Background: The mechanisms by which bone responds to changes in its loading environment are poorly understood.
Results: Mechanical signals induce Hif-1α expression, and mice lacking Hif-1α in bone are more responsive to loading.
Conclusion: Hif-1α is a novel regulator of skeletal mechanotransduction that impinges on Wnt signaling.
Significance: Understanding skeletal mechanotransduction may lead to the development of therapies designed to enhance bone formation.
Keywords: Beta-Catenin, Hypoxia-inducible Factor (HIF), Mechanotransduction, Osteoblasts, Osteocyte
Abstract
Mechanical loads induce profound anabolic effects in the skeleton, but the molecular mechanisms that transduce such signals are still poorly understood. In this study, we demonstrate that the hypoxia-inducible factor-1α (Hif-1α) is acutely up-regulated in response to exogenous mechanical stimuli secondary to prostanoid signaling and Akt/mTOR (mammalian target of rapamycin) activation. In this context, Hif-1α associates with β-catenin to inhibit Wnt target genes associated with bone anabolic activity. Mice lacking Hif-1α in osteoblasts and osteocytes form more bone when subjected to tibia loading as a result of increased osteoblast activity. Taken together, these studies indicate that Hif-1α serves as a negative regulator of skeletal mechanotransduction to suppress load-induced bone formation by altering the sensitivity of osteoblasts and osteocytes to mechanical signals.
Introduction
The ability of bone to serve as an effective weight-bearing structure depends upon its capacity to adapt to its functional environment. As such, skeletal mass is continually added and removed according to physical demands. When mechanical loads exceed those associated with habitual use, new bone is formed via an osteogenic response that is characterized by increased osteoblast proliferation, enhanced osteogenic gene expression, and decreased osteoclast activity (1, 2). Conversely, when routine loads are removed or reduced, osteoblastic activity is suppressed and bone loss ensues (3). In this way, skeletal architecture can be optimized to prevent fracture and minimized to limit the energy expenditure necessary for maintenance.
Previous studies using both in vivo and in vitro models of mechanical loading have begun to identify anabolic signaling mechanisms associated with load-induced bone formation. These mechanisms include the rapid release of autocrine/paracrine factors such as ATP, prostaglandin, and nitric oxide (4–6) and the activation of intracellular calcium and kinase signaling pathways (7, 8). A growing body of work indicates that Wnt signaling is an important regulator of skeletal response to mechanical loading (9, 10). Experimental loading regulates the expression of several Wnt ligands, receptors, and antagonists, which in turn increases β-catenin nuclear translocation (10–12) and the expression of its target genes (13–15).
Other studies suggest the existence of negative feedback mechanisms that operate to modulate skeletal mechanotransduction. For example, during prolonged exposure to fluid flow, osteoblasts exhibit refractory periods in which the activation of anabolic signaling mechanisms is impaired (16, 17). Additionally, intermittent intervals of loading interspersed with periods of rest produce more bone than sustained loading, suggesting the engagement of mechanisms that desensitize bone cells to mechanical signals (18–20). As described below, previous studies from a number of laboratories suggested that the transcription factor, Hif-1α,3 might function as a regulator of bone cell sensitivity to mechanical stimuli.
Hif-1, a transcription factor originally identified as a regulator of the cellular response to molecular oxygen levels (21, 22), activates angiogenic and glycolytic gene programs required for cells to adapt to hypoxia. The cellular levels of Hif-1 and its nuclear translocation are governed by regulated proteolysis. The α-subunit of the molecule contains an oxygen-dependent degradation domain that is subject to prolyl hydroxylation and subsequent E3 ubiquitin ligation by the tumor suppressor protein von Hippel-Lindau (Vhl) and proteasomal degradation. Inhibition of prolyl hydroxylation under hypoxic conditions allows Hif-1α to accumulate and translocate to the nucleus, where it forms a dimer with the Hif-1β subunit (21, 22).
It is increasingly recognized that the Hif-1 pathway also regulates cellular functions independent of those related to hypoxia. A number of growth factors (21, 22) and paracrine factors (23–26) activate the Hif-1 pathway in several cell types under normoxia. In osteoblasts, insulin-like growth factor-1 stabilizes Hifα via a mechanism that involves PI3-kinase/Akt signaling (27), and the expression of Hif-1α is necessary for normal osteoblast proliferation (28). Additionally, mechanical signals prevent the degradation of Hif α-subunits in both skeletal and cardiac muscle (29, 30). In response to these stimuli, Hif-1 activates angiogenic and metabolic responses that are required for the anabolic response.
In the course of analyzing the role of Hif-1α in skeletal development, we created mice that lacked this transcription factor in osteoblasts and osteocytes (31). The immature Hif-1α mutants had deficits in cortical and trabecular bone, which we attributed to impaired development of the skeletal vasculature (31). Surprisingly, as these mice matured, they acquired more cortical bone when compared with controls, suggesting that the loss of Hif-1α caused an enhanced adaptive response in cortical bone architecture. In this study, we show that in vitro fluid flow markedly up-regulates Hif-1α, which partners with β-catenin to inhibit Wnt target genes associated with bone anabolic activity. Consequently, removal of Hif-1α from osteoblasts sensitizes bone to load-induced bone formation in vivo by enhancing the response of osteoblasts to mechanical stimuli. Our findings indicate that Hif-1α functions as a negative regulator of skeletal mechanotransduction to suppress load-induced bone formation.
