Background: Methicillin-resistant Staphylococcus aureus (MRSA) PK has been recently identified as a potential novel antimicrobial drug target.
Results: Screening of a marine extract library led to the identification of several bis-indole alkaloids as novel potent and selective MRSA PK inhibitors.
Conclusion: These results help to understand the mechanism of the antibacterial activities of marine bis-indole alkaloids.
Significance: This study provides the basis for development of potential novel antimicrobials.
Keywords: Enzyme Inhibitors, Natural products, Pyruvate Kinase, Staphylococcus aureus, X-ray Crystallography, Bis-indole Alkaloids, Methicillin-resistant S. aureus (MRSA), Antibacterial Activity
Abstract
Novel classes of antimicrobials are needed to address the emergence of multidrug-resistant bacteria such as methicillin-resistant Staphylococcus aureus (MRSA). We have recently identified pyruvate kinase (PK) as a potential novel drug target based upon it being an essential hub in the MRSA interactome (Cherkasov, A., Hsing, M., Zoraghi, R., Foster, L. J., See, R. H., Stoynov, N., Jiang, J., Kaur, S., Lian, T., Jackson, L., Gong, H., Swayze, R., Amandoron, E., Hormozdiari, F., Dao, P., Sahinalp, C., Santos-Filho, O., Axerio-Cilies, P., Byler, K., McMaster, W. R., Brunham, R. C., Finlay, B. B., and Reiner, N. E. (2011) J. Proteome Res. 10, 1139–1150; Zoraghi, R., See, R. H., Axerio-Cilies, P., Kumar, N. S., Gong, H., Moreau, A., Hsing, M., Kaur, S., Swayze, R. D., Worrall, L., Amandoron, E., Lian, T., Jackson, L., Jiang, J., Thorson, L., Labriere, C., Foster, L., Brunham, R. C., McMaster, W. R., Finlay, B. B., Strynadka, N. C., Cherkasov, A., Young, R. N., and Reiner, N. E. (2011) Antimicrob. Agents Chemother. 55, 2042–2053). Screening of an extract library of marine invertebrates against MRSA PK resulted in the identification of bis-indole alkaloids of the spongotine (A), topsentin (B, D), and hamacanthin (C) classes isolated from the Topsentia pachastrelloides as novel bacterial PK inhibitors. These compounds potently and selectively inhibited both MRSA PK enzymatic activity and S. aureus growth in vitro. The most active compounds, cis-3,4-dihyrohyrohamacanthin B (C) and bromodeoxytopsentin (D), were identified as highly potent MRSA PK inhibitors (IC50 values of 16–60 nm) with at least 166-fold selectivity over human PK isoforms. These novel anti-PK natural compounds exhibited significant antibacterial activities against S. aureus, including MRSA (minimal inhibitory concentrations (MIC) of 12.5 and 6.25 μg/ml, respectively) with selectivity indices (CC50/MIC) >4. We also report the discrete structural features of the MRSA PK tetramer as determined by x-ray crystallography, which is suitable for selective targeting of the bacterial enzyme. The co-crystal structure of compound C with MRSA PK confirms that the latter is a target for bis-indole alkaloids. It elucidates the essential structural requirements for PK inhibitors in “small” interfaces that provide for tetramer rigidity and efficient catalytic activity. Our results identified a series of natural products as novel MRSA PK inhibitors, providing the basis for further development of potential novel antimicrobials.
Introduction
Recent increases in antibiotic resistance among bacterial pathogens such as methicillin-resistant Staphylococcus aureus (MRSA),3 and multidrug-resistant Gram-negative bacilli, coupled with a dearth of new antibiotic development over the past four decades, has created major problems in the clinic. Thus, identification of new targets for antibacterial development based upon novel scaffolds with unique mechanisms of action are critically needed (3). To this end, we recently identified pyruvate kinase (PK), an evolutionarily conserved, highly connected essential hub protein in MRSA, with structural features distinct from the mammalian orthologs, as a novel candidate drug target (1, 4, 5). Using structural modeling, virtual data base screening, and enzyme activity assays in combination with medicinal chemistry, we recently identified low micromolar selective inhibitors of MRSA PK exhibiting strong antibacterial activity against Staphylococci and a range of other Gram-positive bacteria such as Enterococcal and Streptococcal species with low micromolar minimal inhibitory concentration (MIC) values (5).
PK (EC 2.7.1.40) plays a major role in the regulation of carbohydrate metabolism, catalyzing the rate-limiting final step in glycolysis with the irreversible conversion of phosphoenolpyruvate (P-enolpyruvate) to pyruvate with concomitant phosphorylation of ADP to ATP (6). Both the substrates and products of PK feed into a number of biosynthetic pathways, placing it at a pivotal metabolic intersection. The x-ray crystal structures of several PKs from different species (e.g. Escherichia coli, Leishmania mexicana, Bacillus stearothermophilus, cat, rabbit muscle, human erythrocyte, and yeast) reveal a generally conserved architecture (7–11). PKs exist as homo-tetramers of identical subunits, each consisting of three or four domains: N-terminal, A, B, and C domains. The N-terminal helical domain is absent in prokaryotic bacterial PKs. The active site lies at the interface of the A and B domains in each of the four subunits, whereas the binding site for the allosteric effector appears to be located in the C domain (8, 9, 12). Allosteric regulation in particular, provides a mechanism for the tight modulation of PK enzymes. Tetramerization occurs through interactions between A and C domains of adjacent subunits so that bordering A domains constitute the A-A or “large” interface and adjacent C domains form the C-C or “small” interface between monomers (12). Recently it was shown that the allosteric transition between inactive T-state and active R-state involved a symmetrical 6° rigid-body rocking motion of the A- and C-domain cores in each of the four subunits. This also involved the formation of eight essential salt-bridge locks across the small interface that provided tetramer rigidity and a 7-fold increase in enzyme activity (13). Sequence alignment between MRSA and human PKs revealed particular sequence divergence in domain C (e.g. sites for effector binding), which is involved in formation of the small interfaces between the C domains of adjacent PK subunits providing tetramer rigidity and efficient catalytic activity of the enzyme (2, 4).
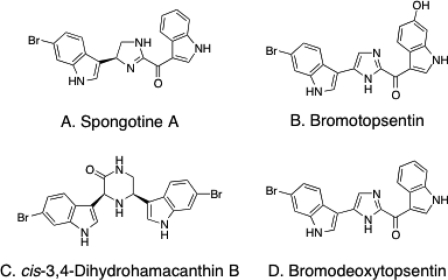
In the present study, as part of our interest in discovering natural marine products that potently and selectively inhibit bacterial PKs, we screened the inhibitory potential of a natural marine product library of 968 crude benthic invertebrate extracts collected from Papua New Guinea, Indonesia, Dominica, Brazil, British Columbia, South Africa, and Norway on MRSA and human recombinant PKs enzymatic activity. Bioactivity-guided fractionation of the an active crude extract (number 11) that exhibited significant selective inhibitory activity (IC50 ≤ 5 μg/ml) toward MRSA PK, using various chromatographic techniques, led to the identification of four known bis-indole alkaloids (A-D) (Fig. 2).
FIGURE 2.
Structures of bis-indole alkaloids with MRSA PK inhibitory activities purified from T. pachastrelloides in this study.
