIGURE 2.
Up-regulated primary miRNAs and microprocessor in round spermatids of GRTH KO compared with wild type (WT) mice. A, quantitative real time qPCR measurements of a panel of primary miRNAs corresponding to their respective mature miRNAs from round spermatids in the GRTH-null (KO) and wild type (WT). Results are the means ± S.E. of three independent experiments in triplicate. B, quantitative real time PCR measurements of Drosha and DGCR8 mRNA from round spermatids of GRTH-null (KO) and wild type (WT) mice. C, Western blot analysis of Drosha protein and DGCR8 isoform (*, NM_022720.6 and 001190326.1) from round spermatids of GRTH-null (KO) and wild type. Signals from three independent experiments were quantified and normalized by β-actin. The KO values (means ± S.E.) are presented relative to WT (100%).
Identification of miR-469 Target Gene
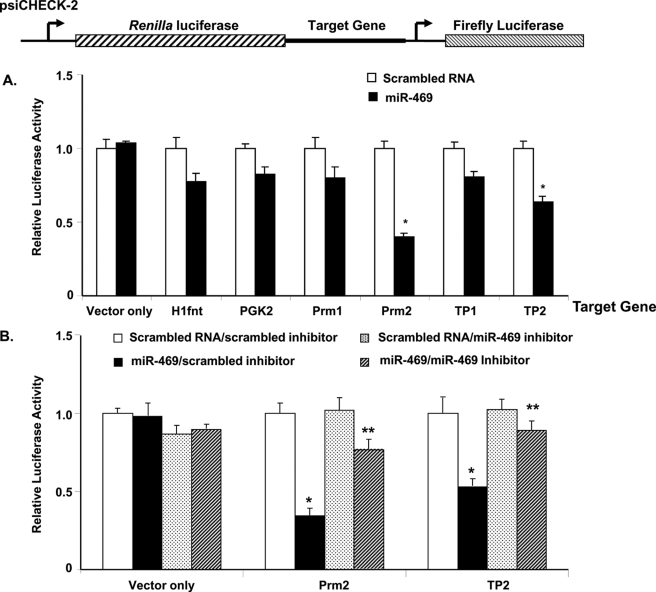
Among the verified up-regulated miRNAs (Fig. 1), the testis-specific miR-469 (16, 17) was selected to focus our effort in this study. This miRNA was found to be testis-specific with expression in germ cells, meiotic spermatocytes, and round spermatids (16, 17) and in Leydig cells of adult testis (supplemental Fig. S1). Because the initial analysis by target scan program did not reveal TP2 and Prm2 as targets for miR-469 function, a panel of proven GRTH-regulated genes of relevance for germ cell development, including transition proteins 1 and 2 (TP1 and TP2) and protamine 1 and 2 (Prm1 and Prm2), were assessed as the putative target genes for miR-469 using dual-luciferase reporter assays. The coding region and 3′-UTR of candidate genes were inserted into the psiCHECK-2 vector directly 3′ downstream of reporter gene Renilla luciferase, and further downstream Firefly luciferase was used as the internal control in the study (Fig. 3, top panel). Results from the dual-luciferase assay showed the normalized level of Renilla luciferase relative to Firefly luciferase activity in the presence of miR-469 significantly decreased in the cells cotransfected with Prm2 and TP2 constructs when compared with scrambled RNA (p < 0.01). The negative control, plasmid lacking the targeting sequence, showed no difference in the luciferase activity between the scrambled and miR-469 group. The luciferase activity for H1fnt, PGK2, Prm1, and TP1 showed minimal inhibition by miR-469 (p > 0.05) (Fig. 3A). Thus, Prm2 and TP2 appeared to be the target gene regulated by miR-469 either via coding region or 3′-UTR. Recovery from miR-469 inhibition of TP2 and Prm2 was evident in cells cotransfected with miR-469 inhibitor (Fig. 3B). As target miR-469 sequences in Prm2 or TP2 mRNA could not be identified initially by the RNA22 program in either untranslated or coding regions of the gene (21), we proceeded with the investigation of functional elements using deletions and mutations utilizing dual-luciferase assays.
FIGURE 3.
Identification of miR-469 target genes. A, panel of GRTH-targeted testicular genes derived from polysome microarray gene expression analysis (40) was selected for the study. miR-469 inhibits the expression of a luciferase construct containing coding region and 3′-UTR of protamine 2 (Prm2) and transition protein 2 (TP2) mRNA *, p < 0.01. No effect on testis-specific histone H1 gene (H1fnt), PGK2, Prm1, and TP1. B, recovery of miR-469 inhibition in Prm2 and TP2 by miR-469 inhibitor. Normalized luciferase activity (Renilla/Firefly) in NIH3T3 cells cotransfected with reporter constructs in the presence of scrambled RNA, miR-469 precursors, scrambled inhibitor, and or miR-469 inhibitor as indicated. Results are the means ± S.E. from three independent experiments with four culture replicates each. miR-469/scrambled inhibitor versus scrambled RNA/scrambled inhibitor in Prm2 and TP2 groups, *, p < 0.01 and miR-469/miR-469 inhibitor versus scrambled RNA/miR-469 inhibitor in Prm2 and TP2 groups; **, p > 0.05. Vector only indicates no differences among groups.
