Background: The role of S100A7 in ERα+ breast cancer is not known.
Results: S100A7 overexpression in ERα+ breast cancer cells inhibits tumorigenesis by decreasing β-catenin and TCF4 expression through activation of GSK3β and E-cadherin.
Conclusion: S100A7 may regulate tumorigenesis through modulation of the β-catenin/TCF4 pathway.
Significance: S100A7 may possess differential activity in ERα+ compared with ERα− cells, as S100A7 has been shown to enhance tumorigenesis in ERα− cells.
Keywords: β-Catenin, Breast Cancer, Estrogen, Oncogene, T Cell Factor (TCF), Tumor Suppressor Gene, S100A7
Abstract
Psoriasin (S100A7) is expressed in several epithelial malignancies including breast cancer. Although S100A7 is associated with the worst prognosis in estrogen receptor α-negative (ERα−) invasive breast cancers, its role in ERα-positive (ERα+) breast cancers is relatively unknown. We investigated the significance of S100A7 in ERα+ breast cancer cells and observed that S100A7 overexpression in ERα+ breast cancer cells, MCF7 and T47D, exhibited decreased migration, proliferation, and wound healing. These results were confirmed in vivo in nude mouse model system. Mice injected with S100A7-overexpressing MCF7 cells showed significant reduction in tumor size compared with mice injected with vector control cells. Further mechanistic studies revealed that S100A7 mediates the tumor-suppressive effects via a coordinated regulation of the β-catenin/TCF4 pathway and an enhanced interaction of β-catenin and E-cadherin in S100A7-overexpressing ERα+ breast cancer cells. We observed down-regulation of β-catenin, p-GSK3β, TCF4, cyclin D1, and c-myc in S100A7-overexpressing ERα+ breast cancer cells. In addition, we observed increased expression of GSK3β. Treatment with GSK3β inhibitor CHIR 99021 increased the expression of β-catenin and its downstream target c-myc in S100A7-overexpressing cells. Tumors derived from mice injected with S100A7-overexpressing MCF7 cells also showed reduced activation of the β-catenin/TCF4 pathway. Therefore, our studies reveal for the first time that S100A7-overexpressing ERα+ breast cancer cells exhibit tumor suppressor capabilities through down-modulation of the β-catenin/TCF4 pathway both in vitro and in vivo. Because S100A7 has been shown to enhance tumorigenicity in ERα− cells, our studies suggest that S100A7 may possess differential activities in ERα+ compared with ERα− cells.
Introduction
S100A7 is a low molecular mass protein (∼11.4 kDa) and a member of the S100 protein family which is characterized by two calcium-binding sites of the helix-loop-helix (EF-hand type) conformation. It is a part of the S100 gene cluster located on human chromosome 1q21, which constitutes the epidermal differentiation complex. This region is of particular interest because it encodes many genes that have been linked to epidermal differentiation and inflammation (1–4). Further, S100A7 has been shown to regulate inflammatory processes by enhancing the chemotaxis of T cells and by modulating the cytokine production in different cell types (5–7). Apart from its role as an inflammatory molecule, S100A7 has been associated with various epithelial malignancies, including breast cancer (8, 9).
S100A7 has been shown to be highly associated with the estrogen receptor (ER)4 α-negative (ERα−) breast cancer and is expressed in ductal carcinoma in situ and invasive carcinomas (10–15). Expression of S100A7 in human breast tumors represents a poor prognostic marker and correlates with lymphocyte infiltration in high grade morphology (16). Furthermore, recent studies have shown that S100A7 down-regulation in ERα− cells inhibits tumor growth in in vivo mouse model systems (11) and EGF-induced migration (14). In addition, S100A7 overexpression in ERα− cells was shown to enhance proliferation and invasion in in vitro conditions and tumor growth and metastasis in vivo (17, 18). S100A7 has been shown to enhance tumor growth in ERα− cells by regulating prosurvival mechanisms, such as NF-κB and phospho-AKT (18). Furthermore, S100A7 has been shown to interact with Jab1 and translocate it to the nucleus that leads to the induction of AP-1-regulated genes and down-regulation of p27kip1 (17, 18). These studies indicate the protumorigenic role of S100A7 in ERα− cells, but the exact role of S100A7 in the ERα+ cells has not been elucidated comprehensively until now.
Hyperactivation of the canonical β-catenin/TCF4 pathway is one of the most frequent signaling abnormalities in many types of cancer (19, 20). The central event in this pathway is the stabilization and nuclear translocation of β-catenin, where it binds to the transcription factors of TCF4/TCF7L2 family and subsequently activates a cluster of genes that ultimately establish the oncogenic phenotype (21, 22). β-Catenin has also been shown to interact with α-catenin and E-cadherin, thereby stabilizing the expression of E-cadherin in the membranes and thus maintaining the epithelial integrity of the cells (23). Further, loss of E-cadherin confers mesenchymal ability to the epithelial cells leading to increased metastasis and migration (24). Stabilization of the β-catenin and overexpression of its target cyclin D1 have been observed in >50% of patients with breast cancer (25). Furthermore, increased β-catenin activity was found to be significantly correlated with poor prognosis of breast cancer patients (26).
We report for the first time that overexpression of S100A7 in ERα+ breast cancer cells inhibits growth and migration in vitro as well as tumor growth in an in vivo mouse model system. We have also shown that S100A7 mediates its tumor-suppressive activities by down-modulating the β-catenin/TCF4 signaling pathway. Further, we show that inhibiting GSK3β activity and TCF4 overexpression reverses the S100A7-mediated inhibitory effects. These studies suggest that S100A7 may have a differential role in ERα+ cells compared with ERα− where it has been shown to enhance growth and metastasis.
EXPERIMENTAL PROCEDURES
Cell Culture, Reagents, and Antibodies
Human breast carcinoma cell lines MCF7 and T47D (obtained originally from ATCC) were cultured as described previously (27). GSK3β inhibitor CHIR 99021 was purchased from Stemgent, MA. Antibodies used were S100A7 (IMGENEX); β-catenin, phospho-β-catenin, phospho-GSK3β, GSK3β, secondary mouse and rabbit antibodies (Cell Signaling); and GAPDH (Santa Cruz Biotechnology); E-cadherin (Abcam); TCF4 and active β-catenin (Millipore); Ki67 (Neomarker), and CD31 (BD Pharmingen).