Indole-containing alkaloids are a class of natural marine products that show unique promise in the development of new drug leads. Bis-indole alkaloids, consisting of two indole moieties connected to each other via heterocyclic units, belong to a rapidly growing group of sponge metabolites that exhibit potent and diverse pharmacological activities including cytotoxic (14–16), antitumor (17, 18), antiviral (17, 18), antimicrobial (17, 19–21), antiparasitic (22), antifungal (14, 15), and anti-inflammatory activities as well as R1 adrenergic inhibitory effects (23), antiserotonin, Ca2+-calmodulin-antagonistic and antitopoisomerase-I activities (24–28). Thus they have received considerable attention as highly attractive potential starting points for drug development.
Despite the potential of bis-indole alkaloids as chemotherapeutics, it is surprising how little is currently known about their mechanism of action underlying their antimicrobial activity. It has been previously reported that the bis-indole alkaloids, hamacanthin A and deoxytopsentin, isolated from the marine sponge Spongosorites sp., exhibited significant in vitro antibacterial activity against S. aureus (minimal inhibitory concentration (MIC) = 3.12–6.35 μg/ml) in addition to a variety of other Gram-positive and Gram-negative bacteria (19, 29). Interestingly, these alkaloids inhibited the recombinant S. aureus sortase A (StrA) (IC50 = 15.7 and 86.3 μg/ml, respectively), a membrane-associated transpeptidase that plays a key role in invasion of host cells by Gram-positive pathogenic bacteria (20). Nevertheless, the molecular target(s) of these bis-indole alkaloids accounting for their in vitro antimicrobial activities is not yet known. However, it is certainly not related to inhibition of StrA, which serves only as a virulence factor. Thus, the results we describe here serve as a starting point for understanding the mechanism accounting for the antibacterial activities of these bis-indole alkaloids.
Furthermore, in this study, we describe for the first time the structural features of S. aureus PK obtained by x-ray crystallography and highlight significant structural differences in the bacterial enzyme compared with human PKs in the small interface located between the adjacent PK subunits. This region was important for the preferential binding of bis-indole alkaloids to MRSA PK during co-crystallization of compound C with MRSA PK. The results of these studies may allow for the design of potent and specific bacterial PK inhibitors.
EXPERIMENTAL PROCEDURES
Bacterial Strains
An epidemic methicillin-resistant S. aureus (MRSA) strain sequenced at the Sanger Centre (MRSA252, NRS71), and S. aureus RN4220 (NCTC8325 NRS144) were obtained from NARSA (Network on Antimicrobial Resistance in S. aureus).
Generation of PK Constructs
N-terminal His-tagged MRSA PK, and human M1, R, and L PKs constructs were generated in pET-28-a(+) vectors as described previously (2).
Expression and Purification of Recombinant His-tagged MRSA and Human PKs
MRSA and human M1, M2, R, and L constructs in pET-28a(+) (courtesy of Dr. L. Cantley, Harvard Medical, School, Boston, MA) were used to express relevant recombinant PK proteins in E. coli BL21(DE3). For enzymatic experiments, the proteins were expressed and purified using nickel-nitrilotriacetic acid-agarose (Qiagen) as described previously (2). For crystallographic studies, cells were resuspended in buffer A (20 mm Tris, 150 mm KCl, 150 mm NaCl, 0.5 mm K2HPO4, 20% glycerol, pH 7.4) plus 20 mm imidazole and lysed with an EmulsiFlex-C5 (Avestin). His-tagged PK was captured from the lysate using a Ni2+-charged 5-ml FF metal chelating Sepharose gravity flow column, washed with buffer A plus 50 mm imidazole, and eluted with buffer A plus 300 mm imidazole. Protein was concentrated and resolved on a Superdex 200 16/60 equilibrated in buffer A prior to flash freezing and storage at −80 °C. For crystallization experiments, protein was buffer exchanged into buffer B (5 mm Tris, pH 7.4, 50 mm KCl) using a Superdex 200 10/30 and concentrated to 8–10 mg/ml.
Measurement of Pyruvate Kinase Activity
Marine extracts and their purified bis-indole alkaloids were assayed for their ability to inhibit enzymatic activities of MRSA and human PKs. PK activity was determined using a continuous assay coupled to lactate dehydrogenase in which the change in absorbance at 340 nm, because of oxidation of NADH, was measured using a Benchmark Plus microplate spectrophotometer (Bio-Rad) (30). The reaction contained 60 mm Na+-HEPES, pH 7.5, 5% glycerol, 67 mm KCl, 6.7 mm MgCl2, 0.24 mm NADH, 5.5 units of l-lactate dehydrogenase from rabbit muscle (Sigma), 2 mm ADP, and 10 mm P-enolpyruvate (i.e. close to the Km of MRSA PK, so that the IC50 values should approximate the Ki) in a total volume of 200 μl. Reactions were initiated by the addition of 15 nm of one of the PK enzymes. PK activity proportional to the rate of change at 340 nm was expressed as specific activity (μmol/min/mg), which is defined as the amount of PK that catalyzes the formation of 1 μmol of either product per minute. Marine extracts or bis-indole compounds were dissolved in DMSO with the final concentration of the solvent never exceeding 1% of the assay volume. IC50 values were calculated by curve fitting on a four-parameter dose-response model with variable slope using GraphPad Prism 5.0 (GraphPad Software Inc., La Jolla, CA). In all studies, less than 10% of total P-enolpyruvate was exhausted during the reaction. Reactions were performed at 30 °C for 5 min. All values determined represent three measurements, each in triplicate.
In Vitro Susceptibility Testing
The antimicrobial activities of PK inhibiting crude marine extracts and bis-indole compounds were determined using the 96-well microtiter standard 2-fold serial broth microdilution method as described using CLSI (formerly National Committee for Clinical Laboratory Standards) recommended protocols (31) with S. aureus strains mentioned above as described previously (2). Each stock solution of crude marine extract or bis-indole compounds in DMSO was diluted with BHI to prepare serial 2-fold dilutions in the range of 100 to 0.5 μg/ml or 500 to 0.06 μm, respectively. We defined the MIC as the lowest concentration of test compound leading to complete inhibition of cell growth in relationship to compound-free control wells as determined by optical density. Erythromycin was used as reference compound. Experiments were replicated at least three times in triplicates to verify reproducibility using the above conditions.
Determination of Mammalian Cytotoxicity
The cytotoxic activities of marine crude extracts and bis-indole compounds were determined for HeLa cells 229 (ATCC: CCL-2-.1) and HEK 293T cells (ATCC:CRL-11268), respectively, in microtiter cultures by measuring dehydrogenase activity using CellTiter 96® AQueous One Solution Cell Proliferation Assay (Promega, Madison, WI), according to the manufacturer's protocol as described previously (2). Growth in compound-free control wells was considered as 100% and percentage of growth inhibition was calculated for each compound concentration. Cytotoxicity was quantified as the CC50, the concentration of compound that inhibited 50% of conversion of 3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) to formazan (32). The “selectivity index” is defined as the ratio of the mammalian cell cytotoxicity to the MIC against S. aureus (i.e. CC50/MIC). Positive control measurements were performed with xanthohumol (HeLa cells, CC50 ≈9 μg/ml; HEK 293T cells, CC50 ≈15 μg/ml). All assays were performed three times in triplicate.
