Abstract
Background
Endophenotypes reflect more proximal effects of genes than diagnostic categories, hence providing a more powerful strategy in searching for genes involved in complex psychiatric disorders. There is strong evidence suggesting the P3 amplitude of the event-related potential (ERP) as an endophenotype for the risk of alcoholism and other disinhibitory disorders. Recent studies demonstrated a crucial role of corticotropin releasing hormone receptor 1 (CRHR1) in the environmental stress response and ethanol self-administration in animal models. The aim of the present study was to test the potential associations between single-nucleotide polymorphisms (SNPs) in the CRHR1 gene and the quantitative trait, P3 amplitude during the processing of visual target signals in an oddball paradigm, as well as alcohol dependence diagnosis.
Methods
We analyzed a sample from the Collaborative Study on the Genetics of Alcoholism (COGA) comprising 1049 Caucasian subjects from 209 families (including 472 alcohol-dependent individuals). Quantitative transmission disequilibrium test (QTDT) and family-based association test (FBAT) were used to test the association, and false discovery rate (FDR) was applied to correct for multiple comparisons.
Results
Significant associations (p < 0.05) were found between the P3 amplitude and alcohol dependence with multiple SNPs in the CRHR1 gene.
Conclusions
Our results suggest that CRHR1 may be involved in modulating the P3 component of the ERP during information processing and in vulnerability to alcoholism. These findings underscore the utility of electrophysiology and the endophenotype approach in the genetic study of psychiatric disorders.
Keywords: P3, Disinhibition, Endophenotype, Stress, Corticotropin Releasing Factor (CRF)
Psychosocial stress has been suggested to be an important risk factor for precipitating the development of dependence on alcohol and other substances of abuse. Preclinical and clinical studies have shown an increase in alcohol drinking following stress (Sillaber et al., 2002; de Wit et al., 2003). The neuroendocrine mechanism of the stress reaction is primarily mediated through the hypothalamic-pituitary-adrenal (HPA) system. Corticotropin releasing hormone (CRH) is released from the hypothalamus when an individual is exposed to stressful signals including alcohol intake and binds to the corticotropin releasing hormone receptor 1 (CRHR1) in the pituitary gland (Bittencourt and Sawchenko, 2000). Activation of the HPA system in response to alcohol may differ among individuals because of family history of alcoholism. For example, lower levels of adrenocorticotropin (ACTH, (Schuckit et al., 1988) and cortisol (Schuckit et al., 1987) were found after ingestion of a high dose of 0.88 g/kg ethanol in sons of alcoholics compared to control males; subjects with a positive family history of alcoholism had an increased cortisol response to the stimulation of the HPA system induced by intravenous injection of the opiate antagonist naloxone (Wand et al., 1999); sons of alcohol-dependent fathers were more sensitive to the alcoholinduced attenuation of ACTH response to psychosocial stress than family history negative controls (Zimmermann et al., 2004).
In addition to the involvement of the HPA system, studies reported that corticotropin releasing hormone (CRH, also known as corticotropin releasing factor, CRF) may play an important role in negative mood states and in the negative affective aspects of drug withdrawal (Koob, 1999). Spontaneous alcohol or cocaine withdrawal has been shown to elevate extracellular CRH levels in the central nucleus of the amygdala, a brain area implicated in anxiety response (Merlo Pich et al., 1995; Richter and Weiss, 1999). Antagonism of CRH receptors decreases anxiety-like behavior associated with withdrawal from alcohol, cocaine, and other drugs of abuse (Heinrichs et al., 1994; Lu et al., 2001; Overstreet et al., 2004). Corticotropin releasing hormone receptor 1 (CRHR1) antagonists have been shown to reduce ethanol self-administration in ethanol-dependent rats (Funk et al., 2007) and to decrease the operant responding for ethanol in rats with a history of ethanol vapor exposure after acute withdrawal (Valdez et al., 2002). Furthermore, genetic polymorphism in the Crhr1 promoter region was found to be associated with a lowered threshold for stress-induced reinstatement of alcohol seeking in a strain of rats that were genetically selected for high alcohol preference (Hansson et al., 2006). In the Crhr1 - knockout mouse model, mice lacking the Crhr1 receptor, but not the highly homologous Crhr2 receptor, exhibited impaired stress response and reduced anxiety under basal conditions and during alcohol withdrawal (Timpl et al., 1998). A recent study in human subjects suggests that treatment with a high-affinity CRHR1 receptor antagonist NBI-34041 did not impair the diurnal HPA secretion but attenuated the neuroendocrine response to psychosocial stress (Ising et al., 2007). These results suggest that there may be an association between the genetic polymorphism in the CRHR1 gene and not only responsiveness to stress but also alcohol dependence or other substance-related disorders.
Event-related potentials (ERPs) provide a noninvasive tool to explore the characteristics of sensory processes and higher cognitive function in the brain. The P3 (or P300) component is possibly the best-studied ERP component. This positive electric potential deflection is elicited approximately 300–500 ms following the occurrence of infrequent stimuli during an oddball experiment paradigm. P3 is highly heritable (Katsanis et al., 1997; O’Connor et al., 1994) and provides quantitative endophenotypes for disinhibitory disorders including alcohol-related or substance-related disorders, conduct disorder, attention-deficit hyperactive disorder (ADHD), and antisocial personality disorder (ASP) (Hesselbrock et al., 2001; Porjesz et al., 2005). Endophenotypes, or intermediate phenotypes, are closer to gene action than diagnostic categories, and therefore they provide a more powerful strategy in searching for genes involved in complex (non-Mendelian) psychiatric diagnoses (Almasy and Blangero, 1998; Gottesman and Gould, 2003; Porjesz et al., 2005). By using this strategy, analysis of brain oscillation data from the Collaborative Study on the Genetics of Alcoholism (COGA) has revealed significant linkage on a number of chromosomes as well as positive association with single-nucleotide polymorphisms (SNPs) within candidate genes for disinhibitory disorders (e.g., Begleiter et al., 1998; Porjesz et al., 2004).
Recent studies using whole genome linkage scans have identified linkage peaks with genome-wide significant LOD scores of 3.5 for resting electroencephalogram (EEG) alpha and beta power in the region of chromosome 5q13-14, where the corticotropin releasing hormone-binding protein (CRH-BP) gene resides at the apex of these convergent linkage peaks (Enoch et al., 2008). Association studies of SNPs in the CRH-BP gene were also found to be significant with EEG alpha power. Moreover, the same SNPs and haplotypes located within the CRH-BP haplotype block were also associated with anxiety disorders in the Plains Indians and alcohol use disorders in the Caucasians (Enoch et al., 2008). These results suggest a possible role for CRH-BP in stress-related alcoholism. The data also indirectly suggest that there may be an association of other genes in the CRH system with EEG and alcohol dependence. The physiological function of CRH-BP is still not clear. CRH-BP is thought to modulate CRH activity, because a large proportion (65–90%) of total CRH forms a complex with CRH-BP, and is therefore unavailable for receptor activation in the baseline conditions (Westphal and Seasholtz, 2006). In addition, a recent study in rhesus macaques (Barr et al., 2008) showed that a functional single-nucleotide polymorphism in the CRH gene that is associated with sensitivity of the CRH promoter to glucocorticoids in vitro and CSF levels of CRH may also be involved in increased alcohol consumption in the social group, an animal model for high-risk alcohol-seeking behavior. Taken together, these results strongly suggest that genetic variations in the CRH system play important roles in alcohol dependence.
