Background: AGR2 is a novel ER protein for which the molecular and cellular functions remain uncharacterized.
Results: AGR2 associates to nascent chains in the ER, and its silencing impacts UPR and ERAD and sensitizes cells to autophagy.
Conclusion: AGR2 plays an important role in the maintenance of ER homeostasis.
Significance: AGR2-mediated control of ER homeostasis could be of importance for cancer development.
Keywords: Endoplasmic Reticulum Stress, Endoplasmic Reticulum (ER), ER-associated Degradation, Protein Secretion, Stress Response, Tumor, Unfolded Protein Response
Abstract
The protein-disulfide isomerase (PDI) family member anterior gradient 2 (AGR2) is reportedly overexpressed in numerous cancers and plays a role in cancer development. However, to date the molecular functions of AGR2 remain to be characterized. Herein we have identified AGR2 as bound to newly synthesized cargo proteins using a proteomics analysis of endoplasmic reticulum (ER) membrane-bound ribosomes. Nascent protein chains that translocate into the ER associate with specific ER luminal proteins, which in turn ensures proper folding and posttranslational modifications. Using both imaging and biochemical approaches, we confirmed that AGR2 localizes to the lumen of the ER and indirectly associates with ER membrane-bound ribosomes through nascent protein chains. We showed that AGR2 expression is controlled by the unfolded protein response and is in turn is involved in the maintenance of ER homeostasis. Remarkably, we have demonstrated that siRNA-mediated knockdown of AGR2 significantly alters the expression of components of the ER-associated degradation machinery and reduces the ability of cells to cope with acute ER stress, properties that might be relevant to the role of AGR2 in cancer development.
Introduction
Nascent secretory and integral membrane proteins are targeted to and translocated into the ER5 lumen where they are folded and assembled (1, 2). To ensure that proteins entering this compartment are properly folded, the ER has evolved a quality control system (ERQC), achieving partial surveillance on the protein folding status (3, 4). Improperly folded or misfolded proteins are not allowed to escape this compartment and remain in the ER where they accumulate (5). These proteins can then be recognized as terminally misfolded and be targeted for degradation by the ERAD machinery and proteasome-dependent mechanisms (6). About one-third of the genome products mature in the ER; however an apparently small number of ERQC and ERAD actors have been identified as responsible for the survey of ER protein conformation. Moreover, the control of protein secretion has been shown to play significant roles in systemic diseases such as diabetes, neurodegenerative disorders, and cancer. This suggests that additional components of ERQC/ERAD might still remain to be identified; this would most likely enhance the specificity of the existing molecular machines or provide selective advantages to diseased cells. Therefore, we designed an experimental strategy to identify additional constituents of these pathways.
Protein folding and quality control take place as early as newly synthesized proteins translocate co-translationally into the ER. We consequently postulated that isolating proteins in the vicinity of the translocation channel (translocon) would allow the identification of potentially novel component of ERQC/ERAD systems. In the past 20 years, studies using classical membrane biochemistry (7), yeast genetics (8, 9), and structural biology (10, 11), have led to the characterization of the translocon. In mammals, during co-translational import, the Sec61 complex, consisting of Sec61α, Sec61β, and Sec61γ (12), binds to ribosomes at the ER membrane, giving the ER its typical “rough” appearance (13). A ribosome-associated protein (RAP) enrichment strategy has revealed the presence of ER membrane proteins in close proximity to the minimal/immature translocon leading to the concept of an extended/mature version of this molecular machinery (14). In this version of the translocon, signal peptidases, the oligosaccharyl transferase complex, BiP, ERp57, the Signal Recogation particle receptor, Signal Recogation particle, p34, and p180 (12) have been shown to associate stably or transiently with the translocation channel. The translocon-associated protein (TRAP) complex (α, β, δ, and γ) has also been shown to be in close proximity to the translocon (15), but its role still remains unclear.
In the present work, we used the RAP enrichment strategy described previously (16) and analyzed this fraction by mass spectrometry sequencing (MS). We identified a total of 205 proteins, among which were found various proteins previously identified as translocon components per se, thereby confirming the validity of the approach. We also were able to identify 12 proteins with uncharacterized functions. We specifically focused our attention on a novel ER protein containing a single-cysteine thioredoxin-like motif, named anterior gradient 2 (AGR2). This protein has been described previously as pro-oncogenic and may thus represent a mechanistic intermediate in the connection between ERAD, ERQC and cancer development.
EXPERIMENTAL PROCEDURES
Antibodies, Cell Lines, and Reagents
Antibodies against calnexin (CNX) and gp25L were described previously (16). Anti-UDP-glucose:glycoprotein glucosyltransferase (UGGT), anti-ERp72, and anti-calreticulin were from Stressgen (Assay Design Inc., Ann Arbor, MI). Anti-Nck was kindly provided by Dr. Larose (McGill University, Montreal, Canada), anti-ribophorin I (RibI) and anti-TRAPα were kindly provided by Dr. Kreibich (New York University), anti-ORP150 was a gift from Dr. Ozawa (Kanazawa University, Japan), and anti-Sec61α antibodies were kindly provided by Dr. Nicchitta (Duke University Medical Center). Antibodies against ERK1, GRP78, IRE1α, JNK, and PERK were obtained from Santa Cruz Biotechnology (Santa Cruz, CA; sc-571, sc-1050, sc-20790, sc-93, and sc-13073, respectively). Anti-α-1 antitrypsin (α1AT) and anti-phospho-eIF2α were purchased from Dako (Glostrup, Denmark) and Invitrogen, respectively. Monoclonal anti-AGR2 antibodies were from Abnova (Abnova Corp., Taiwan). Mouse monoclonal anti-ATF6α was purchased from BioAcademia (Osaka, Japan). Anti-LC3 antibody was from Novus Biologicals (Littleton, CO). Anti-phospho-JNK (Thr-183/Tyr-185) and anti-eIF2α were from Cell Signaling Technology (Danvers, MA). Tunicamycin (Tun) was from Calbiochem (EMD Biosciences Inc., Darmstadt, Germany). Dithiothreitol (DTT), NH4Cl, and thapsigargin were obtained from Sigma. All cell lines used in this study were cultured in DMEM supplemented with 10% FBS (Hyclone, Logan, UT) and antibiotics unless specified. All cell cultures were maintained in a 37 °C incubator containing 5% CO2.
Preparation of Ribosome-associated ER Proteins and Immunoblot Analyses
Dog pancreatic microsomes were prepared as described previously (17). Briefly, a postmitochondrial supernatant (10,000 × g, 10 min) was prepared from freshly excised dog pancreas. This homogenate was then layered over a discontinuous sucrose gradient composed of sucrose 5 ml at 1 m, 5 ml at 1.5 m, 7.5 ml at 1.75 m, and 5 ml at 2 m in buffer A (50 mm triethanolamine-HCl, pH 7.5, 50 mm KCl, and 5 mm MgCl2) and centrifuged at 180,000 × g for 20 h. Rough microsomes collected with a syringe from the 1.75 m sucrose layer were diluted with 1 volume of 0.25 m sucrose in buffer A. These microsomes were then layered over a 5-ml 1.3 m sucrose cushion and centrifuged at 190,000 × g for 1 h. The pellet was aliquoted, frozen in liquid nitrogen, and stored at −80 °C for future use. RAPs were purified as reported previously (16), except that the CHAPS concentration was kept at 1.5% in the 1.5 m sucrose cushion. For MS analyses, four different preparations were used, and the RAP fraction isolated from 500 mg of total ER proteins was analyzed. For preparation of total cell extracts, cells were washed twice with ice-cold PBS and then lysed in radioimmune precipitation assay buffer (25 mm Tris-HCl, pH 7.6, 150 mm NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS). Total ER microsomes (20 mg), the RAP fraction (purified from 200 mg of total ER microsomes), and cell lysates were separated by SDS-PAGE prior to transferring onto a nitrocellulose membrane. Membranes were blocked for 45 min at room temperature with PBS containing 5% skim milk or 3% bovine serum albumin and 0.1% Tween 20 followed by incubation overnight at 4 °C with the indicated primary antibodies. Proteins were detected using the appropriate secondary antibodies coupled to horseradish peroxidase (Dako). Blots were revealed using LumiGLO chemiluminescent substrate system (Kirkegaard & Perry Laboratories, Gaithersburg, MD).