Constructs and Transfections
The open reading frame (ORF) clone of Homo sapiens S100A7 homolog was purchased from OriGene Technologies (Rockville, MD) and subcloned into pIRES2-EGFP (Invitrogen). MCF7 and T47D cells were transfected with pIRES2-EGFP plasmid alone or containing S100A7 with Lipofectamine according to manufacturer's protocol (Invitrogen). After 24 h of transfection, cells were incubated for 3 weeks in medium containing G418 (500 μg/ml) to select the stably overexpressing S100A7 clones. S100A7 expression in cells was analyzed by Western blotting. Vector- and S100A7-transfected ERα+ cells hereafter are termed MCF7/Vec, T47D/Vec, and MCF7/S100A7, T47D/S100A7, respectively. TCF4 was transfected in MCF7/S100A7 in pcDNA3.1 vector using Lipofectamine according to the manufacturer's recommendations. For siRNA studies, MCF7/Vec and MCF7/S100A7 cells were transfected with siRNA smart pool (Dharmacon) against GSK3β using Lipofectamine according to the manufacturer's recommendations. siRNA was used at 100 nm and 200 nm concentrations to observe the dose-dependent effects. Scrambled nontargeting siRNA (200 nm) was used as control. The cells were harvested 48 h after transfection, and GSK3β, phospho-β-catenin, and β-catenin protein levels were determined by immunoblotting. For proliferation studies, cells were harvested after 24 h and seeded for the MTT proliferation assay (Roche Applied Science).
Proliferation, Chemotaxis, and Wound Healing Assay
Proliferation of MCF7/Vec and MCF7/S100A7 was measured using MTT assay as described previously (27). Briefly, 5 × 103 cells were seeded in 96-well tissue culture plates. The proliferation of the cells was measured by MTT assay (Roche Applied Science) at different days as a percentage increase in the absorbance reading at 570 nm with respect to the control (day 0). In the case of GSK3β inhibitor CHIR 99021 (2 μm), cells were serum-starved before adding the inhibitor. Serum-induced chemotaxis assay was measured using 8-μm Transwell plates as described previously (28). The migrated cells were stained using Hema stain and counted. The wound scratch assay was done as previously described (28).
Western Blotting, Co-immunoprecipitation, Confocal Microscopy
70–80% confluent ERα+ cells were cultured for 24 h at 37 °C. After treatment, the cells were washed and lysed, and protein concentration was measured using a Bradford Reagent kit (Bio-Rad). Then 50 μg of protein was loaded in NuPage BisTris 4–12% gradient gels (Invitrogen), and detection was done using ECL reagent (Amersham Biosciences) as described (27). Co-immunoprecipitation was carried out with 500 μg of cell lysate using protein G plus A-agarose beads as described (27). Samples were eluted using 2× Laemmli reducing buffer and subjected to SDS-PAGE and analyzed by Western blotting. Confocal microscopy was done as described previously (27–29). Briefly, cells were cultured in chamber slides, fixed, and treated with mouse anti-E-cadherin and/or rabbit anti-β-catenin antibody overnight at 4 °C, washed, and stained with Alexa Fluor-488-conjugated anti-mouse and Alexa Fluor-594-conjugated secondary IgG antibodies, respectively. The cells were then examined under Olympus FV1000 filter confocal microscope, and the images were acquired and quantified using FV10-ASW2.0 software.
TCF4 Luciferase Reporter Assay
The β-catenin/TCF4 pathway activity was determined using TCF4 luciferase reporter assay. To determine luciferase reporter activity, TCF luciferase constructs (0.5 μg), containing the wild-type (pTOPFLASH) or mutant (pFOPFLASH) TCF binding sites, were transfected into MCF7/Vec and MCF7/S100A7 (2 × 105) cells using Lipofectamine. In addition, the cells were co-transfected with an internal control (0.1 μg of pRL-TK Renilla luciferase vector). The cells were incubated for 48 h after the transfection and were lysed and analyzed for luciferase activity.
Microarray Analysis and Quantitative Real Time PCR (qRT-PCR)
Total RNA from MCF7/Vec and MCF7/S100A7 cells was extracted using TRIzol reagent (Invitrogen), according to the manufacturer's protocol. Microarray analysis was performed at The Ohio State University Medical Center genomics core facility using an Affymetrix Microarray Human Genome U133 chip containing 40,000 genes. Microarray data were further analyzed by Ingenuity Pathway Analysis. Isolated RNA was also subjected to qRT-PCR using RT-PCR kits (Applied Biosystems).
Xenograft Mouse Model
Female nude (nu/nu) mice, obtained from Charles River Laboratories (Wilmington, MA), were housed under specific pathogen-free conditions. The in vivo experiments were performed in accordance with the guidelines of our Institutional Animal Care and Use Committee, University Laboratory Animal Research. MCF7/Vec and MCF7/S100A7 (3 × 106 cells/200 μl of PBS) were injected subcutaneously into the right flank of each mouse. Mice were also injected subcutaneously with 2.5 μg of β-estradiol 17-valerate in 50 μl of sesame oil once a week. Tumor size was assessed once a week, and tumor volume was calculated using digital calipers according to the formula: volume = length × (width)2/2. Tumors were excised and fixed in formalin and paraformaldehyde to obtain frozen and paraffin-embedded sections, which were further subjected to immunohistochemical analyses.
Immunohistochemistry
Immunohistochemistry was performed on 4-μm sections from frozen tumor xenografts of mice as described previously (28, 29) using Ki67 (1:100), CD31 (1:50), cyclin D1 (1:100), E-cadherin (1:100), and β-catenin (1:50) antibodies for 60 min at room temperature or overnight at 4 °C. Slides were developed as described previously (28, 29). The nuclei for Ki67 staining and CD31 expression in vessels was quantitated in three random microscopic (×10) fields/tumor.
Statistical Analyses
The results are expressed as the mean ± S.D. of data obtained from three or four experiments performed in duplicate or triplicate. The statistical significance was determined by Student's t test or one-way ANOVA, and p < 0.05 was considered significant.
RESULTS
S100A7 Overexpression Reduces Proliferation, Chemotaxis, and Wound Healing in ERα+ Breast Cancer Cells
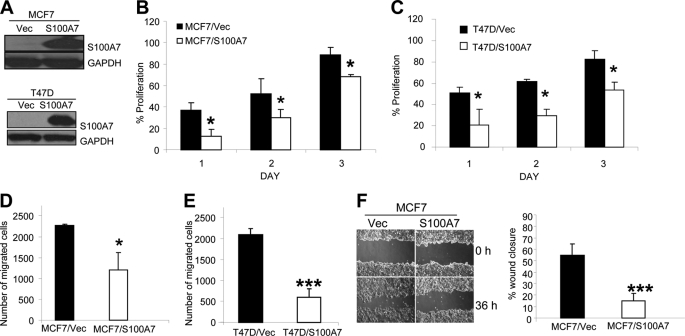
To determine the role of S100A7 on tumorigenic properties of ERα+ cells, we transfected pIRES2-EGFP-S100A7 and pIRES2-EGFP alone in ERα+ MCF7 and T47D cells. High expression of S100A7 was observed in stably S100A7-transfected MCF7 and T47D cells compared with vector control by Western blotting (Fig. 1A). Increased migration, adhesion, and proliferation are important characteristics of tumorigenesis and metastasis of cancer cells. We found that S100A7 overexpression significantly decreased serum-induced proliferation by ∼50% in MCF7 and T47D cells (Fig. 1, B and C). We also observed a significant decrease in chemotactic ability of S100A7-overexpressing MCF7 and T47D cells (Fig. 1, D and E). A wound healing assay also confirmed significantly decreased migratory abilities of S100A7-overexpressing MCF7 cells (Fig. 1F). Furthermore, morphological examination showed the prominent presence of lamellipodia or migratory structures in the MCF7/Vec compared with the MCF7/S100A7 cells (supplemental Fig. 1). These data support that S100A7 overexpression in ERα+ cells significantly inhibits the proliferative and migratory properties of ERα+ cells.