To determine the possible contribution of the CRHR1 gene to alcohol dependence in humans, we investigated a large sample of 1049 Caucasian adults from multiplex families recruited from alcoholic probands in this study. We report here significant associations between SNPs in the CRHR1 gene and the quantitative trait of the P3 amplitude of the event-related potential during processing of target visual signals as well as the diagnosis of alcohol dependence, a dichotomous endpoint phenotype.
METHODS
Subjects
The samples included in this study were recruited and tested as part of the COGA, a large multisite national study implemented with the purpose of identifying genetic loci linked with the predisposition to develop alcoholism and other related disorders. Data from six COGA sites were included in this analysis: SUNY Downstate Medical Center, Brooklyn, New York; University of Connecticut Health Science Center; Indiana University School of Medicine; University of Iowa School of Medicine; University of California School of Medicine, San Diego; and Washington University School of Medicine, St Louis. Ascertainment and assessment procedures have been outlined previously (Begleiter et al., 1995; Foroud et al., 2000; Reich et al., 1998).
All probands were recruited from alcohol or other substance dependence treatment facilities, and met DSM-IIIR criteria for alcohol dependence and Feighner definite criteria (COGA criteria). In addition to the probands, the study required two additional first-degree relatives who were alcohol dependent by the same COGA criteria on direct interview. Semi-Structured Assessment for the Genetics of Alcoholism, SSAGA, a polydiagnostic instrument designed by COGA with well-established reliability (Bucholz et al., 1994) and validity (Hesselbrock et al., 1999), was administered in person to determine psychiatric diagnoses in all family members. Subjects completed a neuropsychological battery and family history questionnaire and EEGs/ERPs were recorded.
A Caucasian-only subset of sample comprising 209 families and 1049 individuals was used in the genetic association analysis (containing 472 individuals diagnosed as alcohol dependent by DSM-IV). Details of the demography of the subjects included in this study were reported in our recent study (Chen et al., 2009) and summarized in Table 1. Briefly, between alcoholic and unaffected groups, there was no significant difference in age (unaffected: 39.30 ± 0.55, mean ± SEM, ranging from 18.09 to 70.41 years old; alcohol dependence: 39.60 ± 0.61, ranging from 17.94 to 73.38), common co-occurring psychiatric disorders, e.g., antisocial personality disorder, major depression, bipolar disorder, obsessive compulsive disorder (Schizophrenia is an exclusion in recruitment.), and other substance dependence except that there was an almost five-time increase in cannabis dependence in alcoholic group. There were slightly more males among the alcoholics (M/F ratio = 237/235 = 1.01) than the unaffected (M/F ratio = 274/303 = 0.90) in this dataset.
Table 1.
Demography of Study Subjects
| Unaffected | Alcohol dependence (by DSM-IV) | Total | ||||
|---|---|---|---|---|---|---|
| n | 577 | 472 | 1049 | |||
| Age (year) | 39.30 ± 0.55 | 39.6 ± 0.61 | 39.45 ± 0.41 | |||
| Gender (M/F) | 274/303 | 0.90 ratio | 237/235 | 1.01 ratio | 511/538 | 0.95 ratio |
| Psychiatric comorbidity (by DSM-IIIR) | ||||||
| ASPD | 49 | 8.49% | 53 | 11.23% | 102 | |
| Major depression | 92 | 15.94% | 84 | 17.80% | 176 | |
| Bipolar I | 3 | 0.52% | 9 | 1.91% | 12 | |
| Bipolar II | 6 | 1.04% | 12 | 2.54% | 18 | |
| OCD | 7 | 1.21% | 14 | 2.97% | 21 | |
| Social phobia | 15 | 2.60% | 23 | 4.87% | 38 | |
| Panic disorder | 13 | 2.25% | 22 | 4.66% | 35 | |
| Cannabis dependence* | 49 | 8.49%* | 191 | 40.47%* | 240 | |
| Cocaine dependence | 99 | 17.16% | 75 | 15.89% | 174 | |
ASPD, antisocial personality disorder; OCD, obsessive compulsive disorder.
p = 1.56E-33.
Prior to administration of the neurophysiology test battery, alcoholic subjects had been detoxified in a 30-day treatment program, and none of the subjects was in the withdrawal phase. Subjects were excluded from the neurophysiology protocol if they manifested any of the following: uncorrected sensory deficits, hepatic encephalopathy/cirrhosis of the liver, history of significant head injury, seizures or neurosurgical procedures, other acute/chronic medical illness, were on medication known to influence brain functioning (e.g., any psychotropic medications), or tested positive for HIV. Subjects were also excluded on the basis of recent (i.e., 5 days) substance and/or alcohol use, based on self-report as well as breath-analyzer and urine screen. In addition, all subjects were screened for cognitive status, using the Mini Mental State Examination (MMSE, Folstein et al., 1975).
Event-Related Potential Data Acquisition and Signal Analysis
Subjects were seated in a comfortable chair located in a dimly lit sound-attenuated RF-shielded room (IAC, Industrial Acoustics, Bronx, NY) in front of the computer monitor placed one meter away. EEG activity was recorded on a Neuroscan system (Version 4.1) (Neurosoft, Inc., El Paso, TX) using a multichannel electrode cap (Electro-cap International, Inc., Eaton, OH), which included 19 electrodes of the 10–20 International System and 42 additional electrode sites (Electrode Position Nomenclature, American Electroencephalographic Association, 1991), as shown previously (Jones et al., 2004). The electrodes were referenced to the tip of the nose, and the ground electrode was at the forehead (frontal midline). Artifact-free data obtained from the 7 posterior channels were included for genetic analyses of visual P3 amplitude.
Details of the visual oddball paradigm for eliciting event-related potential employed in the present study have been previously described (Chen et al., 2007; Porjesz et al., 1998). It consists of presentation of three types of visual stimuli (N = 280), 60 ms duration, subtending a visual angle of 2.5°, with an inter-stimulus interval of 1.625 second. The rare target stimulus (n = 35) was the letter X, to which the subject was required to press a button as quickly as possible; the responding hand was alternated across subjects to counter-balance any laterality effects because of responding. Speed was emphasized, but not at the cost of accuracy. The frequently occurring nontarget stimuli (n = 210) were squares and the novel stimuli (n = 35) consisted of colored geometric polygons that were different on each trial; the subject was not required to respond to the nontarget or novel stimuli.
The visual P3 (VP3) amplitude was measured as the voltage difference between the prestimulus baseline and the largest positive going peak in the latency window 300–600 ms after stimulus onset. For each individual, the amplitude and latency measures were calculated using a semi-automatic peak-picking program, wherein the time window for each component was manually selected in the computer while the peak within the window was automatically detected, measured, and tabulated for each channel. However, operator intervention was possible during the process to ensure that the computer did not make anomalous peak selections. Each subject had a minimum of 20 good trials in each condition. The grand averages were computed and plotted to determine the components and time windows (Kamarajan et al., 2005).
Genotyping
Publicly available databases dbSNP (http://www.ncbi.nlm.nih.gov/SNP/) and HapMap (http://hapmap.ncbi.nlm.nih.gov/) were used to identify SNPs within the CRHR1 gene, which is located on Chromosome 17q12-q22. Genotyping was performed using Sequenom MassArray technology (http://www.sequenom.com, San Diego, CA), homogenous MassEXTEND (hME). PCR primers, termination mixes, and multiplexing were determined with Sequenom MassARRAY Assay Designer software v3.1.2.2. The hME assay was carried out following standard procedures, and genotype spectra were analyzed with the Sequenom SpectroTYPER software v3.4. All genotypic data were checked for Mendelian inheritance of marker alleles with the USERM13 (Boehnke, 1991) option of the MENDEL linkage computer programs, which was then used to estimate marker allele frequencies. Chi-square tests were used to ensure that all SNPs were in Hardy Weinberg equilibrium.