MS Analyses
Following resolution of the RAP fraction by one-dimensional PAGE and Coomassie R-250 staining, each band was excised, dehydrated in acetronitrile, and washed for two 10-min cycles in 100 mm (NH4)2CO3 before the addition of an equal volume of acetonitrile. The destained gel slices were then treated for 30 min with 10 mm DTT to reduce cystinyl residues followed by 20 min with 55 mm iodoacetamide to perform alkylation. After an additional round of (NH4)2CO3 and acetronitrile washes, the slices were extracted with acetonitrile at 37 °C. The samples were then incubated with trypsin (6 ng/ml in 50 mm NH4CO3) for 5 h at 37 °C, and the peptides were first extracted in 1% formic acid, 2% acetronitrile followed by two further extractions with additions of acetonitrile. All treatments were performed robotically using a MassPrep workstation (MicroMass-Waters, Milford, MA). Extracted peptides were applied to a reverse-phase guard column and then eluted in-line on a 10-cm × 75 mm Picofritt column filled with BioBasic C18. Samples were eluted with a linear gradient of 5–70% acetonitrile, 0.1% formic acid. A 2000-V charge was applied to the PicoFrit column such that the eluted peptides were electrosprayed into a cap liquid chromatography quadrupole time-of-flight MS (MicroMass-Waters). When doubly or triply charged ions were detected, the quadrupole was used to select the precursor ion, which was passed into a collision cell where fragmentation was induced by collision with argon gas. For peptide matching analyses, a definition of the Mascot score can be found online.
Complementary DNA Cloning and Plasmid Construction and Transfections
PCR-amplified products of RAP cDNA were inserted into pDONR201 by BP recombination (Invitrogen). The resulting ORF clones were transferred by LR recombination into Gateway®-compatible destination vectors (pdEYFP and pCDNA6.1; Invitrogen). All constructs were analyzed by DNA sequencing. AGR2 cDNA was amplified from the cDNA of HeLa cells and cloned into pcDNNA6 using Gateway® technology (Invitrogen). Wild-type human α1AT was cloned into pCDNA3 vector, and pRep9-α1AT-NHK was given by Dr I. Wada (Fukushima Medical University, Japan). HeLa cells were transiently transfected using Lipofectamine and PLUS reagents (Invitrogen).
Cell Imaging Analyses
For transfection, HEK-293T cells were plated 24 h prior to transfection onto ionized glass coverslips to achieve 80% confluency. Each EYFP fusion plasmid (pEYFP vector) (0.8 μg) and a CFP-300 ER control construct (a kind gift from Prof. J. Simpson, University College Dublin, Ireland) were co-transfected using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. The cells were cultured for a further 24 h prior to fixation in 4% paraformaldehyde and imaging by confocal microscopy as described previously (18). For staining of endogenous AGR2 protein, HuH6 and HuH7 cells were grown in 24-well plates, fixed with 4% paraformaldehyde for 10 min, and permeabilized with 0.1% Triton X-100 for 1 min at room temperature. Cells were then incubated with anti-AGR2 and anti-CNX antibodies for 1 h at room temperature followed by incubation with fluorescently labeled secondary antibodies. Protein subcellular localization was performed using an epifluorescence microscope with a Zeiss 63 × 1.4 oil immersion objective, recorded with a digital camera, and analyzed with Northern Eclipse software (Emprix Imaging, Mississauga, Ontario, Canada) or a LEICA SP5 confocal microscope.
RNA Isolation, Reverse transcriptase-Polymerase Chain Reaction, Semiquantitative RT-PCR, and Quantitative PCR (qPCR)
Total RNA was extracted using TRIzol reagent (Invitrogen). Semiquantitative RT-PCR analysis was carried out as described previously (19). The primers used are listed in supplemental Table S1. The PCR products were applied to 1–3% agarose gels. For qPCR analysis, all reactions were performed with a Stratagene X4000 thermocycler (Stratagene, Amsterdam, The Netherlands) using a SYBR Green PCR Core Reagents kit (Bio-Rad). The primers used are listed in supplemental Table S3. Each sample was normalized using Gapdh mRNA expression as reference.
Small Interfering RNA (siRNA) and Small Hairpin RNA (shRNA)
siRNA targeting AGR2 was designed using Greg Hannon's laboratory web tool. All other siRNAs used in this study are listed in supplemental Table S2. siRNAs were transfected into the cells by using Lipofectamine RNAiMAX (Invitrogen) according to the manufacturer's protocols. Cells were used for the experiments 48 or 72 h posttransfection. shRNA control and targeting AGR2 were purchased from OpenBiosystems (Thermo Inc., Huntsville, AL) and cloned into the pGIPZ vector. Lentiviruses were packaged using the pGIPZ packaging kit (Thermo Inc.) and cellular cultures (HeLa and HuH7) were infected with the resulting viral particles. Stable cell populations were selected using puromycin (2 μg/ml).
Cytotoxicity Assay
A cytotoxicity assay based on sulforhodamine B was carried out using an in vitro toxicology assay kit (Sigma).
[35S]Methionine Metabolic Labeling and Pulse-Chase Experiments
HeLa cells transfected with siRNA were preincubated in methionine- and cysteine-free medium (Invitrogen) for 2 h. Cells were then pulse-labeled with 100 μCi/dish EXPRE35S35S protein labeling mix (PerkinElmer Life Sciences). Labeled cells were then chased for various times in complete medium and lysed with lysis buffer containing 30 mm Tris-HCl, pH 7.5, 150 mm NaCl, and 2% CHAPS (Calbiochem, EMD Bioscience Inc.). Supernatants were recovered following centrifugation at 13,000 rpm for 15 min at 4 °C and incubated overnight with rabbit polyclonal anti-α1AT antibodies. Protein A-Sepharose beads were added to the immune complexes for 45 min at 4 °C with gentle rotation followed by washing five times with buffer containing 0.5% CHAPS. Beads were resuspended in Laemmli buffer containing 0.05% CHAPS. Labeled proteins were resolved on 10% polyacrylamide gels, processed to fluorography, dried, and exposed to x-ray films. Bands on the autoradiograms were quantified by scanning densitometry.
Statistical Analyses
Data are presented as means ± S.D. Statistical significance (p < 0.05) was determined using Student's t test with two-tailed distribution, assuming equal variance for the samples.
RESULTS
Proteomics Analysis of Ribosome-associated Proteins in the ER
The use of rough ER microsomes required an approach that would allow efficient solubilization of the ER membrane with minimal disruption of the binding interactions between ribosomes and RAPs. Consequently, dog pancreatic rough ER microsomes were purified as described under “Experimental Procedures,” and their structures were analyzed by electron microscopy (Fig. 1A). Our preparation showed the characteristic morphology of rough ER microsomes. Then, ribosome and RAP complexes were purified as depicted schematically in Fig. 1B and resolved by SDS-PAGE followed by Coomassie Blue staining. The resulting band profiles from four independent experiments were compared, and this allowed us to establish an average profile of 31 bands (±3) (Fig. 2A). The bands identified ranged from 180 to 4 kDa. Of the 31 bands excised, 205 unique proteins were identified by MS followed by database searches that revealed the presence of >1 protein/gel slice (supplemental Fig. S1). The identified proteins are listed in supplemental Table S3 indicating their respective functional categories, their NCBI Gi number, their predicted and/or published subcellular localization, the presence of predicted (as computed with TMHMM v2.0 (20)) or reported transmembrane domains, peptide numbers (unique, distinct, and total), and finally the number of times the protein was found in the four experimental repeats. Also included are the Mascot score results quantifying the identification probability of the fragment match. Only statistically significant Mascot score results (p < 0.05) were included in supplemental Table S3.
FIGURE 1.
Schematic representation of the ribosome-associated ER proteins enrichment strategy. A, electron micrograph of dog pancreatic rough ER microsomes purified from dog pancreas. Scale bar, corresponds to 1 μm. B, representation of the experimental scheme. Rough ER microsomes were solubilized with 1.5% CHAPS for 30 min on ice. The resulting lysates were centrifuged, and the protein content in the supernatant was spun through a 1.5 m sucrose cushion for 3 h at 95,000 rpm in a Beckman TLA100.2 rotor. RAP pellets were then collected.
