Abstract
An association between the gram-positive anaerobe Filifactor alocis and periodontal disease has recently emerged; however, possible pathogenic mechanisms have not been investigated. In this study we examined the responses of primary cultures of gingival epithelial cells (GECs) to infection with F. alocis. Secretion of the proinflammatory cytokines IL-1β, IL-6 and TNF-α from GECs was stimulated by F. alocis infection. F. alocis also induced apoptosis in GECs through pathways that involved caspase-3 but not caspase-9. Apoptosis was coincident with inhibition of MEK (MAPK kinase) activation. These results show that F. alocis has characteristics in common with established periodontal pathogens and has the potential to contribute to periodontal tissue destruction.
Keywords: periodontal pathogens, virulence, periodontal disease, cytokine, apoptosis
Introduction
Periodontitis, one of the most prevalent diseases throughout the world (Brown et al., 2002; Albandar, 2011), is a chronic bacterial inflammatory infection leading to destruction of the periodontal tissue, and culminating in alveolar bone loss and exfoliation of the teeth. Until recently, research into the etiology of periodontal disease has focused primarily on a small group of bacteria that can be recovered in high numbers from periodontal lesions. On the basis of association, Socransky et al. (1998) proposed that Porphyromonas gingivalis, Tannerella forsythia and Treponema denticola, designated the red complex, were the primary pathogens, with orange complex organisms, including Fusobacterium nucleatum, Prevotella intermedia and Campylobacter rectus, also contributing to disease to a lesser degree. A large number of studies have since revealed the pathogenic properties of these organisms, along with the nature of protective and destructive host responses (Lamont and Jenkinson, 1998; Holt and Ebersole, 2005; Feng and Weinberg, 2006; Frederick et al., 2011; Sharma, 2010; Darveau, 2010).
Approximately 300 bacterial species from the oral cavity have been isolated in culture and formally named; however it is estimated that less than half of the bacterial species present in the oral cavity can be readily cultivated (Wade, 2011). The development of culture-independent techniques such as 16S rRNA sequencing and high throughput sequencing, that allow for the identification of bacterial species directly from DNA, has led to a broader understanding of the diversity of bacterial species present in the oral environment (Dewhirst et al., 2010; Wade, 2011; Griffen et al., 2011). Indeed, a recent study utilizing 16S rRNA sequencing found over 1000 phylogenetically different taxa in the oral cavity, around 400 of which were novel (Dewhirst et al., 2010).
Molecular methods of bacterial identification have facilitated the ability to identify previously overlooked bacteria associated with periodontal disease. One such organism is Filifactor alocis, a gram-positive anaerobic rod. First isolated in 1985 from the gingival sulcus, the bacterium was taxonomically classified as Fusobacterium alocis (Cato et al., 1985), with later phylogenetic analysis leading to its reassignment to Filifactor in 1999 (Jalava and Eerola, 1999). Although cultivable, this organism is slow growing and difficult to detect by conventional culture-based methodologies. However, through molecular approaches it is becoming increasingly apparent that the presence of F. alocis is indicative of a number of oral diseases including caries, endodontic infections and periodontal disease. F. alocis is weakly glycolytic, and children with caries have been shown to have elevated levels of F. alocis in plaque (Dahlen et al., 2010). F. alocis is among the most commonly detected taxa in sites of endodontic infection (Siqueira and Rocas, 2004; Sakamoto et al., 2006), and F. alocis is present in the root canals of teeth with primary apical periodontitis (Siqueira et al., 2009), and in periapical lesions of root filled teeth (Gomes et al., 2008). Several studies have found F. alocis at increased frequency and in higher numbers in periodontal disease sites compared to healthy sites, leading to the proposal that F. alocis should be included as a diagnostic indicator of disease (Kumar et al., 2006; Kumar et al., 2005; Dahlen and Leonhardt, 2006, Colombo et al., 2009). Thus, there is a growing body of evidence supporting the notion that F. alocis may be a key causative agent in the development of oral diseases.
A recent study reported F. alocis forms biofilms in vivo, preferentially colonizing the apical parts of the gingival pocket in close proximity to the soft tissues (Schlafer et al., 2010). We hypothesized, therefore, that F. alocis would exert an influence on gingival epithelial cells that was consistent with the characteristics of a periodontal pathogen. Hence we investigated the ability of F. alocis to induce proinflammatory cytokine secretion and apoptotic cell death in gingival epithelial cells. F. alocis infection leads to the secretion of IL-1β, IL-6 and TNF-α from gingival epithelial cells, and eventually causes apoptotic cell death. Our results begin to establish pathogenic credentials for F. alocis and support a role for the organism in the etiology of periodontal disease.