EXPERIMENTAL PROCEDURES
Generation of Transgenic Mice
The generation of mice lacking Hif-1α in osteoblasts (ΔHif-1α) was described previously (31). Briefly, OC-Cre mice (32) were crossed with mice in which the second exon of Hif-1α is floxed (33). ΔHif-1α;ΔHif-2α mice were generated by crossing ΔHif-1α mice with mice containing Hif-2α floxed alleles (34). BAT-gal mice (35) were obtained from The Jackson Laboratory and crossed with Hif-1α floxed mice to generate Hif-1αflox/flox;BAT-gal+/− mice. Mice containing mTOR floxed alleles were generously provided by Dr. Christopher Lynch (36). All mice were maintained on a C57BL/6 background. PCR analysis from ear biopsies was used to confirm genotypes. The Institutional Animal Care and Use Committees of the Johns Hopkins University School of Medicine and the University of Washington approved all animal procedures.
MicroCT Analysis
The mouse femur was scanned using a desktop microtomographic imaging system (SkyScan 1172, SkyScan, Kontich, Belgium) in accordance with the recommendations of the American Society for Bone and Mineral Research (ASBMR) (37). The femur was scanned at 50 keV and 200 mA using a 0.5-mm aluminum filter with an isotropic voxel size of 10 μm. The resulting two-dimensional cross-sectional images are shown in gray scale. Cortical bone parameters were assessed at the femoral midshaft and represent an average of 50 CT slices (500 μm).
Cell Isolation and Culture
Osteoblasts were isolated from the calvaria of newborn Hif-1α floxed, Hif-1α flox;BAT-gal+/−, and mTOR floxed mice by serial digestion in 1.8 mg/ml collagenase type I and maintained in minimum essential medium α supplemented with 10% FBS and 1% penicillin/streptomycin. To disrupt Hif-1α or mTOR expression, osteoblasts were infected with control adenovirus expressing green fluorescent protein or adenovirus expressing Cre recombinase (Vector Biolabs) at a Multiplicity of Infection 100 (28). MLO-Y4 cells, kindly provided by Dr. Lynda Bonewald, were maintained in minimum essential medium α supplemented with 5% FBS, 5% calf serum, and 1% penicillin/streptomycin.
Application of Fluid Flow
For fluid flow studies, cells were seeded to glass slides (75 × 38 × 1.0 mm) coated with type I collagen (150 μg/ml) 48 h before stimulation. After starving cells overnight in medium containing 0.5% FBS, slides were positioned in parallel plate flow chambers (38) connected to a digital peristaltic pump (Masterflex, Cole-Parmer) via rigid wall tubing in a closed loop. A shear stress of 10 dynes/cm2 was generated using a peak flow rate of 28 ml/min. Untreated, controls were positioned in the flow chamber but not connected to the peristaltic pump. Pharmacological agents were added 30 min prior to the initiation of fluid flow and maintained in the flow medium.
In Vivo Mechanical Loading
The right tibiae of female control and ΔHif-1α mice were non-invasively loaded via cantilever bending (39). Briefly, the right tibia was held proximal to the tibial crest with a brass gripping cup, whereas a brass tine attached to a computer control linear actuator applied the small required forces to the lateral distal tibia (<0.3 newtons). The experimental loading regimen consisted of 100 cycles of loading per bout (1 Hz), 3 days/week for 3 weeks. Given the differing cortical morphology of the ΔHif-1α mice, end loads were adjusted using beam theory applied to midshaft cortical bone morphology obtained by pre-experimental high resolution microCT imaging (vivaCT 40, Scanco, Inc.) so that peak-induced normal strains were equivalent for control and ΔHif-1α mice. The left tibiae served as a non-loaded contralateral control for each mouse. Calcein (15 mg/kg of interperitoneal) was administered on days 10 and 19, and all mice were sacrificed on day 22. Dynamic histomorphometric measurements were performed 2 mm proximal to the tibial-fibular junction according to standard practices (40).
Quantitative Real-time PCR
Total RNA was extracted from osteoblasts using TRIzol (Invitrogen), and 1 μg was reverse-transcribed using the iScript cDNA synthesis system (Bio-Rad). Two μl of cDNA was subjected to PCR amplification using the iQ SYBR Green Supermix (Bio-Rad). Primer sequences were obtained from PrimerBank. Reactions were normalized to endogenous β-actin reference transcript.
Chromatin Immunoprecipitation
ChIP assays were performed using an agarose ChIP kit (Pierce) according to the manufacturer's instructions and a ChIP-qualified antibody specific for β-catenin (Thermo Scientific). For each sample, cells isolated from two slides were pooled to ensure sufficient chromatin yield. Primer sequences for the Axin2 promoter are available upon request.
Protein Isolation and Assays
Protein was extracted from cultured osteoblast and MLO-Y4 cells in 0.1% Triton X-100 containing protease and phosphatase inhibitors. The extracts were separated on 10% SDS/polyacrylamide gels and transferred to PVDF membranes. Antibodies were obtained from Cell Signaling Technologies and Novus Biological. Bound antibodies were visualized using the SuperSignal West Femto substrate (Pierce). Co-immunoprecipitation was performed overnight at 4 °C in a reaction containing 2 μg of antibody specific for Hif-1α (Novus) or β-catenin (Cell Signaling). A β-galactosidase enzyme assay system (Promega) was used to assess activity from the BAT-gal reporter and was normalized to total protein concentration using the BCA method (Pierce).