FIGURE 2.
Isolation of RAPs. A, representative Coomassie Blue-stained gel of rough ER microsome fractions: total solubilized rough ER microsomes (TOT; 50 μg) and RAP (corresponding to 500 μg of starting rough ER microsomes). Each individually stained band was excised and trypsin-digested prior to mass spectrometry sequencing. Results are representative of four independent experiments. B, distribution of functional groups of proteins identified in the RAP fractions. C, distribution of functional groups of proteins identified in the ribosome-associated membrane protein (RAMP) fraction. D, validation of the proteins found in the RAP fraction. Total solubilized rough ER (RER) microsomes (TOT; 20 μg) and the RAP fraction (from 200 μg of total solubilized rough ER microsomes) were resolved by SDS-PAGE and transferred to nitrocellulose prior to immunoblotting analysis using the indicated antibodies. Ib, immunoblot. Results are representative of three independent experiments.
The 205 candidate RAPs were divided into 12 functional categories based on protein prediction analysis, including proteins involved in translocation, folding, translation, and signaling. In addition, cargo, mitochondrial, nuclear, or cytoskeletal proteins as well as those with a different localization (others) and uncharacterized hits are also identified in the graph (Fig. 2B). As expected, the largest proportion (33%) of the proteins found in the RAP fraction was of ribosomal origin, and 7% belonged to the translation machinery. In addition, the RAPs comprised 8% cargo proteins; 12% were nuclear or nucleic acid-binding contaminants, 7% cytoskeletal proteins, and 2% signaling proteins. Interestingly, 25% of the proteins belonged to the translocation/folding machinery, and the remaining 5% corresponded to proteins of uncharacterized function. Of the 205 unique proteins identified, 12 proteins of uncharacterized function (RAP1–12) were identified. Four of these uncharacterized proteins displayed at least one hydrophobic domain. When the ribosome-associated membrane proteins from the initial pool were reclustered into the 12 functional groups, 34.5% of the proteins belonged to the translocation machinery, a percentage that increased to 55.2% when the folding machinery was included (Fig. 2C). One major limitation of our approach was the presence of excessive amounts of ribosomal proteins in the purified fraction, which restricted the detection of low abundance proteins migrating in the 45–10-kDa region of the SDS-gel (supplemental Fig. S1). To validate the presence of the proteins identified through the proteomic approach in the RAP fraction, a subset of those was selected based on two criteria: 1) their known ER localization and 2) their identification in the RAP fraction by MS sequencing, which was confirmed immunoblotting. Interestingly, only the proteins consistently found in the RAP fraction by MS were also identified in that fraction using immunoblotting (Fig. 2D). Immunoblot analysis of the RAP fraction was compared with that of the total ER extract, revealing that ribophorin I (RibI), TRAPα, ORP150, and Gp25L (ordered by decreasing amount in the RAP fraction) were enriched in the RAP fraction and were identified at least twice (in four repeats) in the proteomics approach. In contrast, UDP-glucose:glycoprotein glucosyltransferase and glucosidase II (GII) were identified neither by MS sequencing in the RAP fraction nor by immunoblot analysis. By comparison, ERp72 was found only once (in four repeats) in the proteomics analysis and was not detected by immunoblotting (Fig. 2D).
Computational Analysis of the RAP Fraction
To provide some functional significance to the list of proteins identified in the proteomics screen, we first generated a functional interaction network using the STRING database (21) and Medusa software (22). This led to the building of a network bearing 143 nodes and 4733 edges (Fig. 3A). A complex containing 69 ribosomal proteins was clearly visualized (Fig. 3A, white nodes). The other most important machinery identified included proteins involved in translocation and folding (Fig. 3A, purple and orange nodes). This analysis revealed a highly connected functional network comprising more than 60% of the proteins identified in the proteomics screen. Based on this, we aimed at investigating the functions of the uncharacterized RAPs. We used a computational method called protein or oligonucleotide probability profile (POPP) analysis (23, 24), which allows proteins to be compared using profiles based on similarities in the composition of their amino acids or different sized peptides rather than similarities in their entire sequences. The power of the POPP approach lies in the ability to interrogate a database of POPPs for proteins of known function with similar peptide profiles to query for proteins of unknown function and to cluster similar profiles. By “POPPing” the RAPs of uncharacterized function against 201 proteins out of 211, we were able to cluster them into six functional families based on their POPP homology profile (Fig. 3B). Interestingly, all the RAPs of uncharacterized function showed some similarity with ribosomal proteins (ranging from 15 to 30% to >45% for groups 1 and 4, respectively). This was most likely because of the high representation of ribosomal proteins in our RAP fraction (>35%). Fig. 3C provides a graphic representation of the RAP clusters that were generated.
FIGURE 3.
Computational analysis of the proteomics data. A, network representation of the proteins identified in the proteomics analysis. The functional network, built using the STRING program suite, contains 143 nodes and 4733 edges. White nodes, ribosomal; orange nodes, ER translocation and folding; yellow nodes, translation; green nodes, export (light) and cargo (dark). B, POPP analysis of the RAPs of uncharacterized function against the 213 proteins found in our proteomics approach. C, cluster analysis of the RAPs of uncharacterized functions into six functional families based on their POPP homology profiles.
Subcellular Localization of Novel RAPs and characterization of RAP7
To further characterize the novel RAPs identified through the proteomics screen, we amplified novel RAPs encoding cDNAs from a human liver cDNA library by PCR (supplemental Fig. S2A, left panel). The corresponding cDNA fragments were inserted into the peYFP-C vector using Gateway® recombinational cloning (supplemental Fig. S2A, right panel). We then assessed the localization of nine novel RAP proteins of uncharacterized function, which were expressed as YFP COOH-terminally tagged recombinant proteins in COS-7 cells (supplemental Fig. S2, B and C) and HEK-293T cells (data not shown). A plasmid containing the ER marker VIP36-like fused to cyan fluorescent protein (CFP-300 ER) was co-transfected with the RAP cDNA as the co-localization marker. The results showed that four of the nine RAP proteins localized exclusively in the ER in COS-7 cells (RAPs 1, 6, 7, and 10) (supplemental Fig. S2C). In contrast, RAP3 displayed nuclear localization and RAPs 2, 5, 8, and 12 were ambiguously localized (supplemental Fig. S2B). Similar results were obtained in HEK-293T cells (data not shown), thus confirming the ER-specific localization of RAPs 1, 6, 7, and 10.
RAP7 was then selected for further studies because MS/MS analysis revealed that the sequence of RAP7 is identical to that of AGR2 protein, the human homolog of the Xenopus laevis cement gland protein (XAG2). This protein, which contains a pseudo thioredoxin domain, is encoded by an estrogen receptor-responsive gene. AGR2 is found in various cancers to be overexpressed in precancerous states and is indicative of a bad prognosis (25–30). However the molecular roles played by AGR2 remain unclear. A recent study using germ line Agr2−/− mice showed that this gene plays an essential role in the production of intestinal mucus by goblet cells (31). Moreover, AGR2 was also recently found to be involved in the regulation of ER homeostasis in Paneth and Goblet cells in mice (32). At the structural level, AGR2 contains a thioredoxin-like motif and is similar to the proteins ERp19 and ERp29, two members of the PDI family involved in ER protein folding. AGR2 and ERp19 share 38% identity with major differences at their extremities. However, unlike ERp19 and ERp29 and thioredoxin proteins in general, AGR2 contains only a single cysteine. Alignment of the amino acid sequences of human AGR2, ERp19 and ERp29 is shown in Fig. 4A. Each protein contains a putative NH2-terminal signal sequence and a COOH-terminal ER retention motif (KTEL, EDEL, and KEEL, respectively). Despite only having one cysteine residue, AGR2 is clearly homologous to the other two proteins. Fig. 4B shows a structural model of AGR2 created by 3D-JIGSAW (33) and visualized using Chimera (34) software. The Matchmaker function has been used to align the model structure with Protein Data Bank entry 1SEN, the protein structure determined for ERp19. The AGR2 model cysteine, shown in mauve in Fig. 4B, very closely matches the first cysteine in 1SEN. Fig. 4C depicts the ribbon model of the predicted structure overlaid with the hydrophobicity surface. The cysteine residue lies near the surface of the molecule in a shallow, somewhat hydrophobic pocket (Fig. 4C, shown in orange). Note, however, that the size of the pocket could change markedly, as it is bounded by loops.
