Background: The degradation of promyelocytic leukemia (PML) protein is regulated by phosphorylation-dependent manner.
Results: The inhibition or knockdown of ERK2 increase PML expression level and decrease the interaction between PML and Pin1.
Conclusion: The activation of ERK2 promotes Pin1-mediated degradation of PML in response to EGF.
Significance: Understanding the mechanism by which mitogens promote PML protein turnover will have implication in developing therapeutic agents in treating cancer.
Keywords: Epidermal growth factor (egf), ERK, phosphorylation, Protein degradation, Protein stability, PML, Pin1
Abstract
The promyelocytic leukemia (PML) protein is a tumor suppressor that has an important role in several cellular processes, including apoptosis, viral infection, DNA damage repair, cell cycle regulation, and senescence. PML is an essential component of sub-nuclear structures called PML nuclear bodies (NBs). Our laboratory has previously demonstrated that the peptidyl-prolyl cis-trans isomerase, Pin1, binds and targets PML for degradation in a phosphorylation-dependent manner. To further elucidate the mechanisms underlying Pin1-mediated PML degradation, we aimed to identify one or more factors that promote PML phosphorylation. Here we show that treatment with U0126, an inhibitor of the ERK2 upstream kinases MEK1/2, leads to an increase in PML protein accumulation and an inhibition of the interaction between Pin1 and PML in MDA-MB-231 breast cancer cells. Consistent with this observation, phosphorylated ERK2 partially co-localized with PML NBs. Although U0126 up-regulated exogenous wild-type PML levels, it did not have an effect on the steady-state level of a mutant form of PML that is defective in binding Pin1. In addition, exogenous wild-type, but not Pin1 binding-defective PML protein expression levels were decreased by overexpression of ERK2. In contrast, knockdown of ERK2 by siRNA resulted in an increase in PML protein levels and an increase in the formation of PML NBs. Using phospho-specific antibodies, we identified Ser-403 and Ser-505 as the ERK2 targets that promote Pin1-mediated PML degradation. Finally, we demonstrated that EGF induced activation of ERK and interaction between PML and phosphorylated ERK resulting in a decrease in PML protein levels. Taken together, our results support a model in which Pin1 promotes PML degradation in an ERK2-dependent manner.
Introduction
The promyelocytic leukemia (PML)2 gene was originally identified in acute promyelocytic leukemia patients, as a fusion partner of retinoic acid receptor α due to chromosomal translocation (1, 2). The endogenous PML protein primarily localizes in multiprotein subnuclear structures, termed PML nuclear bodies (NBs), and is essential for the biogenesis of these particles (3). Recent evidence implies that the PML NBs are structurally and functionally heterogeneous, and their formation is intrinsically dynamic in response to stress and environmental stimuli (4). Many proteins have been found to localize to PML NBs partially or temporally, and as a consequence, PML NBs have been implicated in diverse cellular processes such as DNA repair, DNA replication, transcription, repression of messenger RNA nuclear export, and post-translational modification of proteins (5–8). Although the physiological roles of PML and PML NBs are still a matter of debate, many studies have suggested a tumor-suppressive role for PML (9, 10). PML−/− mice are highly susceptible to tumor development in chemical and physical models of carcinogenesis, and PML−/− cells are resistant to lethal doses of gamma irradiation and treatment with Fas antibody (11, 12). A recent report demonstrated that a cytoplasmic form of PML is localized in complex with the inositol 1,4,5-triphosphate receptor, Akt, and protein phosphatase 2a and regulates apoptosis via increasing Ca2+ release from the endoplasmic reticulum (13). Moreover, PML recruits mammalian target of rapamycin to PML NBs during hypoxia and inhibits mammalian target of rapamycin activity; as a result, PML negatively regulates HIF-1α translation and tumor angiogenesis (14). It is known that a decrease or loss of PML protein expression is frequently observed in leukemia and in various solid tumors (15). Therefore, understanding the mechanism by which the expression of PML is regulated is an important issue.
Expression levels of PML can be regulated at the transcriptional and post-translational levels. Type I and II interferons increase both the size and number of PML NBs through STAT- and IRF8-mediated transcriptional induction of PML mRNA (16–19).3 Furthermore, PML is a direct transcriptional target of p53 in mouse embryonic fibroblasts (20). Various mechanisms have been suggested to reduce cellular PML protein levels. Treatment with arsenic trioxide causes SUMOylation of PML and its proteasome-dependent degradation in Chinese hamster ovary (CHO) cells (21–23). Arsenic trioxide binds directly to cysteine residues located within the N-terminal RING finger/B-box/coiled coil (RBCC) domain of PML in a reactive oxygen species-dependent manner, and this binding increases the interaction between PML and SUMO-conjugating enzyme UBC9, resulting in enhanced SUMOylation and degradation of PML (24, 25). Viral infection also results in degradation of PML protein and disruption of the PML NBs (26, 27). We have previously demonstrated that tumor necrosis factor α (TNFα) significantly induces PML accumulation in human umbilical vascular endothelial cells (28) and that HDAC7 is critical for TNFα-induced NB formation by modulating PML SUMOylation (29). We also reported that PML interacts with the peptidyl-prolyl cis-trans isomerase, Pin1, in a phosphorylation-dependent manner and that this interaction promotes PML degradation (30). Additionally we showed that IGF-1 negatively regulates PML protein abundance through a Pin1-dependent mechanism to promote cell migration (31). These observations are consistent with the notion that PML NBs are a dynamic subnuclear structure that undergoes constant re-organization.