Statistical Analysis
Results are expressed as mean ± S.E. All statistical tests were two-sided. A p value less than 0.05 was considered significant. Comparability of two groups of data was assessed using a Student's t test.
RESULTS
Mice Lacking Hif-1α in Osteoblasts Acquire Increased Cortical Bone
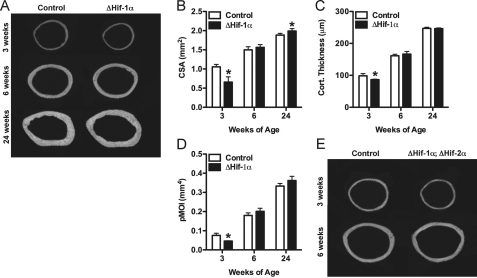
The potential role for Hif-1α in the response to mechanical loading was suggested by the striking changes in cortical bone architecture observed in ΔHif-1α mice. As reported previously (31), female ΔHif-1α mice begin life with narrow bones with thin cortices and a reduced polar moment of inertia at 3 weeks of age (Fig. 1, A–D). However, by 6 weeks of age, cortical bone structure at the femoral mid-diaphysis normalizes, and by 24 weeks of age, ΔHif-1α mice exhibit significantly increased cross-sectional area and increased resistance to torsional loading (polar moment of inertia, p = 0.06) when compared with wild-type littermates. A similar phenomenon was observed in male mice (data not shown) and the trabecular bone compartment (supplemental Fig. 1). We initially thought that this age-related change in cortical bone architecture might be due to compensatory up-regulation of Hif-2α (31), and therefore, we examined cortical bone architecture in mice lacking either Hif-2α alone or double knock-out mice lacking both Hif α-subunits. No alterations in cortical bone structure were seen in the ΔHif-2α mice (data not shown and Ref. 28), and mice lacking both Hif α-subunits in osteoblasts exhibited changes in cortical bone structure that were identical to that seen in ΔHif-1α mice (Fig. 1E). Thus, the increase in cortical bone in ΔHif-1α mice was not due to compensation by Hif-2α.
FIGURE 1.
Mice lacking Hif-1α in osteoblasts acquire increased cortical bone. A, representative microCT images illustrate cortical bone structure at the femoral mid-diaphysis in control and ΔHif-1α mice at 3, 6, and 24 weeks of age. B–D, bar graphs show quantification of the tissue cross-sectional area (CSA) (B), cortical thickness (Cort. Thickness) (C), and polar moment of inertia (pMOI) (D). E, representative microCT images from the femoral mid-diaphysis in control and ΔHif-1α;ΔHif-2α double mutants. Data are plotted mean ± S.E. with at least five mice being examined per genotype. *, p < 0.05
Mechanical Signals Induce Hif-1α Expression in Osteocytes and Osteoblasts
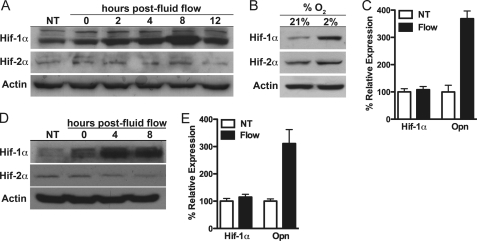
The major determinants of the shape and size of the cortical bone during skeletal maturation are sex hormones and mechanical forces. Because increases in cortical bone were observed in both male and female ΔHif-1α mice, we examined whether Hif-1α influences the response of bone to anabolic loading. We first tested the effect of mechanical signals on Hif-1α expression in vitro by exposing the osteocyte-like cell line MLO-Y4 (41) and primary mouse osteoblasts to fluid flow. Hif-1α protein levels were increased 2 h after fluid flow exposure and reached a peak 8 h after treatment in MLO-Y4 cells, whereas levels of Hif-2α were not affected (Fig. 2A). Hypoxia increased the levels of both isoforms (Fig. 2B). A comparable induction of Hif-1α, but not Hif-2α, expression was observed in primary osteoblasts with Hif-1α protein levels peaking 4–8 h after fluid flow stimulation (Fig. 2D). As expected, the levels of Hif-1α mRNA were not affected by fluid flow stimulation in either cell type (Fig. 2, C and E), indicating that mechanical signals regulate Hif-1α protein levels via a post-transcriptional mechanism.
FIGURE 2.
Mechanical signals induce Hif-1α expression in osteocytes and osteoblasts. A and D, Hif-1α and Hif-2α levels were examined at the indicated times after MLO-Y4 osteocytes (A), and primary osteoblasts (D) were exposed to fluid flow for 1 h or left untreated (NT). B, exposing MLO-Y4 cells to hypoxia (2% O2) for 6 h increased both Hif-1α and Hif-2α protein levels. C and E, Hif-1α and osteopontin (Opn) mRNA levels were examined by quantitative PCR 8 h after MLO-Y4 cells (C) and primary osteoblasts (E) were exposed to fluid flow or left untreated.