Genetic Analyses
Age and gender were regressed before genetic analysis of the P3 amplitude of the ERP. The quantitative transmission disequilibrium test (QTDT) (Abecasis et al., 2000) was used to test population stratification, total association, and within-family association with each of the SNPs and the P3 amplitude of the ERP. QTDT, one of the most popular family-based linkage disequilibrium tests, is able to avoid spurious evidence of association in the presence of population stratification. In the QTDT framework, association effects are partitioned into orthogonal between-family and within-family components. The between-family association can be impacted by population structure. Within-family association, however, is significant only if there is linkage disequilibrium and is robust to population stratification/admixture. The total association tests use all information including within-family and between-family components, and are proper only if there is no population stratification/admixture. The within-family association results were reported if the test of population stratification was significant, otherwise, the total association (i.e., measured genotype; Boerwinkle et al., 1986) results were reported.
Family-based association tests (FBAT) (Rabinowitz and Laird, 2000) was used for genetic association analyses of alcohol dependence diagnosis. FBAT builds also on the original transmission disequilibrium test (TDT) method (Spielman and Ewens, 1996) in which alleles transmitted to affected offspring are compared with the expected distribution of alleles among offspring. The approach compares the genotype distribution observed in the “cases” to its expected distribution under the null hypothesis; in this case, the null hypothesis tested was “no linkage and no association” (Lange and Laird, 2002).
Haplotype frequencies were estimated using a Bayesian approach implemented with the FBAT software, which carries out a family-based test for association and/or linkage between the haplotype locus and any trait influencing gene. These haplotype frequencies closely agreed with results from a maximum likelihood method implemented via an expectation-maximization (EM) algorithm (Long et al., 1995). Haploview V. 2.0.2 (Whitehead Institute for Biomedical Research, Cambridge, MA) was used to produce LD matrices. Haplotype blocks were reconstructed using the pairs of markers with LD greater than 0.8 (Gabriel et al., 2002).
In this study, both a quantitative trait, P3 amplitude of the ERP, and a dichotomous variable, alcohol dependence by DSM-IV criteria, were the phenotypes tested. False discovery rate, FDR (Benjamini and Hochberg, 1995; Storey and Tibshirani, 2003), was calculated to correct for multiple comparisons using the method developed by Storey and Tibshirani (2003).
RESULTS
Among the 10 SNPs that we have genotyped, 1 SNP (rs991246, located upstream of the CRHR1 gene) had a very low minor allele frequency (MAF) 0.0024, and was omitted from analysis. Pair-wise estimates of linkage disequilibrium (Abecasis and Cookson, 2000) between the remaining 9 CRHR1 SNPs in this study are shown by Haploview in Fig. 1. The SNPs showing the strongest association with the visual P3 amplitude and with alcohol dependence phenotypes (Table 2) are in strong linkage disequilibrium (LD) with one another (Fig. 1).
Fig. 1.
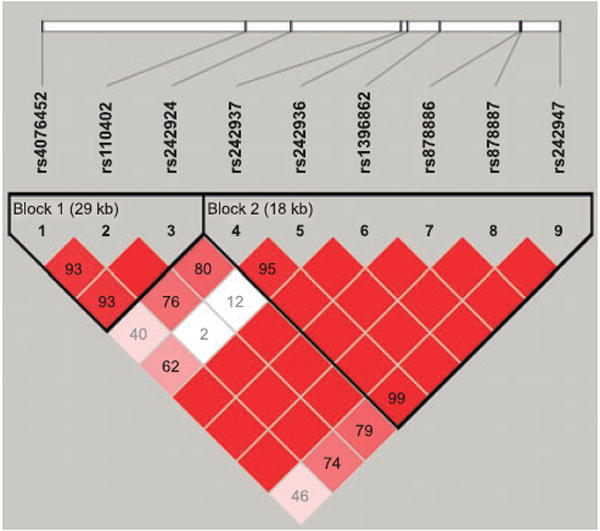
Pair-wise measures of linkage disequilibrium (LD) plot of the 9 CRHR1 SNPs based on subjects included in this study. The Lewontin’s standardized disequilibrium coefficient, D’, value of each SNP pair is shown in the squares. The numbers in the squares are D’ × 100. Empty squares indicate D’ = 1. Squares are colored bright red (dark gray) if the D’ value is high and the confidence in the value of D’ is high as well.
Table 2.
Data Represent the p-Values of the Results of the Total Family Based Association Test of QTDT With the P3 Amplitude of the Event-Related Potential in the Posterior Electrodes and the Results of FBAT With the DSM-IV Diagnoses of Alcohol Dependence.
| CRHR1 SNP | Position | Location | MAF | P7 | P3 | Pz | P4 | P8 | O1 | O2 | DSM-IV |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs4076452 | 41211660 | Upstream | 0.147 | 0.841 | 0.764 | 0.806 | 1 | 0.596 | 0.698 | 0.532 | 0.783 |
| rs110402 | 41235818 | intron 1 | 0.458 | 0.032 | 0.005 | 0.005 | 0.005 | 0.017 | 0.007 | 0.028 | 0.019 |
| rs242924 | 41241147 | intron 2 | 0.459 | 0.039 | 0.009 | 0.008 | 0.008 | 0.023 | 0.008 | 0.029 | 0.010 |
| rs242937 | 41254149 | intorn 3 | 0.411 | 0.080 | 0.067 | 0.059 | 0.039 | 0.104 | 0.037 | 0.083 | 0.003 |
| rs242936 | 41254990 | intron 4 | 0.095 | 0.887 | 1 | 0.887 | 0.718 | 0.764 | 0.862 | 0.841 | 0.539 |
| rs1396862 | 41258778 | intron 4 | 0.206 | 0.005 | 0.018 | 0.043 | 0.009 | 0.008 | 0.003 | 0.011 | 0.051 |
| rs878886 | 41268271 | Exon 13 untranslated | 0.204 | 0.007 | 0.026 | 0.068 | 0.018 | 0.008 | 0.004 | 0.017 | 0.041 |
| rs878887 | 41268363 | Exon 13 untranslated | 0.206 | 0.006 | 0.022 | 0.055 | 0.013 | 0.010 | 0.004 | 0.013 | 0.048 |
| rs242947 | 41272916 | Downstream | 0.428 | 0.047 | 0.070 | 0.060 | 0.031 | 0.054 | 0.035 | 0.054 | 0.005 |
Numbers in bold fond represent significant difference (p < 0.05). MAF, minor allele frequency.
Table 2 summarizes the results of the total association test of QTDT with the P3 amplitude of the event-related potential in the posterior electrodes and the results of FBAT with diagnoses of alcohol dependence. Multiple significant genetic associations (p < 0.05) were identified with the P3 amplitude of ERP and the SNPs in the introns 1, 2, 3, and 4, and untranslated regions of exon 13 of the CRHR1 gene. These positive results were not because of population stratification as examined by the quantitative transmission disequilibrium test (QTDT) program (Abecasis et al., 2000) (p-values for population stratification are all >0.05). Of note, most SNPs that were significantly associated with the quantitative trait phenotype (P3 amplitude) were also associated with alcohol dependence.
False discovery rate (FDR) was calculated to correct for multiple comparisons with the quantitative trait, visual P3 amplitude. We found 38 significant tests, and five false positives are expected under the traditional p-value cutoff criterion (0.05). If the p-value cutoff point is set to 0.01, it yields 17 significant tests, where only 1 positive result is expected by chance.
To confirm that the association between CRHR1 and P3 found in the current study was not attributed to the genetic association with alcohol-dependent subjects, which are known to have low P3 amplitude in previous studies, we performed an additional transmission disequilibrium test (TDT) on the nonaffected subjects only. The results (Table 3) demonstrated that the association between the variations of CRHR1 and P3 amplitude is independent of the state of alcohol dependence.