Identification of Protamine 2 Functionally Responsive Element of miR-469
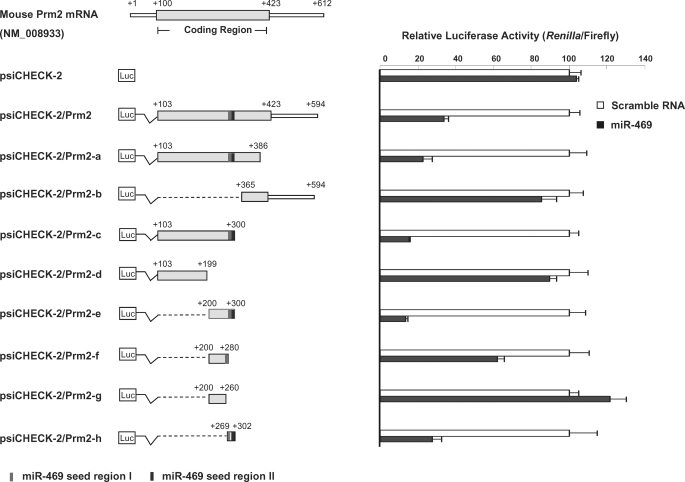
To characterize the mechanisms regulating Prm2 or TP2 expression at the post-transcriptional level by miR-469, serial deletions from the distal and proximal ends of Prm2 or TP2 were generated as illustrated in the left panel of Figs. 4 and 6, respectively. Luciferase assays demonstrated that strong regulatory elements are located in the coding region of both target genes. miR-469 (Fig. 4) significantly decreased luciferase activity in the plasmid containing the full-length coding region (103/423 nt) along with the 172-bp 3′-noncoding region (423/594 nt) of Prm2 (103/594 nt) (psiCHECK-2/Prm2). Similar inhibition was noted in the construct of 3′ deletion from nt 386 (103/386 nt) (psiCHECK-2/Prm2-a) when compared with the negative control group with scrambled RNA. However, miR-469 did not have any effect on the plasmid containing isolated fragment from 365 to 594 nt (psiCHECK-2/Prm2-b). These results suggest the functional MRE(s) is/are located in the coding region of Prm2. A significant further decrease in luciferase activity was observed in the reporter gene containing Prm2 at nt 103–300 (psiCHECK-2/Prm2-c), whereas this inhibition was recovered in the construct-containing fragment of nt 100–199 (psiCHECK-2/Prm2-c versus -d). Location of MREs between 200/300 nt of Prm2 was confirmed by the loss of reporter gene activity in cells transfected with psiCHECK-2/Prm2-e in the presence of miR-469. Detailed evaluation of this region showed that the repression was completely recovered in nt 200–260 (psiCHECK-2/Prm2-g). The partial recovery observed with the nt 200–280 (psiCHECK-2/Prm2-f) indicated that sequences between nt 260 and 300 play a key role in the miR-469-mediated Prm2 repression. This was further confirmed in the construct-containing sequences between nt 269 and 302 (psiCHECK-2/Prm2-h).
FIGURE 4.
Mapping of miR-469 response element(s) in protamine 2. Left panel, diagram of the constructs of mouse Prm2 full-length and deletions in the psi-CHECK2 vector. Coding region with deletion of ATG codon or 3′-UTR of Prm2 was subcloned in the downstream 3′-UTR of Renilla luciferase (Luc). Shaded bar indicates the coding region of Prm2, and solid square indicates two response elements of miR-469. Right panel, normalized luciferase activity (Renilla/Firefly) in NIH3T3 cells cotransfected with deletion constructs in the presence of scrambled RNA or miR-469 precursors, as indicated. Results are the means ± S.E. of three independent experiments performed in quadruplicate.
FIGURE 6.
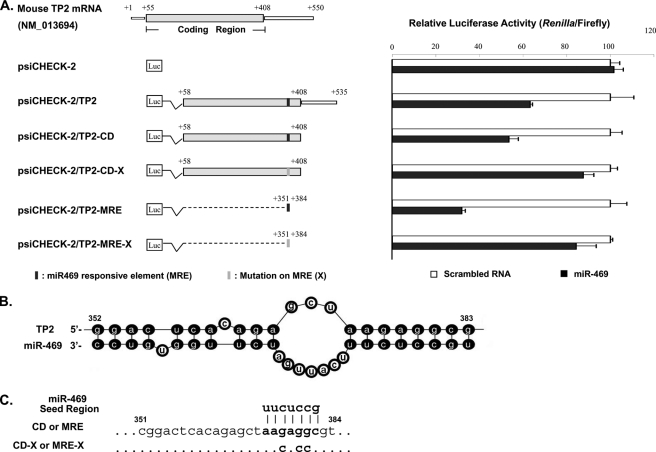
Identification of functional response element of miR-469 in transition protein 2 (TP2) coding region. A, left panel, diagram of the constructs of mouse TP2 deletion or mutation in the psi-CHECK2 vector. Coding region or 3′-UTR of TP2 is located in the downstream 3′-UTR of Renilla luciferase. Shaded bar, coding region of TP2. Solid squares, putative response element of miR-469 predicted by mfold program (B). Right panel, normalized luciferase activity (Renilla/Firefly) in NIH3T3 cells cotransfected with respective deletion or mutation constructs in the presence of scrambled RNA or miR-469 precursors. Results are the means ± S.E. of three independent experiment in quadruplicate. B, duplex structure between miR-469 and TP2 mRNA predicted by mfold program. One binding site located at nt 352–383 of TP2 mRNA contains the perfect match sequence (nt 376–383) to miR-469 5′ seed region. C, diagram of mutation in the TP2 mRNA. Three nucleotide mutations were generated in the coding region (CD) of TP2-binding site to the miR-469 seed region using psi-CHECK2/TP2-CD as the template. Annealed DNA oligonucleotides, wild type (MRE), or mutated mir469-responsive element (MRE-X) was directly subcloned to the vector as psi-CHECK2/TP2-MRE or MRE-X.