Prostanoid/mTOR Signaling Is Required for Induction of Hif-1α in Response to Fluid Flow
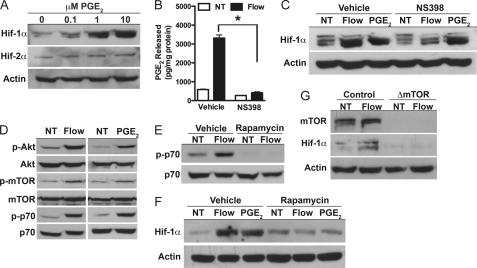
The delay in Hif-1α accumulation following fluid flow suggested the involvement of an intermediary factor in the induction of Hif-1α. Because prostaglandins are known to regulate Hif-1α expression in non-osseous cells (26) and are released by bone cells following mechanical stimulation (4), we examined the effect of PGE2 on Hif-1α expression in MLO-Y4 cells. The addition of PGE2 increased the expression of Hif-1α in a dose-dependent fashion (Fig. 3A), consistent with a possible role in mediating the induction of Hif-1α following fluid flow exposure. Treatment of cells with a COX-2 antagonist NS398, which inhibited PGE2 synthesis in response to fluid flow (Fig. 3B), abolished the effect of fluid flow on Hif-1α expression (Fig. 3C), indicating that prostanoid signaling is required for load-induced expression of Hif-1α.
FIGURE 3.
Prostanoid/mTOR signaling is required for induction of Hif-1α in response to fluid flow. A, Hif-1α, but not Hif-2α, protein levels were dose-dependently increased 8 h after treating MLO-Y4 cells with PGE2. B, NS398 (0.1 μm) was used to inhibit PGE2 synthesis after fluid flow exposure. NT, untreated. C, blocking PGE2 synthesis inhibited the increase in Hif-1α protein levels after exposure to fluid flow, but not 10 μm PGE2. D, both fluid flow and PGE2 induced the activation of Akt/mTOR signals as indicated by increased phosphorylation levels. p-Akt, phosphorylated Akt; p-mTOR, phosphorylated m-TOR; p-p70, phosphorylated p70. E and F, rapamycin (10 nm) abolished mTOR activity (E) and the effect of fluid flow and PGE2 on Hif-1α protein levels (F). G, disrupting mTOR expression abolished the induction of Hif-1α in primary osteoblasts. Data are plotted mean ± S.E. *, p < 0.05
We next probed signaling pathways downstream of prostanoid signaling with a focus on Akt/mTOR signaling as this pathway has previously been linked with the regulation of Hif-1α. In this pathway, Akt stimulates an increase in the activity of mTOR, which then directly enhances the expression of Hif-1α via phosphorylation events that inhibit Hif-1α proteolysis and by increasing Hif-1α protein synthesis (42, 43). Because both fluid flow and PGE2 activated the Akt/mTOR pathway (Fig. 3D), we treated MLO-Y4 cells with rapamycin to determine whether this signaling pathway is required for Hif-1α expression after fluid flow exposure (Fig. 3E). Rapamycin treatment inhibited the effects of both fluid flow and PGE2 on Hif-1α expression (Fig. 3F). Identical effects of mTOR signaling were observed in primary mouse osteoblasts as disrupting mTOR abolished the effect of fluid flow on Hif-1α expression (Fig. 3G). These data suggest a mechanism whereby mechanical signals stimulate the production and release of prostaglandins that in turn activate the Akt/mTOR pathway to regulate Hif-1α expression.
Loss of Hif-1α in Osteoblasts Enhances Responsiveness to Mechanical Loading
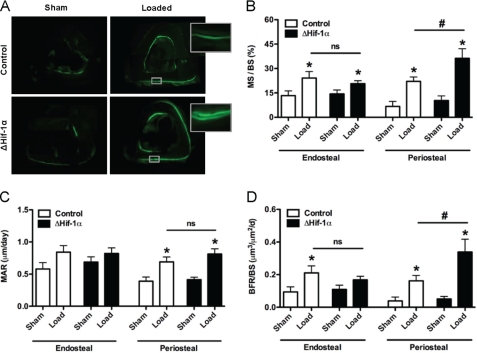
To directly test the role of Hif-1α in the adaptive response of bone to mechanical loading in vivo,we exposed ΔHif-1α mice to a tibia-loading regimen (39). The right tibiae of 21-week-old female control and ΔHif-1α mice were subjected to 100 cycles/day of tibial cantilever bending thrice weekly for a total of 3 weeks. Peak-induced normal strains were similar in both genotypes (1790 ± 70 microstrain for control versus 1745 ± 40 microstrain for ΔHif-1α). At this time point, measures of osteoblast performance in the contralateral tibiae were not significantly affected by the disruption of Hif-1α. As expected, dynamic measures of periosteal bone formation including mineralizing surface, mineral apposition rate, and bone formation rate were significantly increased in the loaded tibiae of both genotypes. However, the mineralizing surface and bone formation rate increased to a greater extent in ΔHif-1α mice when compared with controls (Fig. 4). Specifically, the increase in bone formation rate after loading in ΔHif-1α mice was 161% of that evident in the controls. Measures of osteoblast activity after loading on the endocortical surface were not influenced by the loss of Hif-1α function. These data suggest that loss of Hif-1α increases bone formation in response to a mildly osteogenic mechanical stimulus due to increased activation of resident bone cells and is compatible with the proposed role of Hif-1α as a negative regulator of load-induced bone formation.
FIGURE 4.