Table 3.
Data Represent the p-Values of the Results of the Family Based Association Test of TDT with the P3 Amplitude of the Event-Related Potential in the Posterior Electrodes in the Nonaffected Subjects Only.
| CRHR1 SNP | Position | Location | MAF | P7 | P3 | Pz | P4 | P8 | O1 | O2 |
|---|---|---|---|---|---|---|---|---|---|---|
| rs4076452 | 41211660 | Upstream | 0.159 | 0.964 | 0.067 | 0.694 | 0.648 | 0.382 | 0.622 | 0.360 |
| rs110402 | 41235818 | intron 1 | 0.452 | 0.006 | 0.000 | 0.000 | 0.000 | 0.004 | 0.007 | 0.012 |
| rs242924 | 41241147 | intron 2 | 0.452 | 0.010 | 0.001 | 0.000 | 0.000 | 0.009 | 0.012 | 0.019 |
| rs242937 | 41254149 | intorn 3 | 0.434 | 0.111 | 0.020 | 0.003 | 0.005 | 0.143 | 0.092 | 0.111 |
| rs242936 | 41254990 | intron 4 | 0.120 | 0.966 | 0.823 | 0.638 | 0.845 | 0.965 | 0.879 | 0.881 |
| rs1396862 | 41258778 | intron 4 | 0.235 | 0.026 | 0.013 | 0.013 | 0.009 | 0.045 | 0.048 | 0.074 |
| rs878886 | 41268271 | Exon 13 untranslated | 0.238 | 0.025 | 0.015 | 0.018 | 0.016 | 0.050 | 0.053 | 0.094 |
| rs878887 | 41268363 | Exon 13 untranslated | 0.235 | 0.024 | 0.012 | 0.012 | 0.009 | 0.047 | 0.043 | 0.068 |
| rs242947 | 41272916 | Downstream | 0.460 | 0.059 | 0.017 | 0.002 | 0.003 | 0.075 | 0.079 | 0.059 |
Numbers in bold fond represent significant difference (p < 0.05). MAF, minor allele frequency of the nonaffected subjects only.
The LD pattern as computed with Haploview (Fig. 1) indicates that all 9 SNPs are in fairly strong LD with each other. The positive SNP associations extend over both blocks; however, the LD figure displays two blocks based on a more conservative definition. Therefore, for haplotype analysis, we have derived haplotypes from all 9 SNPs as they appear to be within one haplotype block. Table 4 summarizes the haplotype frequencies (of those 8 haplotypes with frequency greater than 0.01) and the respective p-values resulted from the haplotype analyses with the continuous phenotypes of P3 amplitude in the seven posterior electrodes and with the dichotomous diagnostic phenotype of alcohol dependence by DSM-IV. The most common haplotype, H1 (121111112), is over-transmitted to individuals with reduced P3 amplitude (Table 4 and Fig. 2).
Table 4.
Data Represent the p-Values of the Results of the Haplotype Analyses With the P3 Amplitude of the Event-Related Potential in the Posterior Electrodes and With the DSM-IV Diagnoses of Alcohol Dependence.
| Haplotypes | SNP1 | SNP2 | SNP3 | SNP4 | SNP5 | SNP6 | SNP7 | SNP8 | SNP9 | Haplotype Frequency | P7 | P3 | Pz | P4 | P8 | O1 | O2 | DSM-IV |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 1 | 2 | 0.463 | 0.023 | 0.085 | 0.149 | 0.082 | 0.012 | 0.031 | 0.043 | 0.189 |
| H2 | 1 | 1 | 2 | 2 | 1 | 2 | 2 | 2 | 1 | 0.179 | 0.146 | 0.117 | 0.159 | 0.087 | 0.057 | 0.066 | 0.155 | 0.570 |
| H3 | 1 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 0.121 | 0.376 | 0.525 | 0.692 | 0.778 | 0.722 | 0.332 | 0.443 | 0.843 |
| H4 | 1 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0.064 | 0.661 | 0.612 | 0.166 | 0.163 | 0.891 | 0.672 | 0.743 | 0.120 |
| H5 | 2 | 1 | 2 | 2 | 2 | 1 | 1 | 1 | 1 | 0.047 | 0.495 | 0.750 | 0.953 | 0.977 | 0.373 | 0.505 | 0.714 | 0.066 |
| H6 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 1 | 0.040 | 0.180 | 0.207 | 0.228 | 0.229 | 0.744 | 0.165 | 0.879 | 0.361 |
| H7 | 2 | 1 | 2 | 1 | 1 | 1 | 1 | 1 | 2 | 0.035 | 0.990 | 0.676 | 0.654 | 0.771 | 0.975 | 0.677 | 0.770 | 0.767 |
| H8 | 1 | 2 | 1 | 2 | 2 | 1 | 1 | 1 | 1 | 0.022 | 0.916 | 0.772 | 0.636 | 0.641 | 0.705 | 0.886 | 0.644 | 0.305 |
Numbers in bold fond represent significant difference (p < 0.05). SNPs are allied with the positions shown in Table 2 (i.e., SNP1 = rs4076452, SNP2 = rs110402, etc.) 1 = allele 1; 2 = allele 2.
Fig. 2.
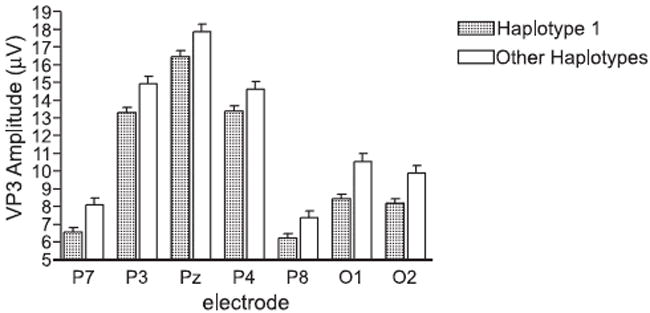
Mean target visual P3 amplitude at the seven posterior electrodes grouped by haplotypes. The bars represent the mean value; the vertical lines issuing from the bars represent the standard error of mean (SEM). Individual p-values are shown in Table 4.
DISCUSSION
The CRHR1 gene contains at least 14 exons spanning 20 kb of genomic DNA, and CRHR1 isoforms originate from the same gene by alternative splicing (Sakai et al., 1998). The findings presented here are the first to identify a significant association of the P3 amplitude, a quantitative trait endophenotype, and alcohol dependence with multiple SNPs from the introns and untranslated exons of the CRHR1 gene.