FIGURE 1.
S100A7 overexpression decreases proliferation, chemotaxis, and motility in ERα+ cells. A, S100A7 expression was analyzed in S100A7-overexpressing MCF7, T47D and vector control cells by Western blotting with anti-S100A7 antibody. GAPDH was used as a loading control. B and C, MCF7/Vec, MCF7/S100A7, T47D/Vec, and T47D/S100A7 cells were subjected to proliferation after seeding 5 × 103 cells in 96-well tissue culture plates. The proliferation of the cells was measured at different days using the MTT assay as a percentage increase in the absorbance reading at 570 nm with respect to the control (day 0). D and E, cells were subjected to serum-induced migration by seeding 2 × 104 cells in the upper chamber of 8-μm Transwell plates. The migrated cells were then stained with Hema stain and counted, and the percentage of migrated cells was calculated. F, wound scratch assay was done as described under “Experimental Procedures.” The bar graph shows the quantitative analyses of wound closure assays and was calculated as described before (45). All experiments were repeated three times and a representative one is shown.*, p < 0.05; ***, p < 0.0001 for all experiments as calculated from Student's t test. Error bars, S.D.
S100A7 Overexpression Inhibits Tumor Growth in Vivo
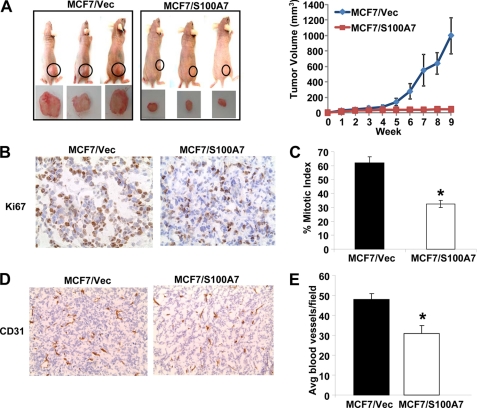
To evaluate the tumor-suppressive effects of S100A7 in vivo, we determined the tumorigenic potential of MCF7/S100A7 cells in a nude mouse model. MCF7/S100A7 and MCF7/Vec cells (3 × 106) were injected subcutaneously into the right flank of female nude mice. We also injected mice with 2.5 μg of β-estradiol 17-valerate weekly after cell injection. Tumor volume was monitored up to 9 weeks, and a dramatic decrease in tumor size was observed in mice injected with MCF7/S100A7 compared with vector control cells (Fig. 2A, left and right). Hematoxylin and eosin (H&E) staining revealed tightly packed cells in MCF7/S100A7 tumors whereas cells were more spread out in MCF7/Vec control-derived tumors. We observed similar cell morphology when cells were grown under in vitro conditions (supplemental Fig. 1). There was also decreased expression of Ki67 (Fig. 2B) and hence, reduced mitotic index in the S100A7-overexpressing tumor (Fig. 2C). Microvasculature was more developed in tumors formed by vector control cells compared with S100A7-overexpressing cells as shown by increased CD31 expression and larger size of microblood vessels (Fig. 2D). The average number of CD31-positive blood vessels/field as counted from three different fields was significantly reduced in S100A7-overexpressing tumors compared with vector control (Fig. 2E). These results extend our in vitro findings of tumor-suppressive capabilities of S100A7 overexpression to an in vivo mouse model system.
FIGURE 2.
S100A7 overexpression inhibits tumor growth in vivo. MCF7/Vec and MCF7/S100A7 cells were injected into the right flank of female nude mice (n = 5). Mice were also injected subcutaneously with 2.5 μg of β-estradiol 17-valerate weekly. Tumors were measured by digital calipers weekly for 9 weeks, and volume was determined using the formula: length × (width)2/2. After 9 weeks, the tumors were harvested, fixed, and evaluated for proliferation and angiogenesis. A, left, representative photographs of mice and tumors 9 weeks after the injection of cells. A, right, increase in tumor volume over time. B and C, Ki67 staining (B) and mitotic index (C) as calculated by the percentage of Ki67-positive cells out of total number of cells in three random fields on the slide. D, CD31 staining of tumors derived from mice. E, average number of microblood vessels/field after counting CD31-positive staining from three random fields on the slide *, p < 0.05 for all experiments as calculated from Student's t test. Error bars, S.D.
Reduced β-Catenin/TCF4 Pathway Activity Is Involved in S100A7-mediated Tumor-suppressive Effects
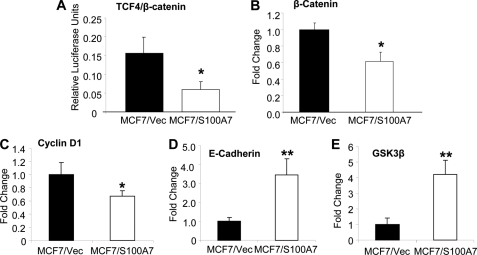
To investigate the mechanism involved in S100A7-mediated suppressive effects we performed microarray analysis by using an Affymetrix gene chip containing 40,000 human genes. Gene ontology data analysis using Ingenuity Pathway Analysis revealed that the S100A7 overexpression significantly affected cellular pathways that are mostly involved in cancer, signifying the importance of S100A7 in breast tumor progression (supplemental Fig. 2A). Further analyses of microarray data revealed that S100A7 overexpression down-regulates genes that are either components of the canonical β-catenin/TCF4 pathway or its downstream targets (supplemental Fig. 2B). Important components of this pathway that were differentially regulated included TCF4 (∼7-fold down-regulated) and GSK3β (∼2.5-fold up-regulated). The downstream targets of the β-catenin/TCF4 pathway that were down-regulated included proto-oncogenes cyclin D1 (2-fold down-regulation) and c-myc (∼3.5-fold down-regulated). Next, we investigated the activity of the β-catenin/TCF4 pathway by using TCF reporter/LEF reporter assay. As shown in Fig. 3A, β-catenin/TCF4 activity in MCF7/S100A7 was significantly reduced by 50% compared with vector control. The candidate targets obtained from microarray analysis were then validated by qRT-PCR. The results showed a significant decrease in the expression of β-catenin and cyclin D1 (Fig. 3, B and C). However, we found an increase in the expression of E-cadherin and GSK3β (Fig. 3, D and E). These results confirm that S100A7 overexpression in ERα+ breast cancer cells down-modulates the β-catenin/TCF4 pathway.