Loss of Hif-1α in osteoblasts enhances responsiveness to mechanical loading. The right tibia of 21-week-old female control and ΔHif-1α mice was subjected to a 3-week tibia loading regime, whereas the left tibia served as an unloaded, internal control (Sham). Both groups experienced equivalent peak periosteal strains. A, representative tissue sections for sham and loaded tibia in which calcein was injected on days 10 and 19 to assess bone formation. Magnified images are shown for the loaded samples. B–D, bar graphs show quantification of the mineralizing surface per bone surface (MS/BS) (B), mineral apposition rate (MAR) (C), and bone formation rate per bone surface (BFR/BS) (D) on both the periosteal and the endosteal surfaces. Data are plotted mean ± S.E. with eight mice being examined per genotype. *, p < 0.05 versus sham limb, # p < 0.05 versus wild-type control. ns, not significant.
Hif-1α Antagonizes Load-induced β-Catenin Signaling
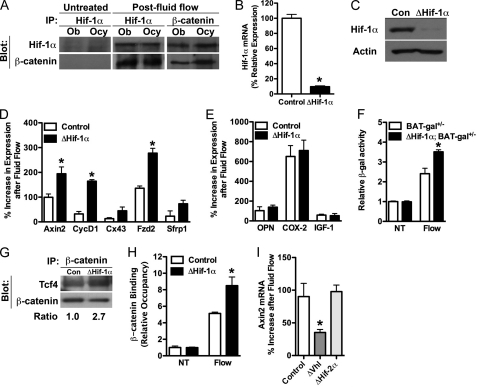
We next sought to determine the molecular basis for the attenuation of load-induced bone formation by Hif-1α. As discussed above, previous studies have demonstrated that mechanical loading of bone is associated with activation of the canonical Wnt/β-catenin pathway (9, 10). The striking up-regulation of Hif-1α by mechanical signals at time points when β-catenin is reported to be active (10, 14, 44) suggested that these two transcription factors might interact to modulate responses to mechanical signals. In accordance with this hypothesis, co-immunoprecipitation studies revealed that Hif-1α and β-catenin directly interact after mechanical stimulation in both primary osteoblasts and MLO-Y4 cells (Fig. 5A). Disruption of Hif-1α in primary osteoblasts (Fig. 5, B and C) greatly increased load-induced expression of β-catenin target genes and the activity of a β-catenin reporter gene (Fig. 5, D and F) but did not affect genes associated with other signaling pathways (Fig. 5E). Moreover, the amounts of Tcf4 that co-immunoprecipitated with β-catenin (Fig. 5G) and the Axin2 promoter occupancy of β-catenin (Fig. 5H) were increased in osteoblasts lacking Hif-1α after exposure to fluid flow. These data, together with the observations that osteoblasts overexpressing Hifs via the disruption of Vhl demonstrated reduced expression of Axin2 after loading and that disruption of Hif-2α was without effect (Fig. 5I), indicate that Hif-1α acts as a negative regulator of load-induced bone formation by specifically interacting with β-catenin and suppressing its activity.
FIGURE 5.
Hif-1α antagonizes load-induced β-catenin signaling. A, co-immunoprecipitation (IP) revealed that Hif-1α and β-catenin interact in both primary osteoblasts (Ob) and MLO-Y4 (Ocy) cells after exposure to fluid flow. B and C, primary osteoblasts isolated from Hif-1α floxed mice were infected with adenovirus expressing Cre recombinase to abolish Hif-1α expression (B) and prevent its induction after fluid flow exposure (C). Con, control. D and E, osteoblasts deficient for Hif-1α (ΔHif-1α) exhibited increased expression levels of β-catenin target genes (D), but not osteopontin (OPN), Cox-2, or IGF-1 (E), after exposure to fluid flow. F, BAT-gal reporter activity was increased in osteoblasts rendered deficient for Hif-1α and exposed to fluid flow. NT, untreated. G and H, an increased association of β-catenin with Tcf4 (G) and increased binding to the Axin2 promoter (H) were evident in ΔHif-1α osteoblasts after exposure to fluid flow in co-immunoprecipitation and ChIP assays, respectively. I, overexpressing Hif-1α by disrupting the expression of Vhl inhibited Axin2 expression after fluid flow exposure, whereas disrupting Hif-2α expression was without effect. Data are plotted mean ± S.E. *, p < 0.05
DISCUSSION
In this study, we show that activation of the transcription factor Hif-1α is a primary response to mechanical loading of bone and that it appears to function in the osteoblast as a negative regulator of load-induced bone formation. Our studies were prompted by observations in mice lacking Hif-1α specifically in osteoblasts and osteocytes, which exhibited dramatic shifts in cortical bone architecture during postnatal development. Thus, deficits in cortical bone seen in immature ΔHif-1α mice, likely due in part to impaired development of the skeletal vasculature (31), were reversed such that the mature mutants had more cortical bone when compared with controls. We hypothesized that this developmental switch in cortical bone architecture might represent enhanced adaptation to mechanical loading associated with increased ambulation during postnatal life. The studies described here were designed to explore the possible role of Hif-1α in this phenomenon.