One of the important underlying mechanisms for developing alcohol abuse and relapse is negative reinforcement whereby alcohol reduces anxiety and other negative feelings, some of which occur in reaction to stress (Sinha, 2001). CRH (CRF) is well known to mediate unconditioned and conditioned anxiety response after exposure to stress, and maladaptive responses to stress increase alcohol consumption and relapse (Koob, 1999; Sinha, 2001; Zimmermann et al., 2004). Given the relationship among stress, alcohol abuse, and the CRH system, we hypothesized that CRHR1 is involved in the development of alcohol dependence as well as the abnormality of brain oscillations that have been shown to be associated with such dependence. In particular, a recent study demonstrated that both CRH and ethanol enhanced GABA-mediated neurotransmission in central amygdala neurons from wild-type and Crhr2 knockout mice, but not Crhr1 knockout mice (Nie et al., 2004). CRHR1 receptor (but not CRHR2) antagonists blocked both CRH and ethanol effects in wild-type mice (Nie et al., 2004). These results suggest a pivotal role of CRHR1 receptor in modulating ethanol enhancement of GABAergic synaptic transmission in the central amygdala and lead us to consider a role for the CRHR1 gene in modulating the brain oscillations and in the predisposition to alcohol dependence. Our hypothesis was further supported by several studies (Brazdil et al., 1999; Watanabe et al., 2002) in intracranial ERPs, which demonstrated that amygdala is one of the major subcortical structures in the generation of P3 component of the ERP. Evidence indicates that dysregulation of the GABA neurotransmitter system may be involved in the development of alcohol dependence, including the fact that polymorphisms in genes encoding GABA receptor subunits have been shown to be associated with brain oscillations (endophenotypes related to alcohol dependence) as well as with the diagnosis of alcohol dependence (Dick et al., 2004; Edenberg et al., 2004; Porjesz et al., 2002, 2004). In addition, medications that target the GABAA receptor system, including benzodiazepines, are often used in the detoxification and treatment of patients who are prone to withdrawal after alcohol binge drinking (Kosten and O’Connor, 2003). The interaction with the GABAergic system may in part explain the mechanisms by which the CRHR1 receptor or the HPA system regulates the brain electrophysiology associated with alcohol dependence (P3 for example). Based on this evidence, we theorized that polymorphisms in the CRHR1 gene that alter the receptor’s actions on GABAergic neurons in response to alcohol contribute to the variation of the endophenotype P3 amplitude of the ERP and to the vulnerability to development of alcohol dependence.
A recent study in individuals of European ancestry who suffer from alcohol dependence and children of high risk failed to detect any significant associations between individual SNPs or haplotypes in the CRHR1 gene with the alcohol-dependent phenotype by the DSM-IV criteria (Treutlein et al., 2006). However, in the same study, an association was found between two SNPs, rs242938 in intron 3 and rs1876831 in intron 6 regions of the CRHR1 gene, and lifetime binge drinking and lifetime prevalence of drunkenness. In two other recent studies on individuals of European ancestry (Dahl et al., 2005; Soyka et al., 2004), no association was found between CRHR1 SNPs and alcohol dependence. Of note, these earlier mentioned studies tested a relatively small population, ranging from 120 to 299 alcohol-dependent individuals with a similar number of unaffected subjects. Two of these three studies analyzed only the clinical diagnosis of alcohol dependence by DSM-IV. Only the Treutlein and colleagues (2006) study, which also had the largest sample size of 299 alcohol-dependent individuals, further characterized the specific patterns of alcohol consumption with which an association of two SNPs in the CRHR1 gene was found. Only two of the SNPs tested in those three studies correspond to the 10 SNPs in our study; rs1396862 in intron 4 and rs878887 in the un-translated region of Exon 13 were included in the studies by Dahl and colleagues (2005) and by Treutlein and colleagues (2006). In addition to the heterogeneity of alcohol dependence, these factors may also contribute to the reasons why these three recent studies failed to identify significant association between SNPs in CRHR1 and alcohol dependence.
Following the identification of associations of two SNPs in intron 3 and intron 6 regions of CRHR1 with binge drinking in adolescents and with high amount of drinking in adults but not with alcohol dependence diagnosis (Treutlein et al., 2006), the same research group recently investigated the interaction between these two SNPs in CRHR1 and negative stressful life events in adolescents at age 15, in an attempt to understand the mechanisms underlying the gene-environment interaction and alcohol drinking (Blomeyer et al., 2008). Significant interactions between one SNP, rs1876831 in intron 6, and life stress on heavy drinking were found, revealing an effect of negative life events on heavy drinking only among individuals carrying a particular genotype of CRHR1. The findings are parallel to recent reports of significant associations of the short allele of the serotonin transporter gene 5-HTTLPR with depression under life stress (Caspi et al., 2003; Dick et al., 2007). For alcohol dependence, some studies (e.g., Parsian and Cloninger, 2001) have reported association with the long allele of the 5-HTTLPR polymorphism, others have found association with the short allele (e.g., Thompson et al., 2000), whereas other studies have found no association with either allele (e.g., Dick et al., 2007; Edenberg et al., 1998). A recent meta-analysis reported that the short allele was significantly associated with alcohol dependence with an odds ratio of 1.18, suggesting a gene of small effect (Feinn et al., 2005). Given the high co-occurrence of depression with alcohol dependence and the influence of stress on both disorders, we speculated a similar moderating effect of CRHR1 genotype on the interaction with depression. We performed additional genetic association tests in the same 1049 subjects with depression (by DSM-IV) only, alcohol dependence and depression, using the same 10 SNPs in the CRHR1 gene we had tested. No significant association (p < 0.05) was found (data not shown). If the clinical diagnosis phenotype tested was “alcohol dependence or depression” (both by DSM-IV criteria), only 4 SNPs that we tested from the CRHR1 gene, rs242924, rs1396862, rs878886, rs878887, resulted in statistical significance (p < 0.05) in the genetic association test. It is plausible that there are different gene variations or different combination of polymorphisms in the CRHR1 gene that moderate the development of depression and alcohol dependence after stressful life events.
In conclusion, the present study demonstrates that variations in the CRHR1 gene are associated with the visual P3 amplitude of the event-related potential and with alcohol dependence. Our findings provide further evidence that brain electrophysiological measures evoked under cognitive conditions as endophenotypes in combination with genetic and other neurobiological information show great promise in deciphering the interaction of the subsystems involved in the pathophysiology of complex neuropsychiatric diseases. We expect that the identification of genes that regulate cognitive processes will be of enormous benefit to the field of psychiatric genetics.
Acknowledgments
The Collaborative Study on the Genetics of Alcoholism (COGA), Co-Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes nine different centers where data collection, analysis, and storage take place. The nine sites and Principal Investigators and Co-Investigators are: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St Louis (L. Bierut, A. Goate, J. Rice); University of California at San Diego (M. Schuckit); Howard University (R. Taylor); Rutgers University (J. Tischfield); Southwest Foundation (L. Almasy). Max Guo serves as the NIAAA Staff Collaborator. This national collaborative study is supported by the NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA). Dr. Andrew C. Chen received support from the American Psychiatric Association/American Psychiatric Institute for Research and Education PMRTP Award.
In memory of Henri Begleiter and Theodore Reich, Principal and Co-Principal Investigators of COGA since its inception; we are indebted to their leadership in the establishment and nurturing of COGA, and acknowledge with great admiration their seminal scientific contributions to the field.
Footnotes
Previous presentation: This work was presented in the 46th annual meeting of the American College of Neuropsychopharmacology (ACNP), December 9–13, 2007 in Boca Raton, Florida.