In the present work, we identify the MAPK ERK2 as a negative PML regulator. We found that treatment with U0126, an inhibitor of the ERK1/2 upstream kinases MEK1/2, increased PML protein levels and the formation of PML NBs. Importantly, U0126 decreased the interaction between PML and Pin1. Furthermore, knockdown of ERK2 increased the expression levels of PML without altering PML mRNA expression, and overexpression of ERK2 promoted the degradation of exogenous wild-type PML protein but had no effect on the levels of PML mutant defective in Pin1 binding. We conclude that ERK2 phosphorylates PML and promotes its interaction with Pin1, leading to PML degradation.
EXPERIMENTAL PROCEDURES
Cell Lines and Medium
MDA-MB-231 (breast cancer cells), HeLa, CV-1, PML−/− mouse embryonic fibroblasts and HEK293 cells were grown at 37 °C in 5% CO2 in 1× Dulbecco's modified Eagle's medium with 4.5 g/liter glucose, l-glutamine, and sodium pyruvate (Cellgro) supplemented with 10% charcoal-stripped fetal bovine serum (FBS), 50 units/ml penicillin G, and 50 μg/ml streptomycin sulfate. PML−/− mouse embryonic fibroblasts were a gift from Dr. Kun-Sang Chang.
Chemicals and Antibodies
U0126 was used at 20 μm and was purchased from Calbiochem. EGF was used at 100 ng/ml and was purchased from Promega (Madison, WI). Antibodies used were as follows: PML (Santa Cruz Biotechnology, H-238 and PG-M3), p-ERK (Santa Cruz Biotechnology, Tyr-204), ERK2 (Santa Cruz Biotechnology, C-14), α-tubulin (Sigma, B-5-1-2), β-actin (Sigma, AC-15), GFP (Santa Cruz Biotechnology, B-2), HA-HRP (Roche Applied Science), and FLAG (Sigma, M5). Anti-Pin1 and anti-phospho-specific PML antibodies were produced according to the manufacturer's protocol (ABR).
Plasmid Construction and Transfection
CMX-HA-PML4, CMX-HA-PML4 (4×), and CMX-GFP have been described previously (30). CMX-FLAG-ERK2 and CMX-FLAG-PML4 were generated by PCR and subcloned into CMX-1F vector (28). HeLa, CV-1, and HEK293 cells were treated with the indicated drug and/or transfected as indicated with ≥5 μg of plasmid DNA using Lipofectamine 2000 following the manufacturer's protocol (Invitrogen).
Immunofluorescence Microscopy
Immunostaining was carried out as described previously (30) with the following changes. Primary antibody incubation with anti-PML (Santa Cruz Biotechnology, PG-M3), anti-p-ERK (Santa Cruz Biotechnology, Tyr-204), or anti-ERK2 (Santa Cruz Biotechnology, C-14) was carried out at room temperature for 2 h. The secondary antibodies used were Alexa Fluor 488 and Alexa Flour 594 (Invitrogen). All images were taken with a Leica Wetzlar Gbmh microscope. Data acquisition was done with a SPOT camera and software (Diagnostic Instruments, Inc.).
Western Blotting and Immunoprecipitation
Whole cell extracts were prepared at the indicated time after U0126 treatment or 48 h after transfection using a radioimmune precipitation assay (RIPA) buffer (1× PBS, 1% Nonidet P-40, 0.5% sodium deoxycholate, and 0.1% SDS) plus protease inhibitors and phosphatase inhibitors (Roche Applied Science). After 90 min of incubation with the RIPA buffer, insoluble components were removed and the resulting cell extracts were separated by SDS-PAGE. Proteins were transferred to PVDF membranes, and products were visualized by immunoblotting. Blocking was performed in 10% nonfat milk in 1× PBS with 0.1% Tween-20 (PBST), and primary and secondary antibodies were incubated in 5% milk/PBST. Primary antibody incubation was carried out overnight at 4 °C. Detection was performed using an ECL detection kit (Pierce). To detect endogenous interactions, MDA-MB-231 cell extracts were prepared using NETN buffer (20 mm Tris-HCl, pH 8.0, 100 mm NaCl, 1 mm EDTA, 10% glycerol, 1 mm dithiothreitol, 0.1% Nonidet P-40) with a mixture of protease inhibitors (Roche Applied Science). The resulting cell extracts were immunoprecipitated with anti-Pin1 antibody in NETN buffer for 4 h. The immunoprecipitated complexes were resolved by SDS-PAGE and immunoblotted with anti-PML and anti-Pin1 antibodies.
GST Pulldown Assay
For glutathione S-transferase (GST) pulldown assays, whole cell lysates were prepared using RIPA buffer and incubated for 60 min on a Nutator at 4 °C with GST-PML4 conjugated glutathione-Sepharose beads in NETN buffer with protease inhibitors and phosphatase inhibitors (Roche Applied Science). After incubation, the beads were washed three times with NETN and collected by centrifugation, and the pulldown fractions were resolved on SDS-PAGE gels and immunoblotted with anti-phospho-ERK antibody.
siRNA
MDA-MB-231 cells were grown to 50–60% confluency in 6-well cell culture plates in the medium noted above without the antibiotics. Cells were treated with either a non-targeting small interfering RNA (siRNA) oligonucleotide (Dharmacon, D-001810-01-50) or siRNA directed against ERK2 (Dharmacon, LQ-003555-00), according to the manufacturer's protocol using the Dharmafect 1 reagent. Each well received 200 pmol of siRNA. Forty-eight hours after transfection, cells were harvested, and immunoprecipitation/immunoblotting was performed. HEK293 cells were transfected with Pin1-targeting siRNA (Dharmacon, J-003291-13) or non-targeting siRNA. Forty-eight hours after transfection with siRNA, cells were transfected with plasmid expressing HA-PML4 and/or FLAG-ERK2, and 24 h after plasmid transfection, cells were harvested and analyzed by immunoblotting.