Preparation of Marine Crude Extracts
1 mg/ml stocks of a library of 968 lyophilized specimens of crude benthic marine invertebrate extracts collected from Papua New Guinea, Indonesia, Dominica, Brazil, British Columbia (Canada), South Africa, and Norway were prepared in DMSO and stored at −20 °C.
Isolation and Extraction of Bis-indole Alkaloid Compounds
Three sponge specimens were collected by hand using scuba (40 m depth) in June, 1994, off the Aliwal Shoal, a large reef system situated 5 km offshore from the village of Umkomaas in southern Kwazulu-Natal, South Africa. The collected samples were a loose association of two sponges: a major yellow colored sponge identified as Topsentia pachastrelloides (Topsent 1892) (Porifera, Demospongiae, Halichondrida, and Halichondriidae), and a minor olive green sponge, Polymastia species (Fig. 1).
FIGURE 1.
T. pachastrelloides collected from South Africa used in this study to purify MRSA PK inhibiting bis-indole alkaloids.
The two sponges were separated, and only T. pachastrelloides was subjected to chemical analysis. The massive rich yellow sponge, 200-mm wide × 120-mm high × 180-mm broad, outer layer, was identified as T. pachastrelloides, which had oxea of various size categories: very small oxea (115 × 2 μm); small oxea (220 × 7 μm), medium oxea (364 × 12 μm), and large oxea (403 × 14 μm). The surface of the body was smooth but rough to the touch and the ectosome can barely be differentiated by the choanosome. The texture was firm, being corky crumbly and compressible. Oscules were not visible in the preserved specimen. The embedded encrusting olive green sponge, 12-mm wide × 9-mm high, with small wart-like papillae on the smooth surface, was identified as Polymastia sp. It has various categories of modified styles with the primary styles being smooth, slightly curved and centrally thickened with very slim heads, distal end fusiform (672 × 14 μm); subectosomal subtylostyles, distally fusiform (460 × 9.6 μm).
The specimens were immediately frozen and kept at −25 °C until chemically investigated. They were lyophilized (dry weight, 420 g) to provide crude marine extract number 11. The lyophilized mixture of these two sponges was extracted with EtOAc (2 × 1 liter) and MeOH (2 × 1 liter) to give 3.4- and 47-g extracts, respectively. Antimicrobial activity was found to reside predominantly in the 60% aqueous MeOH fraction (1.87 g) obtained from the initial polymeric reversed phase separation of 14.8 g of the orange methanolic sponge extract. Exhaustive reversed phase C18 HPLC (aqueous MeOH, 0.05% TFA) of a portion (420 mg) of the 60% aqueous methanol fraction yielded the known bis-indole alkaloids spongotine A (A, 18 mg) (33, 34), bromotopsentin (B, 36 mg) (18, 29), cis-3,4-dihydrohamacanthin (C, 3 mg) (29, 35), and bromodeoxytopsentin (D, 23 mg) (15, 29) (Fig. 2). The structures of A-D were established through recourse to standard spectroscopic techniques including NMR and high-resolution electron impact mass spectrometry data. Spectral data for these compounds were in good agreement with those reported previously (15, 18, 29, 33–35) and are available upon request.
S. aureus StrA Gene Inactivation Using TargeTron Technology
The StrA gene (StrA, 618 bp) was disrupted at target site (254 bp) by insertion of group II intron in the antisense orientation relative to StrA transcription using TargeTron technology. The StrA-TargeTron target fragments of 350 bp were amplified by 3 specific primers (ST-IBS, ST-EBS1d, and ST-EBS2), according to the protocol of the TargeTronTM Gene Knock-out System Kit (Sigma) to retarget the RNA portion of the intron. Primers were designed using the InGex Intron Prediction Program accompanying the TargetTron products, which introduced nucleotide substitutions into the IBS, EBS2, and EBS1d regions of the LtrBL1 intron originating from Lactococcus lactis. S. aureus intron-donor vector pNL9162 was used as a template for PCR to construct StrA-specific TargeTron donor plasmid pST254, containing the StrA-specific IBS-EBS fragments (350 bp) between the HindIII and BsrGI sites inserting LtrBLl TargeTrons into the selected sites in the StrA gene. pST254 were electroporated into S. aureus RN4220. Transformants were grown until early log phase when intron invasion was induced by 10 μm CdCl2 at 37 °C. After overnight induction, cells were isolated on BHI plates containing 10 μg/ml of erythromycin. The intron insertions were identified by colony PCR, using the genomic DNA of these mutants as templates and specific StrA primers flanking the predicted insertion sites in the StrA gene. The amplified fragments were cloned into PUC19 (Sigma) and sequenced to confirm the intron insertion. Attempts were made to cure the StrA TargeTron plasmids by plate-streaking the insertion mutants on BHI (no erythromycin) at 37–40 °C to identify erythromycin-sensitive mutants. Due to temperature sensitivity of the IEP-assisted splicing reaction in the TargeTron system, wild-type cells, transformant having uninduced StrA donor plasmids, and StrA disruptants in the antisense orientation were streaked on plates incubated separately at 32 and 43 °C to access StrA essentiality for growth. The plates were incubated for 24–48 h and photographed. Control parent strain and induced transformants of pST254 were grown in BHI medium overnight at 32 °C, then diluted 1:20 into fresh medium, and incubated for 4 h at 32 °C or for 2 h at 43 °C. RNA was extracted and analyzed by RT-PCR, using StrA-specific primers flanking the StrA-intron inserted in the StrA gene. Primer sequences are available upon request. Controls showed no products if the initial reverse transcription step was omitted indicating that the amplified bands in the experimental samples were derived from cellular RNA. Disruption of hsa, a previously confirmed essential gene in S. aureus was performed as a control (36).
Crystallization, Data Collection, and Refinement
Crystals belonging to space group P1 were obtained using the sitting drop method by mixing equal volumes of protein and reservoir solution containing 15–20% PEG 3350, 0.4 m sodium malonate, pH 7, 0.1 m Bicine, pH 8–8.5. For ligand studies, ligands in 100% DMSO were diluted ×100 in protein solution, centrifuged, and set up as above. Crystals were cryoprotected by supplementing with 20% glycerol and flash-cooled in liquid nitrogen. Data were collected at 100 K at station 08ID-1 of the Canadian Light Source (Saskatoon, Canada) and processed using SCALA (37). Phases were obtained by molecular replacement with the program PHASER (38) using the structure of the related Bacillus geothermophilus PK (39). Refinement was carried out with REFMAC (40), PHENIX (41), and COOT (42) with TLS parameters included in the later stages. See Table 1 for data processing and refinement statistics. Model validation was carried out with MolProbity (43) with 91.1% of residues in the favored region of the Ramachandran map and 2.8% outliers for the apo structure and 90% of residues in the favored and 1.7% outliers for the holo structure. Structure factors and coordinates have been deposited to the Protein Data Bank with codes 3T05 and 3T07.
TABLE 1.
