Abstract
BACKGROUND
Warfarin is a drug with a narrow therapeutic index and large interindividual variability in daily dosing requirements. Patients commencing warfarin treatment are at risk of bleeding due to excessive anticoagulation caused by overdosing. The interindividual variability in dose requirements is influenced by a number of factors, including polymorphisms in genes mediating warfarin pharmacology, co-medication, age, sex, body size and diet.
AIMS
To develop population pharmacokinetic models of both R- and S-warfarin using clinical and genetic factors and to identify the covariates which influence the interindividual variability in the pharmacokinetic parameters of clearance and volume of distribution in patients on long-term warfarin therapy.
METHODS
Patients commencing warfarin therapy were followed up for 26 weeks. Plasma warfarin enantiomer concentrations were determined in 306 patients for S-warfarin and in 309 patients for R-warfarin at 1, 8 and 26 weeks. Patients were also genotyped for CYP2C9 variants (CYP2C9*1,*2 and *3), two single-nucleotide polymorphisms (SNPs) in CYP1A2, one SNP in CYP3A4 and six SNPs in CYP2C19. A base pharmacokinetic model was developed using NONMEM software to determine the warfarin clearance and volume of distribution. The model was extended to include covariates that influenced the between-subject variability.
RESULTS
Bodyweight, age, sex and CYP2C9 genotype significantly influenced S-warfarin clearance. The S-warfarin clearance was estimated to be 0.144 l h−1 (95% confidence interval 0.131, 0.157) in a 70 kg woman aged 69.8 years with the wild-type CYP2C9 genotype, and the volume of distribution was 16.6 l (95% confidence interval 13.5, 19.7). Bodyweight and age, along with the SNPs rs3814637 (in CYP2C19) and rs2242480 (in CYP3A4), significantly influenced R-warfarin clearance. The R-warfarin clearance was estimated to be 0.125 l h−1 (95% confidence interval 0.115, 0.135) in a 70 kg individual aged 69.8 years with the wild-type CYP2C19 and CYP3A4 genotypes, and the volume of distribution was 10.9 l (95% confidence interval 8.63, 13.2).
CONCLUSIONS
Our analysis, based on exposure rather than dose, provides quantitative estimates of the clinical and genetic factors impacting on the clearance of both the S- and R-enantiomers of warfarin, which can be used in developing improved dosing algorithms.
Keywords: NONMEM software, pharmacogenetics, population pharmacokinetics, warfarin
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
A number of pharmacokinetic studies have focused on S-warfarin. These have shown that demographic factors, such as bodyweight, genetic factors, such as CYP2C9 genotype, and interacting medicines, particularly amiodarone, contribute to the interindividual estimates of clearance.
WHAT THIS STUDY ADDS
This study not only reinforces what has previously been learned about S-warfarin, but also provides an insight into the pharmacokinetics of R-warfarin. The study also focuses on individuals who are on long-term warfarin therapy, which is more reflective of clinical practice.
Introduction
Warfarin is an oral anticoagulant commonly used in the treatment of individuals with venous thromboembolism, atrial fibrillation and heart valve replacement. It is a racemic mixture of R- and S-enantiomers; the anticoagulant effect is mainly attributed to the S-isomer, which is thought to have three to five times the potency of the R-form [1]. Warfarin has a narrow therapeutic range [2–4], exhibits a complex dose–response relationship [5] and is characterized by high interindividual variability in daily dose requirements [6]. At one extreme are individuals who require very high doses to attain a therapeutic international normalization ratio, while at the other are those who suffer serious adverse events, including haemorrhages [1, 7]. Warfarin toxicity has been reported to account for over 10% of all adverse drug reactions leading to hospital admission [8].