The contrasting skeletal phenotypes observed as the ΔHif-1α mice mature suggest that Hif-1 exerts distinct functions at different times during skeletal development. During early development, the accumulation of both trabecular and cortical bone in the axial skeleton of ΔHif-1α mice is reduced, likely due to a decrease in vascularization critical for the initial specification and differentiation of bone forming osteoblasts. As the mouse matures and increases its ambulatory activity, it is possible that Hif-1α-generated signals impinge on other cellular functions including those involved in adaptation of bone to mechanical loads. In this regard, Hif-1α is known to be regulated by mechanical events in other tissues. For example, loading of the rat extensor digitorum longus muscle induces the accumulation of Hif α-subunits (29). Likewise, hemodynamic loading or stretching of cardiac tissue by expanding an intraventricular balloon stimulated Hif-1α accumulation in the cardiac muscle (30). In this context, Hif-1α-generated signals likely participate in mechanisms that enable muscle to adapt to increased mechanical forces as well as the heightened demand for oxygen delivery evidenced by up-regulation of angiogenic gene expression in the stretched myocardium. This situation differs from the exogenous mechanical stimuli implemented in the current study, which are not associated with significant hypoxia or increased angiogenesis (45), but rather, are considered anabolic. On the other hand, application of much more intensive loads that induce bone fatigue with significant tissue damage is associated with increased skeletal vascularity likely due to tissue hypoxia (46, 47).
An important conceptual conclusion from our studies is that Hif-1α normally functions to attenuate anabolic responses to exogenous loads. As mentioned above, intermittent application of mechanical loading with interspersed periods of rest is a more effective regimen for increasing bone formation than uninterrupted sustained loading (18–20). Unregulated anabolic signaling in loaded osteoblasts would increase oxidative, metabolic, and genetic stress and ultimately limit their performance. Interestingly, reactive oxygen species, which are generated by mechanical loading in other tissues (48), stabilize Hif α-subunits (49, 50), and in turn, Hif-1-generated signals inhibit reactive oxygen species production (51, 52). These observations suggest that Hif-1α may function transiently as a negative regulator of load-induced bone formation to ensure periods of quiescence during which time cells can recover from the potentially damaging effects of cellular stressors.
Several lines of evidence support the conclusion that Hif-1α attenuates load-induced bone formation by interfering with β-catenin, a critical regulator of osteoblast specification and function (53, 54). First, Hif-1α directly interacted with β-catenin. Second, osteoblasts deficient in Hif-1α exhibited enhanced activation of a β-catenin reporter gene, enhanced target gene expression, and increased association with Tcf4 after exposure to fluid flow. Conversely, osteoblasts overexpressing Hif-1α had diminished measures of β-catenin activity. Moreover, previous work in other cell types support our findings. For example, inhibition of Wnt/β-catenin signaling has been reported in colon cancer and non-small lung cancer cell lines exposed to hypoxia (55, 56). In these studies, Hif-1α interacted with β-catenin via its NH2 terminus and thereby inhibited the interaction of β-catenin with TCF4. However, the regulation of Wnt/β-catenin signaling may be cell- and differentiation stage-specific as Hif-1 was recently reported to enhance canonical Wnt signaling in embryonic stem cells but to have no effect on the pathway in neuroprogenitor cells (57). Finally, it should be noted that other factors including FoxO1 (58) and NMP4-CIZ (12) have also been shown to modulate β-catenin activity in osteoblasts, with the actions of the latter being responsive to experimental loading. Whether or to what extent these factors cooperate with Hif-1α to regulate mechanical responses to loading in bone remains to be determined but fully supports the importance of controlling β-catenin activity.
In summary, we have identified Hif-1α as a negative regulator of load-induced bone formation. Our results suggest a model in which loading activates the canonical Wnt/β-catenin pathway, which induces bone anabolic pathways in osteoblasts. Concomitant up-regulation of Hif-1α attenuates the magnitude of Wnt/β-catenin activity by sequestration of β-catenin, thereby enabling periods of quiescence during which time cells can recover from the potentially damaging effects of cellular stressors. Our findings advance the understanding of the mechanisms by which bone cells perceive and respond to changes in their mechanical environment and may lead to the development of strategies designed to increase bone mass via an anabolic response.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AR049410 (to T. L. C.). This work was also supported by a Career Development Award from the Veterans Administration (to R. C. R).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Fig. 1.
- Hif
- hypoxia-inducible factor
- Vhl
- Von Hippel-Lindau
- mTOR
- mammalian target of rapamycin
- CT
- computer tomography
- microCT
- micro-computer tomography
- PGE2
- prostaglandin E2.