Data collection and refinement statistics
| PK apo | PK + compound C | |
|---|---|---|
| Data collection | ||
| Wavelength (Å) | 0.9803 | 0.9795 |
| Space group | P1 | P1 |
| Cell dimensions | ||
| a, b, c (Å) | 84.02, 111.26, 111.15 | 83.96, 111.91, 112.03 |
| α, β, γ (°) | 85.36, 80.15, 70.46 | 86.05, 72.14, 80.88 |
| Resolution (Å) | 3.05 (3.21–3.05)a | 3.3(3.48–3.3) |
| Rsym | 0.079 (0.984) | 0.059 (0.689) |
| I/σI | 11.1 (1.5) | 10.2 (1.5) |
| Completeness (%) | 97.7 (97.7) | 93.4 (90.1) |
| Redundancy | 3.9 (3.9) | 3.1 (2.9) |
| Refinement | ||
| Resolution (Å) | 3.05 | 3.3 |
| No. reflections (unique) | 69,583 (10,187) | 53,839 (7,594) |
| Rwork/Rfree | 0.236/0.264 | 0.198/0.226 |
| No. atoms | ||
| Protein | 17,628 | 17,628 |
| Ligand/ion | 20 | 74 |
| Water | 0 | 0 |
| B-factors | ||
| Protein | 54.56 | 111.19 |
| Ligand/ion | 127.37 | 147.35 |
| Water | ||
| Root mean square deviations | ||
| Bond lengths (Å) | 0.013 | 0.011 |
| Bond angles (°) | 1.408 | 1.413 |
a Values in parentheses are for highest resolution shell.
RESULTS
In Vitro Screening of the Marine Extract Library
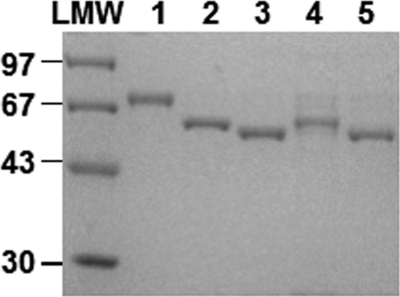
During the course of our search for potent and selective MRSA PK inhibitors from marine organisms, purified recombinant MRSA PK (Fig. 3) was used as a target enzyme to examine a total of 968 crude benthic marine invertebrate extracts collected from Papua New Guinea, Indonesia, Dominica, Brazil, British Columbia, South Africa, and Norway. For initial screening, all extracts were used at a concentration of 5 μg/ml, with a substrate concentration of 10 mm P-enolpyruvate, which is close to the MRSA PK Km (e.g. 6.6 mm) (4), so that the IC50 values should approximate the Ki. Seventy-four (7.6%) extracts tested exhibited a robust effect (≥90% inhibition) on MRSA PK enzyme activity at 5 μg/ml. Subsequently, these extracts were screened at a lower concentration (1 μg/ml) against MRSA PK as well as human PK isoforms (M1, M2, R, and L) (Fig. 3) in search for extract(s) that potently and selectively inhibit MRSA PK. This resulted in the identification of 11 crude marine extracts that contained potent and selective inhibitors of MRSA PK (Table 2). These 11 crude marine extracts were also tested for in vitro antibacterial activities. Among them, only extract number 11 isolated from T. pachastrelloides from South Africa (Fig. 1) demonstrated strong antibacterial activity (MIC of 25 μg/ml for S. aureus strain RN4220). The other extracts exhibited poor antibacterial activity most likely due to lack of cellular penetration. Subsequently, crude marine extract number 11 was examined against human HeLa 229 cells to test for specificity. The results indicated that crude marine extract number 11 had no significant growth inhibitory effects on HeLa 229 cells (CC50 >100 μg/ml). Therefore, extract number 11 was characterized further to identify the active component with selective anti-PK activity and antibacterial activity.
FIGURE 3.
SDS-PAGE of purified MRSA PK and human PK isoforms. 5-μg aliquots of MRSA PK, human M1, M2, R, and L PK proteins purified to homogeneity through the nickel-nitrilotriacetic acid chromatography step as described under “Experimental Procedures” were applied to lanes 1–5, respectively. Low molecular weight (LMW) standards (GE Healthcare) were applied as size markers. Proteins were stained with Coomassie Brilliant Blue dye.
TABLE 2.
High-throughput screening of natural marine products identifies several inhibitory extracts for MRSA PK
Shown are mean ± S.D. of inhibitory effects (%) of 1 μg/ml of most promising crude marine extracts on the activities of MRSA PK and human PK isoforms from 3 independent experiments each performed in triplicate.
| Marine extract No. | Inhibition of PK activity |
||||
|---|---|---|---|---|---|
| MRSA PK | Human M1 | Human M2 | Human R | Human L | |
| % | |||||
| 1 | 79 ± 8 | −11 ± 2 | −9 ± 3 | −9 ± 9 | −13 ± 7 |
| 2 | 91 ± 4 | −4 ± 1 | 1 ± 10 | −7 ± 6 | −7 ± 6 |
| 3 | 81 ± 8 | −1.7 ± 0.8 | 1 ± 3 | −16 ± 2 | −16 ± 8 |
| 4 | 96 ± 10 | −10 ± 14 | −7 ± 4 | −14 ± 1 | −19 ± 8 |
| 5 | 96 ± 2 | −2.6 ± 0.1 | −1 ± 2 | 16.5 ± 0.6 | 40 ± 4 |
| 6 | 90 ± 3 | −6 ± 4 | −12 ± 8 | −29 ± 3 | −15.3 ± 0.7 |
| 7 | 92 ± 2 | −2 ± 1 | −4 ± 6 | −10 ± 5 | −17 ± 7 |
| 8 | 91 ± 3 | −16 ± 4 | −8 ± 5 | −10 ± 6 | −10 ± 1 |
| 9 | 93 ± 2 | −6 ± 7 | −2 ± 5 | −10 ± 9 | −2.3 ± 0.1 |
| 10 | 69 ± 4 | −7 ± 10 | −3 ± 5 | −2 ± 5 | −4 ± 12 |
| 11 | 91 ± 2 | −1 ± 3 | −2 ± 4 | 12 ± 4 | 33 ± 4 |
Identification of Bis-indole Alkaloids as Bacterial PK Inhibitors
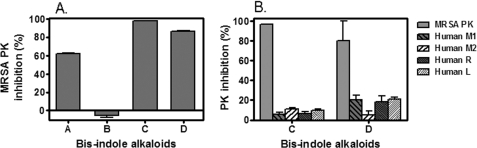
To identify the active compound(s) with potent and selective inhibitory activity against MRSA PK, based upon the results of combined spectral analyses and comparison of spectral data with those of known compounds, antibacterial activity-guided fractionation of marine extract number 11 as described under “Experimental Procedures” led to the identification of known bis-indole alkaloids (A-D) (Fig. 2), including spongotine A (A) (33, 34), bromotopsentin (B) (18, 29), cis-3,4-dihydrohamacanthin B (C) (29, 35), and bromodeoxytopsentin (D) (15, 29). Compounds A-D were evaluated for their potent inhibitory activities toward MRSA PK (Fig. 4A). The results demonstrated that three bis-indole alkaloids (A, C, and D) robustly inhibited MRSA PK enzymatic activity. Interestingly, compound B did not show any anti-PK inhibitory activity at the same concentration tested (10 μm) (Fig. 4A). Notably, hydroxylation of the indole residue of bromotopsentin (B) led to a total loss of MRSA PK inhibitory activity. These results suggest that the MRSA PK inhibitory activities of the topsentins are greatly affected by substitution on the indole rings. To determine whether the bis-indole-containing compounds were capable of acting selectively against MRSA PK, the inhibitory effects of the two most potent compounds (e.g. C and D) were tested against human M1, M2, R, and L PK isoforms in single-enzyme catalytic assays (Fig. 4B). The results demonstrated that compounds C and D displayed marked selectivity for bacterial PK.