References
- Abecasis GR, Cookson WO. GOLD–graphical overview of linkage disequilibrium. Bioinformatics. 2000;16(2):182–183. doi: 10.1093/bioinformatics/16.2.182. [DOI] [PubMed] [Google Scholar]
- Abecasis GR, Cookson WO, Cardon LR. Pedigree tests of transmission disequilibrium. Eur J Hum Genet. 2000;8(7):545–551. doi: 10.1038/sj.ejhg.5200494. [DOI] [PubMed] [Google Scholar]
- Almasy L, Blangero J. Multipoint quantitative-trait linkage analysis in general pedigrees. Am J Hum Genet. 1998;62(5):1198–1211. doi: 10.1086/301844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- American Electroencephalographic Association. American Electroencephalographic Society guidelines for standard electrode position nomenclature. J Clin Neurophysiol. 1991;8:200–202. [PubMed] [Google Scholar]
- Barr CS, Dvoskin RL, Yuan Q, Lipsky RH, Gupte M, Hu X, Zhou Z, Schwandt ML, Lindell SG, McKee M, Becker ML, Kling MA, Gold PW, Higley D, Heilig M, Suomi SJ, Goldman D. CRH haplotype as a factor influencing cerebrospinal fluid levels of corticotropin-releasing hormone, hypothalamic-pituitary-adrenal axis activity, temperament, and alcohol consumption in rhesus macaques. Arch Gen Psychiatry. 2008;65(8):934–944. doi: 10.1001/archpsyc.65.8.934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Reich T, Edenberg HJ, Goate A, Blangero J, Almasy L, Foroud T, Van Eerdewegh P, Polich J, Rohrbaugh J, Kuperman S, Bauer LO, O’Connor SJ, Chorlian DB, Li TK, Conneally PM, Hesselbrock V, Rice JP, Schuckit MA, Cloninger R, Nurnberger J, Jr, Crowe R, Bloom FE. Quantitative trait loci analysis of human event-related brain potentials: P3 voltage. Electroencephalogr Clin Neurophysiol. 1998;108(3):244–250. doi: 10.1016/s0168-5597(98)00002-1. [DOI] [PubMed] [Google Scholar]
- Begleiter H, Porjesz B, Wang W. Event-related brain potentials differentiate priming and recognition to familiar and unfamiliar faces. Electroencephalogr Clin Neurophysiol. 1995;94(1):41–49. doi: 10.1016/0013-4694(94)00240-l. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. J R Stat Soc Series B Stat Methodol. 1995;57(1):289–300. [Google Scholar]
- Bittencourt JC, Sawchenko PE. Do centrally administered neuropeptides access cognate receptors?: an analysis in the central corticotropin-releasing factor system. J Neurosci. 2000;20(3):1142–1156. doi: 10.1523/JNEUROSCI.20-03-01142.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blomeyer D, Treutlein J, Esser G, Schmidt MH, Schumann G, Laucht M. Interaction between CRHR1 Gene and Stressful Life Events Predicts Adolescent Heavy Alcohol Use. Biol Psychiatry. 2008;63(2):146–151. doi: 10.1016/j.biopsych.2007.04.026. [DOI] [PubMed] [Google Scholar]
- Boehnke M. Allele frequency estimation from data on relatives. Am J Hum Genet. 1991;48(1):22–25. [PMC free article] [PubMed] [Google Scholar]
- Boerwinkle E, Chakraborty R, Sing CF. The use of measured genotype information in the analysis of quantitative phenotypes in man. I. Models and analytical methods. Ann Hum Genet. 1986;50(Pt 2):181–194. doi: 10.1111/j.1469-1809.1986.tb01037.x. [DOI] [PubMed] [Google Scholar]
- Brazdil M, Rektor I, Dufek M, Daniel P, Jurak P, Kuba R. The role of frontal and temporal lobes in visual discrimination task-depth ERP studies. Neurophysiol Clin. 1999;29(4):339–350. doi: 10.1016/s0987-7053(99)90047-3. [DOI] [PubMed] [Google Scholar]
- Bucholz KK, Cadoret R, Cloninger CR, Dinwiddie SH, Hesselbrock VM, Nurnberger JI, Jr, Reich T, Schmidt I, Schuckit MA. A new, semi-structured psychiatric interview for use in genetic linkage studies: a report on the reliability of the SSAGA. J Stud Alcohol. 1994;55(2):149–158. doi: 10.15288/jsa.1994.55.149. [DOI] [PubMed] [Google Scholar]
- Caspi A, Sugden K, Moffitt TE, Taylor A, Craig IW, Harrington H, McClay J, Mill J, Martin J, Braithwaite A, Poulton R. Influence of life stress on depression: moderation by a polymorphism in the 5-HTT gene. Science. 2003;301(5631):386–389. doi: 10.1126/science.1083968. [DOI] [PubMed] [Google Scholar]
- Chen AC, Porjesz B, Rangaswamy M, Kamarajan C, Tang Y, Jones KA, Chorlian DB, Stimus AT, Begleiter H. Reduced frontal lobe activity in subjects with high impulsivity and alcoholism. Alcohol Clin Exp Res. 2007;31(1):156–165. doi: 10.1111/j.1530-0277.2006.00277.x. [DOI] [PubMed] [Google Scholar]
- Chen AC, Tang Y, Rangaswamy M, Wang JC, Almasy L, Foroud T, Edenberg HJ, Hesselbrock V, Nurnberger J, Jr, Kuperman S, O’Connor SJ, Schuckit MA, Bauer LO, Tischfield J, Rice JP, Bierut L, Goate A, Porjesz B. Association of single nucleotide polymorphisms in a glutamate receptor gene (GRM8) with theta power of event-related oscillations and alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):359–368. doi: 10.1002/ajmg.b.30818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl JP, Doyle GA, Oslin DW, Buono RJ, Ferraro TN, Lohoff FW, Berrettini WH. Lack of association between single nucleotide polymorphisms in the corticotropin releasing hormone receptor 1 (CRHR1) gene and alcohol dependence. J Psychiatr Res. 2005;39(5):475–479. doi: 10.1016/j.jpsychires.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Dick DM, Edenberg HJ, Xuei X, Goate A, Kuperman S, Schuckit M, Crowe R, Smith TL, Porjesz B, Begleiter H, Foroud T. Association of GABRG3 with alcohol dependence. Alcohol Clin Exp Res. 2004;28(1):4–9. doi: 10.1097/01.ALC.0000108645.54345.98. [DOI] [PubMed] [Google Scholar]
- Dick DM, Plunkett J, Hamlin D, Nurnberger J, Jr, Kuperman S, Schuckit M, Hesselbrock V, Edenberg H, Bierut L. Association analyses of the serotonin transporter gene with lifetime depression and alcohol dependence in the Collaborative Study on the Genetics of Alcoholism (COGA) sample. Psychiatr Genet. 2007;17(1):35–38. doi: 10.1097/YPG.0b013e328011188b. [DOI] [PubMed] [Google Scholar]
- Edenberg HJ, Dick DM, Xuei X, Tian H, Almasy L, Bauer LO, Crowe RR, Goate A, Hesselbrock V, Jones K, Kwon J, Li TK, Nurnberger JI, Jr, O’Connor SJ, Reich T, Rice J, Schuckit MA, Porjesz B, Foroud T, Begleiter H. Variations in GABRA2, encoding the alpha 2 subunit of the GABA(A) receptor, are associated with alcohol dependence and with brain oscillations. Am J Hum Genet. 2004;74(4):705–714. doi: 10.1086/383283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edenberg HJ, Reynolds J, Koller DL, Begleiter H, Bucholz KK, Conneally PM, Crowe R, Goate A, Hesselbrock V, Li TK, Nurnberger JI, Jr, Porjesz B, Reich T, Rice JP, Schuckit M, Tischfield JA, Foroud T. A family-based analysis of whether the functional promoter alleles of the serotonin transporter gene HTT affect the risk for alcohol dependence. Alcohol Clin Exp Res. 1998;22(5):1080–1085. [PubMed] [Google Scholar]