FIGURE 3.
S100A7 overexpression down-regulates expression of genes involved in the β-catenin/TCF4 pathway. A, TCF4 luciferase reporter activity was calculated as described under “Experimental Procedures.” B–E, total RNA from MCF7/Vec and MCF7/S100A7 cells was extracted using TRIzol reagent (Invitrogen) according to the manufacturer's protocol. cDNA was subjected to SYBR Green qRT-PCR for β-catenin, cyclin D1, E-cadherin, and GSK3β. The primers used are listed in supplemental Table 1. *, p < 0.05; **, p < 0.005 for all experiments as calculated from Student's t test. Error bars, S.D. qRT-PCR experiments were repeated four times, and a representative one is shown.
S100A7 Overexpression in ERα+ Cells Down-modulates the β-Catenin/TCF4 Pathway by Regulating GSK3β and TCF4
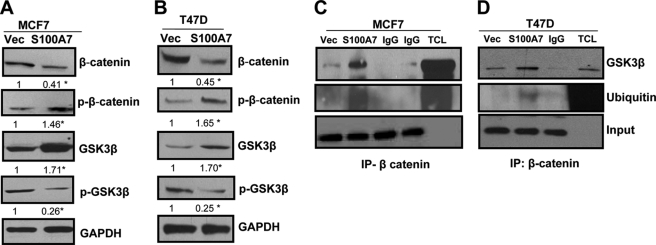
It has been shown that GSK3β phosphorylates β-catenin which is important for the ubiquitination and proteasomal-mediated degradation of β-catenin. Increased phosphorylation of GSK3β reduces the β-catenin-phosphorylating activity of GSK3β (30, 31). Our studies indicate a significant increase in phosphorylation of β-catenin and decrease in β-catenin expression in S100A7-overexpressing MCF7 and T47D cells (Fig. 4, A and B). We also observed increased GSK3β expression in S100A7-overexpressing MCF7 and T47D cells as well as decreased expression of inactive phospho-GSK3β (Fig. 4, A and B). Because GSK3β phosphorylates β-catenin, we determined the interaction between GSK3β and β-catenin through co-immunoprecipitation. The results showed that S100A7 overexpression enhanced the interaction of GSK3β with β-catenin in both MCF7 and T47D cells (Fig. 4, C and D). Because phosphorylation of β-catenin leads to ubiquitination-mediated degradation, we analyzed the ubiquitination level of β-catenin and observed enhanced ubiquitination of β-catenin in S100A7-overexpressing cells (Fig. 4, C and D). These studies indicate that S100A7 may regulate GSK3β which in turn enhances β-catenin phosphorylation that leads to increased ubiquitination and degradation.
FIGURE 4.
S100A7 overexpression reduces the β-catenin/TCF4 pathway in ERα+ cells. A and B, 50 μg of cell lysates obtained from MCF7/Vec, MCF7/S100A7, T47D/Vec, and T47D/S100A7 cells were subjected to Western blotting using β-catenin, phospho-β-catenin (p-β-catenin), GSK3β, or phospho-GSK3β (p-GSK3β) antibodies. GAPDH was used as a loading control. C and D, 500 μg of cell lysates from MCF7 (left) or T47D (right) vector control or S100A7-overexpressing cells were subjected to immunoprecipitation and probed with anti-GSK3β or ubiquitin antibody. The bottom panel shows the IgG band for input. IgG, immunoglobulin control; TCL, total cell lysate; IP, immunoprecipitation. All experiments were repeated three times, and a representative one is shown. The values below the blots show relative intensity obtained after densitometric analyses. *, p < 0.05 as calculated from Student's t test.
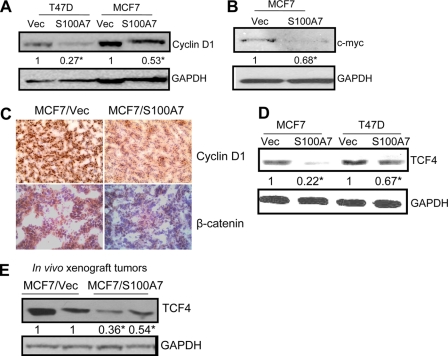
Stabilized β-catenin has been shown to translocate to the nucleus where it interacts with transcription factors of the TCF/LEF family, leading to the increased expression of genes, such as cyclin D1 and c-myc (30, 31). These genes have been shown to play an important role in tumor development (32). We observed decreased expression of downstream targets of the β-catenin/TCF4 pathway such as cyclin D1 (Fig. 5A) and c-myc (Fig. 5B) in S100A7-overexpressing cells compared with vector controls. These results were also confirmed through immunohistochemical analysis in tumors derived from MCF7/S100A7- and MCF7/Vec-injected mice. As shown in Fig. 5C, decreased expression of cyclin D1 and β-catenin was observed in S100A7-overexpressing tumors. β-Catenin has also been shown to modulate expression of TCF4, which has been shown to promote tumorigenesis (19). Because our initial microarray analysis revealed that S100A7-overexpressing MCF7 cells exhibit decreased expression of TCF4, we analyzed the expression of TCF4 by immunoblotting and observed that its expression was significantly inhibited in MCF7/S100A7 and T47D/S100A7 cells (Fig. 5D). Similarly, significantly reduced TCF4 expression was observed in MCF7/S100A7 tumor lysates compared with vector control (Fig. 5E). These results suggest that S100A7 overexpression may down-modulate the β-catenin/TCF4 pathway by regulating the expression of GSK3β and TCF4.
FIGURE 5.
S100A7 overexpression reduces expression of downstream targets of the β-catenin/TCF4 pathway in ERα+ cells. A and B, 50 μg of cell lysates obtained from T47D/Vec, T47D/S100A7, MCF7/Vec, and MCF7/S100A7 cells were subjected to Western blotting using cyclin D1 (A) or c-myc (B) antibodies. C, tumors derived from mice injected with MCF7/Vec or MCF7/S100A7 cells were subjected to immunohistochemical staining for cyclin D1 or β-catenin. D, 50 μg of cell lysates were subjected to immunoblot analysis with TCF4 antibody or GAPDH. E, 50 μg of tumor lysates were subjected to Western blotting with TCF-4 antibody. GAPDH served as a loading control in all blots. All experiments were repeated three times, and a representative one is shown. The values below the blots show relative intensity obtained after densitometric analyses. *, p < 0.05 as calculated from Student's t test.