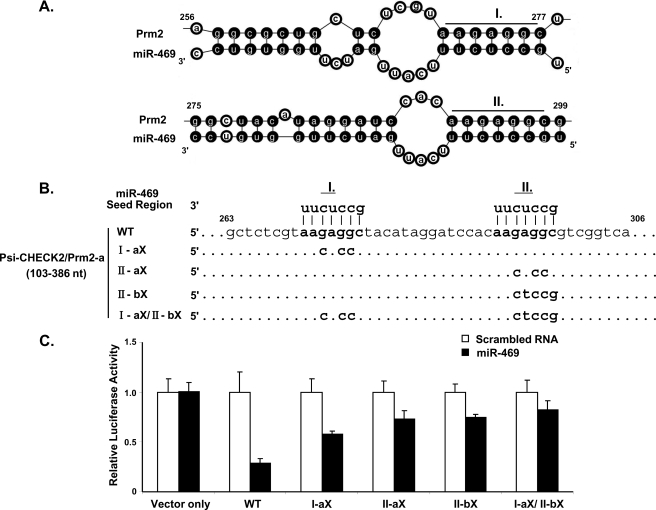
Computational and bio-informational approaches were then utilized to predict potential Prm2 mRNA targets between nt 256 and 299 of miR-469 (Fig. 5A). The structure between Prm2 mRNA and miR-469 predicted by mfold program) revealed the presence of two critical binding sites with complete match to the seed region (nt 2–8) of miR-469 located in nt 271–277 and 292–299 of Prm2 mRNA (Fig. 5). To investigate which target site(s) confer the specificity of interaction between miR-469 and Prm2 mRNA, four different mutations were created in the predicted Prm2-binding sites of miR-469 (Fig. 5B). Results showed that luciferase activities were partially recovered in either singly mutated binding site compared with WT. Inhibition was only abolished by mutations of both target sites (I-aX/II-bx). Collectively, these results demonstrate miR-469 regulates Prm2 expression through binding to two functional response elements in the coding region.
FIGURE 5.
Verification of functional response element of miR-469 in Prm2 coding region by mutagenic analysis. A, duplex structure between miR-469 and Prm2 mRNA predicted by mfold program. Sequences located at nt 271–277 (I) and 292–299 (II) of Prm2 mRNA (nt 256–299) were perfect match to the 5′ seed region of miR-469 (nt 2–8 of miR-469). B, diagram of mutation sites in the Prm2 mRNA. Three or five nucleotides mutations were generated in the Prm2 I and/or II binding sites to miR-469 seed region using psi-CHECK2/Prm2-a (Fig. 3) as the template. C, normalized luciferase activity (Renilla/Firefly) in NIH3T3 cells cotransfected with wild type (WT) or mutation constructs in the presence of scrambled RNA or miR-469 precursors. Relative luciferase activity was recovered gradually in the I and/or II mutation construct compared with the WT. Results are the means ± S.E. from three independent experiments in quadruplicate.
Identification of Transition Protein 2 Response Element of miR-469
The functional MRE(s) in TP2 was also determined by dual-luciferase assay with combined deletions and mutagenesis approach (Fig. 6). The sequence containing the coding region alone (psiCHECK-2/TP2-CD) exhibits comparable loss of luciferase activity as the constructs with additional 3′-UTR sequences (psiCHECK-2/TP2) (Fig. 6A). MRE was then proposed to be localized in the coding region rather at 3′-UTR. mRNA-fold program confirmed the formation of a duplex structure of TP2 mRNA (nt 352–383) with a perfect match at nt 376–383 to the 5′ seed region of miR-469 (Fig. 6B). Significant loss of luciferase activity was observed in the construct containing the MRE sequence alone (psiCHECK-2/TP2-MRE). Mutation of the proposed MRE site in the psiCHECK-2 vector containing either coding region of TP2 (psiCHECK-2/TP2-CD-X) or the isolated MRE (psiCHECK-2/TP2-MRE-X) completely recovered the luciferase activity in the presence of miR-469. Negative control with vector alone showed no difference in luciferase activity in the presence or absence of miR-469. Similar to the Prm2, these results demonstrated the miR-469 regulated TP2 expression through its binding to the coding region of TP2 but not the 3′-UTR.