Materials and Methods
Bacterial and eukaryotic cell culture
F. alocis strain ATCC 38596 and was routinely cultured anaerobically at 37°C on Brucella agar plates containing hemin and menadione (Sigma) and supplemented with 5% sheep’s blood. Primary cultures of gingival epithelial cells (GECs) were generated as described previously (Mao et al., 2007). Briefly, healthy gingival tissue was collected from patients undergoing surgery for removal of impacted third molars and following Institutional Review Board Guidelines. Basal epithelial cells were separated and cultured in Keratinocyte Growth Medium (DermaLife Basal Medium; Lifeline) in the absence of antibiotics. Eukaryotic cells were cultured at 37°C in 5% CO2.
Confocal microscopy
For examination of F. alocis GEC association, GECs were seeded at 1 × 105 cells on glass coverslips in 12-well plates and grown until ≈40% confluent. Cells were infected with Syto 17 (Invitrogen) labeled F. alocis at MOI 20 for 1 h. Coverslips were washed 4 times in phosphate-buffered saline (PBS) and fixed for 10 min in 4% paraformaldehyde. Following a 20 min block in 10% goat serum, actin was labeled using 1:100 FITC-phalloidin (Sigma) for 40 min at room temperature. After 4 washes in PBS, coverslips were mounted using ProLong Gold with DAPI mounting medium (Invitrogen). Images were acquired on an Olympus DSU Spinning Disk Confocal Scanner mounted on an Olympus IX81 inverted microscope, using a 60x water immersion objective. Z-stacks were obtained (1 μm between layers, 20 layers/stack from base to top of cells) through the z-axis of cells (3 z-stacks/coverslip), and numbers of associated F. alocis/cell were enumerated using means of bacteria associated with ≈50 cells/assay (3 coverslips/group, 3 z-stacks/coverslip, average of 6 GECs/field).
For apoptosis assays, GECs were cultured on glass coverslips until ≈40% confluent and infected with F. alocis at MOI 100 for 24 h. Coverslips were washed 4 times in phosphate-buffered saline (PBS) and fixed for 10 min in 4% paraformaldehyde. Permeabilization was with 0.2% Triton X-100 for 10 min at room temperature, prior to blocking in 10% goat serum for 20 min. Caspases were detected by reacting with primary active caspase-3 or caspase-9 antibodies (Sigma) at 1:100 dilution for 1 h, followed by Alexa-647-conjugated anti-rabbit secondary antibody (1:200) for 1 h in the dark. After 4 washes in PBS, coverslips were mounted using ProLong Gold with DAPI mounting medium (Invitrogen). Images were acquired on a Leica DM IRE2 inverted fluorescent microscope, with a Leica TCS SP2 AOBS spectral confocal scanner, using a 63x water immersion HCX PL APO WCORR objective. Z-stacks were obtained (10 layers/stack, 2 μm between layers) through the z-axis of cells (3 z-stacks/coverslip), and maximum projections obtained using Leica LCS Software.
ELISA
GECs were cultured to 80% confluence and infected with F. alocis (MOI 100) for 6 h, 24 h or 48 h. Supernatants were collected and centrifuged at 4000 g for 10 min to remove bacteria. Secretion of IL-1β, IL-6, IL-8, and TNF-α, was assessed using Quantikine kits (R & D Systems), according to the manufacturer’s instructions.
Western immunoblotting
F. alocis infected GECs were lysed in SDS-PAGE buffer, separated by SDS-PAGE, and transferred onto nitrocellulose membranes by electroblotting. Membranes were blocked in 10% skimmed dry milk in Tris-buffered saline (TBS) overnight at 4°C. Primary antibody was rabbit anti-MEK1/2, rabbit anti-phospho-MEK1/2 (Cell Signaling) or rabbit anti-GAPDH (Cell Signaling), 1:1000 for 2 h at room temperature. Antigen-antibody binding was detected using horseradish peroxidase-conjugated species-specific secondary antibodies followed by ECL Western Blotting detection reagents (Perkin-Elmer). Densiometric analysis was performed and p-MEK:MEK ratios calculated following normalization to GAPDH.
Annexin V/Sytox Green flow cytometry assay
GECs were infected with F. alocis (MOI 100) or treated with 10 μM campthothecin (apoptosis control) or 0.3% H2O2 (necrosis control). Cells were harvested by trypsinization, and a PE Annexin V/Dead cell Apoptosis kit for Flow Cytometry (Invitrogen) was employed according to the manufacturer’s instructions. Briefly, cell pellets were resuspended in Annexin-binding buffer to wash, centrifuged, and stained with Annexin V and Sytox Green in the dark at 37°C in 5% CO2 for 15 min prior to flow cytometry analysis.
Caspase-3 and caspase-9 luminosity assay
GECs were infected with F. alocis (MOI 100) for 24 h. Cells were then incubated with Caspase-Glo assay substrates for caspase-3 and caspase-9 (Promega), at room temperature in the dark for 1h. Luminosity was measured using a Wallac Victor3 1420 Multilabel Counter Luminometer (Waltham).