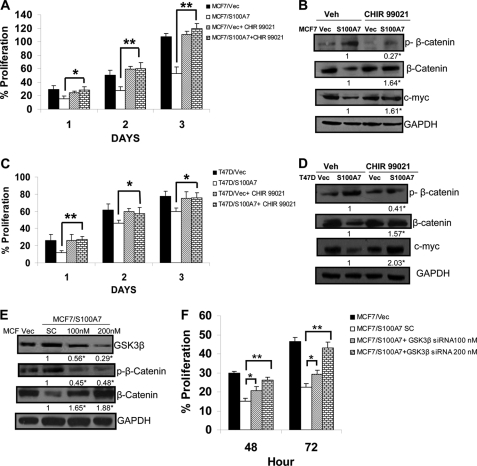
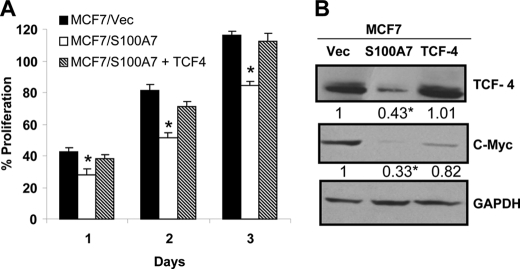
To test whether activity of GSK3β and TCF4 is responsible for decreased activation of the β-catenin/TCF4 pathway, we hypothesized that inhibiting GSK3β activity and restoration of TCF4 expression may reverse the S100A7-mediated effects in ERα+ breast cancer cells. Therefore, we treated S100A7-overexpressing MCF7 and T47D cells with CHIR 99021 (highly specific GSK3β inhibitor) and observed that CHIR 99021 treatment, compared with vehicle control, reversed the S100A7 overexpression-induced decrease in proliferation of MCF7 and T47D cells (Fig. 6, A and C), respectively. Further, immunoblotting experiments showed increased expression of β-catenin and its downstream target, c-myc in inhibitor-treated S100A7-overexpressing MCF7 and T47D cells (Fig. 6, B and D). We also observed decreased phosphorylation of β-catenin in CHIR 99021-treated S100A7-overexpressing cells which may be due to the inhibition of GSK3β activity (Fig. 6, B and D). These results were further confirmed by an siRNA approach. As shown in Fig. 6E, siRNA-mediated down-regulation of GSK3β increased the expression of β-catenin compared with the scrambled control in MCF7/S100A7 cells. Furthermore, the phosphorylation of β-catenin was decreased in GSK3β-down-regulated cells (Fig. 6E). Knockdown of GSK3β also significantly increased the proliferation in MCF7/S100A7 cells compared with the cells transfected with scrambled control (Fig. 6F). Further, to determine whether restoration of TCF4 expression in S100A7-overexpressing cells may reverse the S100A7-mediated inhibitory effects, we transfected TCF4 into S100A7-overexpressing MCF7 cells. The results showed that the restoration of TCF4 expression increased the proliferation of S100A7-overexpressing cells (Fig. 7A). In addition, increased expression of c-myc, a downstream target of the β-catenin/TCF4 pathway, was also observed in TCF4-transfected S100A7-overexpressed MCF7 cells (Fig. 7B). These results indicate that TCF4 down-regulation along with increased activation of GSK3β may be responsible for the tumor-suppressive effects of S100A7 in ERα+ breast cancer cells.
FIGURE 6.
S100A7 regulates the β-catenin/TCF4 pathway through GSK3β. A, MCF7/Vec and MCF7/S100A7 cells were serum-starved for 5 h and then treated with CHIR 99021 (2 μm), and proliferation was measured using the MTT assay as described under “Experimental Procedures.” B, 50 μg of cell lysates were subjected to immunoblot analysis with phospho-β-catenin, β-catenin, or c-myc antibodies. C, proliferation of T47D/Vec and T47D/S100A7 cells was measured as described above for MCF-7 cells. D, 50 μg of cell lysates were subjected to immunoblot analysis with phospho-β-catenin, β-catenin, or c-myc antibodies. GAPDH served as a loading control in all the blots. E, MCF7/S100A7 cells were transiently transfected with 100 nm and 200 nm GSK3β siRNAs using Lipofectamine according to the manufacturer's recommendations. 200 nm scrambled nontargeting siRNA served as the control. Cell lysates were prepared after 48 h of transfection, and levels of GSK3β, phospho-β-catenin, and β-catenin were analyzed using the respective antibodies. F, the proliferation of MCF7/S100A7 cells transfected with either GSK3β-specific or scrambled siRNA was measured using the MTT assay as described under “Experimental Procedures.” The values below the blots show relative intensity obtained after densitometric analyses. *, p < 0.05; **, p < 0.005 as calculated from Student's t test. Veh, vehicle. Error bars, S.D.
FIGURE 7.
TCF4 overexpression in S100A7-overexpressing cells restores the oncogenic properties. A, TCF4 was transiently transfected in MCF7/S100A7 in pcDNA3.1 vector using Lipofectamine according to the manufacturer's recommendations. 24 h after transfection, 5 × 103 cells were seeded in a 96-well tissue culture plate, and proliferation was measured using the MTT assay as described under “Experimental Procedures.” B, 50 μg of cell lysates of TCF4 and vector-transfected MCF7/S100A7 were subjected to Western blotting with anti-TCF4 or c-myc antibodies. GAPDH served as a loading control in all blots. The values below the blots show relative intensity obtained after densitometric analyses. *, p < 0.05 as calculated from one-way ANOVA. Error bars, S.D.
S100A7 Overexpression Enhances the Co-localization and Interaction of β-Catenin and E-cadherin
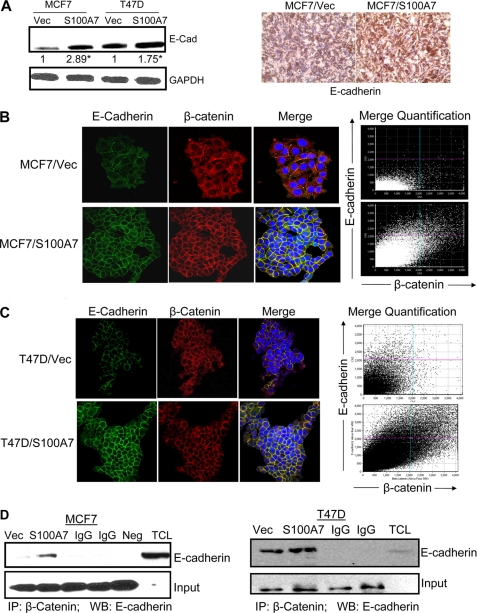
It is known that β-catenin controls E-cadherin-mediated cell adhesion at the plasma membrane and regulates adherens junction molecules within the actin cytoskeletal system (33, 34). We observed increased E-cadherin expression both in vitro in S100A7-overexpressing MCF7 and T47D cells (Fig. 8A, left) as well as tumor samples (Fig. 8A, right). We also observed enhanced expression of E-cadherin in S100A7-overexpressing MCF7 and T47D cells by immunofluorescence (Fig. 8, B and C, left). Further analysis revealed increased co-localization of β-catenin with E-cadherin in the membranes of MCF7/S100A7 and T47D/S100A7 cells compared with vector controls by immunofluorescence (Fig. 8, B and C, right). Quantification analysis of merged E-cadherin and β-catenin showed significantly enhanced co-localization as shown by the linear increase in the merged intensities of E-cadherin and β-catenin (Fig. 8, B and C, right). In addition, we observed enhanced association of β-catenin with E-cadherin in the cell lysates obtained from MCF7/S100A7 (Fig. 8D, left) and T47D/S100A7 (Fig. 8D, right) compared with vector control. These data indicate that S100A7 overexpression might enhance cell-cell adhesion and decreased migration by regulation of β-catenin and E-cadherin in ERα+ breast cancer cells.