Effect of miR-469 on Endogenous Prm2 and TP2 Expression
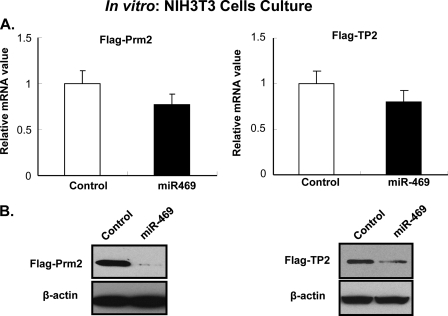
To verify the relevance of the coding regions of Prm2 and TP2 as targets of miR-469 function, mammalian expression constructs (pCMV-FLAG) containing Prm2 and TP2 that excluded 3′-UTR were utilized. In NIH3T3 cells transfected with either PCMV-Prm2 or -TP2 plasmid, mRNA of Prm2 or TP2 was not altered by the overexpression of miR-469 (p > 0.05) (Fig. 7A). However, the level of protein expression in either case was significantly decreased (Fig. 7B) by overexpression of miR-469. These results indicate that Prm2 and TP2 protein expression was repressed by miR-469 at the translation level but not via mRNA degradation.
FIGURE 7.
miR-469 represses Prm2 and TP2 protein expression with minor effect on mRNA level. A, quantitative real time qPCR measurements of Prm2 and TP2 mRNA from transfected NIH3T3 cell line. Cells were cotransfected pCMV-FLAG-Prm2 or pCMV-FLAG-TP2 construct with either scrambled RNA (control, open bar) or miR-469 precursor (solid bar). B, Prm2 and TP2 protein expression level in the transfected NIH3T3 cells. Western blot analyses were performed using anti-FLAG antibody and anti-β-actin as an internal control. Results presented are from three independent studies. Data are presented as the means ± S.E.
DISCUSSION
This study has demonstrated a subset of microRNAs that were differentially up-regulated in round spermatids of GRTH KO mice, including testis-specific miR-469, testis-preferred (miR-34c and miR-470), ubiquitous let-7 family members, and others that might participate in germ cell development (Fig. 1). The primary miRNAs of interest were also increased along with the enzyme complex (Drosha-DGCR8) that promotes the processing of primary miRNA transcripts. Our studies provide evidence that miR-469 specifically targets chromatin remodeling gene products TP2 and Prm2, which are essential for germ cell development, and further understanding of the GRTH regulatory function during spermatogenesis.
It is interesting to note in the present microarray analysis the presence of solely differentially up-regulated microRNAs (Fig. 1, I and II) covering a wide range cellular actions that could participate in germ cell development. This type of up-regulation was similarly observed in the absence of Tdrd6 gene expression where the majority of differentially expressed miRNAs (91%) were up-regulated. Tdrd6 is a Tudor domain containing protein that exhibits the same pattern of expression as GRTH and is also required for the architecture of the chromatoid and the progress of spermiogenesis (9, 22). Only the miRNAs miR-29c, miR-34a, miR-34c, and miR-470 were commonly up-regulated in both null mice models. Target scan predicted DNA-modifying enzymes (DNA methyltransferase and histone deacetylase), RNA-processing enzyme (Dicer), and eukaryotic translational initiation factors as the miRNA target genes. This suggests a common control mechanism that might be shared by both GRTH and Tdrd6. Some aspects of the specificity of GRTH action differing from Tdrd6 rely on the unique regulation of miRNAs like miR-469 through its association with the coding region of chromatin remodeling genes TP2 and Prm2 (Figs. 3–6). Other not yet identified miRNAs differentially regulated by GRTH could also target TP2 and Prm2 in this mouse model. Neither target scan nor RNA22 program revealed any other miRNA candidates in the panel of differentially expressed species (WT versus KO) targeting TP2 and Prm2 in this study. The interaction between miRNAs and target genes seems to play an important factor in the regulation of stage-specific expression of germ cell genes.
In addition to the fact that both GRTH and miR-469 are expressed specifically in the testis, the striking similarity of their increased expression from puberty to adulthood (3, 16), and the cell-specific expression in the late stage of meiosis and round spermatids (4, 16, 17) indicate their functional importance during testicular spermatogenesis. Evidence for the presence of mature miR-469 in round spermatids and its functional role in the regulation of genes (TP2/Prm2) required for chromatin remodeling during spermatogenesis have been shown in this study. The finding of an increased miR-469 level in the absence of GRTH (Fig. 1) led us to propose that GRTH might have an intrinsic regulatory role to prevent overexpression of miR-469 and possible others shown to be up-regulated in this study.
miR-34c, is also known to be highly expressed in the spermatocytes and round spermatids (23). Although the RNA22 program predicted few potential low energy binding sites in the target genes of interest (Fig. 3) governed by GRTH (7), none of these genes were proven as miR-34c targets by the dual-luciferase analysis (data not shown). However, it is conceivable that the miR-34c target Transforming growth factor inhibitor 2 (TGIF2), a known inhibitor of the TGFβ receptor/signaling pathway for its regulation during spermatogenesis (24, 25), might be linked to GRTH regulation. Other evolutionarily conserved members of miR-34 family, miR-34a and -34b, known to have a role in determining cell fate, provide a new avenue for the GRTH action in the germ cells.