Results
F. alocis associates with GEC surfaces
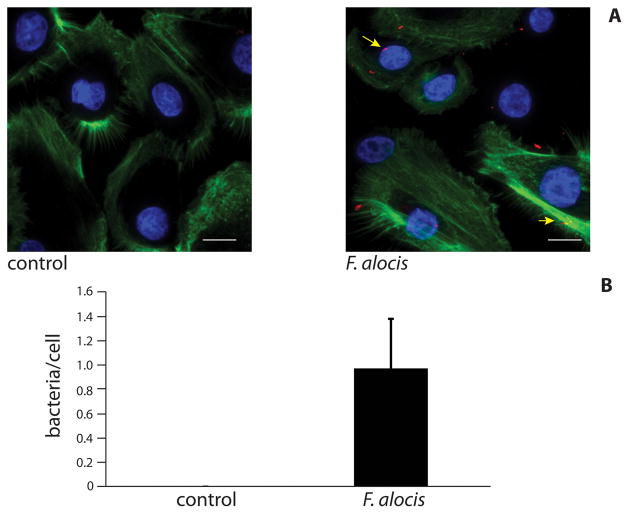
Initially, we undertook fluorescent image analysis to investigate whether F. alocis associates with gingival cells and F. alocis was observed to adhere to the surface of GECs (Fig 1A, arrows). Examination of the z-stacks through layers indicated that F. alocis was located within the cells, as bacteria were visible within the cytoplasmic region through central layers of the stack; however, this requires further investigation. Similarly, clinical isolates of F. alocis have been shown to invade epithelial cells (H. Fletcher, personal communication). F. alocis associated with cells were enumerated (Fig. 1B), and there was 1 bacterium/cell at an MOI of 20.
Figure 1. F. alocis localizes to gingival epithelial cells.
A) GECs were infected with F. alocis (MOI 20) for 1 h and analyzed by confocal microscopy. Control was uninfected GECs. F. alocis (red) was labeled with Syto 17 prior to infection, actin (green) was stained with FITC-phalloidin, and nuclei (blue) stained with DAPI. Magnification x60. Results are representative of two independent assays. Data shown are maximum projections of z-stacks (20 slices/z stack, 3 coverslips/group). B) Levels of F. alocis associated with gingival epithelial cells. Numbers of bacteria co-localized with host cells were counted throughout z-stacks (20 slices/stack; 3 coverslips/group). Results are representative of two independent assays. Data are means of bacteria associated with ≈50 cells/assay (3 coverslips/group, 3 z-stacks/coverslip, average of 6 GEC/field), and error bars indicate standard deviations. Scale bar = 5 μm.
Proinflammatory cytokine secretion is stimulated in F. alocis infected GECs
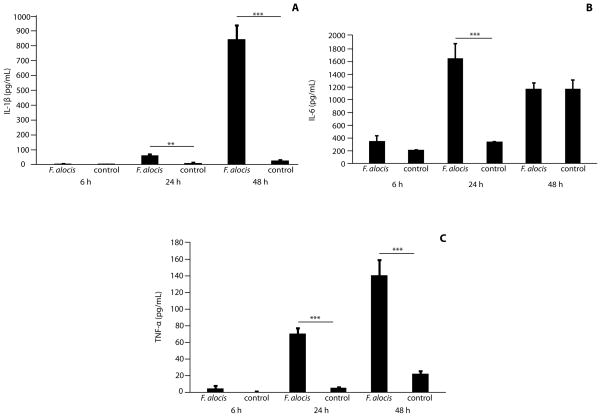
Next, we sought to determine the cytokine responses of GECs to F. alocis. Levels of IL-1β, IL-6 and TNF-α in GEC culture supernatants were quantified by ELISA. In response to F. alocis, IL-1β levels showed a slight increase 24 h post-infection (p < 0.01), and a more substantial increase following 48 h infection (p <0.001) (Fig 2A). The amount of IL-6 secreted from infected cells was comparable to uninfected controls at 6 h; however, after 24 h IL-6 levels were elevated more than 4-fold in F. alocis infected GECs (p < 0.001) (Fig 2B). At 48 h, IL-6 levels were comparable between control and infected conditions. F. alocis infection caused a significant increase in TNF-α secretion (Fig 2C) following 24 h incubation (p <0.001), and secretion levels continued to increase up to 48 h (p <0.001). In contrast, IL-8 levels were unchanged following F. alocis infection at all time periods (not shown). These results indicate that F. alocis selectively induces a proinflammatory cytokine response from gingival epithelial cells.
Figure 2. F. alocis induces secretion of IL-1β, IL-6 and TNF-α from GECs.
Supernatants were obtained from F. alocis-infected GECs, or uninfected controls, and analyzed by ELISA. A) IL-6, B) IL-1β and C) TNF-α. Data are means and error bars indicate standard deviation (n=3). Data are representative of three independent experiments. **, p <0.01 ***, p <0.001 by Tukey-Kramer Multiple Comparison test.