Warfarin undergoes extensive metabolism by the cytochrome P450 isoforms. CYP2C9 is responsible for the metabolism of S-warfarin, while R-warfarin is metabolized by CYP1A2, CYP2C19 and CYP3A4[9]. To date, most attention has focused on the role of CYP2C9 and its allelic variants in the metabolism of S-warfarin [10]. The most common allele is designated CYP2C9*1 and is considered the wild-type allele. Two variants (CYP2C9*2 and CYP2C9*3) with functional consequences have been identified in the general population [11]. Several studies have demonstrated that the disposition of S-warfarin is influenced by CYP2C9 genotype [12–15]; however, little work has been undertaken with R-warfarin.
In addition to CYP2C9 genotypes, several other factors affect interindividual variability in the response to warfarin therapy. These include demographic factors, such as age or bodyweight, clinical factors, including hepatic dysfunction, interacting medications (for example, enzyme inhibitors), vitamin K intake and allelic variation in the pharmacodynamic target for warfarin, VKORC1[16, 17]. Indeed, recent data suggest that at least 50% of the variance in dose requirements can be accounted for by age, bodyweight (or body mass index) and genetic polymorphisms in VKORC1 and CYP2C9[18]. However, the evidence is in relation to dose, rather than plasma drug concentration, which may be more reliable as a measure of exposure that takes into account pharmacokinetics and non-adherence.
The pharmacokinetics of warfarin has been studied extensively [1, 19]. However, population analyses quantifying the influence of genetic polymorphism on the pharmacokinetics of enantiomeric forms of warfarin are limited to those reported by Hamberg et al. [5, 20]. In their analysis of 150 patients from two studies, polymorphism in CYP2C9 was found to affect S-warfarin clearance, but not R-warfarin clearance. Genes coding for other candidate CYP450 enzymes have not been assessed.
As part of a planned interim analysis of 354 patients recruited to a prospective study of genetic and environmental factors determining clinical outcomes in patients commencing warfarin therapy [21], we undertook a population pharmacokinetic analysis of R-warfarin and S-warfarin. The aim of the study was to establish appropriate models to identify the factors that contribute to interindividual variability in the pharmacokinetic parameters and, as a consequence, the differences in exposure to inform warfarin dose requirements in patients on long-term therapy.
Methods
Patients
Data from patients who had started warfarin treatment, irrespective of indication, between November 2004 and March 2006 were included in the analysis after having obtained written informed consent. A total of 354 patients were recruited from the Royal Liverpool and Broadgreen University Hospital NHS Trust and University Hospital Aintree, Liverpool, UK. All patients were informed of the nature of the study prior to being enrolled, and the only exclusion criterion was the refusal to give written informed consent. The study was approved by Birmingham South Research Ethics Committee.
The study design was observational. Patients received the usual clinical care, with the individual warfarin loading doses and subsequent maintenance doses determined by in-house hospital guidelines. Each patient had four study visits scheduled, the first at the time of initiation of warfarin therapy, then at 1, 8 and 26 weeks after the commencement of warfarin [21].
Genotyping
Blood samples for DNA extraction were taken at baseline prior to the commencement of the warfarin regimen. The DNA was extracted by the Sanger Institute and genotyped for 29 genes using the mass extended method of Sequenom. Individual single-nucleotide polymorphism (SNP) loci were amplified using the polymerase chain reaction, which provides a template for allele-specific primer extension. Allele-specific products were detected using MALDI-TOF mass spectrometry. All SNPs were checked to ensure that they were in Hardy–Weinberg equilibrium. Full details are provided by Jorgensen et al. [21].
Determination of plasma warfarin enantiomer concentrations
Using a sparse sampling approach, plasma concentrations of R- and S-warfarin were measured after 1, 8 and 26 weeks following the onset of warfarin therapy when possible, but the number of plasma concentrations per individual varied between one and three. The blood samples used to determine the plasma concentrations were taken approximately 16 h after each individual's last dose of warfarin. Sampling times were recorded accurately. Unbound plasma levels of both R- and S-warfarin were measured simultaneously in all patients using the chiral high-performance liquid chromatography method described by Naidong and Lee [22]. Intra- and interday coefficients of variation were less than 6% for both R- and S-warfarin. The inter- and intra-assay accuracy (percentage bias) for all quality control concentrations was within 15% for both R- and S-warfarin. The assay allowed for the quantification of enantiomers of warfarin over a wide concentration range (100–5000 ng ml−1). The limit of quantification was set at 100 ng ml−1 for each warfarin enantiomer, which is sufficiently low to enable patient samples to be analysed with good accuracy and precision.