Quantitative RT-PCR
MDA-MB-231 cells were transfected with siRNA as described previously. Forty-eight hours after transfection, cells were harvested, and total RNA was prepared using PrepEase RNA Spin kits (USB). cDNA was generated using Superscript III reverse transcriptase with oligo-dT according to the manufacturer's protocol (Invitrogen). For quantitative real-time PCR, reactions were performed using iQ SYBR Green supermix (Bio-Rad) and analyzed as previously described (32). Primers used were as follows: PML forward, 5′-GAATCAACGGAATGAATGGCT-3′; PML reverse, 5′-CCAGGGACTCAGAATACAGG-3′; ERK2 forward, 5′-TACACCAACCTCTCGTACATCG-3′; ERK2 reverse, 5′-CATGTCTGAAGCGCAGTAAGATT-3′; GAPDH forward, 5′-GAAGGTGAAGGTCGGAGT-3′; and GAPDH reverse, 5′-GAAGATGGTGATGGGATTTC-3′.
RESULTS
U0126 Up-regulates PML Protein Levels
We have previously demonstrated that the peptidyl-prolyl cis-trans isomerase, Pin1, binds and targets PML for degradation in a phosphorylation-dependent manner (30). To further elucidate the mechanisms underlying Pin1-mediated PML degradation, we aimed to identify one or more factors that promote PML phosphorylation. It has been previously suggested that arsenic trioxide (As2O3) induces ERK2-mediated phosphorylation of PML (33). To examine whether modulation of the MEK/ERK pathway influences the expression level of PML protein, we examined changes in PML NBs and PML protein levels using the inhibitor of ERK upstream kinases MEK1/2, U0126, by immunofluorescence and immunoblotting, respectively. We found that U0126 treatment resulted in an increase in PML NB size and number in MDA-MB-231 cells (Fig. 1A) and in HeLa cells (Fig. 1B), whereas there was no apparent change in PML localization. In HeLa cells, the number of PML NBs increased ∼3-fold after 1 h of U0126 treatment. In addition, we found that PML and phosphorylated ERK1/2 partially co-localized in the nucleus through 1 h of treatment (Fig. 1A, panel f). The increase in PML NB number correlated with increased PML protein levels (Fig. 1C, upper panels). To determine whether Pin1 plays a role in U0126-mediated increase in PML protein levels, we examined the interaction between PML and Pin1 in U0126-treated cells by immunoprecipitation. Indeed, the interaction between PML and Pin1 was significantly decreased in U0126-treated samples (Fig. 1C, lower panels). Similarly, U0126 induced PML protein accumulation and decreased Pin1-PML interaction in HeLa cells (supplemental Fig. S1). These results suggest that U0126 up-regulates PML protein abundance by inhibiting its interaction with Pin1.
FIGURE 1.
Effects of U0126 on the formation of PML NBs and PML expression levels. A, MDA-MB-231 were treated with or without 20 μm U0126 for 2 h. Cells were immunostained with anti-PML and anti-phospho-ERK antibodies, and images were taken by fluorescence microscopy. DAPI-stained nuclei (a and g); phosphorylated ERK1/2 (b and h); endogenous PML (c and i); merged staining of phosphorylated ERK1/2 and DAPI (d and j); merged staining of endogenous PML and DAPI (e and k); merged staining of endogenous PML and phosphorylated ERK1/2 (f and l). The arrows mark cells in which PML and phosphorylated ERK2 co-localize. B, HeLa cells were treated with or without 20 μm U0126 for 1 h. Cells were immunostained with anti-PML antibodies followed by fluorescence microscopy. Left panel: DAPI-stained nuclei (a and d); endogenous PML (b and e); merged staining of endogenous PML and DAPI (c and f). Right panel: numbers of PML NBs in each cell were scored. Over 100 cells in duplicated experiments were scored. Error bars indicate ±S.D. C, MDA-MB 231 cells were treated with U0126 for the times indicated, and whole cell extracts were prepared as described under “Experimental Procedures.” Upper panel: whole lysates were analyzed by Western blotting with anti-PML, anti-phospho-ERK, and anti-Pin1 antibodies. Anti-α-tubulin was used as a loading control. Lower panel: whole cell extracts were immunoprecipitated with anti-Pin1 antibody and analyzed by immunoblotting with anti-PML and anti-Pin1 antibodies.
The Formation of PML NBs and Expression Levels of PML Protein Were Increased in ERK2 Knockdown Cells
To further investigate the mechanism underlying U0126-induced formation of PML NBs and protein levels, we examined changes in the formation of PML NBs and expression levels of PML protein in ERK2 knockdown cells and found that the size and number of PML NBs were increased in ERK2 knockdown HeLa cells (Fig. 2A, left panels) and MDA-MB-231 cells (Fig. 2A, right panels). The expression level of PML protein was increased in ERK2 knockdown MDA-MB-231 cells (Fig. 2B). In contrast, PML mRNA level remains unchanged in ERK2 knockdown cells (Fig. 2C). These results suggested that ERK2 negatively regulates PML expression at the protein level.
FIGURE 2.
ERK2 knockdown increases the expression of PML and formation of PML NBs. A, non-targeting siRNA or ERK2-targeting siRNAs were transfected into HeLa (left panels) or MDA-MB-231 (right panels) cells. Transfected cells were immunostained with anti-PML and anti-ERK2 antibodies, and images were taken by fluorescence microscopy. DAPI shows staining for DNA. B, MDA-MB-231 cells were transfected with non-targeting siRNA or ERK2 targeting siRNA, and whole cell extracts were analyzed by immunoblotting with anti-PML, anti-ERK2, and anti-β-actin antibodies. C, MDA-MB-231 cells were transfected with non-targeting siRNA or ERK2-targeting siRNAs, and total RNA was isolated and analyzed by real-time quantitative RT-PCR to determine the mRNA expression levels of PML and ERK2. GAPDH was used as an internal control. Error bars indicate ±S.D.