FIGURE 8.
S100A7 overexpression enhances the co-localization and interaction between β-catenin and E-cadherin in ERα+ breast cancer cells. A, left, 50 μg of cell lysates subjected to immunoblot analysis with anti-E-cadherin antibody and GAPDH. A, right, representative immunohistochemical staining for anti-E-cadherin in tumors derived from mice injected with MCF7/Vec or MCF7/S100A7 cells. B and C, localization of E-cadherin and β-catenin in MCF7/Vec, MCF7/S100A7 (B), or T47D/Vec, T47D/S100A7 (C) as determined by confocal microscopy. These cells were cultured in chamber slides, fixed, and treated with various antibodies, as described under “Experimental Procedures.” The slides were then mounted using Vectashield medium with 4′,6-diamidino-2-phenylindole (DAPI) and examined under an Olympus FV1000 filter confocal microscope. Graph shows the quantification of merged E-cadherin and β-catenin as analyzed by FV10-ASW 2.0 software (B and C, right). D, 500 μg of cell lysates from MCF7 (left) or T47D (right) vector control or S100A7-overexpressing cells were subjected to immunoprecipitation (IP) and probed with anti-E-cadherin antibody. The bottom panel shows the IgG band for input. IgG, immunoglobulin control for both vector and S100A7-overexpressing cells; Neg, negative control; TCL, total cell lysate; IP, immunoprecipitation; WB, Western blotting. All experiments were repeated three times, and a representative one is shown. *, p < 0.05 as calculated from Student's t test.
DISCUSSION
S100A7 expression has been shown to be associated mainly with high grade ductal carcinoma in situ and highly invasive ERα− breast cancers (10–18). In addition, S100A7 down-modulation inhibits tumor growth and EGF-induced migration in ERα− cells (11, 14). Moreover, overexpression of S100A7 in ERα− breast cancer cells has been shown to enhance proliferation, migration, and invasion in vitro as well as tumor growth in vivo (13, 16, 17). However, not much is known about its role in ERα+ breast cancer cells. In the present study, we characterized the role and mechanism of S100A7 in ERα+ breast cancer cells.
We have shown for the first time that S100A7 possesses tumor-suppressive activity in S100A7-overexpressing ERα+ MCF7 and T47D breast cancer cells. We observed reduced serum-induced proliferation, migration, and wound healing activity in S100A7-overexpressing cells. We also observed a significant reduction in tumor formation of estradiol-treated MCF7/S100A7 compared with MCF7/Vec control in nude mice. Although S100A7 overexpression has been shown to enhance tumor growth both in vivo and in vitro in ERα− cells (17), one report has shown that S100A7 overexpression inhibits cell proliferation in vitro and tumor growth/invasion in vivo in cells of squamous cell carcinoma of the oral cavity (36). Several previous studies have demonstrated that regular treatment of MCF7 cells with estrogen sustains and enhances the tumorigenic effect of MCF7 cells in nude mice (37). Our results in xenografted nude mice indicate that S100A7 overexpression significantly overcomes the tumor-enhancing effects of estrogen by exhibiting a marked reduction in tumor size even in the presence of estrogen. We hypothesize that S100A7 may be inhibiting tumor growth in vivo by modulating estrogen and growth factor-mediated signaling. It was recently reported that IL-6 and oncostatin M treatment enhance S100A7 expression that leads to down-regulation of expression of the ERα gene (38). We have also observed that S100A7 overexpression down-regulates ERα expression in both MCF7 and T47D cells (supplemental Fig. 3).
Upon further analyses of the mechanisms that may mediate the tumor-suppressive function of S100A7 in ERα+ breast cancers, we observed a decreased β-catenin/TCF4 pathway in MCF7/S100A7 cells compared with MCF7/Vec cells. Increased levels of β-catenin have been observed in various cancers including breast and have also been linked with poor prognosis in human adenocarcinoma and invasive ductal breast cancers (39–42). Recently, it was also shown that S100A7 and β-catenin may negatively regulate each other in squamous cell carcinoma of the oral cavity (36).
In our studies, we observed reduced expression and increased phosphorylation of β-catenin at Thr-41, Ser-37, and/or Ser-33 phosphorylation sites in MCF7/S100A7 cells. These sites are phosphorylated by GSK3β which are then recognized by the ubiquitin ligase complex leading to β-catenin degradation (43). We observed increased levels of GSK3β which may be phosphorylating β-catenin, leading to its reduced expression and degradation. Also, the phosphorylated level of GSK3β, which is the inactive form of GSK3β, was also reduced in S100A7-overexpressing cells. GSK3β is phosphorylated by kinases like AKT, and we also observed decreased activation of AKT in S100A7-overexpressing cells (data not shown). Therefore, it is possible that S100A7 may be having some direct effects on upstream kinases which phosphorylate GSK3β.
We have also observed decreased expression of TCF4 which binds to β-catenin and regulates the transcription of downstream targets of the β-catenin/TCF4 pathway. Additionally, our luciferase gene reporter assay revealed inhibition of β-catenin/TCF4 transcriptional activity in the S100A7-overexpressing cells. Upon further analyses of the expression of various downstream targets of the β-catenin/TCF4 pathway, we found decreased expression of cyclin D1 and c-myc in the S100A7-overexpressing cells. These genes are proto-oncogenes and considered as critical mediators of proliferation, invasion, and tumor progression.
β-Catenin has also been shown to affect the cell adhesion and cytoskeleton properties by binding to E-cadherin (39, 41). S100A7 overexpression increased E-cadherin expression which is a component of adherens junctions and a known marker of reverse epithelial-mesenchymal transition and epithelial integrity (44). Its increased expression may support reduction in migration of S100A7-overexpressing cells. We also demonstrated increased co-localization of E-cadherin and β-catenin in S100A7-overexpressing cells. Furthermore, there was enhanced interaction between E-cadherin and β-catenin. Although there is increased β-catenin degradation observed in S100A7-overexpressing cells, the increased interaction between E-cadherin and β-catenin may be explained by the significantly increased expression of E-cadherin which may sequester nonphosphorylated β-catenin in S100A7-overexpressing cells, allowing it to escape GSK3β-mediated degradation.