FIGURE 4.
Inhibition of MRSA and human PK isoforms enzymatic activities by bis-indole alkaloids. A, bis-indoles (A, C, and D) exhibited potent inhibitory activity against MRSA PK at 10 μm. B, two bis-indoles (C and D) exhibited potent selective inhibitory activity against MRSA PK versus human PK isoforms at 5 μm. PK enzymatic activity was assayed as described under “Experimental Procedures.” The data shown are mean ± S.D. of inhibitory effects (%) from three independent experiments each performed in triplicate.
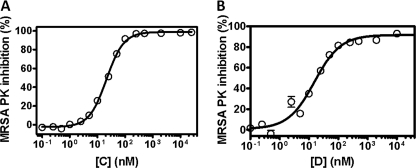
Inhibitory potencies, expressed as IC50 values, were next determined for compounds C and D (Fig. 5). cis-3,4-Dihydrohamacanthin B (C) and bromodeoxytopsentin (D) showed robust inhibition of bacterial PK (Fig. 5) as demonstrated by the nanomolar IC50 values (0.016 ± 0.009 and 0.06 ± 0.003 μm, respectively). These values compared very favorably to that of another reference compound, NSK5-15, a recently identified MRSA PK inhibitor with an IC50 value of 0.185 μm (2). Therefore, bis-indole-containing PK compounds appeared to be a suitable starting point for the development of highly specific PK inhibitors.
FIGURE 5.
Potency of inhibition of catalytic activity of MRSA PK by cis-3,4-dihyrohamacanthin B (left) and bromodeoxytopsentin (right). Potency of inhibition of MRSA PK was determined as described under “Experimental Procedures” with 10 μm P-enolpyruvate and 2 mm ADP substrates. The data are derived from one of three experiments with similar results each performed in triplicate and analyzed with Prism GraphPad software (single-site model). Error bars indicate the range of values within the single experiment shown. The final concentration of MRSA PK protein used in these studies was 15 nm.
Structural Analysis of the PK-Inhibitor Complex
To identify the ligand-binding site and characterize the mechanism of action, and facilitate future rational compound design, we sought to solve the novel apo- and inhibitor-bound structure of S. aureus PK.
The novel apo PK crystal structure was solved to 3.05-Å resolution and refined to Rwork/Rfree of 0.236/0.26 (see Table 1). The refined crystal structure consisted of one tetramer in the asymmetric unit and belonged to space group P1 with unit cell dimensions of a = 84.02 Å, b = 111.26 Å, c = 111.15 Å, α = 85.36°, β = 80.15°, γ = 70.46°. All monomers had the same conformation with an intra-molecular backbone root mean square deviation of <1 Å. The overall structure is conserved when compared with the closely related B. geothermophilus structure (39) (backbone root mean square deviation = 1.3 Å) including the presence and structure of the extra C-terminal sequence, which only presents in a small subset of PKs.
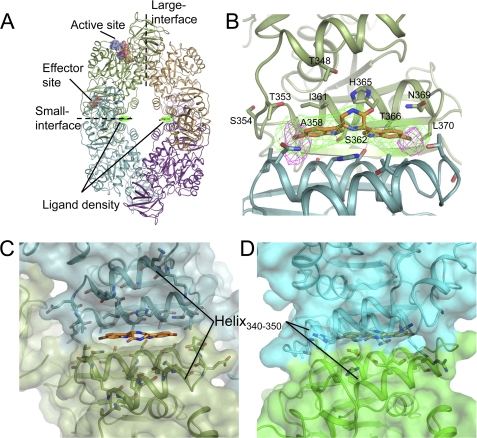
The holo structure with compound C was obtained by co-crystallization in the native condition and solved to 3.3-Å resolution with refined Rwork/Rfree of 0.198/0.226. The monomeric structure was conserved between apo and holo forms (root mean square deviation = 0.56 Å) although inhibitor binding was accompanied by a small but significant conformational rearrangement of the monomers in the tetrameric structure (see supplemental Movie 1). No evidence of bound ligand was observed in either active or effector sites but surprisingly a region of strong density was observed at the small interface (e.g. C-C domains interface) consistent with the size and shape of the ligand (Fig. 8A). In light of the resolution of the data, the anomalous properties of the brominated ligand were exploited to unambiguously confirm the binding of the compound in this interface pocket (Fig. 8B).
FIGURE 8.
Structure of S. aureus PK in complex with compound C. A, tetrameric structure of PK illustrating the ligand density at the small interface. Position of active and effector sites are for reference only. B, interface binding pocket. Refined ligand position shown with ligand omit map (green). Anomalous map calculated from data collected at bromine edge (purple) indicates the position of the ligand bromine atoms. Residues lining the binding pocket from chain A are labeled. Hydrogen bonds between Ser362 and indole nitrogens are marked in red. C and D, comparison of S. aureus (C) and representative human PK structure (PDB 1ZJH) (D) illustrating closer packing of helix 340–350 and differences in the primary structure of this helix and the interface binding pocket, which contribute to selectivity.
The binding pocket was found to be located at the small interface of the PK tetramer and was formed by the anti-parallel interaction of helix 357–370 from the respective subunits (Fig. 8C). The symmetric pocket was lined by 10 residues from each subunit: Thr348, Thr353, Ser354, Ala358, Ile361, Ser362, His365, Thr366, Asn369, and Leu370. The ligand dimensions were complimentary to the binding pocket (∼25 Å in length and 10 Å width) and the ligand filled the majority of the pocket. Prominent were two central histidine residues (His365) that showed side chain rearrangement in the presence of the ligand and sandwiched the central piperazine-2-one ring. Anchoring the ligand were symmetric hydrogen bonds between Ser362 from chains A and B, respectively, and the indole nitrogens. Hydrophobic interactions were prominent, with the indole moieties bound between Ile361 and His365 of opposite chains, whereas the two bromine atoms occupied a hydrophobic pocket formed by Thr353, Ser354, Ala358, and Leu370. The symmetric nature of the binding pocket was mirrored by the pseudo-symmetrical properties of the ligand.
Structural comparison to eukaryotic homologues provides an explanation for the observed selectivity the bis-indole compounds exhibited toward the S. aureus enzyme. Analysis of the Leishmania mexicana inactive and active structures (13) showed the presence of a similar binding pocket at the small interface, the nature of which changes dependent on the enzymatic state (see below). However, analysis of all the higher eukaryotic structures, regardless of apparent state, showed a much tighter dimeric packing around the small interface, which drastically alters the nature of the pocket, and also the closer interaction of a helix 340–350 above the interface, thereby limiting access to the pocket (Fig. 8, C and D). In addition, the primary structure of the residues lining the binding pocket was also poorly conserved in higher eukaryotes resulting in a loss of specific interactions notably for Ser362, which demonstrate hydrogen binding to the ligand indole nitrogens.