A PML Mutant Defective in Binding Pin1 Is Resistant to ERK2-mediated Degradation
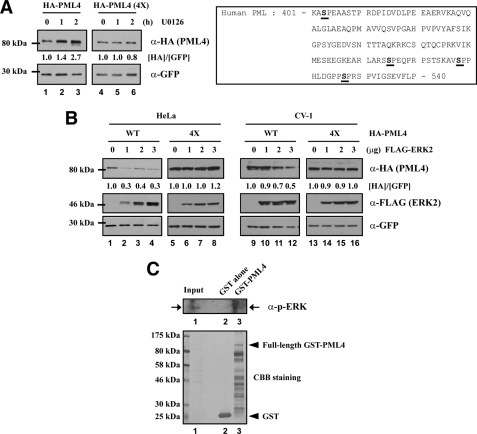
Because we have previously demonstrated that the four putative phosphorylated serines (Ser-403, Ser-505, Ser-518, and Ser-527) of PML (Fig. 3A, right panel) are required for interaction with Pin1 (30), we investigated the effect of U0126 on wild-type and a Pin1 binding-defective mutant PML (4×). HeLa cells were co-transfected with expression plasmids containing HA-tagged wild-type (WT) or the PML4 (4×) mutant and GFP (as an internal control) followed by U0126 treatment. We prepared whole cell extracts and performed Western blotting to examine the expression levels of wild-type and 4× mutant PML probed with anti-HA and anti-GFP antibodies. The protein expression level of transfected wild-type PML4 was increased by U0126 treatment, but there was no change in the expression level of PML4 (4×) observed (Fig. 3A, left panels). Next, to determine the effect of ERK2 overexpression on exogenous expression of wild-type and mutant PML, we co-transfected HeLa or CV-1 cells with varying amounts of expression plasmids encoding HA-tagged wild-type or the PML (4×) mutant in the absence or presence of FLAG-tagged ERK2. The exogenous expression of ERK2 decreased the expression of wild-type PML in dosage-dependent manner, but not PML (4×) in both cell lines (Fig. 3B). Furthermore, to determine whether phosphorylated ERK2 physically interacts with PML4, immobilized GST-PML4 fusion proteins were incubated with HeLa whole cell extracts expressing phosphorylated ERK2. We found that GST-PML4 but not GST alone was capable of pulling down phosphorylated ERK2 (Fig. 3C). These results suggest that activated ERK2 might bind to and phosphorylate PML.
FIGURE 3.
ERK2 overexpression promotes PML degradation. A, HeLa cells were transfected with wild-type or mutant HA-PML expression plasmids and treated with U0126 for the time indicated. Whole cell extracts were prepared in RIPA buffer and analyzed by immunoblotting with anti-HA and anti-GFP antibodies (left panels). GFP was used as a transfection and loading control. The four serine residues mutated to alanine in PML4 (4×) are indicated (right panel). B, HeLa and CV-1 cells were transfected with expression plasmids for HA-PML, GFP, and FLAG-ERK2 as indicated. Whole cell extracts were analyzed by immunoblotting with anti-HA, anti-FLAG, and anti-GFP antibodies. GFP was used as a transfection and loading control. C, whole cell extracts of HeLa cells were prepared for pulldown assays with immobilized GST-PML4 proteins. The pulldown fractions were analyzed by immunoblotting with anti-phospho-ERK antibody. 0.5% of the input is shown.
U0126 Treatment and ERK2 Knockdown Decrease Phosphorylation of PML at Ser-403 and Ser-505
It was previously reported that ERK2 phosphorylates PML in vitro (33). These data along with our observation that U0126 increases PML protein levels suggest that ERK2 enhances PML phosphorylation in mammalian cells. To test this, we determined whether U0126 treatment alters phosphorylation levels at these four residues of PML. We first generated phospho-specific antibodies that recognize PML-phosphorylated serine residues (pS403, pS505, pS518, or pS527). As shown in Fig. 4A, these antibodies recognize wild-type PML, but not mutant PML (4×) (Fig. 4A). Furthermore, calf intestine phosphatase treatment significantly reduced the ability of these antibodies to recognize PML (Fig. 4B). To examine the effect of U0126 on phosphorylation of PML at Ser-403, Ser-505, Ser-518, and Ser-527, HeLa cells transfected with HA-PML4 were treated with vehicle or U0126, whole cell extracts prepared and immunoprecipitated with anti-HA antibodies followed by immunoblotting with phospho-specific anti-PML antibodies. We found that the phosphorylation level at Ser-403 and Ser-505 of PML was decreased by ∼70% in U0126-treated cells. In contrast, phosphorylation at Ser-518 was only slightly decreased and phosphorylation at Ser-527 was not changed (Fig. 4C and supplementary Fig. S2). To further confirm the role of ERK2 on PML phosphorylation, we determined whether knockdown of ERK2 or Pin1 alters PML phosphorylation. HeLa cells were transfected with ERK2 or Pin1-targeting siRNAs followed by transfection with HA-PML4. Whole cell extracts were prepared and immunoprecipitated with anti-HA antibody followed by immunoblotting with phospho-specific anti-PML antibodies. Knockdown of ERK2 did not alter Pin1 protein accumulation. Similarly, knockdown of Pin1 did not affect ERK2 protein abundance (Fig. 4D). We found that the phosphorylation level of Ser-403 and Ser-505 of PML was decreased in ERK2 knockdown cells but remained unchanged in Pin1 knockdown cells (Fig. 4E). Based on these results, we conclude that ERK2 is capable of PML phosphorylation at Ser-403 and Ser-505 in mammalian cells.