Pharmacokinetic analysis
The pharmacokinetic (PK) models were constructed using NONMEM version 7.0 (ICON Development Solutions, Ellicott City, MA, USA). The first-order conditional estimation with interaction method was used to estimate the population PK parameters and the individual-level random effects. Scatter plots and box and whisker plots were generated using SPSS version 15 (SPSS Inc., Chicago, IL, USA) to visually assess relationships between covariates and random effects. The significance of the covariates for entry into the final model was assessed using the objective function provided by NONMEM. A variable was considered for inclusion in the final model if it reduced the objective function value (OFV) by more than 6.63 (1% significance level assuming a χ2 on 1 degree of freedom). The subset of variables which met this criterion were then entered stepwise into the multivariate model, with the variable that had the biggest impact on the OFV being entered first and subsequent variables being added according to their impact on the OFV. Again, variables that reduced the OFV by more than 6.63 were retained in the model. A second model was then constructed with all covariates included. Covariates were then removed one at time, starting with the covariates that had least impact on the OFV. Using both these approaches produced the same final model, which is reported in this manuscript.
Base models
A single-compartment model with first-order absorption and a block covariance structure was identified as the optimal base model for both R- and S-warfarin. A fixed absorption rate of 1.66 h−1 had to be assumed because the sparse sampling did not allow this value to be estimated from the data. This was based on a previously reported value [23]. Other values, ranging from 1 to 5 h−1, were tested in a sensitivity analysis, but had no impact on the NONMEM objective function or other parameter estimates. Likewise, because of the sparse sampling, there was no evidence of a two-compartment model for S-warfarin as reported in some other studies [5]. It should also be noted that Hamberg et al. [20] also identified a one-compartment model with first-order absorption and elimination. The models were parameterized for clearance (CL; in litres per hour) and volume of distribution (V; in litres) using NONMEM subroutines ADVAN2 and TRANS2. For the base model, intersubject variability was estimated for both CL and V. Subsequently, both intersubject and interoccasion variability were added to the pharmacokinetic parameters. Consequently, the parameters consist of a fixed effect (population average θ*) and a random effect which was log additive:
| (1) |
| (2) |
Where θCL and θV are the population averages for clearance and volume of distribution, respectively, and ηi and κi,j are the interindividual and interoccasion variability parameters, respectively, for individual i and occasion j. An additive and proportional error model was used to describe residual variability.
Covariate selection and models
Demographic information included in the analysis was age (in years), sex (1 for a man and 0 for a woman), bodyweight (in kilograms), body surface area, calculated using the Mosteller formula [24], and height (in metres). The CYP2C9 genotype was included as a covariate in the S-warfarin model. The CYP2C9 genotype was included in the PK modelling as a categorical variable, with the wild-type *1/*1 used as the reference category. The remaining genotypes (*1/*2, *1/*3, *2/*2, *2/*3 and *3/*3) were compared against the reference wild-type (*1/*1); a category was also included for missing genotype information. In the R-warfarin model, three SNPs relating to the CYP1A2 genotype, one SNP relating to the CYP3A4 genotype and six SNPs relating to CYP2C19 genotype were also included in the modelling process, along with the CYP2C9 genotype. These SNPs were coded as wild-type, heterozygote or variant homozygotes.