- Enoch MA, Shen PH, Ducci F, Yuan Q, Liu J, White KV, Albaugh B, Hodgkinson CA, Goldman D. Common genetic origins for EEG, alcoholism and anxiety: the role of CRH-BP. PLoS ONE. 2008;3(10):e3620. doi: 10.1371/journal.pone.0003620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinn R, Nellissery M, Kranzler HR. Meta-analysis of the association of a functional serotonin transporter promoter polymorphism with alcohol dependence. Am J Med Genet B Neuropsychiatr Genet. 2005;133(1):79–84. doi: 10.1002/ajmg.b.30132. [DOI] [PubMed] [Google Scholar]
- Folstein MF, Folstein SE, McHugh PR. “Mini-mental state” A practical method for grading the cognitive state of patients for the clinician. J Psychiatr Res. 1975;12(3):189–198. doi: 10.1016/0022-3956(75)90026-6. [DOI] [PubMed] [Google Scholar]
- Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, Li TK, Hesselbrock V, Crowe R, Schuckit M, Porjesz B, Begleiter H, Reich T. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24(7):933–945. [PubMed] [Google Scholar]
- Funk CK, Zorrilla EP, Lee MJ, Rice KC, Koob GF. Corticotropin-releasing factor 1 antagonists selectively reduce ethanol self-administration in ethanol-dependent rats. Biol Psychiatry. 2007;61(1):78–86. doi: 10.1016/j.biopsych.2006.03.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D. The structure of haplotype blocks in the human genome. Science. 2002;296(5576):2225–2229. doi: 10.1126/science.1069424. [DOI] [PubMed] [Google Scholar]
- Gottesman II, Gould TD. The endophenotype concept in psychiatry: etymology and strategic intentions. Am J Psychiatry. 2003;160(4):636–645. doi: 10.1176/appi.ajp.160.4.636. [DOI] [PubMed] [Google Scholar]
- Hansson AC, Cippitelli A, Sommer WH, Fedeli A, Bjork K, Soverchia L, Terasmaa A, Massi M, Heilig M, Ciccocioppo R. Variation at the rat Crhr1 locus and sensitivity to relapse into alcohol seeking induced by environmental stress. Proc Natl Acad Sci USA. 2006;103(41):15236–15241. doi: 10.1073/pnas.0604419103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs SC, Menzaghi F, Pich EM, Baldwin HA, Rassnick S, Britton KT, Koob GF. Anti-stress action of a corticotropin-releasing factor antagonist on behavioral reactivity to stressors of varying type and intensity. Neuropsychopharmacology. 1994;11(3):179–186. doi: 10.1038/sj.npp.1380104. [DOI] [PubMed] [Google Scholar]
- Hesselbrock V, Begleiter H, Porjesz B, O’Connor S, Bauer L. P300 event-related potential amplitude as an endophenotype of alcoholism–evidence from the collaborative study on the genetics of alcoholism. J Biomed Sci. 2001;8(1):77–82. doi: 10.1007/BF02255974. [DOI] [PubMed] [Google Scholar]
- Hesselbrock M, Easton C, Bucholz KK, Schuckit M, Hesselbrock V. A validity study of the SSAGA–a comparison with the SCAN. Addiction. 1999;94(9):1361–1370. doi: 10.1046/j.1360-0443.1999.94913618.x. [DOI] [PubMed] [Google Scholar]
- Ising M, Zimmermann US, Kunzel HE, Uhr M, Foster AC, Learned-Coughlin SM, Holsboer F, Grigoriadis DE. High-Affinity CRF(1) Receptor Antagonist NBI-34041: Preclinical and Clinical Data Suggest Safety and Efficacy in Attenuating Elevated Stress Response. Neuropsychopharmacology. 2007;32(9):1941–1949. doi: 10.1038/sj.npp.1301328. [DOI] [PubMed] [Google Scholar]
- Jones KA, Porjesz B, Almasy L, Bierut L, Goate A, Wang JC, Dick DM, Hinrichs A, Kwon J, Rice JP, Rohrbaugh J, Stock H, Wu W, Bauer LO, Chorlian DB, Crowe RR, Edenberg HJ, Foroud T, Hesselbrock V, Kuperman S, Nurnberger J, Jr, O’Connor SJ, Schuckit MA, Stimus AT, Tischfield JA, Reich T, Begleiter H. Linkage and linkage disequilibrium of evoked EEG oscillations with CHRM2 receptor gene polymorphisms: implications for human brain dynamics and cognition. Int J Psychophysiol. 2004;53(2):75–90. doi: 10.1016/j.ijpsycho.2004.02.004. [DOI] [PubMed] [Google Scholar]
- Kamarajan C, Porjesz B, Jones KA, Choi K, Chorlian DB, Padmanabhapillai A, Rangaswamy M, Stimus AT, Begleiter H. Alcoholism is a disinhibitory disorder: neurophysiological evidence from a Go/No-Go task. Biol Psychol. 2005;69(3):353–373. doi: 10.1016/j.biopsycho.2004.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsanis J, Iacono WG, McGue MK, Carlson SR. P300 event-related potential heritability in monozygotic and dizygotic twins. Psychophysiology. 1997;34(1):47–58. doi: 10.1111/j.1469-8986.1997.tb02415.x. [DOI] [PubMed] [Google Scholar]
- Koob GF. Stress, corticotropin-releasing factor, and drug addiction. Ann N Y Acad Sci. 1999;897:27–45. doi: 10.1111/j.1749-6632.1999.tb07876.x. [DOI] [PubMed] [Google Scholar]
- Kosten TR, O’Connor PG. Management of drug and alcohol withdrawal. N Engl J Med. 2003;348(18):1786–1795. doi: 10.1056/NEJMra020617. [DOI] [PubMed] [Google Scholar]
- Lange C, Laird NM. Power calculations for a general class of family-based association tests: dichotomous traits. Am J Hum Genet. 2002;71(3):575–584. doi: 10.1086/342406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long JC, Williams RC, Urbanek M. An E-M algorithm and testing strategy for multiple-locus haplotypes. Am J Hum Genet. 1995;56(3):799–810. [PMC free article] [PubMed] [Google Scholar]
- Lu L, Liu D, Ceng X. Corticotropin-releasing factor receptor type 1 mediates stress-induced relapse to cocaine-conditioned place preference in rats. Eur J Pharmacol. 2001;415(2-3):203–208. doi: 10.1016/s0014-2999(01)00840-8. [DOI] [PubMed] [Google Scholar]
- Merlo Pich E, Lorang M, Yeganeh M, Rodriguez de Fonseca F, Raber J, Koob GF, Weiss F. Increase of extracellular corticotropin-releasing factor-like immunoreactivity levels in the amygdala of awake rats during restraint stress and ethanol withdrawal as measured by microdialysis. J Neurosci. 1995;15(8):5439–5447. doi: 10.1523/JNEUROSCI.15-08-05439.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie Z, Schweitzer P, Roberts AJ, Madamba SG, Moore SD, Siggins GR. Ethanol augments GABAergic transmission in the central amygdala via CRF1 receptors. Science. 2004;303(5663):1512–1514. doi: 10.1126/science.1092550. [DOI] [PubMed] [Google Scholar]
- O’Connor S, Morzorati S, Christian JC, Li TK. Heritable features of the auditory oddball event-related potential: peaks, latencies, morphology and topography. Electroencephalogr Clin Neurophysiol. 1994;92(2):115–125. doi: 10.1016/0168-5597(94)90052-3. [DOI] [PubMed] [Google Scholar]
- Overstreet DH, Keeney A, Hogg S. Antidepressant effects of citalopram and CRF receptor antagonist CP-154,526 in a rat model of depression. Eur J Pharmacol. 2004;492(2-3):195–201. doi: 10.1016/j.ejphar.2004.04.010. [DOI] [PubMed] [Google Scholar]
- Parsian A, Cloninger CR. Serotonergic pathway genes and subtypes of alcoholism: association studies. Psychiatr Genet. 2001;11(2):89–94. doi: 10.1097/00041444-200106000-00006. [DOI] [PubMed] [Google Scholar]