FIGURE 4.
Characterization of AGR2. A, amino acid sequence alignment of AGR2 with two members of the PDI superfamily, ERp19 and ERp29 (GenBankTM accession numbers: NM006408, NM015913, and NM006817, respectively). An amino acid sequence comparison was performed using the ClustalW algorithm. Vertical alignments between the sequences for identical and similar amino acids are highlighted in different colors. Gaps were introduced as represented by dots to optimize the alignment. Green boxes indicate the signal peptide, red boxes indicate thioredoxin-like domains, and yellow boxes indicate putative ER retrieval motifs. B, comparison of a predicted model for the AGR2 structure version (Protein Data Bank code 1SEN, the structure determined for ERp19). C, superposition of the hydropobicity surface on the predicted structure for AGR2 (ribbon). D, validation by immunoblotting analysis of AGR2 in the RAP fraction as described in C. E, evaluation of the presence of AGR2 in ribosomal pellet in conditions where the ribosome pulldown was performed under control conditions (Ctl), in the presence of 5 mm EDTA (middle blots), or following treatment with puromycin (Puro). Ib, immunoblot. RER, rough ER. The scheme shows the experimental strategy used. F, immunofluorescence analysis of endogenous AGR2 protein by confocal microscopy. HuH6 cells were costained using anti-AGR2 (middle panels) and anti-CNX antibodies (left panels). Merged images are shown on the right panels.
To further confirm that AGR2 localized in the RAP fraction, ribosome pulldown was performed (as carried out in Fig. 2) followed by immunoblotting analysis (Fig. 4, D and E). ER microsomes were incubated either in the presence or absence (Fig. 4E, control (Ctl)) of 5 mm EDTA for 1 h on ice to promote ribosome detachment from the ER membrane or following treatment with puromycin (1 mm, 10 min at 30 °C) to favor the detachment of translationally engaged ribosomes. As expected, ribosomal protein L4 was detected in the pellets collected after centrifugation through a 1.5 m sucrose cushion under both experimental conditions. By contrast, AGR2 was not detected in the ribosomal pellet when microsomes were treated with EDTA, thus indicating that the association between AGR2 and the ribosome is dependent on an ER membrane component (Fig. 4E). Even more interesting, AGR2 was not detected in the ribosomal pellet following puromycin treatment, thereby suggesting that its association with the ribosomes occurred through nascent protein chains (Fig. 4E).
To evaluate the AGR2 expression pattern in different human cell lines, RT-PCR with AGR2 specific oligonucleotide primers and immunoblots with antibodies against AGR2 were performed on total RNA or protein, respectively (supplemental Fig. S3). AGR2 was expressed in all cell lines tested, with a higher expression found in HuH6 (human hepatocellular carcinoma) HT29 (human colon cancer), and mzCHA1 (human cholangiocarcinoma) cells (supplemental Fig. S3). To further confirm the ER localization of endogenous AGR2, HuH6 cells were subjected to immunofluorescence analysis with antibodies against AGR2 and CNX using confocal microscopy. The immunofluorescence data from HuH6 cells showed that AGR2 was confined to the reticular structure around the nucleus and co-localized with the ER marker CNX as illustrated in the merged image in Fig. 4F.
AGR2 Expression Is Regulated by the UPR in Mammalian Cells
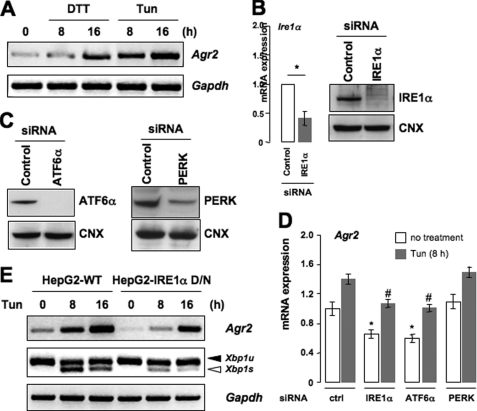
Members of the PDI family are thought to be for the most part stress-inducible and to participate in the control of ER homeostasis under challenging conditions. Therefore we investigated whether AGR2 expression could be induced in response to ER stress. HeLa (Fig. 5A) and HepG2 were treated for 8 or 16 h with Tun and DTT, two well characterized UPR inducers that prevent N-linked glycosylation and disulfide bond formation, respectively. RT-PCR analysis revealed that both stressors significantly enhanced the expression of Agr2 transcripts when normalized to the expression of Gapdh, which did not vary throughout the treatment (Fig. 5A). These results indicate that Agr2 expression is up-regulated upon ER stress. To further dissect the regulation of AGR2 by the UPR, IRE1α, ATF6α, and PERK were silenced in HeLa cells using siRNA. We first confirmed that IRE1α siRNA-mediated silencing effectively down-regulated Ire1α mRNA levels (Fig. 5B, left panel) and IRE1α protein expression (Fig. 5B, right panel). ATF6α and PERK protein expression was also attenuated upon treatment of the cells with the respective siRNAs (Fig. 5C). These experiments were also carried out in HepG2 cells with similar results (not shown). The effects of IRE1α and ATF6α silencing were further confirmed by measuring the expression of their cognate target genes, Edem1 mRNA and BiP protein, by RT-PCR and immunoblotting, respectively, in response to Tun treatment (data not shown). HepG2 cells transfected previously with siRNAs against either of the three UPR sensors were treated with Tun for 0 or 8 h, as this was the strongest AGR2 mRNA inducer (Fig. 5A), and AGR2 mRNA levels were quantified by RT-PCR (Fig. 5D). Our results indicated that both IRE1α and ATF6α knockdowns resulted in decreased basal Agr2 mRNA expression and also prevented Agr2 induction upon Tun treatment when compared with nontransfected cells (Fig. 5D). In contrast, PERK knockdown had no effect on Agr2 expression (Fig. 5D). The results indicating that Agr2 expression was partly under the control of the IRE1α arm of the UPR were confirmed by treating HepG2 cells expressing a dominant negative form of IRE1α (35) with Tun for 8 or 16 h (Fig. 5E). As expected, Xbp1 mRNA splicing was attenuated in IRE1α dominant negative expressing cells (Fig. 5E).
FIGURE 5.
AGR2 expression is controlled by the UPR in mammalian cells. A, HeLa cells were treated with one of two ER stressors, Tun (5 μg/ml) or DTT (1 mm), for 8 or 16 h. Total mRNA was isolated, and Agr2 mRNA expression was analyzed by semiquantitative RT-PCR using the specific primers against Agr2 and Gapdh. B, IRE1α expression was silenced in HeLa cells for 48 h by siRNA. RT-PCR (left graph) and immunoblotting (right panels) analyses were carried out using the primers against Ire1α and Gapdh or antibodies against IRE1α or CNX. Results represent the mean ± S.D. from three independent experiments (*, p < 0.05, as compared with control siRNA transfection). C, HeLa cells were transfected with ATF6α or PERK siRNA. At 48 h post-siRNA transfection, cells were lysed in radioimmune precipitation assay buffer and analyzed by immunoblotting using anti-ATF6α or PERK antibodies. CNX expression was used as a loading control. D, HepG2 cells were transfected with control siRNAs (ctrl) or siRNA targeting of IRE1α, ATF6α, or PERK followed by treatment with Tun for 8 h or left untreated. RT-PCR analysis was performed as described in A. Three independent experiments were performed, and the results are presented as means ± S.D. (*, p < 0.05, as compared with control; #, p < 0.03, as compared with control siRNA transfection upon Tun treatment). E, wild-type HepG2 (HepG2-WT) or HepG2 cells expressing a dominant negative IRE1α (HepG2-IRE1α D/N) were treated with Tun for 8 or 16 h. Agr2 mRNA expression and splicing of Xbp1 mRNA were analyzed by semiquantitative RT-PCR. The results are representative of three independent experiments.