REFERENCES
- 1. Hillam R. A., Skerry T. M. (1995) J. Bone Miner. Res. 10, 683–689 [DOI] [PubMed] [Google Scholar]
- 2. Pead M. J., Skerry T. M., Lanyon L. E. (1988) J. Bone Miner. Res 3, 647–656 [DOI] [PubMed] [Google Scholar]
- 3. Morey E. R., Baylink D. J. (1978) Science 201, 1138–1141 [DOI] [PubMed] [Google Scholar]
- 4. Bakker A. D., Soejima K., Klein-Nulend J., Burger E. H. (2001) J. Biomech. 34, 671–677 [DOI] [PubMed] [Google Scholar]
- 5. Genetos D. C., Geist D. J., Liu D., Donahue H. J., Duncan R. L. (2005) J. Bone Miner. Res. 20, 41–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pead M. J., Lanyon L. E. (1989) Calcif. Tissue Int. 45, 34–40 [DOI] [PubMed] [Google Scholar]
- 7. Riddle R. C., Taylor A. F., Genetos D. C., Donahue H. J. (2006) Am. J. Physiol. Cell Physiol. 290, C776–C784 [DOI] [PubMed] [Google Scholar]
- 8. You J., Reilly G. C., Zhen X., Yellowley C. E., Chen Q., Donahue H. J., Jacobs C. R. (2001) J. Biol. Chem. 276, 13365–13371 [DOI] [PubMed] [Google Scholar]
- 9. Sawakami K., Robling A. G., Ai M., Pitner N. D., Liu D., Warden S. J., Li J., Maye P., Rowe D. W., Duncan R. L., Warman M. L., Turner C. H. (2006) J. Biol. Chem. 281, 23698–23711 [DOI] [PubMed] [Google Scholar]
- 10. Case N., Ma M., Sen B., Xie Z., Gross T. S., Rubin J. (2008) J. Biol. Chem. 283, 29196–29205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Santos A., Bakker A. D., Zandieh-Doulabi B., de Blieck-Hogervorst J. M., Klein-Nulend J. (2010) Biochem. Biophys. Res. Commun. 391, 364–369 [DOI] [PubMed] [Google Scholar]
- 12. Yang Z., Bidwell J. P., Young S. R., Gerard-O'Riley R., Wang H., Pavalko F. M. (2010) J. Cell Physiol. 223, 435–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Xia X., Batra N., Shi Q., Bonewald L. F., Sprague E., Jiang J. X. (2010) Mol. Cell. Biol. 30, 206–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Armstrong V. J., Muzylak M., Sunters A., Zaman G., Saxon L. K., Price J. S., Lanyon L. E. (2007) J. Biol. Chem. 282, 20715–20727 [DOI] [PubMed] [Google Scholar]
- 15. Robling A. G., Niziolek P. J., Baldridge L. A., Condon K. W., Allen M. R., Alam I., Mantila S. M., Gluhak-Heinrich J., Bellido T. M., Harris S. E., Turner C. H. (2008) J. Biol. Chem. 283, 5866–5875 [DOI] [PubMed] [Google Scholar]
- 16. Donahue S. W., Donahue H. J., Jacobs C. R. (2003) J. Biomech. 36, 35–43 [DOI] [PubMed] [Google Scholar]
- 17. Hung C. T., Pollack S. R., Reilly T. M., Brighton C. T. (1995) Clin. Orthop. Relat. Res. 256–269 [PubMed] [Google Scholar]
- 18. Srinivasan S., Ausk B. J., Poliachik S. L., Warner S. E., Richardson T. S., Gross T. S. (2007) J. Appl. Physiol. 102, 1945–1952 [DOI] [PubMed] [Google Scholar]
- 19. Robling A. G., Burr D. B., Turner C. H. (2001) J. Exp. Biol. 204, 3389–3399 [DOI] [PubMed] [Google Scholar]
- 20. Srinivasan S., Agans S. C., King K. A., Moy N. Y., Poliachik S. L., Gross T. S. (2003) Bone 33, 946–955 [DOI] [PubMed] [Google Scholar]
- 21. Semenza G. L. (2000) Genes Dev. 14, 1983–1991 [PubMed] [Google Scholar]
- 22. Semenza G. L. (2001) Cell 107, 1–3 [DOI] [PubMed] [Google Scholar]
- 23. Treins C., Giorgetti-Peraldi S., Murdaca J., Semenza G. L., Van Obberghen E. (2002) J. Biol. Chem. 277, 27975–27981 [DOI] [PubMed] [Google Scholar]
- 24. Shi Y. H., Wang Y. X., Bingle L., Gong L. H., Heng W. J., Li Y., Fang W. G. (2005) J. Pathol. 205, 530–536 [DOI] [PubMed] [Google Scholar]
- 25. Phillips R. J., Mestas J., Gharaee-Kermani M., Burdick M. D., Sica A., Belperio J. A., Keane M. P., Strieter R. M. (2005) J. Biol. Chem. 280, 22473–22481 [DOI] [PubMed] [Google Scholar]
- 26. Liu X. H., Kirschenbaum A., Lu M., Yao S., Dosoretz A., Holland J. F., Levine A. C. (2002) J. Biol. Chem. 277, 50081–50086 [DOI] [PubMed] [Google Scholar]
- 27. Akeno N., Robins J., Zhang M., Czyzyk-Krzeska M. F., Clemens T. L. (2002) Endocrinology 143, 420–425 [DOI] [PubMed] [Google Scholar]
- 28. Shomento S. H., Wan C., Cao X., Faugere M. C., Bouxsein M. L., Clemens T. L., Riddle R. C. (2010) J. Cell Biochem. 109, 196–204 [DOI] [PubMed] [Google Scholar]
- 29. Milkiewicz M., Doyle J. L., Fudalewski T., Ispanovic E., Aghasi M., Haas T. L. (2007) J. Physiol. 583, 753–766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kim C. H., Cho Y. S., Chun Y. S., Park J. W., Kim M. S. (2002) Circ. Res. 90, E25–33 [DOI] [PubMed] [Google Scholar]