Interacting concomitant medications were initially coded as none, enzyme inhibitor or enzyme inducer. A second analysis was conducted in which amiodarone, the most commonly used P450 enzyme inhibitor in the cohort of patients, was included as a separate variable, coded as one for an individual taking amiodarone and zero otherwise. Also included in the data set was the recorded dosing history of each individual. This included the loading doses (given over a 3 day period), followed by a daily prescribed maintenance dose taken at 18.00 h the previous evening before the sampling in the morning; however, it had to be assumed that each individual was fully adherent to their dosing regimen.
Seven covariates (age, bodyweight, body surface area, height, sex, co-medication and CYP2C9 genotype) were assessed to determine whether they had any significant impact on reducing the unexplained variability in the PK parameters CL and V of S-warfarin. The same demographic covariates and CYP2C9 genotype were assessed in the R-warfarin model, along with the SNPs in CYP1A2, CYP3A4 and CYP2C19. These covariates were first explored graphically and each potential covariate individually added to the base model if graphical trends were shown. If the objective function decreased by more than 3.84 points (P≤ 0.05, d.f. = 1) with the addition of each parameter, then the covariate was retained. In the backward elimination step, each covariate was removed separately from the model; if its removal caused an increase in objective function of at least 6.63 points (P≤ 0.01, d.f. = 1), then it was retained for the final model.
Results
Patient characteristics
From the 354 patients initially recruited into the study, 306 were included in the pharmacokinetic (PK) study for S-warfarin and 309 for R-warfarin. A total of 759 S-warfarin and 739 R-warfarin plasma concentrations were available for the PK modelling. There was complete information on bodyweight, height, age, sex and co-medication, but genotyping information was not available for 14 participants in the case of CYP2C9, 61 patients for CYP2C19 and 38 patients for CYP3A4. The demographic information about the participants is presented in Table 1.
Table 1.
Participant demographics
| Number of participants | |
| S-warfarin | 306 |
| R-warfarin | 309 |
| S-warfarin plasma concentrations | 739 |
| R-warfarin plasma concentrations | 759 |
| Men S-warfarin | 178 (58%) |
| Men R-warfarin | 181 (59%) |
| Age, mean (range) (years) | 66.4 (19–95) |
| Bodyweight, mean (range) (kg) | 80.7 (36–172) |
| Co-medication (S-warfarin) | |
| Amiodarone Yes | 20 (6.5%) |
| Amiodarone No | 286 (93.5%) |
| CYP2C9 genotype* | |
| *1/*1 | 195 (63.7%) |
| *1/*2 | 59 (19.3%) |
| *1/*3 | 29 (9.5%) |
| *2/*2 | 1 (0.3%) |
| *2/*3 | 6 (2.0%) |
| *3/*3 | 2 (0.6%) |
| Missing | 14 (4.6%) |
| CYP2C19 genotype* (SNP rs3814637) | |
| Homozygote | 219 (70.9%) |
| Heterozygote | 25 (8.1%) |
| Mutant-type – homozygote | 4 (1.3%) |
| Missing | 61 (19.7%) |
| CYP3A4 genotype* (SNP rs2242480) | |
| Homozygote | 226 (73.1%) |
| Heterozygote | 42 (13.6%) |
| Mutant-type – homozygote | 3 (1.0%) |
| Missing | 38 (12.3%) |
CYP2C9 frequencies are for S-warfarin, whilst CYP2C19 and CYP3A4 frequencies apply to R-warfarin. Frequencies for nonsignificant SNPs are available but not reported.
S-Warfarin models
The initial base model for S-warfarin (Table 2) had parameter values of 0.149 l h−1 for CL (95% confidence interval 0.140, 0.158) and 15.2 l for V (95% confidence interval 12.0, 18.4), with the unexplained intersubject variability being 49.3 and 38.6%, respectively. The proportional component of the residual error was 30.1% and the additive component was 45.8 µg l−1.
Table 2.