Apoptosis is induced in F. alocis-infected GECs
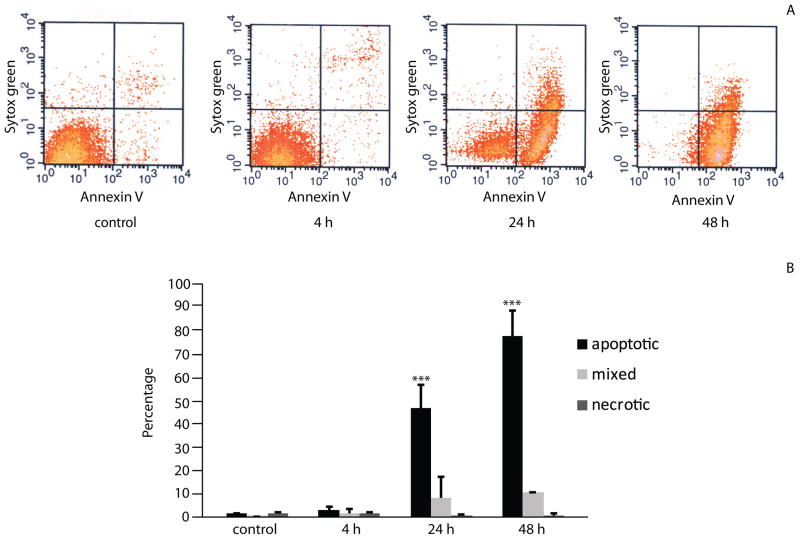
To investigate whether F. alocis may affect cell viability, we examined the levels of apoptotic and necrotic cells following infection. Flow cytometry plots (Fig 3A) revealed that after 4 h incubation, levels of apoptosis in infected cells and uninfected controls were comparable. However, after 24 h greater than 50% of infected cells were apoptotic (p <0.001), increasing to 88% apoptotic following 48 h infection (p <0.001) (Fig 3B). Uninfected controls showed no higher than 4% apoptosis. No significant necrosis was detected in either group. This result provides the first evidence that F. alocis induces apoptosis in primary gingival epithelial cells.
Figure 3. F. alocis stimulates apoptosis in GECs.
F. alocis-infected GECs were stained with Annexin V and Sytox Green. A) FACS profiles showing apoptotic (lower right quadrants), necrotic (upper left) or mixed apoptotic and necrotic (upper right) cells. B) Percentages of cells undergoing apoptosis/necrosis. Data are means and error bars indicate standard deviation. Results shown are the average from two independent assays. ***, p <0.001 compared to control by Tukey-Kramer Multiple Comparison test.
F. alocis activates an extrinsic apoptosis pathway in GECs
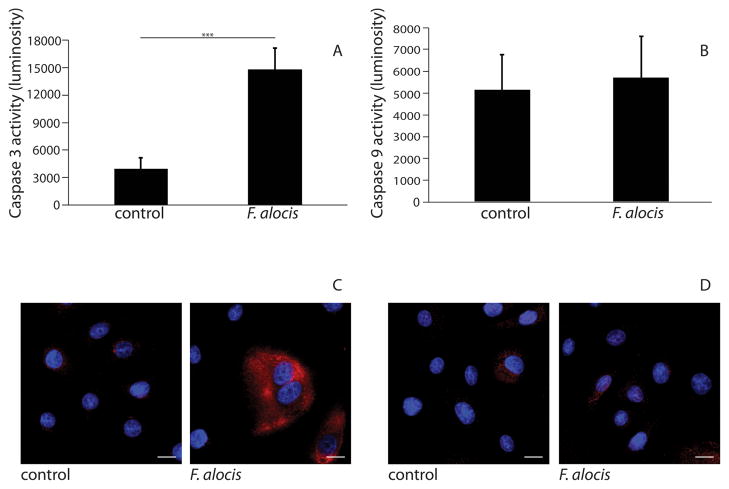
To begin to address whether apoptosis induction occurred through intrinsic or extrinsic pathways, activation of caspase-3 and caspase-9 in infected cells was determined. Following 24 h infection, caspase-3 activity increased compared with uninfected controls (p <0.001), (Fig 4A). In contrast, no caspase-9 activation was detected (Fig 4B). Caspase-3 and caspase-9 activation levels were also examined by confocal microscopy. As shown in Figure 4C, caspase-3 activation was increased in cells infected with F. alocis as compared to uninfected controls. Caspase-9 activation was not detectable in either infected or uninfected cells (Fig 4D). As activation of caspase-9 is indicative of intrinsic, mitochondrial-induced apoptosis, these results support the concept that F. alocis activates an extrinsic apoptotic pathway in GECs.
Figure 4. Caspase-3 is activated in F. alocis-infected GECs.