Bis-indole PK Inhibitors Exhibit Antibacterial Activity
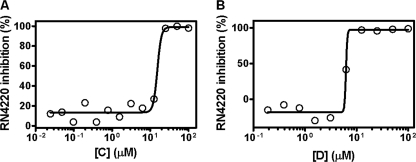
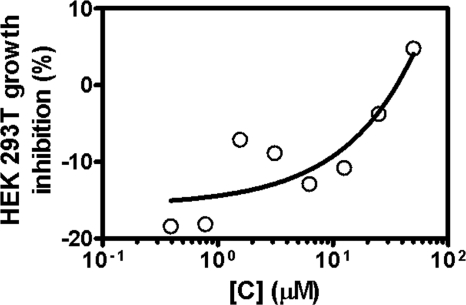
To investigate the antibacterial properties of bis-indole inhibitors, the three most potent compounds were tested in vitro for their antibacterial activities against methicillin-susceptible (e.g. RN4220, ATCC25923) (Fig. 6) and methicillin-resistant (e.g. MRSA252) S. aureus strains (Table 3). Results in Table 3 indicate that bis-indole compounds showed potent in vitro antibacterial activity against both methicillin-sensitive and -resistant strains of S. aureus tested (MIC 6.25–12.5 μg/ml, equal to 15.4–25.6 μm). Toxicity for mammalian cells for compound C was also determined using HEK 293T cells as described under “Experimental Procedures.” Fig. 7 shows that compound C exhibited lower cytotoxicity toward mammalian cells than bacterial cells (i.e. 24% of cell death at 50 μg/ml, when compared with the internal control of the assay) with CC50 values of >50 μg/ml. Thus, the selectivity indice (CC50/MIC) for compound C evaluated in relationship to their potent antibacterial activity against methicillin-sensitive S. aureus and MRSA (Fig. 6, Table 3) was determined to be >4, indicating selectivity of these compounds for bacterial versus mammalian cells. Taken together, these findings showed that bis-indole-containing compounds C and D exhibited the most potent and selective inhibitory activity against bacterial PK enzyme with IC50 and antibacterial activities (MIC) in the low nanomolar and micromolar range, respectively. Data obtained from these studies are summarized in Table 3.
FIGURE 6.
Antibacterial activities of two bis-indole compounds C (left) and D (right) with potent PK inhibitory activities against S. aureus strain RN4220.
TABLE 3.
Inhibition of MRSA PK activity and cell growth by bis-indole alkaloids
MIC was determined against S. aureus strains RN4220 and MRSA252, as determined by the broth-microdilution method; NSK5–15 is the MRSA PK inhibitor described previously (2).
| Compound | IC50 | MIC (μg/ml) |
|
|---|---|---|---|
| S. aureus RN4220 | S. aureus MRSA252 | ||
| μm | μm | ||
| A | 1–10 | 12.5 (30.6) | NDa |
| C | 0.016 | 12.5 (25.6) | 12.5 (25.6) |
| D | 0.06 | 6.25 (15.4) | 6.25 (15.4) |
| NSK5–15 | 0.185 | 1.4 (3.0) | 2.9 (6.0) |
| Erythromycin | NAb | 0.5 (0.7) | >10 (>13.6) |
a ND, not determined.
b NA, not applicable.
FIGURE 7.
Cytotoxicity assessment of the PK inhibiting bis-indole compound against HEK 293T cells. Cytotoxicity of compound C was determined using HEK 293T cells through a 1-day incubation assay as described under “Experimental Procedures.” The data presented are representative of three experiments performed in triplicate.
DISCUSSION
The development of resistance to clinically important antibiotics by bacterial pathogens and potential biowarfare agents pose major threats to public health (3, 44–46). Consequently, the development of new antibacterial compounds, possessing novel scaffolds and unique mechanisms of action, are urgently needed to combat the serious challenge of antimicrobial drug resistance in the clinic (3). Potential target genes should be essential for either bacterial viability, pathogenesis or both (47). In this study, we screened a marine extract library of 968 benthic marine invertebrates against recombinant MRSA PK. The latter was recently identified as a potential novel drug target based upon it being a highly connected, essential hub in the MRSA interactome. Library screening led to the identification of several marine bis-indole alkaloids (compounds A, C, and D) (Fig. 2) as potent and selective MRSA PK inhibitors with nanomolar IC50 values (i.e. 16–60 nm) for MRSA PK and at least 166- to 600-fold selectivity toward the bacterial enzyme (Figs. 4 and 5). Because the hubs (e.g. MRSA PK) in bacterial interactomes are critical to network integrity and stability, they should be ideal, novel targets for new antimicrobials.
In addition, PKs have recently been drawing attention as novel targets for anti-trypanosomal, anti-leishmania, anti-malaria, and anti-cancer drugs (30, 48–50). The results of this study have particular relevance, because the high prevalence of multidrug-resistant organisms such as methicillin-resistant S. aureus (MRSA) has generated an urgent need for alternative targets for the development of new antimicrobials.
It has been known that marine bis-alkaloids exhibit antibacterial activity (19, 20, 29). However, the molecular bases underlying their antibacterial activity is not yet elucidated. Oh and colleagues (19) have previously shown that these bis-alkaloids inhibit StrA, a known bacterial virulence factor in S. aureus. To examine whether the in vitro antibacterial activities of bis-indole-containing compounds were due to their inhibitory effect on StrA, the TargeTron system was used to knock-out the StrA gene. Our results provided direct evidence that S. aureus StrA is not essential for growth and viability of this key Gram-positive pathogen (data not shown). This is based upon the finding that inactivation of chromosomal StrA by insertion of group II intron at the antisense strand of StrA created a viable phenotype (data not shown). In contrast, PK was identified as an essential gene through independent chromosomal gene knock-down experiments using TargeTron technology (4). Using allelic replacement mutagenesis, PK has recently been identified as an absolutely essential gene for survival of other bacteria such as Haemophilus influenzae, Streptococcus pneumoniae, and Mycobacterium tuberculosis (51–54). Importantly, specific StrA inhibitors did not impair microbial growth in cell culture (55), suggesting that in vitro antibacterial activity of bis-indole may be rather due to the inhibition of MRSA PK enzymatic activity. Taken together, our findings showing that StrA is not an essential gene, coupled with the direct inhibition of MRSA PK enzymatic activity by bis-indole alkaloids suggests that MRSA and MSSA PK are the likely targets of these marine compounds leading to bacterial cell death.
Importantly, in this study we showed that the crystal structure of MRSA PK revealed structural differences between human and bacterial PK (Fig. 8), which could potentially be exploited for drug development thereby avoiding toxicity to host cells. In light of the fact that bis-indole compound C did not bind to either the active or effector sites, the question remains how ligand binding at a remote site resulted in enzyme inhibition. An attractive hypothesis is that this represents a form of allosteric inhibition with ligand binding at the small interface preventing the enzyme adopting the active state, effectively stalling enzymatic activity. Indeed, binding of compound C is accompanied by a conformational rearrangement of the PK tetramer toward a more compressed and elongated form (see supplemental Movie 1). In support of this, a recent study from Walkinshaw and colleagues (13) presented the first comparison of a series of substrate and effector-bound PK structures from the same species and proposed a novel mechanism for allosteric control. Central to this mechanism was the increased rigidity across the small interface in the active conformation achieved by the formation of a series of salt bridges in the region immediately in the vicinity of the compound C binding pocket. Concomitant with the increased rigidity is a conformational change across the small interface, tightening the packing between helix 357 and 370 (S. aureus numbering). Thus, by interfering with the packing of the small interface, the bis-indole compounds could achieve allosteric inhibition by preventing the PK tetramer from adopting the conformational state required for effective enzymatic activity.