- 31. Wang Y., Wan C., Deng L., Liu X., Cao X., Gilbert S. R., Bouxsein M. L., Faugere M. C., Guldberg R. E., Gerstenfeld L. C., Haase V. H., Johnson R. S., Schipani E., Clemens T. L. (2007) J. Clin. Invest. 117, 1616–1626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang M., Xuan S., Bouxsein M. L., von Stechow D., Akeno N., Faugere M. C., Malluche H., Zhao G., Rosen C. J., Efstratiadis A., Clemens T. L. (2002) J. Biol. Chem. 277, 44005–44012 [DOI] [PubMed] [Google Scholar]
- 33. Schipani E., Ryan H. E., Didrickson S., Kobayashi T., Knight M., Johnson R. S. (2001) Genes Dev. 15, 2865–2876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Gruber M., Hu C. J., Johnson R. S., Brown E. J., Keith B., Simon M. C. (2007) Proc. Natl. Acad. Sci. U.S.A. 104, 2301–2306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Maretto S., Cordenonsi M., Dupont S., Braghetta P., Broccoli V., Hassan A. B., Volpin D., Bressan G. M., Piccolo S. (2003) Proc. Natl. Acad. Sci. U.S.A. 100, 3299–3304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lang C. H., Frost R. A., Bronson S. K., Lynch C. J., Vary T. C. (2010) Am. J. Physiol. Endocrinol. Metab. 298, E1283–1294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Bouxsein M. L., Boyd S. K., Christiansen B. A., Guldberg R. E., Jepsen K. J., Müller R. (2010) J. Bone Miner. Res. 25, 1468–1486 [DOI] [PubMed] [Google Scholar]
- 38. Frangos J. A., McIntire L. V., Eskin S. G. (1988) Biotechnol. Bioeng. 32, 1053–1060 [DOI] [PubMed] [Google Scholar]
- 39. Gross T. S., Srinivasan S., Liu C. C., Clemens T. L., Bain S. D. (2002) J. Bone Miner. Res. 17, 493–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parfitt A. M., Drezner M. K., Glorieux F. H., Kanis J. A., Malluche H., Meunier P. J., Ott S. M., Recker R. R. (1987) J. Bone Miner. Res. 2, 595–610 [DOI] [PubMed] [Google Scholar]
- 41. Kato Y., Windle J. J., Koop B. A., Mundy G. R., Bonewald L. F. (1997) J. Bone Miner. Res. 12, 2014–2023 [DOI] [PubMed] [Google Scholar]
- 42. Land S. C., Tee A. R. (2007) J. Biol. Chem. 282, 20534–20543 [DOI] [PubMed] [Google Scholar]
- 43. Hudson C. C., Liu M., Chiang G. G., Otterness D. M., Loomis D. C., Kaper F., Giaccia A. J., Abraham R. T. (2002) Mol. Cell. Biol. 22, 7004–7014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hens J. R., Wilson K. M., Dann P., Chen X., Horowitz M. C., Wysolmerski J. J. (2005) J. Bone Miner. Res. 20, 1103–1113 [DOI] [PubMed] [Google Scholar]
- 45. Piekarski K., Munro M. (1977) Nature 269, 80–82 [DOI] [PubMed] [Google Scholar]
- 46. McKenzie J. A., Silva M. J. (2011) Bone 48, 250–258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Matsuzaki H., Wohl G. R., Novack D. V., Lynch J. A., Silva M. J. (2007) Calcif. Tissue Int. 80, 391–399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Powers S. K., Jackson M. J. (2008) Physiol. Rev. 88, 1243–1276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Simon M. C. (2006) Adv. Exp. Med. Biol. 588, 165–170 [DOI] [PubMed] [Google Scholar]
- 50. Brunelle J. K., Bell E. L., Quesada N. M., Vercauteren K., Tiranti V., Zeviani M., Scarpulla R. C., Chandel N. S. (2005) Cell Metab. 1, 409–414 [DOI] [PubMed] [Google Scholar]
- 51. Kim J. W., Tchernyshyov I., Semenza G. L., Dang C. V. (2006) Cell Metab. 3, 177–185 [DOI] [PubMed] [Google Scholar]
- 52. Papandreou I., Cairns R. A., Fontana L., Lim A. L., Denko N. C. (2006) Cell Metab. 3, 187–197 [DOI] [PubMed] [Google Scholar]
- 53. Holmen S. L., Zylstra C. R., Mukherjee A., Sigler R. E., Faugere M. C., Bouxsein M. L., Deng L., Clemens T. L., Williams B. O. (2005) J. Biol. Chem. 280, 21162–21168 [DOI] [PubMed] [Google Scholar]
- 54. Hill T. P., Später D., Taketo M. M., Birchmeier W., Hartmann C. (2005) Dev. Cell 8, 727–738 [DOI] [PubMed] [Google Scholar]
- 55. Kaidi A., Williams A. C., Paraskeva C. (2007) Nat. Cell Biol. 9, 210–217 [DOI] [PubMed] [Google Scholar]
- 56. Lim J. H., Chun Y. S., Park J. W. (2008) Cancer Res. 68, 5177–5184 [DOI] [PubMed] [Google Scholar]
- 57. Mazumdar J., O'Brien W. T., Johnson R. S., LaManna J. C., Chavez J. C., Klein P. S., Simon M. C. (2010) Nat. Cell Biol. 12, 1007–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Almeida M., Han L., Martin-Millan M., O'Brien C. A., Manolagas S. C. (2007) J. Biol. Chem. 282, 27298–27305 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.