GECs were infected with F. alocis (MOI 100) for 24 h and reacted proluminescent substrates for A) Caspase-3, B) Caspase-9. Luminosity (arbitrary units) data are means and error bars indicate standard deviation (n=3). Data are representative of three independent experiments. ***, p <0.001 by t-test. C) & D) F. alocis-infected cells or uninfected controls were labeled with C) caspase-3 antibodies or D) caspase-9 antibodies (red) and nuclei (blue) stained with DAPI. Cells were subsequently analyzed by CSLM. Magnification x63. Results are representative of three independent assays. Data shown are maximum projections of z-stacks (10 slices/z stack, 3 coverslips/group). Scale bar = 5 μm.
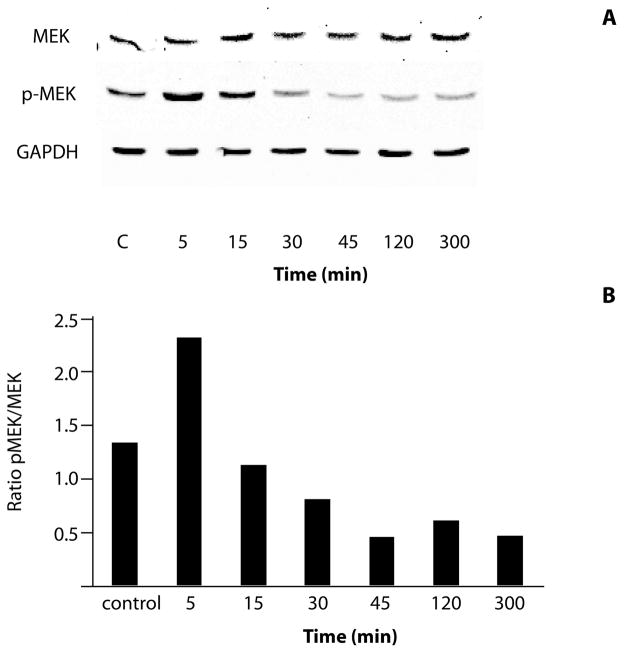
F. alocis modifies MEK signaling in GECs
Inhibition of MEK activity can induce apoptosis, and can impact both the intrinsic and extrinsic pathways (Meng et al., 2010; Wang et al., 2007; Dai et al., 2003; Liu et al., 2006). Therefore, the impact of F. alocis on MEK phosphorylation was investigated by western blotting with specific MEK1/2 and phospho(p)-MEK1/2 antibodies (Fig 5A). While F. alocis caused transient phosphorylation of MEK1/2 after 5 min of bacterial challenge, levels of phospho-MEK1/2 were reduced compared with uninfected controls after 30 min and for up to 5h. Densiometric analysis of bands showed an approximately 70% reduction in the ratio of p-MEK to MEK after 45 min of F. alocis infection (Fig 5B).
Figure 5. MEK1/2 activity is modulated by F. alocis.
A) Lysates of F. alocis infected (inf) or uninfected control (C) GECs were examined by Western blotting with antibodies to MEK1/2 or phospho(p)-MEK1/2. GAPDH was used as a loading control. B) Scanning densitometry showing ratio of p-MEK to MEK. Data are representative of three independent experiments.
Discussion
Periodontal diseases ensue from the disruption of the balance between the host and the complex polymicrobial community that colonizes the gingival crevice. As the periodontal pathogens are also frequently present in the absence of disease, the identities of the organisms associated with the initiation and progression of disease are difficult to determine with certainty. Criteria that are used to impute pathogenic potential to periodontal bacteria include: an increase in number at disease sites; a reduction in number after treatment; pathogenicity in animal models; and display of appropriate virulence factors (Socransky and Haffajee, 1992). These criteria have been very successful in indentifying key components of the pathogenic microbial communities in periodontal disease, and the virulence of organisms such as P. gingivalis, Tannerella forsythia and Treponema denticola is now well established. With the development and successful implementation of culture-independent identification technology it is now possible to more accurately catalogue the complete range of organisms present in health and disease. F. alocis has emerged as an organism that increases in number in diseased periodontal sites in comparison to healthy sites (Dahlen and Leonhardt, 2006; Kumar et al., 2005; Kumar et al., 2006). In addition, cessation of smoking reduces the prevalence of F. alocis, along with other bacterial species associated with periodontal disease (Delima et al., 2010). In terms of association with disease, therefore, F. alocis exhibits characteristics of a periodontal pathogen. We undertook this study to begin to investigate the pathogenic profile of F. alocis.
Epithelial cells that line the gingival crevice are among the first host cells encountered by periodontal bacteria. In addition to providing a mechanical barrier to microbial intrusion, gingival epithelial cells also produce effectors of innate immunity, such as cytokines, and act as sensors of infection by signaling to immune cells in the underlying periodontal tissues (Tribble and Lamont, 2010; Kagnoff and Eckmann, 1997). Successful periodontal pathogens often can disrupt cytokine networks and also impact apoptotic cell death in gingival epithelial cells. We thus examined the interaction between F. alocis and primary cultures of gingival epithelial cells (GECs) in the context of cytokine responses and apoptosis.