Two related and important points to address are: 1) the evidence that the bis-indole compounds are actually acting through inhibition of PK and, 2) the mechanism by which inhibition of PK leads to bacterial killing. In regard to the latter point, PK is required to generate the product pyruvate for the Krebs cycle and inhibition of the enzyme would be expected to compromise ATP levels resulting in disruption of the bacterial metabolism. Our results show three lines of evidence that PK is the target of the bis-indole alkaloids. These include: (a) the essential nature of S. aureus PK (1), (b) the strong correlation between MICs (Table 3) and IC50 values of bis-indole compounds for growth inhibition of staphylococcal strains and inhibition of MRSA PK enzymatic activity, respectively, and (c) the atomic resolution structure of bis-indole alkaloids complexed with MRSA PK (Fig. 8). Moreover, the results of the co-crystallization experiments provided significant insight into the binding determinants for selectivity and potency. This information is important and may be fundamental for the design of novel antibacterial agents that are not compromised by existing resistance mechanisms. As PK is an essential hub in the MRSA protein-protein interaction network, it may be less prone to genetic mutation and subsequent resistance due to the network centrality-lethality rules (5). In addition, modeling based upon the crystal structure of PK will help to determine more comprehensively the actual potential for targeting bacterial PKs more widely.
The possibility that bis-indole alkaloids might have another target(s) other than PK cannot be completely ruled out at this point. However, the binding mode of bis-indole compounds with MRSA PK enzyme defined in co-crystal studies is strong evidence supporting the specific mechanism of action. In fact, most compounds generally hit more than one target to some degree (56–59). Therefore, the possibility of distinct effects of bis-alkaloids may be worth exploring.
In summary, we have identified several promising natural bis-indole alkaloids with robust antibacterial activities that potently and selectively inhibit bacterial PK enzymatic activity. These low, nanomolar range inhibitors may be considered as potential starting points for the development of novel antimicrobials. The results of the co-crystal structure defined in this study also provide insight into the mechanism of action of these PK inhibitors. Future studies will use structure-based approaches to develop compounds with even greater potency and improved selectivity indices (CC50/MIC).
Supplementary Material
Acknowledgments
We thank Drs. Lewis C. Cantley and Matthew G. Vander Heiden, Department of Systems Biology, Harvard Medical School, for providing M2, R, and L human PK constructs. We thank the Canadian Light Source CMCF for access to synchrotron x-ray radiation and the CFI and BCKDF for infrastructure support.
This work was supported by grants from Genome Canada and Genome British Columbia, Vancouver General Hospital & University of British Columbia Hospital Foundation, The Natural Sciences and Engineering Research Council of Canada, SARS Accelerated Vaccine Initiative to the PRoteomics for Emerging PAthogen REsponse (PREPARE) Project, and the South African National Research Foundation and Department of Environmental Affairs through the SeaChange research program.

The on-line version of this article (available at http://www.jbc.org) contains supplemental Movie 1.
The atomic coordinates and structure factors (codes 3T05 and 3T07) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- MRSA
- methicillin-resistant Staphylococcus aureus
- PK
- pyruvate kinase
- P-enolpyruvate
- phosphoenolpyruvate
- CC50/MIC
- ratio of the mammalian cell cytotoxicity to the MIC against S. aureus
- MIC
- minimal inhibitory concentration
- BHI
- brain-heart infusion broth
- StrA
- sortase A
- Bicine
- N,N-bis(2-hydroxyethyl)glycine.
REFERENCES
- 1. Cherkasov A., Hsing M., Zoraghi R., Foster L. J., See R. H., Stoynov N., Jiang J., Kaur S., Lian T., Jackson L., Gong H., Swayze R., Amandoron E., Hormozdiari F., Dao P., Sahinalp C., Santos-Filho O., Axerio-Cilies P., Byler K., McMaster W. R., Brunham R. C., Finlay B. B., Reiner N. E. (2011) J. Proteome Res. 10, 1139–1150 [DOI] [PubMed] [Google Scholar]
- 2. Zoraghi R., See R. H., Axerio-Cilies P., Kumar N. S., Gong H., Moreau A., Hsing M., Kaur S., Swayze R. D., Worrall L., Amandoron E., Lian T., Jackson L., Jiang J., Thorson L., Labriere C., Foster L., Brunham R. C., McMaster W. R., Finlay B. B., Strynadka N. C., Cherkasov A., Young R. N., Reiner N. E. (2011) Antimicrob. Agents Chemother. 55, 2042–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Pathania R., Brown E. D. (2008) Biochem. Cell Biol. 86, 111–115 [DOI] [PubMed] [Google Scholar]
- 4. Zoraghi R., See R. H., Gong H., Lian T., Swayze R., Finlay B. B., Brunham R. C., McMaster W. R., Reiner N. E. (2010) Biochemistry 49, 7733–7747 [DOI] [PubMed] [Google Scholar]
- 5. Fraser H. B., Hirsh A. E., Steinmetz L. M., Scharfe C., Feldman M. W. (2002) Science 296, 750–752 [DOI] [PubMed] [Google Scholar]
- 6. Valentini G., Chiarelli L., Fortin R., Speranza M. L., Galizzi A., Mattevi A. (2000) J. Biol. Chem. 275, 18145–18152 [DOI] [PubMed] [Google Scholar]
- 7. Stuart D. I., Levine M., Muirhead H., Stammers D. K. (1979) J. Mol. Biol. 134, 109–142 [DOI] [PubMed] [Google Scholar]
- 8. Larsen T. M., Laughlin L. T., Holden H. M., Rayment I., Reed G. H. (1994) Biochemistry 33, 6301–6309 [DOI] [PubMed] [Google Scholar]
- 9. Jurica M. S., Mesecar A., Heath P. J., Shi W., Nowak T., Stoddard B. L. (1998) Structure 6, 195–210 [DOI] [PubMed] [Google Scholar]
- 10. Speranza M. L., Valentini G., Iadarola P., Stoppini M., Malcovati M., Ferri G. (1989) Biol. Chem. Hoppe Seyler 370, 211–216 [DOI] [PubMed] [Google Scholar]
- 11. Rigden D. J., Phillips S. E., Michels P. A., Fothergill-Gilmore L. A. (1999) J. Mol. Biol. 291, 615–635 [DOI] [PubMed] [Google Scholar]
- 12. Tulloch L. B., Morgan H. P., Hannaert V., Michels P. A., Fothergill-Gilmore L. A., Walkinshaw M. D. (2008) J. Mol. Biol. 383, 615–626 [DOI] [PubMed] [Google Scholar]