From the 13 let-7 miRNAs family members evolutionarily conserved from Drosophila to human, let-7a/d, -b, and -e–g were up-regulated in GRTH KO mice. let-7 members have either activator or repressor function depending on specific cellular stages. let-7 is known to target endonuclease Dicer within its coding region as a mechanism for a miRNA/Dicer autoregulatory negative feedback loop (26). This appears not to be the case in the GRTH KO mice model where no change in Dicer protein expression was observed (8) despite the marked increases noted in several members of the let-7 family (Fig. 1I, panel B). High level of let-7 expression has been proposed as a unique property of testis-derived male stem cell germ line for primordial germ cells' proliferation and development (27–29). In contrast, in many human cancers, let-7 expression is deregulated, and let-7 is widely viewed as a tumor suppressor (30). In pituitary adenomas, the negative regulation of high mobility group protein A2 (HMGA2) by let-7 is believed to contribute to the pathogenesis of the disease (31). The HMGA2 is a non-histone chromatin protein that binds DNA in AT regions and can regulate transcription by altering chromatin structure and consequently promoting the assembly of transcription factor complexes (32). Its expression is highest in pachytene spermatocytes and early spermatids, and disruption of the HMGA2 gene results in a block of spermatogenesis with testis showing degenerating spermatocytes and few spermatids (33). In the GRTH KO mice, the HMGA2 protein is completely absent, but its mRNA level was unchanged compared with the WT (7). let-7 increases in the GRTH KO could effectively silence the HMGA2 mRNA and prevent its expression, and its deficiency could contribute to the phenotype observed in the GRTH KO mice. Thus, let-7 regulation of germ cell target induced by GRTH regulation during spermatogenesis could be envisioned.
Identification of putative functional response element (MRE) sites localized only within the coding region of TP2 and Prm2 provides evidence for a nonclassical regulation of these target genes by miR-469 (Figs. 4–6). It was further demonstrated that the regulation occurred through the repression of protein translation with no effect on mRNA degradation (Fig. 7). Most of the reported functional mammalian microRNAs were found to target 3′-UTR sequences, and few studies have revealed 5′-UTR participation. More recently, evidence of elements within the coding region of mRNA have been found to be targeted by miRNAs (34). Our studies have clearly shown exclusive participation of functional binding sites in the coding region and excluded cooperating 3′-UTR sequences for miR-469. The presence of functional binding sites for both genes was demonstrated in their mRNA coding region (two for Prm2 and one for TP2) through assessment by deletions and mutagenesis. The RNA folding program further confirmed those functional motifs by their complete complementarity with miR-469 5′ seed sequence (at bp 2–8 of the microRNA) to form stable RNA-miRNA duplex with low energy (Figs. 4–6) (35). The functional importance of miRNA targeting coding regions and mechanism of miRNA-mediated translational repression are not well understood compared with the 3′-UTR (14). We speculate that the presence of two functional miR-469 MREs in Prm2 versus one in TP2 might contribute to the higher degree of down-regulation in the protein level of Prm2 compared with TP2 observed in the dual-luciferase assay (Fig. 3) and in vitro expression studies (Fig. 7).
Progress has been gradually unveiled in understanding the function of miRNAs during male germ cell development. The base pairing complementarity of the microRNA with imperfect duplexes guides the endoribonuclease Argonaute (Ago) to repress the translational level or alternatively to degrade the target mRNA directly (36). Previous evidence relates to the degradation of target gene mRNA message through mRNA deadenylation, such as down-regulation of TP2 by miR-122a (16), TGIF2, and NOTCH2 by miR-34c (23), and heat shock factor 2 (HSF2) by miR-18 (37). miR-122a reported to specifically associate with the 3′-UTR of TP2 (16) was not identified in the present differential microarray panel (GRTH WT versus KO). In the case of miR-469-mediated down-regulation of Prm2 and TP2, our studies point to the repression of both Prm2 and TP2 at the translational level with minor effect on mRNA degradation. This is in contrast to a recent study indicating the predominant destabilization of targets with at least one 3′-UTR site by microRNAs (38). In our case, two target sites for miR-469 in each Prm2 and TP2 mRNA were located within their coding region (Figs. 4–6).
Gene expression such as Prm2 and TP2 in haploid spermatids requires temporal uncoupling of transcription and translation. These mRNAs destined for translation during the late spermiogenesis phase (elongating and elongated spermatids, steps 9–16) are actively transcribed prior to the step 9 of round spermatids. It has been long postulated that those messages are temporarily stored in the chromatoid body to await the specific time to be translated (10, 11). Translation followed by chromatin condensation and nuclear elongation ultimately progresses to the generation of mature germ cells. Expression profiling array in the mouse testis (17) showed a high level of miR-469 expression coincident with the phase of classic active transcription and suppressed TP2/Prm2 translation in the meiotic and early haploid germ cells. This is followed by its decreased expression in elongating spermatids with active TP2/Prm2 protein production. The direct evidence on miR-469 targeting these messages to repress the translation observed in this study (Fig. 7) support the physiological relevance of miR-469 in germ cell development under GRTH regulation.