FIGURE 4.
The effect of ERK2 on phosphorylation status of PML. A, immortalized PML−/− mouse embryonic fibroblasts were transfected with either HA-PML4 or HA-PML4 (4×). Whole cell extracts were prepared in RIPA buffer, which contained the phosphatase inhibitors sodium fluoride and sodium orthovanadate. The resulting whole cell extracts were analyzed by immunoprecipitation with anti-HA antibody followed by immunoblotting with either anti-HA antibody or anti-phospho-specific PML antibodies as indicated. B, HeLa whole cell extracts expressing wild-type FLAG-PML4 were immunoprecipitated with anti-FLAG antibodies. The immunoprecipitates were treated with or without calf intestinal phosphatase for 30 min at 30 °C and then analyzed by immunoblotting with anti-FLAG or anti-phospho-specific PML antibodies as indicated. C, HeLa cells were transfected with HA-PML4 and treated with U0126 for the indicated time. Whole cell extracts were prepared and were immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-HA and anti-phospho-PML-specific antibodies as indicated. D, knockdown of ERK2 or Pin1 in HeLa cells. E, HeLa cells were transfected with HA-PML4 following transient transfection with a non-targeting siRNA, ERK2-targeting siRNAs (left panel), or a Pin1-targeting siRNA (right panel).as described in D. Whole cell extracts were prepared and followed by immunoprecipitation with anti-HA antibody and immunoblotting with anti-HA and anti-phospho-PML-specific antibodies as indicated.
MAPK ERK2 Promotes PML Degradation via Pin1-dependent Pathway
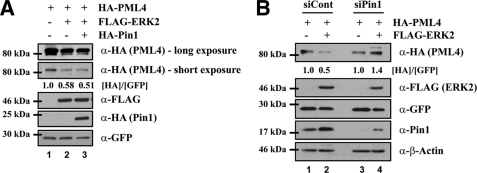
As shown in Fig. 1C, inhibition of ERK2 activity by U0126 resulted in a reduced interaction between PML and Pin1. Next, to determine the effect of Pin1 on ERK2-mediated degradation of PML protein, HEK293 cells were co-transfected with combinations of HA-tagged PML, FLAG-tagged ERK2, and HA-tagged Pin1. The expression of PML was decreased by ∼40% in ERK2-overexpressing cells and by ∼50% in ERK2- and Pin1-overexpressing cells (Fig. 5A). We further investigated ERK2-mediated degradation of PML in Pin1 knockdown cells. HEK293 cells were transfected with non-targeting siRNA or Pin1-targeting siRNA followed by co-transfection of HA-tagged PML with or without FLAG-tagged ERK2 (Fig. 5B). We found that overexpressed ERK2 failed to decrease PML protein levels in Pin1 knockdown cells. Taken together, these results indicate that ERK2 promotes Pin1-dependent degradation of PML.
FIGURE 5.
ERK2 promotes the degradation of PML in Pin1-dependent pathway. A, HEK293 cells were transfected with plasmids encoding HA-PML, FLAG-ERK2, HA-Pin1, and GFP, and whole cell extracts were prepared and analyzed by immunoblotting with anti-HA, anti-FLAG, and anti-GFP antibodies. B, HEK293 cells were transfected with HA-PML, FLAG-ERK2, and GFP, 48 h after cells had been transfected with Pin1-targeting siRNA as indicated. Whole cell extracts were analyzed by immunoblotting with anti-HA, anti-FLAG, anti-GFP, anti-Pin1, and anti-β-actin. GFP was used as transfection and loading control. β-Actin was used as an internal control for protein loading.
EGF Down-regulates the Expression of PML in an ERK2-dependent Manner
The canonical ERK pathway is stimulated by extracellular growth factors such as EGF, VEGF, IGF, and PDGF binding to their cognate receptors (34). To determine whether EGF had an effect on PML protein accumulation, we transiently transfected HA-tagged PML and a GFP expression plasmids (as an internal control) into HEK293 cells followed by EGF treatment for the times indicated. We used HEK293 cells because of its low basal level of activated ERK compared with other cancer cell lines. Whole cell extracts were prepared, and immunoblotting was performed to determine the expression level of PML and phospho-ERK. The activation of ERK1/2 peaked at 15 min while the exogenous PML protein didn't show a significant decrease until 1 h after EGF treatment (Fig. 6A). This suggests that EGF signaling promoted the PML degradation and that activation of ERK preceded PML degradation. In contrast, there was no significant change in the expression levels of ERK2 and GFP (Fig. 6A). To further investigate whether EGF-mediated ERK activation results in PML degradation, we transfected HEK293 cells with HA-tagged PML and GFP expression plasmid and treated the cells with EGF in the presence or absence of U0126 for the times indicated. We found that the EGF-induced decrease in PML protein accumulation was blocked by U0126 (Fig. 6B). Next, we determined the phosphorylation status of PML at Ser-403 and Ser-505 in EGF in U0126-treated cells. Whole cell extracts were prepared from HeLa cells transfected with HA-tagged PML followed by EGF treatment in the presence or absence of U0126. We performed immunoprecipitation with anti-HA antibody-conjugated beads after normalizing HA-PML expression levels and immunoblotting using anti-phospho-specific PML antibodies. We observed that phosphorylation of PML at Ser-403 and Ser-505 was induced by EGF treatment, and this increase was abolished by U0126 treatment (Fig. 6C, 0.8–0.9 versus 1.4). Next, to investigate whether EGF-mediated PML degradation is Pin1-dependent, we examined whether knockdown of Pin1 has an effect on EGF-mediated PML degradation. HEK293 cells were transfected with a control or Pin1 siRNA followed by transfection with HA-PML4 and GFP expression plasmids. Cells were treated with EGF for the indicated times, and whole cell extracts were prepared. We found that PML protein accumulation was decreased by EGF treatment in cells transfected with control siRNA. In contrast, knockdown of Pin1 abolished the ability of EGF to down-regulate PML accumulation (Fig. 6D). This observation was accompanied by the finding that EGF increased the interaction between PML and Pin1 in HEK293 cells (Fig. 6E and supplemental Fig. S3) and the interaction between PML and phospho-ERK (Fig. 6F). Taken together, we conclude that EGF promotes PML protein degradation via an ERK2-Pin1 axis.