F. alocis induced the secretion of the proinflammatory cytokines IL-1β, IL-6 and TNF-α, but not IL-8, from GECs. In terms of relevance to periodontal disease, IL-1β, IL-6 and TNF-α are capable of upregulation of pathways that stimulate osteoclasts and increase alveolar bone resorption (Preshaw and Taylor, 2011). IL-1β, IL-6 and TNF-α can also contribute to tissue degradation through the induction of MMPs and other inflammatory mediators (Birkedal-Hansen, 1993; Graves and Cochran, 2003; Graves, 2008). Moreover, a number of studies have demonstrated increased IL-1β, IL-6 and TNF-α levels in periodontitis patients (Howells, 1995; Okada and Murakami, 1998), and the application of antagonists to IL-1 and TNF reduces the severity of experimental periodontitis (Graves and Cochran, 2003). Much of the tissue destruction in periodontal disease is thus thought to result from disruption of cytokine homeostasis (Preshaw and Taylor, 2011). Interestingly, the GEC cytokine responses to F. alocis bear a remarkable resemblance to those of the consensus periodontal pathogen, P. gingivalis. In response to P. gingivalis infection GECs produce IL-1β, TNF-α and IL-6, but not IL-8 (Stathopoulou et al., 2010; Darveau et al., 1998). In addition, P. gingivalis can antagonize production of IL-8 in response to stimulation with other oral bacteria (Darveau et al., 1998). Suppression of the neutrophil chemokine IL-8 contributes localized immune suppression and may allow overgrowth of other destructive bacteria. The ability of F. alocis to antagonize IL-8 production remains to be investigated.
Epithelial cell apoptosis can be demonstrated in periodontal lesions (Vitkov et al., 2005; Tonetti et al., 1998), and apoptosis may be the direct result of bacterial action or the indirect result of proinflammatory cytokine secretion. F. alocis was capable of inducing apoptosis in GECs, and apoptosis was associated with the activation of caspase-3 but not caspase-9. The absence of caspase-9 activation would tend to suggest F. alocis-induced apoptosis occurs through the extrinsic pathway. In contrast to the concordance between P. gingivalis and F. alocis in cytokine expression, P. gingivalis does not induce apoptosis in GECs (Mao et al., 2007); however other periodontal pathogens such as Treponema denticola can cause epithelial cell apoptosis (Leung et al., 2002).
MEK1/2 is a member of the dual specificity protein kinase family, which lies upstream of the MAP kinases (extracellular signal-regulated kinases or ERKs). MEK1/2 can activate MAPK pathways upon stimulation by variety of extra- and intracellular signals, and MAPK signaling can control cell proliferation and differentiation. F. alocis caused a transient activation of MEK1/2, and a longer term inhibition of MEK activity. Apoptosis induction resulting from the inhibition of MEK1/2 has been reported in several cell types, and can impact both the intrinsic and extrinsic pathways (Meng et al., 2010; Wang et al., 2007; Dai et al., 2003; Liu et al., 2006; Lunghi et al., 2008; Pellicano et al., 2011). Thus, the pro-apoptotic effect of F. alocis may be related to its ability to suppress MEK activity. It is also possible that the proinflammatory cytokines induced by F. alocis may play a role in apoptosis, and the matter requires further investigation.
In conclusion, we have begun the characterization of the virulence properties of the recently recognized periodontal pathogen F. alocis. This organism can induce the secretion of proinflammatory cytokines from GECs. In addition, F. alocis causes apoptosis in GECs coincident with the suppression of MEK1/2 activation. The proinflammatory, pro-apoptotic phenotype of F. alocis may have relevance to the pathogenesis of periodontal disease.