Base models for S- and R-warfarin
| S-Warfarin | R-Warfarin | |||||
|---|---|---|---|---|---|---|
| Objective function | 8975 | 9425 | ||||
| Parameter | Estimate | Standard error | 95% Confidence interval | Estimate | Standard error | 95% Confidence interval |
| CL (l h−1) | 0.149 | 0.00465 | (0.140, 0.158) | 0.132 | 0.00381 | (0.125, 0.139) |
| V (l) | 15.2 | 1.64 | (12.0, 18.4) | 9.11 | 0.934 | (7.28, 10.9) |
| Ka | 1.66 | Fixed | – | 1.66 | Fixed | – |
| IIV CL* | 49.3% | (43.7%, 54.3%) | 47.3% | (41.1%, 52.8%) | ||
| IIV V* | 38.6% | (5.46%, 54.3%) | 56.5% | (38.3%, 74.7%) | ||
| Proportional error* | 30.1% | (25.3%, 34.2%) | 31.9% | (29.2%, 34.5%) | ||
| Additive error (µg ml−1) | 45.8 | (–40.8, 76.6) | 1 | Fixed | – | |
Interindividual variability (IIV) and residual proportional error are expressed as an approximate coefficient of variation (square root of the variance). Ka, absorption rate.
The univariate analysis showed that in the S-warfarin model the bodyweight, body surface area, sex, age and CYP2C9 genotype were significantly related to CL, but no covariates were related to V. When the five individual covariates were included stepwise in a multivariate model for CL, bodyweight, age, sex and CYP2C9 genotype were shown to be significant. Following a confirmatory backward stepping approach, these were the only covariates shown to have an impact on CL. Interestingly, interacting medication was identified by the univariate analysis as not being statistically significant. Likewise, when amiodarone alone was included in the univariate analysis as a separate variable it was also shown to have no significant impact on the OFV. The ETA shrinkage values for CL and V were 10.7 and 49.4%, respectively. As these are relatively high values, post hoc plots were only used for guidance and all covariate effects were tested formally using the likelihood ratio test. The influence of interoccasion variability was found to be nonsignificant; however, it was possible to estimate a covariance between the random intersubject effects on CL and V. The additive term in the residual variability model dropped out of the model and was set to a low fixed value (1). The proportional residual variability was 31.6%.
The final equations for CL and V in the S-warfarin model were as follows:
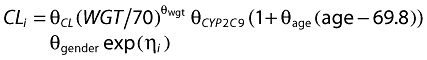 |
(3) |
| (4) |
where θwgt is the exponent on weight (WGT), θCYP2C9, θgender and θage are the coefficients for CYP2C9 genotype, sex and age, respectively, and i indicates the patient.
In the final multivariate S-warfarin model (Table 3), four covariates (bodyweight, age, sex and CYP2C9 genotype) were shown to have a significant impact on the objective function, reducing it from 8975 to 8899 (P < 0.01). With the covariates added, the proportion of unexplained variability in CL was reduced from 49.3 to 41.8%.
Table 3.