AGR2 Knockdown Affects ER Homeostasis
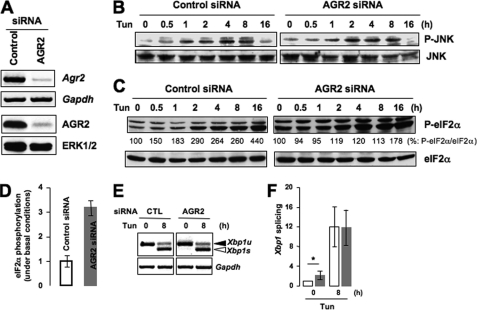
To explore the biological function(s) of AGR2, we used a siRNA knockdown strategy in cultured cells. Given that AGR2 is an ER protein (Fig. 4) and that its expression is induced upon ER stress (Fig. 5A), we examined the effect of AGR2 silencing on the ER stress response. We first transfected HeLa cells with siRNA against AGR2 for 72 h and examined the knockdown efficiency using RT-PCR and immunoblotting. The abundance of Agr2 mRNA and AGR2 protein was significantly attenuated (∼80%) in AGR2 knockdown cells when compared with cells transfected with a control siRNA (Fig. 6A). AGR2 siRNA-transfected cells were treated with Tun for various periods of time, and the phosphorylation of JNK and that of the translation initiation factor eIF2α were monitored by immunoblot. This revealed that the activation of JNK upon Tun treatment remained identical in control and AGR2 siRNA-treated cells (Fig. 6B). On the other hand, although basal eIF2α phosphorylation was increased by 3-fold in AGR2 knockdown cells compared with control cells (Fig. 6, C and D), Tun-induced phosphorylation of eIF2α increased by ∼2-fold in the former and more than 4-fold in the latter (Fig. 6C). Finally, Xbp1 mRNA splicing was found to be significantly up-regulated in AGR2-silenced cells under basal conditions (Fig. 6, E and F), but no difference from the control cells was observed upon Tun-induced stress (Fig. 6, E and F).
FIGURE 6.
AGR2 silencing moderately affects ER stress signals under basal conditions. A, AGR2 expression was knocked down in HeLa cells for 72 h. RT-PCR analysis of Agr2 mRNA expression was performed using mRNAs isolated from control or AGR2 siRNA-transfected cells (upper panels). Gapdh mRNA expression was used as internal control. Protein extracts from AGR2-transfected cells were prepared and analyzed by immunoblotting (lower panels). ERK1/2 was used as a protein loading control. B, HeLa cells were transfected with control or AGR2 siRNA for 72 h and treated with Tun for the indicated periods of time. Cell lysates were prepared and analyzed by immunoblotting using antibodies against JNK and phosphorylated JNK (P-JNK). C, HeLa cells were transfected and treated as described in B. Cells were lysed and analyzed by immunoblotting using anti-eIF2α and P-eIF2α antibodies. Bands were quantified by densitometry; the ratio of P-eIF2α to eIF2α is shown below each band. D, quantification of the levels of eIF2α phosphorylation in control and AGR2-knocked down cells under the non-stress conditions shown in C. E, HeLa cells were transfected as described in B and treated with Tun for 8 h. Total RNA was isolated, and the splicing of Xbp-1 mRNA was analyzed by semiquantitative RT-PCR. Gapdh was used as an internal control (CTL). F, quantification of Xbp1 splicing obtained from three independent experiments as represented in E. Results represent means ± S.D. (*, p < 0.05, as compared with control).
These data indicate that the absence of AGR2 caused a mild ER stress under the basal conditions in our experimental system, manifested by increased eIF2α phosphorylation and XBP1 mRNA splicing but not JNK1 phosphorylation. This might either suggest a dominance of AGR2 over those two pathways or an insufficient stress to reach JNK activation. Moreover, AGR2 expression is under the control of both ATF6 and IRE1α arms of the UPR, which might suggest tight maintenance of AGR2 levels in the cell.
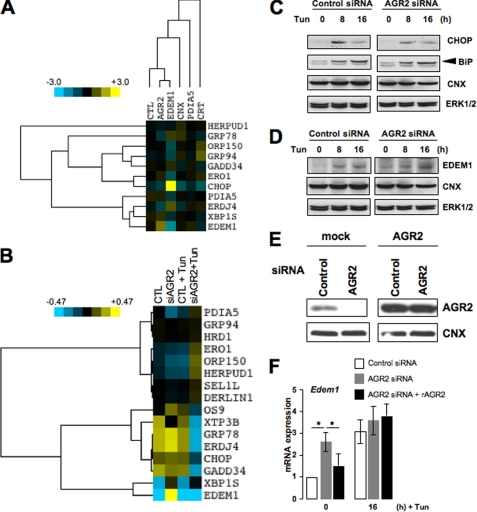
Then to ensure that AGR2 silencing in HeLa cells resulted in a specific response, we compared the basal expression profiles of 11 ER stress-responsive genes in cells silenced for various ERQC components including EDEM1, CNX, PDIA5, and CRT. This analysis revealed that AGR2 silencing resulted in a basal expression profile significantly different from those obtained in Edem1-, calnexin-, PDIA5-, or calreticulin-silenced cells (Fig. 7A). We then investigated the ER stress response in AGR2-silenced cells. Using qPCR, the induction of PERK (supplemental Fig. S4, Chop/Gadd153 and Gadd34), ATF6α (supplemental Fig. S3B, BiP, Ero1Lb, Grp94, Orp150, and Pdia5), and IRE1α (spliced form of Xbp1; supplemental Fig. S4, Xbp1s, Edem1, and ERdj4) target genes was measured in control and AGR2-silenced cells under basal conditions and upon Tun treatment (Fig. 7B and supplemental Figs. S4 and S5). Hierarchical clustering of these data (Fig. 7B) showed that AGR2 silencing induced Edem1 mRNA expression under basal conditions. As EDEM1 is a key component of ERAD, we next monitored the expression of different genes encoding ERAD components in AGR2 knockdown cells compared with control cells under basal conditions or upon Tun treatment. Os-9, Xtp3-b (two lectins of the ERAD system), Hrd1, Derlin1, Herpud1, and Sel1l mRNA expression was analyzed using qPCR, revealing that only the expression of Os-9 and Xtp3-b was significantly altered in AGR2 knockdown cells (Fig. 7B and supplemental Fig. S5). Indeed, although the expression of both mRNAs was induced upon Tun treatment, it was 30 to 50% lower in AGR2 knocked down cells than in control cells. These data suggest that transient AGR2 silencing slightly affects UPR signaling but has a strong impact on the expression of ERAD components. These results were confirmed by immunoblotting (Fig. 7, C and D). To further confirm the impact of AGR2 on EDEM1 expression, we rescued the expression of AGR2 in AGR2-silenced HeLa cells using siRNA-resistant AGR2 cDNA, which led to high expression levels of AGR2 in silenced cells (Fig. 7E). These cells were then treated with Tun for 16 h, and the expression of Edem1 mRNA was monitored using qPCR. Under basal conditions, AGR2 rescue led to the restoration of Edem1 mRNA expression to levels similar to control cells (Fig. 7F). These results indicate that AGR2 expression might control the basal expression of EDEM1, which is also consistent with the basal splicing of Xbp1 mRNA observed in AGR2-silenced cells (Fig. 6, E and F).
FIGURE 7.