FIGURE 6.
EGF promotes the degradation of PML via ERK2-Pin1 axis. A, HEK293 cells were transfected with HA-PML and GFP as indicated and treated with 100 ng/ml EGF for the times specified. Whole cell extracts were prepared and analyzed by immunoblotting with anti-HA, anti-phospho-ERK, anti-ERK2, anti-Pin1, anti-β-actin, and anti-GFP antibodies as indicated. B, HEK293 cells were transfected with HA-PML and GFP as indicated and followed by EGF treatment in the presence or absence of 20 μm U0126 for the times specified. The resulting whole cell extracts were analyzed by immunoblotting with anti-HA, anti-phospho-ERK, anti-ERK2, and anti-GFP antibodies as indicated. C, HEK293 cells were treated with EGF in presence or absence of U0126 for 1h, 48 h after cells had been transfected with HA-PML4. The resulting whole cell extracts were immunoprecipitated with anti-HA antibodies and analyzed by immunoblotting with anti-HA and anti-phospho-PML antibodies as indicated. D, HEK293 cells were transfected with HA-PML4 and GFP following transfection with a control or Pin1-targeting siRNA. Transfected cells were sup-cultured to three wells for each siRNA and were serum-starved for 24 h, before cells were treated with 100 ng/ml EGF at the indicated times. Whole cell extracts were prepared and analyzed by immunoblotting with anti-HA, anti-phospho-ERK, anti-ERK2, anti-Pin1, anti-β-actin, and anti-GFP antibodies. E, HEK293 cells were transfected with FLAG-PML4 and HA-Pin1, followed by treatment with 100 ng/ml EGF for the indicated times 24 h after serum-starvation. The resulted whole cell extracts were immunoprecipitated with anti-HA antibody followed by immunoblotting with anti-FLAG and anti-HA antibodies. F, HEK293 cells were transfected with FLAG-PML4 and serum-starved for 24 h followed by treatment with 100 ng/ml EGF for 30 min. The expression levels of FLAG-PML4 and phospho-ERK in whole cell extracts was shown after normalizing the expression level of FLAG-PML4 for immunoprecipitation (upper panel). The resulted whole cell extracts were immunoprecipitated with anti-phospho-ERK antibody followed by immunoblotting with anti-FLAG and phospho-ERK antibodies. The arrowhead indicates phospho-ERK2. The asterisk indicates IgG (lower panel).
DISCUSSION
In this study, we investigated the mechanisms by which the PML protein accumulation is regulated by phosphorylation and identified a kinase responsible for PML phosphorylation. Our data show that U0126, an inhibitor of the ERK1/2 upstream kinase MEK1/2, increased the expression of PML and the formation of PML NBs via inhibition of Pin1-dependent degradation (Fig. 1). Furthermore, knockdown of ERK2 increased the expression of PML protein, and overexpression of ERK2 decreased the expression of PML (Figs. 2 and 3). The expression of the PML 4× mutant, which is mutated at four serine residues (S403/505/518/527A) that are potential sites of phosphorylation and is resistant to Pin1-dependent degradation, was not changed by the treatment of U0126 and overexpression of ERK2 (Fig. 3). In addition, we verified that U0126 treatment decreased PML phosphorylation at Ser-403 and Ser-505 (Fig. 4C). We have previously reported that Pin1 promotes the degradation of PML protein in a phosphorylation-dependent manner (30). We further show that reducing Pin1 levels blocks ERK2-mediated PML down-regulation (Fig. 5). Taken together, our present study supports a model in which activated ERK2 decreases PML protein accumulation via phosphorylating PML at Ser-403 and Ser-505 and promoting its interaction with Pin1.
ERK1/2 are kinases that phosphorylate serines and threonines of target protein substrates and regulate a wide range of cellular processes: gene expression, cell growth, differentiation, meiosis, mitosis, cell motility, metabolism, apoptosis, and embryogenesis (35, 36). Oncogenic Ras constitutively activates the ERK1/2 pathways, which contributes to the increased proliferation rate of tumor cells (37). Because growth factors generally activate the Ras-MEK1/2-ERK1/2 pathway, our results suggest that Ras is involved in EGF/ERK-dependent PML phosphorylation (Fig. 6). For this reason, it is possible that activated ERK2 enhances cell growth by promoting the degradation of the tumor-suppressive PML protein in cancer cells. This notion is consistent with the observation that ERK2 is often found highly activated in cancer cells, whereas PML protein levels are low in cancerous tissues comparing to adjacent normal tissues (15, 38). Mitogenic stimulation triggers the translocation of ERK1/2 from the cytoplasm to the nucleus (39). Our observation that PML and activated ERK2 partially co-localize in the nucleus suggests that EGF promotes the activation and translocation of ERK2 into nucleus and ERK2-mediated PML phosphorylation. Indeed, U0126 inhibits EGF-induced PML phosphorylation and down-regulation in HEK293 cells (Fig. 6). Several reports have also suggested that PML is phosphorylated at different residues by ERK, although we did not detect phosphorylation at these residues by mass spectrometry in HeLa cells (30, 33, 40). Furthermore, BMK1/ERK5, the latest mammalian MAP kinase discovered, phosphorylates PML at Ser-403 and Thr-409 in an in vitro kinase assay (41). These data support a model that PML protein abundance is regulated by multiple phosphorylation-dependent mechanisms in response to extracellular stimuli. By doing so, each mechanism is responsible for distinct physiological conditions to control PML protein stability or protein-protein interactions and therefore cell proliferation, differentiation, and survival.