The chromatoid body of haploid round spermatids has been viewed as the center of miRNA-mediated mRNA control to coordinate post-transcriptional gene expression during spermiogenesis (10, 11). GRTH does not associate with any of the components of the RNA-induced silencing complex or other RNA helicase like Mvh involved in this regulation, but it is required for the preservation of the structural integrity of the CB and as transport protein to deliver mRNAs for silencing or processing (8). The up-regulated miRNAs found in the GRTH−/− mice most likely function at cytoplasmic sites other than the CB because this organelle was strikingly condensed and highly decreased in size in GRTH-null mice (9). Moreover, the relevant proteins of the RNA-induced silencing complex that normally reside in the CB of the WT mice are excluded from this organelle in the GRTH KO (8). Dual actions of GRTH in controlling at the nuclear level both primary miRNA (pri-miRNA) of interest and enzymes (Drosha and DGCR8) point to its importance in germ cell miRNA biogenesis (Fig. 2). A connection between testis-specific GRTH, Drosha/DGCR8, mature miR-469, and Prm2/TP2 has been implied indirectly from our comparative studies in GRTH wild type versus GRTH KO mice. However, direct demonstration of this microRNA biogenesis and its impact in Prm2/TP2 protein expression must await the development of suitable germ cell cultures of spermatocytes and round spermatids that would permit optimal silencing of multiple messages (i.e. Drosha/DGCR8/Dicer) and quantitative evaluation of protein expression.
Because both pri-mRNA and enzyme complexes are localized in the nucleus, the 56-kDa nuclear form of GRTH protein is likely to regulate their expression at the level of transcription and/or message stability through a not yet identified mechanism. Our recent studies (not shown) have demonstrated comparable half-lives of Drosha and DGCR8 messages in GRTH wild type and GRTH KO mice. This indicates that GRTH could directly or indirectly regulate members of the microprocessor complex at the transcriptional level. Unlike members of DEAD-box protein family, p68 and p72, which associate Drosha-DGCR8 microprocessor complex (39), GRTH does not associate with this complex as was determined by immunoprecipitation. Also, it was not associated with mRNAs of Drosha and DGCR8 in combined immunoprecipitation/RT-PCR studies (data not shown).
It is conceivable that during early stages of the haploid phase (round spermatids) miR-469 represses TP2 and Prm2 protein expression by silencing message translation in these germ cells. In elongated spermatids, decreased miR-469 expression (17) could release the translational repression of TP2 and Prm2 to proceed with the mRNA processing in a sequential manner. In the case of GRTH-null mice, TP2 and Prm2 mRNA translations were repressed due to the aberrant miR-469 expression level. Moreover, transport of relevant messages to polysomal sites by GRTH and its proposed function in translation is curtailed in GRTH KO mice (5–7). Increased miRNAs in the GRTH-null mice could silence the protein expression of a number of genes post-transcriptionally and thus cause spermatogenesis arrest at the round spermatid stage 7–8 and failure to elongate (Fig. 8).
FIGURE 8.
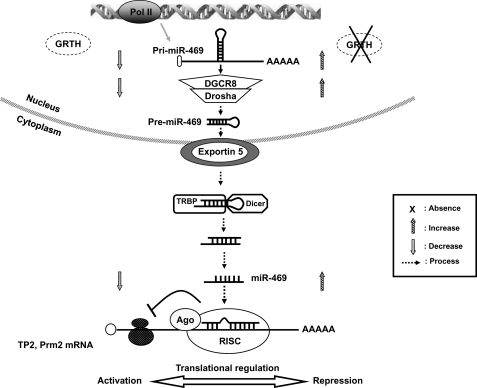
Proposed model: mechanism of GRTH regulation on miR-469 biogenesis that represses TP2 and Prm2 gene translation during spermatogenesis. GRTH plays a critical role to maintain homeostasis of a subset of mature miRNAs by regulating miRNA biogenesis via Drosha-DGCR8 microprocessor complex activation. Absence of GRTH up-regulates pri-miR-469 and Drosha-DGCR8 complex. pre-miR-469 transfers to cytoplasmic sites where it is further processed via Dicer-dependent pathways to generate mature miR-469. miRNA-469 silences TP2 and Prm2 by its association at the coding region through RNA-induced silencing complex. This regulation ultimately controls the temporal progression of spermatogenesis via miR-469 inhibitory action on TP2/Prm2 protein expression, which is essential for their timely expression leading to its chromatin compactation in spermatids and the progress of spermatogenesis.
In summary, we conclude that a negative regulation of GRTH via the route of miRNA biogenesis/action may play an important role during spermatogenesis (Fig. 8). These regulatory aspects provide insights into a novel molecular mechanism of GRTH function during male germ cell development.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Intramural Program through the Eunice Kennedy Shriver National Institute of Child Health and Human Development.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Tables 1–4 and Fig. S1.
- GRTH
- gonadotropin-regulated testicular RNA helicase
- nt
- nucleotide
- qRT
- quantitative RT
- miRNA
- microRNA
- CB
- chromatoid body
- MRE
- miR-469 response element
- CD
- coding region
- pri-miRNA
- primary miRNA.