In the present investigation, we demonstrate for the first time that S100A7 overexpression inhibits proliferation, migration, and wound healing in ERα+ MCF7 and T47D breast cancer cell lines. Furthermore, we have confirmed that S100A7 induces tumor-suppressive activity in MCF7 cells in an in vivo mouse model system. In addition, we showed that S100A7 mediates its tumor-suppressive effects via a novel mechanism through a coordinated regulation of the β-catenin/TCF4 as summarized in Fig. 9. These results suggest that S100A7 may possess differential activity in ERα+ and ERα− breast cancer and therefore may play an important role in breast cancer progression and metastasis. These results may also have implications in developing novel treatments for ERα+ and tamoxifen-resistant breast tumors.
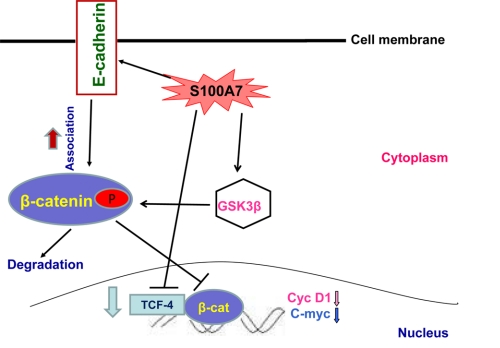
FIGURE 9.
Schematic representation of S100A7-mediated signaling that inhibits proliferation and motility in ERα+ breast cancer. S100A7 overexpression in ERα+ cells may down-regulate the β-catenin/TCF4 pathway by regulating the expression and phosphorylation of GSK3β. The β-catenin/TCF4 pathway may in turn down-regulate the expression of downstream targets such as cyclin D1 and c-myc in S100A7-overexpressing ERα+ cells. Therefore, the β-catenin/TCF4 pathway may be responsible for decreased proliferation and tumor growth in S100A7-overexpressing cells. In addition, S100A7 may modulate migration through enhanced co-localization and interaction of β-catenin and E-cadherin in the membranes of ERα+ cells.
Supplementary Material
Acknowledgments
We thank Drs. Debanjan Chakroborty and Konstantin Shilo for help with immunohistochemical analysis and Dr. Pawan Kumar for critical reading of the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA109527 and R01CA153490 and Department of Defense Grants W81XWH-11-1-0279 and W8XWH-11-1-0275 (to R. K. G.).

The on-line version of this article (available at http://www.jbc.org) contains supplemental Table 1 and Figs. 1–3.
- ER
- estrogen receptor
- BisTris
- bis(2-hydroxyethyl)iminotris(hydroxymethyl)methane
- GSK3β
- glycogen synthase kinase 3β
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- qRT-PCR
- quantitative real time PCR
- TCF4
- T cell factor 4
- Vec
- vector.
REFERENCES
- 1. de Cid R., Riveira-Munoz E., Zeeuwen P. L., Robarge J., Liao W., Dannhauser E. N., Giardina E., Stuart P. E., Nair R., Helms C., Escaramís G., Ballana E., Martín-Ezquerra G., den Heijer M., Kamsteeg M., Joosten I., Eichler E. E., Lázaro C., Pujol R. M., Armengol L., Abecasis G., Elder J. T., Novelli G., Armour J. A., Kwok P. Y., Bowcock A., Schalkwijk J., Estivill X. (2009) Nat. Genet. 41, 211–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Mischke D., Korge B. P., Marenholz I., Volz A., Ziegler A. (1996) J. Invest. Dermatol. 106, 989–992 [DOI] [PubMed] [Google Scholar]
- 3. South A. P., Cabral A., Ives J. H., James C. H., Mirza G., Marenholz I., Mischke D., Backendorf C., Ragoussis J., Nizetic D. (1999) J. Invest. Dermatol. 112, 910–918 [DOI] [PubMed] [Google Scholar]
- 4. Zhang X. J., Huang W., Yang S., Sun L. D., Zhang F. Y., Zhu Q. X., Zhang F. R., Zhang C., Du W. H., Pu X. M., Li H., Xiao F. L., Wang Z. X., Cui Y., Hao F., Zheng J., Yang X. Q., Cheng H., He C. D., Liu X. M., Xu L. M., Zheng H. F., Zhang S. M., Zhang J. Z., Wang H. Y., Cheng Y. L., Ji B. H., Fang Q. Y., Li Y. Z., Zhou F. S., Han J. W., Quan C., Chen B., Liu J. L., Lin D., Fan L., Zhang A. P., Liu S. X., Yang C. J., Wang P. G., Zhou W. M., Lin G. S., Wu W. D., Fan X., Gao M., Yang B. Q., Lu W. S., Zhang Z., Zhu K. J., Shen S. K., Li M., Zhang X. Y., Cao T. T., Ren W., Zhang X., He J., Tang X. F., Lu S., Yang J. Q., Zhang L., Wang D. N., Yuan F., Yin X. Y., Huang H. J., Wang H. F., Lin X. Y., Liu J. J. (2009) Nat. Genet. 41, 205–210 [DOI] [PubMed] [Google Scholar]
- 5. Boniface K., Diveu C., Morel F., Pedretti N., Froger J., Ravon E., Garcia M., Venereau E., Preisser L., Guignouard E., Guillet G., Dagregorio G., Pène J., Moles J. P., Yssel H., Chevalier S., Bernard F. X., Gascan H., Lecron J. C. (2007) J. Immunol. 178, 4615–4622 [DOI] [PubMed] [Google Scholar]
- 6. Jinquan T., Vorum H., Larsen C. G., Madsen P., Rasmussen H. H., Gesser B., Etzerodt M., Honoré B., Celis J. E., Thestrup-Pedersen K. (1996) J. Invest. Dermatol. 107, 5–10 [DOI] [PubMed] [Google Scholar]
- 7. Wolf R., Mascia F., Dharamsi A., Howard O. M., Cataisson C., Bliskovski V., Winston J., Feigenbaum L., Lichti U., Ruzicka T., Chavakis T., Yuspa S. H. (2010) Sci. Transl. Med. 2, 61ra90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Leygue E., Snell L., Hiller T., Dotzlaw H., Hole K., Murphy L. C., Watson P. H. (1996) Cancer Res. 56, 4606–4609 [PubMed] [Google Scholar]
- 9. Moog-Lutz C., Bouillet P., Régnier C. H., Tomasetto C., Mattei M. G., Chenard M. P., Anglard P., Rio M. C., Basset P. (1995) Int. J. Cancer 63, 297–303 [DOI] [PubMed] [Google Scholar]
- 10. Enerbäck C., Porter D. A., Seth P., Sgroi D., Gaudet J., Weremowicz S., Morton C. C., Schnitt S., Pitts R. L., Stampl J., Barnhart K., Polyak K. (2002) Cancer Res. 62, 43–47 [PubMed] [Google Scholar]