We show that inactivating ERK2 with U0126 disrupted the interaction between PML (Fig. 1C) and Pin1 and the ability of EGF to promote PML-Pin1 interaction (Fig. 6E and supplemental Fig. S3), PML-ERK2 interaction (Fig. 6F), and PML protein down-regulation (Fig. 6B). Furthermore, knockdown of Pin1 blocked the ability of ERK2 to promote PML protein degradation (Fig. 5B) and the ability of EGF to induce PML degradation (Fig. 6D). These data indicate that ERK2-mediated PML down-regulation depends on Pin1. Consistent with this notion, we found that inactivating ERK2 decreased phosphorylation of Ser-403 and Ser-505 and treatment with EGF increased this phosphorylation (Figs. 4C and 6C). In contrast, we found that phosphorylation of Ser-518 and Ser-527 remained unchanged, suggesting that phosphorylation of these residues likely regulates the interaction of PML with Pin1 in response to alternative kinase pathways as well. Indeed, a recent report indicated that Cdk2 phosphorylates Ser-518 and promotes PML degradation (42). Taken together, our data support a model in which the ERK2-Pin1 axis mediates EGF-induced PML protein turnover. It will be interesting to know whether such regulation is adopted by other mitogens and whether this axis acts upon other tumor-suppressor proteins.
Supplementary Material
Acknowledgment
We thank Dr. David Samols for his comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK078965 and HL093269 (to H.-Y. K.). This work was also supported by the Pardee Foundation (to H.-Y. K.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S3.
J. H. Lim, Y. Liu, E. Reineke, and H.-Y. Kao, submitted for publication.
- PML
- promyelocytic leukemia
- NB
- nuclear body
- SUMO
- small ubiquitin-like modifier
- RIPA
- radioimmune precipitation assay buffer.
REFERENCES
- 1. de Thé H., Lavau C., Marchio A., Chomienne C., Degos L., Dejean A. (1991) Cell 66, 675–684 [DOI] [PubMed] [Google Scholar]
- 2. Kakizuka A., Miller W. H., Jr., Umesono K., Warrell R. P., Jr., Frankel S. R., Murty V. V., Dmitrovsky E., Evans R. M. (1991) Cell 66, 663–674 [DOI] [PubMed] [Google Scholar]
- 3. Shen T. H., Lin H. K., Scaglioni P. P., Yung T. M., Pandolfi P. P. (2006) Mol. Cell 24, 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bernardi R., Pandolfi P. P. (2007) Nat. Rev. Mol. Cell Biol. 8, 1006–1016 [DOI] [PubMed] [Google Scholar]
- 5. Borden K. L. (2002) Mol. Cell Biol. 22, 5259–5269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Maul G. G., Negorev D., Bell P., Ishov A. M. (2000) J. Struct. Biol. 129, 278–287 [DOI] [PubMed] [Google Scholar]
- 7. Salomoni P., Ferguson B. J., Wyllie A. H., Rich T. (2008) Cell Res. 18, 622–640 [DOI] [PubMed] [Google Scholar]
- 8. Reineke E. L., Kao H. Y. (2009) Int. J. Biol. Sci. 5, 366–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Salomoni P., Pandolfi P. P. (2002) Cell 108, 165–170 [DOI] [PubMed] [Google Scholar]
- 10. Reineke E. L., Kao H. Y. (2009) Cancer Ther. 7, 219–226 [PMC free article] [PubMed] [Google Scholar]
- 11. Wang Z. G., Delva L., Gaboli M., Rivi R., Giorgio M., Cordon-Cardo C., Grosveld F., Pandolfi P. P. (1998) Science 279, 1547–1551 [DOI] [PubMed] [Google Scholar]
- 12. Wang Z. G., Ruggero D., Ronchetti S., Zhong S., Gaboli M., Rivi R., Pandolfi P. P. (1998) Nat. Genet. 20, 266–272 [DOI] [PubMed] [Google Scholar]
- 13. Giorgi C., Ito K., Lin H. K., Santangelo C., Wieckowski M. R., Lebiedzinska M., Bononi A., Bonora M., Duszynski J., Bernardi R., Rizzuto R., Tacchetti C., Pinton P., Pandolfi P. P. (2010) Science 330, 1247–1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bernardi R., Guernah I., Jin D., Grisendi S., Alimonti A., Teruya-Feldstein J., Cordon-Cardo C., Simon M. C., Rafii S., Pandolfi P. P. (2006) Nature 442, 779–785 [DOI] [PubMed] [Google Scholar]
- 15. Gurrieri C., Capodieci P., Bernardi R., Scaglioni P. P., Nafa K., Rush L. J., Verbel D. A., Cordon-Cardo C., Pandolfi P. P. (2004) J. Natl. Cancer Inst. 96, 269–279 [DOI] [PubMed] [Google Scholar]
- 16. Chelbi-Alix M. K., Pelicano L., Quignon F., Koken M. H., Venturini L., Stadler M., Pavlovic J., Degos L., de Thé H. (1995) Leukemia 9, 2027–2033 [PubMed] [Google Scholar]
- 17. Lavau C., Marchio A., Fagioli M., Jansen J., Falini B., Lebon P., Grosveld F., Pandolfi P. P., Pelicci P. G., Dejean A. (1995) Oncogene 11, 871–876 [PubMed] [Google Scholar]