AGR2 silencing slightly affects ER stress signals under basal conditions. A, heat map representation of the expression of IRE1α (Xbp1s, Edem1 and ERdj4), ATF6α (BiP/Grp78, Ero1lb, Grp94, Orp150, and Pdia5) or PERK (Chop and Gadd34) target genes and components of ERAD (Herpud1) in HeLa cells silenced for AGR2, EDEM1, calnexin, PDIA5, or calreticulin under basal conditions. B, heat map representation of the expression of IRE1α (Xbp1s, Edem1 and ERdj4), ATF6α (BiP/Grp78, Ero1lb, Grp94, Orp150 and Pdia5), or PERK (Chop and Gadd34) target genes and components of ERAD (Derlin1, Hrd1, Os-9, Sel1l, Herpud1, and Xtp3-b) in HeLa non-transfected (CTL, control) and silenced for AGR2 upon basal or Tun-induced stress conditions. C, HeLa cells were transfected with control or AGR2 siRNA for 72 h followed by treatment with Tun for 8 or 16 h. Cells were lysed and immunoblotted with anti-CHOP or anti-BiP antibodies. Anti-CNX or ERK1/2 antibodies were used as loading controls. D, AGR2 was silenced in HeLa cells, and Tun was added to the cells as described in B. Cell lysates were analyzed by immunoblotting using antibodies against EDEM1, CNX, or ERK1/2. E, HeLa cells were transfected with control or AGR2 siRNA. After 72 h of siRNA transfection, cells were further transfected with pcDNA6-AGR2 plasmid. Twenty-four hours later, cells were lysed, and the lysates were immunoblotted using anti-AGR2 and anti-CNX antibodies. F, HeLa cells were transfected with AGR2 siRNA and AGR2 cDNA as described in D. Total RNAs from the cells were used for semiquantitative RT-PCR using the primers for Edem1. Quantification of the amplified PCR products was done using three independent experiments (*, p < 0.05, as compared with AGR2 siRNA-transfected cells).
AGR2 Silencing Enhances Sensitivity to ER Stress
To examine whether AGR2 expression has an impact on cell growth under stress conditions, HeLa cells were silenced for AGR2 followed by treatment with Tun or DTT for 36 h. The cell number was determined, and it showed that AGR2-silenced cells were significantly less resistant to Tun or DTT stress than control cells (Fig. 8A). In an attempt to understand the role of AGR2 on the ER and its impact on cell resistance to stress, we first investigated the influence of AGR2 expression levels on protein secretion. To this end, the maturation of wild-type (WT) α1AT and of its terminally misfolded null Hong-Kong (NHK) variant, which is recognized by ERAD, were used as a readout. WT α1AT and NHK were expressed in HeLa cells silenced or not for AGR2 (Fig. 8B), and their cellular fate was investigated using [35S]methionine pulse-chase experiments followed by anti-α1AT immunoprecipitation, SDS-PAGE electrophoresis, and radioautography. This revealed that although AGR2 silencing did not affect the maturation and secretion of WT α1AT, it slightly shortened newly synthesized NHK intracellular half-life (Fig. 8, C and D). This result was consistent with the increased EDEM1 expression observed earlier. However, unexpectedly when both EDEM1 and AGR2 were silenced, this led to an even further reduction in the NHK half-life, whereas it remained unchanged when EDEM1 alone was silenced (Fig. 8D). This suggested that blocking NHK access to ERAD in AGR2-silenced cells might lead to the activation of an alternative degradation pathway. To test this hypothesis, we monitored autophagy using the posttranslational modification of LC3 as a reporter (Fig. 9, A and B). The LC3-II/(LC3-I+II) ratio was determined under conditions of silencing AGR2, EDEM1, or both (Fig. 9, A and B). This revealed that autophagy was activated only when AGR2 and EDEM1 were silenced simultaneously (Fig. 9B). Therefore we hypothesized that AGR2 silencing could sensitize cells to autophagy and consequently enhance their susceptibility to death under conditions where the ERAD was saturated. To test this hypothesis, we generated HeLa cells with stable silencing of AGR2. These cells presented properties that were similar to those observed upon transient AGR2 silencing, as illustrated by AGR2 silencing and eIF2α phosphorylation (supplemental Fig. S6). The results presented in Fig. 9, A and B, suggest that the autophagic fluxes might be increased in AGR2-silenced cells compared with control cells. To test this, we treated HeLa control and AGR2 stably silenced cells with Tun in the presence or absence of 20 mm ammonium chloride (NH4Cl) and monitored the modification of LC3 protein (Fig. 9C). These results revealed that the autophagic flux was accelerated in AGR2-silenced cells upon ER stress. This observation is consistent with the enhanced sensitivity to ER stress displayed by AGR2-silenced cells as well as their reduced quality control/folding abilities.
FIGURE 8.
AGR2 silencing alters the ability of cells to process terminally misfolded proteins. A, 72 h after control or AGR2 siRNA transfection, HeLa cells were treated with Tun (5 μg/ml) or DTT (0.1 mm) for 36 h. Cell viability was measured using the sulforhodamine B assay system. Results represent the mean ± S.D. (*, p < 0.05, **, p < 0.01 as compared with control cells upon ER stressor treatment). B, HeLa cells were transfected with a control siRNA (ctrl) or one targeting AGR2 for 48 h followed by transient transfection with a plasmid expressing α1AT-NHK or α1AT-WT. Expression of AGR2 or α1AT (NHK or WT) was analyzed by immunoblotting, and CNX was used as a loading control. C, control HeLa cells or HeLa cells silenced for AGR2 (siAGR2) were transiently transfected with a plasmid to express α1AT-NHK or α1AT-WT and then pulse-labeled for 15 min with [35S]methionine/cysteine followed by chase for the period indicated as described under “Experimental Procedures.” Shown are the autoradiograms of newly synthesized [35S]methionine-radiolabeled intracellular and secreted α1AT-WT and α1AT-NHK immunoprecipitated from HeLa cell lysates from cells subjected to siRNA with irrelevant siRNA (siCTL or GL2) or against AGR2 (siAGR2). D, HeLa cells were transfected with AGR2 or EDEM1 siRNA or co-transfected with AGR2 and EDEM1 siRNAs. At 72 h after siRNA transfection, cells were further transfected with α1AT-NHK. The next day, cells were metabolically labeled with [35S]methionine for 15 min. Labeled cells were then chased for 1, 2, 4, 6, and 8 h. Intracellular NHK was immunoprecipitated with anti-α1AT antibodies and resolved by SDS-PAGE. The percentage of newly synthesized α1-AT proteins at 6 h in silenced cells was calculated by densitometry and compared with that of control cells. Results represent means ± S.D. of three independent experiments (*, p < 0.05; #, p < 0.02, as compared with control).
FIGURE 9.
AGR2 silencing sensitizes cells to ER stress-induced autophagy. A, AGR2 and/or EDEM1 was silenced by siRNA in HeLa cells for 72 h. Cell lysates were prepared and analyzed by immunoblotting using anti-LC3 antibodies. CNX antibodies were used as a protein loading control. B, quantification of the ratio of LC3-II relative to total LC3 (LC3-I + LC3-II) was performed on the immunoblots shown in A by densitometry. Data are represented as means ± S.D. of three independent experiments. Asterisks indicate statistical significance (*, p < 0.03, compared with control siRNA transfection). C, HeLa cells stably expressing a control shRNA (shCTL) or targeting AGR2 were subjected to tunicamycin treatment (5 μg/ml) for 8 h in the presence or absence of 20 mm NH4Cl. Lysates were analyzed by immunoblotting with antibodies to LC3 and CNX.
DISCUSSION
In this study, we used a suborganellar proteomics approach to identify rough ER proteins associated with membrane-bound ribosomes. This approach was motivated by the fact that proteins in the vicinity of the translocon may be involved in ER protein folding and quality control. We identified and characterized a protein named AGR2, for which the molecular functions were so far unknown. We confirmed that AGR2 localizes in the lumen of the ER and associates with ribosomes through nascent translocating polypeptides.
AGR2 and Protein Degradation in the ER
We showed that AGR2 expression is up-regulated upon ER stress and that AGR2 basal expression levels are controlled mainly by the IRE1α and ATF6α arms of the UPR (Fig. 5). In addition, POPP analysis of AGR2 (Fig. 3C, RAP7) suggests that this protein is related to the ER folding machinery. Moreover, we demonstrated that when AGR2 expression is attenuated using siRNA (or shRNA) in HeLa cells, the expression of EDEM1, a protein involved in ERAD, is significantly increased under non-stress (basal) conditions (Fig. 7). Furthermore, upon Tun-induced ER stress, the expression of two other ERAD lectins, OS-9 and XTP3-B, is affected in AGR2-silenced cells (Fig. 7). These experiments suggest that AGR2 may be an ER-localized protein closely associated with the translocon and may function in the regulation of ER folding/quality control/ERAD machinery.