- Porjesz B, Almasy L, Edenberg HJ, Wang K, Chorlian DB, Foroud T, Goate A, Rice JP, O’Connor SJ, Rohrbaugh J, Kuperman S, Bauer LO, Crowe RR, Schuckit MA, Hesselbrock V, Conneally PM, Tischfield JA, Li TK, Reich T, Begleiter H. Linkage disequilibrium between the beta frequency of the human EEG and a GABAA receptor gene locus. Proc Natl Acad Sci USA. 2002;99(6):3729–3733. doi: 10.1073/pnas.052716399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porjesz B, Begleiter H, Reich T, Van Eerdewegh P, Edenberg HJ, Foroud T, Goate A, Litke A, Chorlian DB, Stimus A, Rice J, Blangero J, Almasy L, Sorbell J, Bauer LO, Kuperman S, O’Connor SJ, Rohrbaugh J. Amplitude of visual P3 event-related potential as a phenotypic marker for a predisposition to alcoholism: preliminary results from the COGA Project. Collaborative Study on the Genetics of Alcoholism. Alcohol Clin Exp Res. 1998;22(6):1317–1323. [PubMed] [Google Scholar]
- Porjesz B, Jones K, Begleiter H. The genetics of oscillations in the human brain. Suppl Clin Neurophysiol. 2004;57:441–449. doi: 10.1016/s1567-424x(09)70382-4. [DOI] [PubMed] [Google Scholar]
- Porjesz B, Rangaswamy M, Kamarajan C, Jones KA, Padmanabhapillai A, Begleiter H. The utility of neurophysiological markers in the study of alcoholism. Clin Neurophysiol. 2005;116(5):993–1018. doi: 10.1016/j.clinph.2004.12.016. [DOI] [PubMed] [Google Scholar]
- Rabinowitz D, Laird N. A unified approach to adjusting association tests for population admixture with arbitrary pedigree structure and arbitrary missing marker information. Hum Hered. 2000;50(4):211–223. doi: 10.1159/000022918. [DOI] [PubMed] [Google Scholar]
- Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, Porjesz B, Li TK, Conneally PM, Nurnberger JI, Jr, Tischfield JA, Crowe RR, Cloninger CR, Wu W, Shears S, Carr K, Crose C, Willig C, Begleiter H. Genomewide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81(3):207–215. [PubMed] [Google Scholar]
- Richter RM, Weiss F. In vivo CRF release in rat amygdala is increased during cocaine withdrawal in self-administering rats. Synapse. 1999;32(4):254–261. doi: 10.1002/(SICI)1098-2396(19990615)32:4<254::AID-SYN2>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- Sakai K, Yamada M, Horiba N, Wakui M, Demura H, Suda T. The genomic organization of the human corticotropin-releasing factor type-1 receptor. Gene. 1998;219(12):125–130. doi: 10.1016/s0378-1119(98)00322-9. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Gold E, Risch C. Plasma cortisol levels following ethanol in sons of alcoholics and controls. Arch Gen Psychiatry. 1987;44(11):942–945. doi: 10.1001/archpsyc.1987.01800230022005. [DOI] [PubMed] [Google Scholar]
- Schuckit MA, Risch SC, Gold EO. Alcohol consumption, ACTH level, and family history of alcoholism. Am J Psychiatry. 1988;145(11):1391–1395. doi: 10.1176/ajp.145.11.1391. [DOI] [PubMed] [Google Scholar]
- Sillaber I, Rammes G, Zimmermann S, Mahal B, Zieglgansberger W, Wurst W, Holsboer F, Spanagel R. Enhanced and delayed stress-induced alcohol drinking in mice lacking functional CRH1 receptors. Science. 2002;296(5569):931–933. doi: 10.1126/science.1069836. [DOI] [PubMed] [Google Scholar]
- Sinha R. How does stress increase risk of drug abuse and relapse? Psychopharmacology (Berl) 2001;158(4):343–359. doi: 10.1007/s002130100917. [DOI] [PubMed] [Google Scholar]
- Soyka M, Preuss UW, Koller G, Zill P, Hesselbrock V, Bondy B. No association of CRH1 receptor polymorphism haplotypes, harm avoidance and other personality dimensions in alcohol dependence: results from the Munich gene bank project for alcoholism. Addict Biol. 2004;9(1):73–79. doi: 10.1080/13556210410001674121. [DOI] [PubMed] [Google Scholar]
- Spielman RS, Ewens WJ. The TDT and other family-based tests for linkage disequilibrium and association. Am J Hum Genet. 1996;59(5):983–989. [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100(16):9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson MD, Gonzalez N, Nguyen T, Comings DE, George SR, O’Dowd BF. Serotonin transporter gene polymorphisms in alcohol dependence. Alcohol. 2000;22(2):61–67. doi: 10.1016/s0741-8329(00)00105-1. [DOI] [PubMed] [Google Scholar]
- Timpl P, Spanagel R, Sillaber I, Kresse A, Reul JM, Stalla GK, Blanquet V, Steckler T, Holsboer F, Wurst W. Impaired stress response and reduced anxiety in mice lacking a functional corticotropin-releasing hormone receptor 1. Nat Genet. 1998;19(2):162–166. doi: 10.1038/520. [DOI] [PubMed] [Google Scholar]
- Treutlein J, Kissling C, Frank J, Wiemann S, Dong L, Depner M, Saam C, Lascorz J, Soyka M, Preuss UW, Rujescu D, Skowronek MH, Rietschel M, Spanagel R, Heinz A, Laucht M, Mann K, Schumann G. Genetic association of the human corticotropin releasing hormone receptor 1 (CRHR1) with binge drinking and alcohol intake patterns in two independent samples. Mol Psychiatry. 2006;11(6):594–602. doi: 10.1038/sj.mp.4001813. [DOI] [PubMed] [Google Scholar]
- Valdez GR, Roberts AJ, Chan K, Davis H, Brennan M, Zorrilla EP, Koob GF. Increased ethanol self-administration and anxiety-like behavior during acute ethanol withdrawal and protracted abstinence: regulation by corticotropin-releasing factor. Alcohol Clin Exp Res. 2002;26(10):1494–1501. doi: 10.1097/01.ALC.0000033120.51856.F0. [DOI] [PubMed] [Google Scholar]
- Wand GS, Mangold D, Ali M, Giggey P. Adrenocortical responses and family history of alcoholism. Alcohol Clin Exp Res. 1999;23(7):1185–1190. [PubMed] [Google Scholar]
- Watanabe N, Hirai N, Maehara T, Kawai K, Shimizu H, Miwakeichi F, Uchida S. The relationship between the visually evoked P300 event-related potential and gamma band oscillation in the human medial and basal temporal lobes: an electrocorticographic study. Neurosci Res. 2002;44(4):421–427. doi: 10.1016/s0168-0102(02)00159-1. [DOI] [PubMed] [Google Scholar]
- Westphal NJ, Seasholtz AF. CRH-BP: the regulation and function of a phylogenetically conserved binding protein. Front Biosci. 2006;11:1878–1891. doi: 10.2741/1931. [DOI] [PubMed] [Google Scholar]
- de Wit H, Soderpalm AH, Nikolayev L, Young E. Effects of acute social stress on alcohol consumption in healthy subjects. Alcohol Clin Exp Res. 2003;27(8):1270–1277. doi: 10.1097/01.ALC.0000081617.37539.D6. [DOI] [PubMed] [Google Scholar]
- Zimmermann U, Spring K, Kunz-Ebrecht SR, Uhr M, Wittchen HU, Holsboer F. Effect of ethanol on hypothalamic-pituitary-adrenal system response to psychosocial stress in sons of alcohol-dependent fathers. Neuropsychopharmacology. 2004;29(6):1156–1165. doi: 10.1038/sj.npp.1300395. [DOI] [PubMed] [Google Scholar]