- 18. Stadler M., Chelbi-Alix M. K., Koken M. H., Venturini L., Lee C., Saïb A., Quignon F., Pelicano L., Guillemin M. C., Schindler C., et al. (1995) Oncogene 11, 2565–2573 [PubMed] [Google Scholar]
- 19. Dror N., Rave-Harel N., Burchert A., Azriel A., Tamura T., Tailor P., Neubauer A., Ozato K., Levi B. Z. (2007) J. Biol. Chem. 282, 5633–5640 [DOI] [PubMed] [Google Scholar]
- 20. de Stanchina E., Querido E., Narita M., Davuluri R. V., Pandolfi P. P., Ferbeyre G., Lowe S. W. (2004) Mol. Cell 13, 523–535 [DOI] [PubMed] [Google Scholar]
- 21. Lallemand-Breitenbach V., Zhu J., Puvion F., Koken M., Honoré N., Doubeikovsky A., Duprez E., Pandolfi P. P., Puvion E., Freemont P., de Thé H. (2001) J. Exp. Med. 193, 1361–1371 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Tatham M. H., Geoffroy M. C., Shen L., Plechanovova A., Hattersley N., Jaffray E. G., Palvimo J. J., Hay R. T. (2008) Nat. Cell Biol. 10, 538–546 [DOI] [PubMed] [Google Scholar]
- 23. Lallemand-Breitenbach V., Jeanne M., Benhenda S., Nasr R., Lei M., Peres L., Zhou J., Zhu J., Raught B., de Thé H. (2008) Nat. Cell Biol. 10, 547–555 [DOI] [PubMed] [Google Scholar]
- 24. Zhang X. W., Yan X. J., Zhou Z. R., Yang F. F., Wu Z. Y., Sun H. B., Liang W. X., Song A. X., Lallemand-Breitenbach V., Jeanne M., Zhang Q. Y., Yang H. Y., Huang Q. H., Zhou G. B., Tong J. H., Zhang Y., Wu J. H., Hu H. Y., de Thé H., Chen S. J., Chen Z. (2010) Science 328, 240–243 [DOI] [PubMed] [Google Scholar]
- 25. Jeanne M., Lallemand-Breitenbach V., Ferhi O., Koken M., Le Bras M., Duffort S., Peres L., Berthier C., Soilihi H., Raught B., de Thé H. (2010) Cancer Cell 18, 88–98 [DOI] [PubMed] [Google Scholar]
- 26. Ling P. D., Tan J., Sewatanon J., Peng R. (2008) J. Virol. 82, 8000–8012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. El McHichi B., Regad T., Maroui M. A., Rodriguez M. S., Aminev A., Gerbaud S., Escriou N., Dianoux L., Chelbi-Alix M. K. (2010) J. Virol. 84, 11634–11645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gao C., Cheng X., Lam M., Liu Y., Liu Q., Chang K. S., Kao H. Y. (2008) Mol. Biol. Cell 19, 3020–3027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Gao C., Ho C. C., Reineke E., Lam M., Cheng X., Stanya K. J., Liu Y., Chakraborty S., Shih H. M., Kao H. Y. (2008) Mol. Cell Biol. 28, 5658–5667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Reineke E. L., Lam M., Liu Q., Liu Y., Stanya K. J., Chang K. S., Means A. R., Kao H. Y. (2008) Mol. Cell Biol. 28, 997–1006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Reineke E. L., Liu Y., Kao H. Y. (2010) J. Biol. Chem. 285, 9485–9492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang H., Stavnezer E. (2009) J. Biol. Chem. 284, 2867–2879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Hayakawa F., Privalsky M. L. (2004) Cancer Cell 5, 389–401 [DOI] [PubMed] [Google Scholar]
- 34. Ramos J. W. (2008) Int. J. Biochem. Cell Biol. 40, 2707–2719 [DOI] [PubMed] [Google Scholar]
- 35. Chen Z., Gibson T. B., Robinson F., Silvestro L., Pearson G., Xu B., Wright A., Vanderbilt C., Cobb M. H. (2001) Chem. Rev. 101, 2449–2476 [DOI] [PubMed] [Google Scholar]
- 36. Johnson G. L., Lapadat R. (2002) Science 298, 1911–1912 [DOI] [PubMed] [Google Scholar]
- 37. Calvo F., Agudo-Ibáñez L., Crespo P. (2010) Bioessays 32, 412–421 [DOI] [PubMed] [Google Scholar]
- 38. Sebolt-Leopold J. S., Herrera R. (2004) Nat. Rev. Cancer 4, 937–947 [DOI] [PubMed] [Google Scholar]
- 39. Brunet A., Roux D., Lenormand P., Dowd S., Keyse S., Pouysségur J. (1999) EMBO J. 18, 664–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ji H., Wang J., Nika H., Hawke D., Keezer S., Ge Q., Fang B., Fang X., Fang D., Litchfield D. W., Aldape K., Lu Z. (2009) Mol. Cell 36, 547–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yang Q., Deng X., Lu B., Cameron M., Fearns C., Patricelli M. P., Yates J. R., 3rd, Gray N. S., Lee J. D. (2010) Cancer Cell 18, 258–267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Yuan W. C., Lee Y. R., Huang S. F., Lin Y. M., Chen T. Y., Chung H. C., Tsai C. H., Chen H. Y., Chiang C. T., Lai C. K., Lu L. T., Chen C. H., Gu D. L., Pu Y. S., Jou Y. S., Lu K. P., Hsiao P. W., Shih H. M., Chen R. H. (2011) Cancer Cell 20, 214–228 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.