In our model, the absence of AGR2 would lead to an increased amount of EDEM1 client proteins, thus leading to the compensatory increase in EDEM1 expression (Fig. 5). Increased EDEM1 expression would then be sufficient to process misfolded protein further into the ERAD pathway. The idea is not absolutely excluded that high expression levels of EDEM1 may also promote the bypass of glycan-dependent glycoprotein delivery to ERAD as demonstrated previously (36).
Germ line Agr2−/− mice were used previously to demonstrate that AGR2 is involved in intestinal homeostasis and ER stress (32). In this study, the authors show that BiP protein levels and sXbp1, Chop, Pdia3, and Perk mRNA expression are dramatically increased in germ line Agr2−/− small intestine (32). However, another study also using germ line Agr2−/− mice (31) showed only modest evidence for activation of the ER stress response in this model. In our experimental system, siRNA-mediated knockdown of AGR2 induced a modest perturbation of the expression of ER stress markers under basal conditions (phosphorylation of eIF2α, Xbp1 mRNA splicing, and Edem1 expression; see Fig. 5). In addition, we observed only minor differences in the expression of other canonical UPR markers upon Tun-induced ER stress (Fig. 4). The differences observed in these three reports could be because of the cell line/model used, the kinetics of the experiments or the capacity of the cells to adapt or compensate for AGR2 knockdown. Interestingly, studies using Agr2−/− mice show that AGR2 might play an essential role for in vivo production of the intestinal mucus protein mucin 2 (MUC2), a large cysteine-rich glycoprotein. Park et al. (31) show that AGR2 co-immunoprecipitates with MUC2 and that this association depends on a conserved cysteine residue in the AGR2 thioredoxin-like domain. Moreover, the surprising observation that AGR2 expression might control the balance between ERAD and autophagy in the processing of misfolded proteins could constitute a physical link between protein folding in the ER and autophagosome formation (Fig. 10).
FIGURE 10.
Schematic representation of AGR2 function in the ER. The functional AGR2 network was built using the STRING program suite and drawn using the Cytoscape program. Pink nodes represent the translocation machinery, green nodes stand for the membrane traffic component of the network, blue nodes correspond to the UPR machinery, and orange nodes correspond to the ERQC/folding machinery. White nodes are also indicated but were not found in the ARG2 functional network in our study. A standard cargo is presented in blue and AGR2 in yellow. The STING-based interactions are shown in gray. The regulatory interactions found in our study are presented in green (positive) or red (negative). A connection between AGR2 and the autophagic machinery is also shown (black dotted line). Finally, the interaction of AGR2 with cargo molecules is shown in blue. The ER membrane is symbolized in gray.
Relevance of AGR2 Functions to Cancer
The expression of AGR2 in normal tissues has been investigated systematically; by the Human Protein Atlas initiative one of the most striking observations is the correlated tissue distribution with mucins 1, 2, 3A, 5B, 12, and 15, which might be indicative of a function for AGR2 in the secretion/folding of these glycoproteins based on data obtained with KO mouse models and on findings presented in the present work. Recently, numerous studies demonstrated the elevated expression of AGR2 in various human preneoplasic states (30) and cancers including those of the esophagus (37), prostate (38–41), pancreas (42), lung (43), breast (25, 27, 44), and biliary system (26), thus showing that it is a widely overexpressed protein in human carcinomas. This might indicate that several experimental and pathophysiological systems could be necessary to tackle the precise molecular and cellular functions of AGR2.
AGR2 has also been proposed as a putative metastasis marker in breast, prostate, and colorectal cancers, and it can be detected in the blood of ovarian cancer patients (27, 28, 45). Furthermore, in vivo and in vitro studies in cancer model systems revealed that overexpression or suppression of AGR2 could affect cell proliferation, adhesion and migration, metastasis, and tumor growth (42, 46). The molecular mechanisms at the origin of these phenotypes are not yet clearly understood, although it was recently shown that AGR2 is present at the cell surface of pancreatic cancer cells (47), a phenomenon similar to that previously reported in tumor immunity (48, 49). However, our work might therefore provide indications on how AGR2 expression could correlate with and impact cancer phenotypes. As shown for other ER-resident proteins (50), increased expression of AGR2, as a member of the PDI chaperone family involved in ERQC, could be predicted to enhance ER folding capacity, allowing cancer cells to cope with the increased protein production and secretion (Fig. 8A). This phenomenon also could be reconciled with the recent observation that AGR2 may control the expression of amphiregulin through the Hippo pathway (51) as well as with the increased mucin secretion observed in the above mentioned cancers. More generally, it can be proposed that AGR2, by regulating the ER capacity to adapt (Fig. 10), might have an impact on cell survival and the nature of the cell secretome under non-stressed and stressed conditions, consequently contributing to the development and progression of cancers.
Supplementary Material
Acknowledgment
Frédéric Delom and mass spectrometry sequencing were supported in part by an operating grant from Genome Quebec/Canada to the Montreal Proteomics Network (Cell Map Project).
This work was supported by grants from INSERM (Avenir) and Institut National du Cancer (INCa), Ligue Nationale contre le Cancer, Fondation pour la Recherche Médicale, Association de Recherche contre le Cancer, and Conseil Régional d'Aquitaine (to E. C.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S6 and Tables S1–S3.
- ER
- endoplasmic reticulum
- AGR2
- anterior gradient 2
- CNX
- calnexin
- EDEM1
- ER degradation-enhancing α-mannosidase-like protein-1
- ERAD
- ER-associated degradation
- ERQC
- ER quality control system
- PDI
- protein-disulfide isomerase
- POPP
- protein or oligonucleotide probability profile
- RAP
- ribosome-associated protein
- TRAP
- translocon-associated protein
- Tun
- tunicamycin
- UPR
- unfolded protein response
- EYFP
- enhanced yellow fluorescent protein
- qPCR
- quantitative PCR
- NHK
- null Hong-Kong
- PERK
- PKR-like ER kinase.
REFERENCES
- 1. Chevet E., Cameron P. H., Pelletier M. F., Thomas D. Y., Bergeron J. J. (2001) Curr. Opin. Struct. Biol. 11, 120–124 [DOI] [PubMed] [Google Scholar]
- 2. Mori K. (2000) Cell 101, 451–454 [DOI] [PubMed] [Google Scholar]
- 3. Kleizen B., Braakman I. (2004) Curr. Opin. Cell Biol. 16, 343–349 [DOI] [PubMed] [Google Scholar]
- 4. Ruddock L. W., Molinari M. (2006) J. Cell Sci. 119, 4373–4380 [DOI] [PubMed] [Google Scholar]
- 5. Määttänen P., Gehring K., Bergeron J. J., Thomas D. Y. (2010) Semin. Cell Dev. Biol. 21, 500–511 [DOI] [PubMed] [Google Scholar]
- 6. Vembar S. S., Brodsky J. L. (2008) Nat. Rev. Mol. Cell Biol. 9, 944–957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Walter P., Gilmore R., Blobel G. (1984) Cell 38, 5–8 [DOI] [PubMed] [Google Scholar]