Acknowledgments
We thank NIH/NIDCR for support through DE11111, DE12505 and DE17921
References
- Albandar JM. Underestimation of periodontitis in NHANES surveys. J Periodontol. 2011;82:337–341. doi: 10.1902/jop.2011.100638. [DOI] [PubMed] [Google Scholar]
- Birkedal-Hansen H. Role of cytokines and inflammatory mediators in tissue destruction. J Periodontal Res. 1993;28:500–510. doi: 10.1111/j.1600-0765.1993.tb02113.x. [DOI] [PubMed] [Google Scholar]
- Brown LJ, Johns BA, Wall TP. The economics of periodontal diseases. Periodontol 2000. 2002;29:223–234. doi: 10.1034/j.1600-0757.2002.290111.x. [DOI] [PubMed] [Google Scholar]
- Cato EP, Moore LVH, Moore WEC. Fusobacterium alocis sp. nov. and Fusobacterium sulci sp. nov. from the human gingival sulcus. Int J Syst Bacteriol. 1985;35:475–477. [Google Scholar]
- Colombo AP, Boches SK, Cotton SL, Goodson JM, Kent R, Haffajee AD, et al. Comparisons of subgingival microbial profiles of refractory periodontitis, severe periodontitis, and periodontal health using the human oral microbe identification microarray. J Periodontol. 2009;80:1421–1432. doi: 10.1902/jop.2009.090185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahlen G, Konradsson K, Eriksson S, Teanpaisan R, Piwat S, Carlen A. A microbiological study in relation to the presence of caries and calculus. Acta Odontol Scand. 2010;68:199–206. doi: 10.3109/00016351003745514. [DOI] [PubMed] [Google Scholar]
- Dahlen G, Leonhardt A. A new checkerboard panel for testing bacterial markers in periodontal disease. Oral Microbiol Immunol. 2006;21:6–11. doi: 10.1111/j.1399-302X.2005.00243.x. [DOI] [PubMed] [Google Scholar]
- Dai Y, Dent P, Grant S. Tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) promotes mitochondrial dysfunction and apoptosis induced by 7-hydroxystaurosporine and mitogen-activated protein kinase kinase inhibitors in human leukemia cells that ectopically express Bcl-2 and Bcl-xL. Mol Pharmacol. 2003;64:1402–1409. doi: 10.1124/mol.64.6.1402. [DOI] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Darveau RP, Belton CM, Reife RA, Lamont RJ. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect Immun. 1998;66:1660–1665. doi: 10.1128/iai.66.4.1660-1665.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delima SL, McBride RK, Preshaw PM, Heasman PA, Kumar PS. Response of subgingival bacteria to smoking cessation. J Clin Microbiol. 2010;48:2344–2349. doi: 10.1128/JCM.01821-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner AC, Yu WH, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Weinberg A. Role of bacteria in health and disease of periodontal tissues. Periodontol 2000. 2006;40:50–76. doi: 10.1111/j.1600-0757.2005.00148.x. [DOI] [PubMed] [Google Scholar]
- Frederick JR, Sarkar J, McDowell JV, Marconi RT. Treponema denticola, molecular signaling mechanisms of the periopathogens. J Dent Res. 2011 doi: 10.1177/0022034511402994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes BP, Pinheiro ET, Jacinto RC, Zaia AA, Ferraz CC, Souza-Filho FJ. Microbial analysis of canals of root-filled teeth with periapical lesions using polymerase chain reaction. J Endod. 2008;34:537–540. doi: 10.1016/j.joen.2008.01.016. [DOI] [PubMed] [Google Scholar]
- Graves D. Cytokines that promote periodontal tissue destruction. J Periodontol. 2008;79:1585–1591. doi: 10.1902/jop.2008.080183. [DOI] [PubMed] [Google Scholar]
- Graves DT, Cochran D. The contribution of interleukin-1 and tumor necrosis factor to periodontal tissue destruction. J Periodontol. 2003;74:391–401. doi: 10.1902/jop.2003.74.3.391. [DOI] [PubMed] [Google Scholar]
- Griffen AL, Beall CJ, Firestone ND, Gross EL, Difranco JM, Hardman JH, et al. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One. 2011;6:e19051. doi: 10.1371/journal.pone.0019051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72–122. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Howells GL. Cytokine networks in destructive periodontal disease. Oral Dis. 1995;1:266–270. doi: 10.1111/j.1601-0825.1995.tb00192.x. [DOI] [PubMed] [Google Scholar]
- Jalava J, Eerola E. Phylogenetic analysis of Fusobacterium alocis and Fusobacterium sulci based on 16S rRNA gene sequences: proposal of Filifactor alocis (Cato, Moore and Moore) comb. nov. and Eubacterium sulci (Cato, Moore and Moore) comb. nov. Int J Syst Bacteriol. 1999;49:1375–1379. doi: 10.1099/00207713-49-4-1375. [DOI] [PubMed] [Google Scholar]
- Kagnoff MF, Eckmann L. Epithelial cells as sensors for microbial infection. J Clin Invest. 1997;100:6–10. doi: 10.1172/JCI119522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Griffen AL, Moeschberger ML, Leys EJ. Identification of candidate periodontal pathogens and beneficial species by quantitative 16S clonal analysis. J Clin Microbiol. 2005;43:3944–3955. doi: 10.1128/JCM.43.8.3944-3955.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665–3673. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamont RJ, Jenkinson HF. Life below the gum line: pathogenic mechanisms of Porphyromonas gingivalis. Microbiol Mol Biol Rev. 1998;62:1244–1263. doi: 10.1128/mmbr.62.4.1244-1263.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung WK, Wu Q, Hannam PM, McBride BC, Uitto VJ. Treponema denticola may stimulate both epithelial proliferation and apoptosis through MAP kinase signal pathways. J Periodontal Res. 2002;37:445–455. doi: 10.1034/j.1600-0765.2002.01007.x. [DOI] [PubMed] [Google Scholar]