- 11. Krop I., März A., Carlsson H., Li X., Bloushtain-Qimron N., Hu M., Gelman R., Sabel M. S., Schnitt S., Ramaswamy S., Kleer C. G., Enerbäck C., Polyak K. (2005) Cancer Res. 65, 11326–11334 [DOI] [PubMed] [Google Scholar]
- 12. Al-Haddad S., Zhang Z., Leygue E., Snell L., Huang A., Niu Y., Hiller-Hitchcock T., Hole K., Murphy L. C., Watson P. H. (1999) Am. J. Pathol. 155, 2057–2066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Emberley E. D., Niu Y., Leygue E., Tomes L., Gietz R. D., Murphy L. C., Watson P. H. (2003) Cancer Res. 63, 1954–1961 [PubMed] [Google Scholar]
- 14. Paruchuri V., Prasad A., McHugh K., Bhat H. K., Polyak K., Ganju R. K. (2008) PLoS One 3, e1741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Petersson S., Bylander A., Yhr M., Enerbäck C. (2007) BMC Cancer 7, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Emberley E. D., Niu Y., Njue C., Kliewer E. V., Murphy L. C., Watson P. H. (2003) Clin. Cancer Res. 9, 2627–2631 [PubMed] [Google Scholar]
- 17. Emberley E. D., Alowami S., Snell L., Murphy L. C., Watson P. H. (2004) Breast Cancer Res. 6, R308–315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Emberley E. D., Niu Y., Curtis L., Troup S., Mandal S. K., Myers J. N., Gibson S. B., Murphy L. C., Watson P. H. (2005) Cancer Res. 65, 5696–5702 [DOI] [PubMed] [Google Scholar]
- 19. Ravindranath A., O'Connell A., Johnston P. G., El-Tanani M. K. (2008) Curr. Mol. Med. 8, 38–50 [DOI] [PubMed] [Google Scholar]
- 20. Saadeddin A., Babaei-Jadidi R., Spencer-Dene B., Nateri A. S. (2009) Mol. Cancer Res. 7, 1189–1196 [DOI] [PubMed] [Google Scholar]
- 21. Iwai S., Yonekawa A., Harada C., Hamada M., Katagiri W., Nakazawa M., Yura Y. (2010) Int. J. Oncol. 37, 1095–1103 [DOI] [PubMed] [Google Scholar]
- 22. Neth P., Ries C., Karow M., Egea V., Ilmer M., Jochum M. (2007) Stem Cell Rev. 3, 18–29 [DOI] [PubMed] [Google Scholar]
- 23. Morin P. J., Weeraratna A. T. (2003) Cancer Treat. Res. 115, 169–187 [DOI] [PubMed] [Google Scholar]
- 24. Moon R. T., Kohn A. D., De Ferrari G. V., Kaykas A. (2004) Nat. Rev. Genet. 5, 691–701 [DOI] [PubMed] [Google Scholar]
- 25. Gillett C., Fantl V., Smith R., Fisher C., Bartek J., Dickson C., Barnes D., Peters G. (1994) Cancer Res. 54, 1812–1817 [PubMed] [Google Scholar]
- 26. López-Knowles E., Zardawi S. J., McNeil C. M., Millar E. K., Crea P., Musgrove E. A., Sutherland R. L., O'Toole S. A. (2010) Cancer Epidemiol. Biomarkers Prev. 19, 301–309 [DOI] [PubMed] [Google Scholar]
- 27. Prasad A., Paruchuri V., Preet A., Latif F., Ganju R. K. (2008) J. Biol. Chem. 283, 26624–26633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Preet A., Qamri Z., Nasser M. W., Prasad A., Shilo K., Zou X., Groopman J. E., Ganju R. K. (2011) Cancer Prev. Res. 4, 65–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Qamri Z., Preet A., Nasser M. W., Bass C. E., Leone G., Barsky S. H., Ganju R. K. (2009) Mol. Cancer Ther. 8, 3117–3129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Papkoff J., Rubinfeld B., Schryver B., Polakis P. (1996) Mol. Cell. Biol. 16, 2128–2134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Rubinfeld B., Albert I., Porfiri E., Fiol C., Munemitsu S., Polakis P. (1996) Science 272, 1023–1026 [DOI] [PubMed] [Google Scholar]
- 32. Behrens J. (2000) Ann. N.Y. Acad. Sci. 910, 21–33 [DOI] [PubMed] [Google Scholar]
- 33. Nagafuchi A. (2001) Curr. Opin. Cell Biol. 13, 600–603 [DOI] [PubMed] [Google Scholar]
- 34. Vasioukhin V., Fuchs E. (2001) Curr. Opin. Cell Biol. 13, 76–84 [DOI] [PubMed] [Google Scholar]
- 35.Deleted in proof [Google Scholar]
- 36. Zhou G., Xie T. X., Zhao M., Jasser S. A., Younes M. N., Sano D., Lin J., Kupferman M. E., Santillan A. A., Patel V., Gutkind J. S., Ei-Naggar A. K., Emberley E. D., Watson P. H., Matsuzawa S. I., Reed J. C., Myers J. N. (2008) Oncogene 27, 3527–3538 [DOI] [PubMed] [Google Scholar]
- 37. Ray G., Banerjee S., Saxena N. K., Campbell D. R., Van Veldhuizen P., Banerjee S. K. (2005) Oncol. Rep. 13, 445–448 [PubMed] [Google Scholar]
- 38. West N. R., Watson P. H. (2010) Oncogene 29, 2083–2092 [DOI] [PubMed] [Google Scholar]
- 39. Crawford H. C., Fingleton B. M., Rudolph-Owen L. A., Goss K. J., Rubinfeld B., Polakis P., Matrisian L. M. (1999) Oncogene 18, 2883–2891 [DOI] [PubMed] [Google Scholar]
- 40. Hatsell S., Rowlands T., Hiremath M., Cowin P. (2003) J. Mammary Gland Biol. Neoplasia 8, 145–158 [DOI] [PubMed] [Google Scholar]
- 41. Tetsu O., McCormick F. (1999) Nature 398, 422–426 [DOI] [PubMed] [Google Scholar]
- 42. Wielenga V. J., Smits R., Korinek V., Smit L., Kielman M., Fodde R., Clevers H., Pals S. T. (1999) Am. J. Pathol. 154, 515–523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Amit S., Hatzubai A., Birman Y., Andersen J. S., Ben-Shushan E., Mann M., Ben-Neriah Y., Alkalay I. (2002) Genes Dev. 16, 1066–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schmalhofer O., Brabletz S., Brabletz T. (2009) Cancer Metastasis Rev. 28, 151–166 [DOI] [PubMed] [Google Scholar]
- 45. Yue P. Y., Leung E. P., Mak N. K., Wong R. N. (2010) J. Biomol. Screen. 15, 427–433 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.