- 8. Deshaies R. J., Sanders S. L., Feldheim D. A., Schekman R. (1991) Nature 349, 806–808 [DOI] [PubMed] [Google Scholar]
- 9. Deshaies R. J., Schekman R. (1987) J. Cell Biol. 105, 633–645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Beckmann R., Spahn C. M., Eswar N., Helmers J., Penczek P. A., Sali A., Frank J., Blobel G. (2001) Cell 107, 361–372 [DOI] [PubMed] [Google Scholar]
- 11. Beckmann R., Spahn C. M., Frank J., Blobel G. (2001) Cold Spring Harb. Symp. Quant. Biol. 66, 543–554 [DOI] [PubMed] [Google Scholar]
- 12. Johnson A. E., van Waes M. A. (1999) Annu. Rev. Cell Dev. Biol. 15, 799–842 [DOI] [PubMed] [Google Scholar]
- 13. Kalies K. U., Görlich D., Rapoport T. A. (1994) J. Cell Biol. 126, 925–934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dejgaard K., Theberge J. F., Heath-Engel H., Chevet E., Tremblay M. L., Thomas D. Y. (2010) J. Proteome Res. 9, 1763–1771 [DOI] [PubMed] [Google Scholar]
- 15. Hartmann E., Görlich D., Kostka S., Otto A., Kraft R., Knespel S., Bürger E., Rapoport T. A., Prehn S. (1993) Eur. J. Biochem. 214, 375–381 [DOI] [PubMed] [Google Scholar]
- 16. Chevet E., Wong H. N., Gerber D., Cochet C., Fazel A., Cameron P. H., Gushue J. N., Thomas D. Y., Bergeron J. J. (1999) EMBO J. 18, 3655–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Blobel G., Dobberstein B. (1975) J. Cell Biol. 67, 852–862 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jenna S., Caruso M. E., Emadali A., Nguyên D. T., Dominguez M., Li S., Roy R., Reboul J., Vidal M., Tzimas G. N., Bossé R., Chevet E. (2005) Mol. Biol. Cell 16, 1629–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nguyên D. T., Kebache S., Fazel A., Wong H. N., Jenna S., Emadali A., Lee E. H., Bergeron J. J., Kaufman R. J., Larose L., Chevet E. (2004) Mol. Biol. Cell 15, 4248–4260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Krogh A., Larsson B., von Heijne G., Sonnhammer E. L. (2001) J. Mol. Biol. 305, 567–580 [DOI] [PubMed] [Google Scholar]
- 21. Snel B., Lehmann G., Bork P., Huynen M. A. (2000) Nucleic Acids Res. 28, 3442–3444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hooper S. D., Bork P. (2005) Bioinformatics 21, 4432–4433 [DOI] [PubMed] [Google Scholar]
- 23. Wise M. J. (2002) Bioinformatics 18, Suppl. 1, S38–S45 [DOI] [PubMed] [Google Scholar]
- 24. Wise M. J., Tunnacliffe A. (2004) Trends Plant Sci. 9, 13–17 [DOI] [PubMed] [Google Scholar]
- 25. Fritzsche F. R., Dahl E., Pahl S., Burkhardt M., Luo J., Mayordomo E., Gansukh T., Dankof A., Knuechel R., Denkert C., Winzer K. J., Dietel M., Kristiansen G. (2006) Clin. Cancer Res. 12, 1728–1734 [DOI] [PubMed] [Google Scholar]
- 26. Lepreux S., Bioulac-Sage P., Chevet E. (2011) Liver Int. 31, 322–328 [DOI] [PubMed] [Google Scholar]
- 27. Liu D., Rudland P. S., Sibson D. R., Platt-Higgins A., Barraclough R. (2005) Cancer Res. 65, 3796–3805 [DOI] [PubMed] [Google Scholar]
- 28. Thompson D. A., Weigel R. J. (1998) Biochem. Biophys. Res. Commun. 251, 111–116 [DOI] [PubMed] [Google Scholar]
- 29. Hrstka R., Nenutil R., Fourtouna A., Maslon M. M., Naughton C., Langdon S., Murray E., Larionov A., Petrakova K., Muller P., Dixon M. J., Hupp T. R., Vojtesek B. (2010) Oncogene 29, 4838–4847 [DOI] [PubMed] [Google Scholar]
- 30. Pohler E., Craig A. L., Cotton J., Lawrie L., Dillon J. F., Ross P., Kernohan N., Hupp T. R. (2004) Mol. Cell. Proteomics 3, 534–547 [DOI] [PubMed] [Google Scholar]
- 31. Park S. W., Zhen G., Verhaeghe C., Nakagami Y., Nguyenvu L. T., Barczak A. J., Killeen N., Erle D. J. (2009) Proc. Natl. Acad. Sci. U.S.A. 106, 6950–6955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhao F., Edwards R., Dizon D., Afrasiabi K., Mastroianni J. R., Geyfman M., Ouellette A. J., Andersen B., Lipkin S. M. (2010) Dev. Biol. 338, 270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Bates P. A., Kelley L. A., MacCallum R. M., Sternberg M. J. (2001) Proteins Suppl. 5, 39–46 [DOI] [PubMed] [Google Scholar]
- 34. Pettersen E. F., Goddard T. D., Huang C. C., Couch G. S., Greenblatt D. M., Meng E. C., Ferrin T. E. (2004) J. Comput. Chem. 25, 1605–1612 [DOI] [PubMed] [Google Scholar]
- 35. Cameron P. H., Chevet E., Pluquet O., Thomas D. Y., Bergeron J. J. (2009) J. Biol. Chem. 284, 34570–34579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Ron E., Shenkman M., Groisman B., Izenshtein Y., Leitman J., Lederkremer G. Z. (2011) Mol. Biol. Cell 22, 3945–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hao Y., Triadafilopoulos G., Sahbaie P., Young H. S., Omary M. B., Lowe A. W. (2006) Gastroenterology 131, 925–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kovalev L. I., Shishkin S. S., Khasigov P. Z., Dzeranov N. K., Kazachenko A. V., Toropygin IIu., Mamykina S. V. (2006) Prikl. Biokhim. Mikrobiol. 42, 480–484 [PubMed] [Google Scholar]
- 39. Kristiansen G., Pilarsky C., Wissmann C., Kaiser S., Bruemmendorf T., Roepcke S., Dahl E., Hinzmann B., Specht T., Pervan J., Stephan C., Loening S., Dietel M., Rosenthal A. (2005) J. Pathol. 205, 359–376 [DOI] [PubMed] [Google Scholar]
- 40. Zhang J. S., Gong A., Cheville J. C., Smith D. I., Young C. Y. (2005) Genes Chromosomes Cancer 43, 249–259 [DOI] [PubMed] [Google Scholar]
- 41. Zhang Y., Forootan S. S., Liu D., Barraclough R., Foster C. S., Rudland P. S., Ke Y. (2007) Prostate Cancer Prostatic Dis. 10, 293–300 [DOI] [PubMed] [Google Scholar]
- 42. Wang Z., Hao Y., Lowe A. W. (2008) Cancer Res. 68, 492–497 [DOI] [PubMed] [Google Scholar]
- 43. Zhu H., Lam D. C., Han K. C., Tin V. P., Suen W. S., Wang E., Lam W. K., Cai W. W., Chung L. P., Wong M. P. (2007) Cancer Lett. 245, 303–314 [DOI] [PubMed] [Google Scholar]
- 44. Persson S., Rosenquist M., Knoblach B., Khosravi-Far R., Sommarin M., Michalak M. (2005) Mol. Phylogenet. Evol. 36, 734–740 [DOI] [PubMed] [Google Scholar]
- 45. Smirnov D. A., Zweitzig D. R., Foulk B. W., Miller M. C., Doyle G. V., Pienta K. J., Meropol N. J., Weiner L. M., Cohen S. J., Moreno J. G., Connelly M. C., Terstappen L. W., O'Hara S. M. (2005) Cancer Res. 65, 4993–4997 [DOI] [PubMed] [Google Scholar]
- 46. Ramachandran V., Arumugam T., Wang H., Logsdon C. D. (2008) Cancer Res. 68, 7811–7818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dumartin L., Whiteman H. J., Weeks M. E., Hariharan D., Dmitrovic B., Iacobuzio-Donahue C. A., Brentnall T. A., Bronner M. P., Feakins R. M., Timms J. F., Brennan C., Lemoine N. R., Crnogorac-Jurcevic T. (2011) Cancer Res., in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Obeid M., Tesniere A., Ghiringhelli F., Fimia G. M., Apetoh L., Perfettini J. L., Castedo M., Mignot G., Panaretakis T., Casares N., Métivier D., Larochette N., van Endert P., Ciccosanti F., Piacentini M., Zitvogel L., Kroemer G. (2007) Nat. Med. 13, 54–61 [DOI] [PubMed] [Google Scholar]
- 49. Panaretakis T., Joza N., Modjtahedi N., Tesniere A., Vitale I., Durchschlag M., Fimia G. M., Kepp O., Piacentini M., Froehlich K. U., van Endert P., Zitvogel L., Madeo F., Kroemer G. (2008) Cell Death Differ. 15, 1499–1509 [DOI] [PubMed] [Google Scholar]
- 50. Moenner M., Pluquet O., Bouchecareilh M., Chevet E. (2007) Cancer Res. 67, 10631–10634 [DOI] [PubMed] [Google Scholar]
- 51. Dong A., Gupta A., Pai R. K., Tun M., Lowe A. W. (2011) J. Biol. Chem. 286, 18301–18310 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.