- 13. Morgan H. P., McNae I. W., Nowicki M. W., Hannaert V., Michels P. A., Fothergill-Gilmore L. A., Walkinshaw M. D. (2010) J. Biol. Chem. 285, 12892–12898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sakemi S., Sun H. H. (1991) J. Org. Chem. 56, 4304–4307 [Google Scholar]
- 15. Shin J., Seo Y., Cho K. W., Rho J. R., Sim C. J. (1999) J. Nat. Prod. 62, 647–649 [DOI] [PubMed] [Google Scholar]
- 16. Morris S. A., Anderson R. J. (1990) Tetrahedron 46, 715–720 [Google Scholar]
- 17. Wright A. E., Pomponi S. A., Cross S. S., McCarthy P. (1992) J. Org. Chem. 57, 4772–4775 [Google Scholar]
- 18. Tsujii S., Rinehart K. L., Gunasekera S. P., Kashman Y., Cross S. S., Lui M. S., Pomponi S. A., Diaz M. C. (1988) J. Org. Chem. 53, 5446–5453 [Google Scholar]
- 19. Oh K. B., Mar W., Kim S., Kim J. Y., Oh M. N., Kim J. G., Shin D., Sim C. J., Shin J. (2005) Bioorg. Med. Chem. Lett. 15, 4927–4931 [DOI] [PubMed] [Google Scholar]
- 20. Oh K. B., Mar W., Kim S., Kim J. Y., Lee T. H., Kim J. G., Shin D., Sim C. J., Shin J. (2006) Biol. Pharm. Bull. 29, 570–573 [DOI] [PubMed] [Google Scholar]
- 21. Gupta L., Talwar A., Chauhan P. M. (2007) Curr. Med. Chem. 14, 1789–1803 [DOI] [PubMed] [Google Scholar]
- 22. Gupta L., Talwar A., Nishi, Palne S., Gupta S., Chauhan P. M. (2007) Bioorg. Med. Chem. Lett. 17, 4075–4079 [DOI] [PubMed] [Google Scholar]
- 23. Phifea D. W., Ramosa R. A., Fenga M., Kinga I., Gunasekerai S. P., Wrighti A., Patela M., Pachtera J. A., Coval S. J. (1996) Bioorg. Med. Chem. Lett. 6, 2103–2106 [Google Scholar]
- 24. Mayer A. M., Rodríguez A. D., Berlinck R. G., Hamann M. T. (2009) Biochim. Biophys. Acta 1790, 283–308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mayer A. M., Rodríguez A. D., Berlinck R. G., Hamann M. T. (2007) Comp. Biochem. Physiol. C Toxicol. Pharmacol. 145, 553–581 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mayer A. M., Hamann M. T. (2005) Comp. Biochem. Physiol. C Toxicol. Pharmacol. 140, 265–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mayer A. M., Hamann M. T. (2004) Mar. Biotechnol. 6, 37–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mayer A. M., Hamann M. T. (2002) Comp. Biochem. Physiol. C Toxicol. Pharmacol. 132, 315–339 [DOI] [PubMed] [Google Scholar]
- 29. Bao B., Sun Q., Yao X., Hong J., Lee C. O., Sim C. J., Im K. S., Jung J. H. (2005) J. Nat. Prod. 68, 711–715 [DOI] [PubMed] [Google Scholar]
- 30. Christofk H. R., Vander Heiden M. G., Harris M. H., Ramanathan A., Gerszten R. E., Wei R., Fleming M. D., Schreiber S. L., Cantley L. C. (2008) Nature 452, 230–233 [DOI] [PubMed] [Google Scholar]
- 31. Watts J. L., Shryock T. R., Apley M., Bade D. J., Brown S. D., Gray J. T., Heine H., Hunter R. P., Mevius D. J., Papich M. G., Silley P., Zurenk G. E. (2008) Clinical and Laboratory Standards Institute, M31-A3, Vol. 28, No. 8 [Google Scholar]
- 32. Marshall N. J., Goodwin C. J., Holt S. J. (1995) Growth Regul. 5, 69–84 [PubMed] [Google Scholar]
- 33. Bao B., Sun Q., Yao X., Hong J., Lee C. O., Cho H. Y., Jung J. H. (2007) J. Nat. Prod. 70, 2–8 [DOI] [PubMed] [Google Scholar]
- 34. Murai K., Morishita M., Nakatani R., Kubo O., Fujioka H., Kita Y. (2007) J. Org. Chem. 72, 8947–8949 [DOI] [PubMed] [Google Scholar]
- 35. Casapullo A., Bifulco G., Bruno I., Riccio R. (2000) J. Nat. Prod. 63, 447–451 [DOI] [PubMed] [Google Scholar]
- 36. Yao J., Zhong J., Fang Y., Geisinger E., Novick R. P., Lambowitz A. M. (2006) RNA 12, 1271–1281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Evans P. (2006) Acta Crystallogr. D Biol. Crystallogr. 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 38. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Suzuki K., Ito S., Shimizu-Ibuka A., Sakai H. (2008) J. Biochem. 144, 305–312 [DOI] [PubMed] [Google Scholar]
- 40. Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D Biol. Crystallogr. 50, 760–76315299374 [Google Scholar]
- 41. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Alanis A. J. (2005) Arch. Med. Res. 36, 697–705 [DOI] [PubMed] [Google Scholar]
- 45. Barrett C. T., Barrett J. F. (2003) Curr. Opin. Biotechnol. 14, 621–626 [DOI] [PubMed] [Google Scholar]
- 46. Monroe S., Polk R. (2000) Curr. Opin. Microbiol. 3, 496–501 [DOI] [PubMed] [Google Scholar]
- 47. García-Lara J., Masalha M., Foster S. J. (2005) Drug Discov. Today 10, 643–651 [DOI] [PubMed] [Google Scholar]
- 48. Opperdoes F. R., Michels P. A. (2001) Int. J. Parasitol. 31, 482–490 [DOI] [PubMed] [Google Scholar]
- 49. Nowicki M. W., Tulloch L. B., Worralll L., McNae I. W., Hannaert V., Michels P. A., Fothergill-Gilmore L. A., Walkinshaw M. D., Turner N. J. (2008) Bioorg. Med. Chem. 16, 5050–5061 [DOI] [PubMed] [Google Scholar]
- 50. Chan M., Tan D. S., Sim T. S. (2007) Travel Med. Infect. Dis. 5, 125–131 [DOI] [PubMed] [Google Scholar]
- 51. Sassetti C. M., Boyd D. H., Rubin E. J. (2003) Mol. Microbiol. 48, 77–84 [DOI] [PubMed] [Google Scholar]
- 52. Song J. H., Ko K. S., Lee J. Y., Baek J. Y., Oh W. S., Yoon H. S., Jeong J. Y., Chun J. (2005) Mol. Cells 19, 365–374 [PubMed] [Google Scholar]
- 53. Zhang R., Ou H. Y., Zhang C. T. (2004) Nucleic Acids Res. 32, D271–272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Akerley B. J., Rubin E. J., Novick V. L., Amaya K., Judson N., Mekalanos J. J. (2002) Proc. Natl. Acad. Sci. U.S.A. 99, 966–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Suree N., Yi S. W., Thieu W., Marohn M., Damoiseaux R., Chan A., Jung M. E., Clubb R. T. (2009) Bioorg. Med. Chem. 17, 7174–7185 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Shaffer C. (2005) Drug Discov. Today 10, 1489. [DOI] [PubMed] [Google Scholar]
- 57. Xie L., Li J., Xie L., Bourne P. E. (2009) PLoS Comput. Biol. 5, e1000387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Scheiber J., Chen B., Milik M., Sukuru S. C., Bender A., Mikhailov D., Whitebread S., Hamon J., Azzaoui K., Urban L., Glick M., Davies J. W., Jenkins J. L. (2009) J. Chem. Inf. Model. 49, 308–317 [DOI] [PubMed] [Google Scholar]
- 59. Scheiber J., Jenkins J. L., Sukuru S. C., Bender A., Mikhailov D., Milik M., Azzaoui K., Whitebread S., Hamon J., Urban L., Glick M., Davies J. W. (2009) J. Med. Chem. 52, 3103–3107 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.