- Liu Y, Borchert GL, Surazynski A, Hu CA, Phang JM. Proline oxidase activates both intrinsic and extrinsic pathways for apoptosis: the role of ROS/superoxides, NFAT and MEK/ERK signaling. Oncogene. 2006;25:5640–5647. doi: 10.1038/sj.onc.1209564. [DOI] [PubMed] [Google Scholar]
- Lunghi P, Giuliani N, Mazzera L, Lombardi G, Ricca M, Corradi A, et al. Targeting MEK/MAPK signal transduction module potentiates ATO-induced apoptosis in multiple myeloma cells through multiple signaling pathways. Blood. 2008;112:2450–2462. doi: 10.1182/blood-2007-10-114348. [DOI] [PubMed] [Google Scholar]
- Mao S, Park Y, Hasegawa Y, Tribble GD, James CE, Handfield M, et al. Intrinsic apoptotic pathways of gingival epithelial cells modulated by Porphyromonas gingivalis. Cell Microbiol. 2007;9:1997–2007. doi: 10.1111/j.1462-5822.2007.00931.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng J, Fang B, Liao Y, Chresta CM, Smith PD, Roth JA. Apoptosis induction by MEK inhibition in human lung cancer cells is mediated by Bim. PLoS One. 2010;5:e13026. doi: 10.1371/journal.pone.0013026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada H, Murakami S. Cytokine expression in periodontal health and disease. Crit Rev Oral Biol Med. 1998;9:248–266. doi: 10.1177/10454411980090030101. [DOI] [PubMed] [Google Scholar]
- Pellicano F, Simara P, Sinclair A, Helgason GV, Copland M, Grant S, Holyoake TL. The MEK inhibitor PD184352 enhances BMS-214662-induced apoptosis in CD34+ CML stem/progenitor cells. Leukemia. 2011 doi: 10.1038/leu.2011.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preshaw PM, Taylor JJ. How has research into cytokine interactions and their role in driving immune responses impacted our understanding of periodontitis? J Clin Periodontol. 2011;38(Suppl 11):60–84. doi: 10.1111/j.1600-051X.2010.01671.x. [DOI] [PubMed] [Google Scholar]
- Sakamoto M, Rocas IN, Siqueira JF, Jr, Benno Y. Molecular analysis of bacteria in asymptomatic and symptomatic endodontic infections. Oral Microbiol Immunol. 2006;21:112–122. doi: 10.1111/j.1399-302X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- Schlafer S, Riep B, Griffen AL, Petrich A, Hubner J, Berning M, et al. Filifactor alocis--involvement in periodontal biofilms. BMC Microbiol. 2010;10:66. doi: 10.1186/1471-2180-10-66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A. Virulence mechanisms of Tannerella forsythia. Periodontol 2000. 2010;54:106–116. doi: 10.1111/j.1600-0757.2009.00332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siqueira JF, Jr, Rocas IN. Simultaneous detection of Dialister pneumosintes and Filifactor alocis in endodontic infections by 16S rDNA-directed multiplex PCR. J Endod. 2004;30:851–854. doi: 10.1097/01.don.0000132300.13023.5d. [DOI] [PubMed] [Google Scholar]
- Siqueira JF, Jr, Rocas IN, Alves FR, Silva MG. Bacteria in the apical root canal of teeth with primary apical periodontitis. Oral Surg Oral Med Oral Pathol Oral Radiol Endod. 2009;107:721–726. doi: 10.1016/j.tripleo.2009.01.042. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. The bacterial etiology of destructive periodontal disease: current concepts. J Periodontol. 1992;63:322–331. doi: 10.1902/jop.1992.63.4s.322. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–144. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Stathopoulou PG, Benakanakere MR, Galicia JC, Kinane DF. Epithelial cell pro-inflammatory cytokine response differs across dental plaque bacterial species. J Clin Periodontol. 2010;37:24–29. doi: 10.1111/j.1600-051X.2009.01505.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonetti MS, Cortellini D, Lang NP. In situ detection of apoptosis at sites of chronic bacterially induced inflammation in human gingiva. Infect Immun. 1998;66:5190–5195. doi: 10.1128/iai.66.11.5190-5195.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribble GD, Lamont RJ. Bacterial invasion of epithelial cells and spreading in periodontal tissue. Periodontol 2000. 2010;52:68–83. doi: 10.1111/j.1600-0757.2009.00323.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitkov L, Krautgartner WD, Hannig M. Surface morphology of pocket epithelium. Ultrastruct Pathol. 2005;29:121–127. doi: 10.1080/01913120590916832. [DOI] [PubMed] [Google Scholar]
- Wade WG. Has the use of molecular methods for the characterization of the human oral microbiome changed our understanding of the role of bacteria in the pathogenesis of periodontal disease? J Clin Periodontol. 2011;38(Suppl 11):7–16. doi: 10.1111/j.1600-051X.2010.01679.x. [DOI] [PubMed] [Google Scholar]
- Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P. Apoptosis induction in human melanoma cells by inhibition of MEK is caspase-independent and mediated by the Bcl-2 family members PUMA, Bim, and Mcl-1. Clin Cancer Res. 2007;13:4934–4942. doi: 10.1158/1078-0432.CCR-07-0665. [DOI] [PubMed] [Google Scholar]