REFERENCES
- 1. Steger K. (2001) Anat. Embryol. 203, 323–334 [DOI] [PubMed] [Google Scholar]
- 2. Eddy E. M. (2002) Recent Prog. Horm. Res. 57, 103–128 [DOI] [PubMed] [Google Scholar]
- 3. Tang P. Z., Tsai-Morris C. H., Dufau M. L. (1999) J. Biol. Chem. 274, 37932–37940 [DOI] [PubMed] [Google Scholar]
- 4. Sheng Y., Tsai-Morris C. H., Dufau M. L. (2003) J. Biol. Chem. 278, 27796–27803 [DOI] [PubMed] [Google Scholar]
- 5. Dufau M. L., Tsai-Morris C. H. (2007) Trends Endocrinol. Metab. 18, 314–320 [DOI] [PubMed] [Google Scholar]
- 6. Tsai-Morris C. H., Sheng Y., Gutti R. K., Tang P. Z., Dufau M. L. (2010) J. Androl. 31, 45–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sheng Y., Tsai-Morris C. H., Gutti R., Maeda Y., Dufau M. L. (2006) J. Biol. Chem. 281, 35048–35056 [DOI] [PubMed] [Google Scholar]
- 8. Sato H., Tsai-Morris C. H., Dufau M. L. (2010) Biochim. Biophys. Acta 1803, 534–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tsai-Morris C. H., Sheng Y., Lee E., Lei K. J., Dufau M. L. (2004) Proc. Natl. Acad. Sci. U.S.A. 101, 6373–6378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kotaja N., Bhattacharyya S. N., Jaskiewicz L., Kimmins S., Parvinen M., Filipowicz W., Sassone-Corsi P. (2006) Proc. Natl. Acad. Sci. U.S.A. 103, 2647–2652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kotaja N., Sassone-Corsi P. (2007) Nat. Rev. Mol. Cell Biol. 8, 85–90 [DOI] [PubMed] [Google Scholar]
- 12. Han J., Lee Y., Yeom K. H., Kim Y. K., Jin H., Kim V. N. (2004) Genes Dev. 18, 3016–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Han J., Lee Y., Yeom K. H., Nam J. W., Heo I., Rhee J. K., Sohn S. Y., Cho Y., Zhang B. T., Kim V. N. (2006) Cell 125, 887–901 [DOI] [PubMed] [Google Scholar]
- 14. Krol J., Loedige I., Filipowicz W. (2010) Nat. Rev. Genet. 11, 597–610 [DOI] [PubMed] [Google Scholar]
- 15. Miyoshi K., Miyoshi T., Siomi H. (2010) Mol. Genet. Genomics 284, 95–103 [DOI] [PubMed] [Google Scholar]
- 16. Yu Z., Raabe T., Hecht N. B. (2005) Biol. Reprod. 73, 427–433 [DOI] [PubMed] [Google Scholar]
- 17. Ro S., Park C., Sanders K. M., McCarrey J. R., Yan W. (2007) Dev. Biol. 311, 592–602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bucci L. R., Brock W. A., Johnson T. S., Meistrich M. L. (1986) Biol. Reprod. 34, 195–206 [DOI] [PubMed] [Google Scholar]
- 19. Mestdagh P., Feys T., Bernard N., Guenther S., Chen C., Speleman F., Vandesompele J. (2008) Nucleic Acids Res. 36, e143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chen C., Ridzon D. A., Broomer A. J., Zhou Z., Lee D. H., Nguyen J. T., Barbisin M., Xu N. L., Mahuvakar V. R., Andersen M. R., Lao K. Q., Livak K. J., Guegler K. J. (2005) Nucleic Acids Res. 33, e179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Miranda K. C., Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., Lim B., Rigoutsos I. (2006) Cell 126, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 22. Vasileva A., Tiedau D., Firooznia A., Müller-Reichert T., Jessberger R. (2009) Curr. Biol. 19, 630–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Bouhallier F., Allioli N., Lavial F., Chalmel F., Perrard M. H., Durand P., Samarut J., Pain B., Rouault J. P. (2010) RNA 16, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Damestoy A., Perrard M. H., Vigier M., Sabido O., Durand P. (2005) Reprod. Biol. Endocrinol. 3, 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Itman C., Loveland K. L. (2008) Dev. Dyn. 237, 97–111 [DOI] [PubMed] [Google Scholar]
- 26. Forman J. J., Legesse-Miller A., Coller H. A. (2008) Proc. Natl. Acad. Sci. U.S.A. 105, 14879–14884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Jung Y. H., Gupta M. K., Shin J. Y., Uhm S. J., Lee H. T. (2010) Mol. Hum. Reprod. 16, 804–810 [DOI] [PubMed] [Google Scholar]
- 28. Hayashi K., Chuva de Sousa Lopes S. M., Kaneda M., Tang F., Hajkova P., Lao K., O'Carroll D., Das P. P., Tarakhovsky A., Miska E. A., Surani M. A. (2008) PLoS One 3, e1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. West J. A., Viswanathan S. R., Yabuuchi A., Cunniff K., Takeuchi A., Park I. H., Sero J. E., Zhu H., Perez-Atayde A., Frazier A. L., Surani M. A., Daley G. Q. (2009) Nature 460, 909–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Boyerinas B., Park S. M., Hau A., Murmann A. E., Peter M. E. (2010) Endocr. Relat. Cancer 17, F19–F36 [DOI] [PubMed] [Google Scholar]
- 31. Qian Z. R., Asa S. L., Siomi H., Siomi M. C., Yoshimoto K., Yamada S., Wang E. L., Rahman M. M., Inoue H., Itakura M., Kudo E., Sano T. (2009) Mod. Pathol. 22, 431–441 [DOI] [PubMed] [Google Scholar]
- 32. Sgarra R., Rustighi A., Tessari M. A., Di Bernardo J., Altamura S., Fusco A., Manfioletti G., Giancotti V. (2004) FEBS Lett. 574, 1–8 [DOI] [PubMed] [Google Scholar]
- 33. Chieffi P., Battista S., Barchi M., Di Agostino S., Pierantoni G. M., Fedele M., Chiariotti L., Tramontano D., Fusco A. (2002) Oncogene 21, 3644–3650 [DOI] [PubMed] [Google Scholar]
- 34. Lee E. K., Gorospe M. (2011) RNA Biol. 8, 44–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Brennecke J., Stark A., Russell R. B., Cohen S. M. (2005) PLoS Biol. 3, e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Shyu A. B., Wilkinson M. F., van Hoof A. (2008) EMBO J. 27, 471–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Björk J. K., Sandqvist A., Elsing A. N., Kotaja N., Sistonen L. (2010) Development 137, 3177–3184 [DOI] [PubMed] [Google Scholar]
- 38. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010) Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fukuda T., Yamagata K., Fujiyama S., Matsumoto T., Koshida I., Yoshimura K., Mihara M., Naitou M., Endoh H., Nakamura T., Akimoto C., Yamamoto Y., Katagiri T., Foulds C., Takezawa S., Kitagawa H., Takeyama K., O'Malley B. W., Kato S. (2007) Nat. Cell Biol. 9, 604–611 [DOI] [PubMed] [Google Scholar]
- 40. Sato H., Gutti R., Dufau M. L., Tsai-Morris C. H. (2008) The Endocrine Society 90th Annual Meeting, San Francisco, CA, June 15–18, 2008, P1–660, The Endocrine Society, Chevy Chase, MD [Google Scholar]
- 41. He L., He X., Lim L. P., de Stanchina E., Xuan Z., Liang Y., Xue W., Zender L., Magnus J., Ridzon D., Jackson A. L., Linsley P. S., Chen C., Lowe S. W., Cleary M. A., Hannon G. J. (2007) Nature 447, 1130–1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Tay Y., Zhang J., Thomson A. M., Lim B., Rigoutsos I. (2008) Nature 455, 1124–1128 [DOI] [PubMed] [Google Scholar]
- 43. Melton C., Judson R. L., Blelloch R. (2010) Nature 463, 621–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yu F., Yao H., Zhu P., Zhang X., Pan Q., Gong C., Huang Y., Hu X., Su F., Lieberman J., Song E. (2007) Cell 131, 1109–1123 [DOI] [PubMed] [Google Scholar]
- 45. Medina P. P., Nolde M., Slack F. J. (2010) Nature 467, 86–90 [DOI] [PubMed] [Google Scholar]
- 46. Jennewein C., von Knethen A., Schmid T., Brüne B. (2010) J. Biol. Chem. 285, 11846–11853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Y., Rathinam R., Walch A., Alahari S. K. (2009) J. Biol. Chem. 284, 23094–23106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Park S. Y., Lee J. H., Ha M., Nam J. W., Kim V. N. (2009) Nat. Struct. Mol. Biol. 16, 23–29 [DOI] [PubMed] [Google Scholar]
- 49. Long J., Wang Y., Wang W., Chang B. H., Danesh F. R. (2011) J. Biol. Chem. 286, 11837–11848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Miyaki S., Sato T., Inoue A., Otsuki S., Ito Y., Yokoyama S., Kato Y., Takemoto F., Nakasa T., Yamashita S., Takada S., Lotz M. K., Ueno-Kudo H., Asahara H. (2010) Genes Dev. 24, 1173–1185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Cordes K. R., Sheehy N. T., White M. P., Berry E. C., Morton S. U., Muth A. N., Lee T. H., Miano J. M., Ivey K. N., Srivastava D. (2009) Nature 460, 705–710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Esau C., Kang X., Peralta E., Hanson E., Marcusson E. G., Ravichandran L. V., Sun Y., Koo S., Perera R. J., Jain R., Dean N. M., Freier S. M., Bennett C. F., Lollo B., Griffey R. (2004) J. Biol. Chem. 279, 52361–52365 [DOI] [PubMed] [Google Scholar]
- 53. Jordan S. D., Krüger M., Willmes D. M., Redemann N., Wunderlich F. T., Brönneke H. S., Merkwirth C., Kashkar H., Olkkonen V. M., Böttger T., Braun T., Seibler J., Brüning J. C. (2011) Nat. Cell Biol. 13, 434–446 [DOI] [PubMed] [Google Scholar]
- 54. Kent O. A., Chivukula R. R., Mullendore M., Wentzel E. A., Feldmann G., Lee K. H., Liu S., Leach S. D., Maitra A., Mendell J. T. (2010) Genes Dev. 24, 2754–2759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Yi R., Poy M. N., Stoffel M., Fuchs E. (2008) Nature 452, 225–229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Bueno M. J., Pérez de Castro I., Gómez de Cedrón M., Santos J., Calin G. A., Cigudosa J. C., Croce C. M., Fernández-Piqueras J., Malumbres M. (2008) Cancer Cell 13, 496–506 [DOI] [PubMed] [Google Scholar]
- 57. Eiring A. M., Harb J. G., Neviani P., Garton C., Oaks J. J., Spizzo R., Liu S., Schwind S., Santhanam R., Hickey C. J., Becker H., Chandler J. C., Andino R., Cortes J., Hokland P., Huettner C. S., Bhatia R., Roy D. C., Liebhaber S. A., Caligiuri M. A., Marcucci G., Garzon R., Croce C. M., Calin G. A., Perrotti D. (2010) Cell 140, 652–665 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.