Final covariate models for S- and R-warfarin
| S-Warfarin | R-Warfarin | |||||
|---|---|---|---|---|---|---|
| Objective function | 8899 | 9334 | ||||
| Parameter | Estimate | Standard error | 95% Confidence interval | Estimate | Standard error | 95% Confidence interval |
| CL (l h−1) | 0.144 | 0.00647 | (0.131, 0.157) | 0.125 | 0.00528 | (0.115, 0.135) |
| V (l) | 16.6 | 1.57 | (13.5, 19.7) | 10.9 | 1.16 | (8.63, 13.2) |
| Ka | 1.66 | Fixed | – | 1.66 | Fixed | – |
| θwgt | 0.321 | 0.145 | (0.037, 0.605) | 0.650 | 0.132 | (0.391, 0.909) |
| θage | −0.00816 | 0.00206 | (–0.0122, −0.00412) | −0.00657 | 0.00223 | (–0.0109, −0.00220) |
| θgender | ||||||
| Female | 1.00 | |||||
| Male | 1.12 | 0.0705 | (0.982, 1.26) | |||
| θCYP2C9 | ||||||
| *1/*1 (wild-type) | 1.00 | |||||
| *1/*2 | 0.855 | 0.0629 | (0.732, 0.978) | |||
| *2/*2 | 0.672 | 0.0568 | (0.561, 0.783) | |||
| *1/*3 | 0.454 | 0.0505 | (0.355, 0.553) | |||
| *2/*3 | 0.496 | 0.121 | (0.259, 0.733) | |||
| *3/*3 | 0.286 | 0.0324 | (0.222, 0.350) | |||
| Missing | 0.782 | 0.0899 | (0.606, 0.958) | |||
| θCYP2C19 | ||||||
| Homozygote | 1.00 | |||||
| Heterozygote | 0.761 | 0.0576 | (0.648, 0.874) | |||
| Mutant – homozygote | 0.494 | 0.191 | (0.120, 0.868) | |||
| Missing | 0.804 | 0.0523 | (0.701, 0.907) | |||
| θCYP3A4 | 1.00 | |||||
| Homozygote | 1.32 | 0.0978 | (1.13, 1.51) | |||
| Heterozygote | 1.06 | 0.172 | (0.723, 1.40) | |||
| Mutant – homozygote | 0.937 | 0.0845 | ().771, 1.10) | |||
| IIV CL* | 41.8% | (37.3%, 45.9%) | 43.0% | (38.6%, 47.0%) | ||
| IIV V* | 35.8% | (18.0%, 47.3%) | 38.3% | (20.2%, 50.3%) | ||
| Covariance (CL, V)† | 0.422 | 0.352 | ||||
| Proportional error* | 31.6% | (28.5%, 34.5%) | 31.9% | (29.2%, 34.5%) | ||
| Additive error (µg ml−1) | 1 | Fixed | 1 | Fixed | ||
Interindividual variability (IIV) and residual proportional error are expressed as an approximate coefficient of variation (square root of the variance).
Covariance is expressed as a correlation coefficient. Ka, absorption rate.
The CYP2C9 genotype was shown to play a significant role in reducing CL. For a 69.8-year-old woman, weighing 70 kg, the population CL was reduced by 71% (from 0.144 to 0.0412 l h−1) for a women with the *3/*3 genotype compared with a women with the wild-type *1/*1 genotype. For a woman with *1/*3, *2/*2 or *2/*3 genotype, the population CL is reduced by approximately half. Individual CL values were increased with increased bodyweight. For example, for a woman weighing 100 kg, of median age and with the wild-type CYP2C9 genotype, the population CL increased to 0.161 l h−1, and for a woman weighing 120 kg, it increased to 0.171 l h−1. The results also showed that the CL rate was approximately 12% higher in men compared with women and that CL decreased with age.
Figures 1 and 2 show some diagnostic plots for the fitted model for S-warfarin, indicating that the model gives an adequate description of the data. Figure 3 shows some plots of the data, stratified by covariate grouping, together with a visual predictive check.
Figure 1.
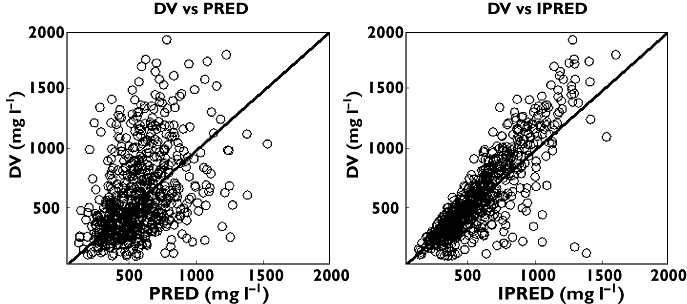
S-Warfarin observed plasma concentration (DV) against population predicted (PRED) and individual predicted concentrations (IPRED)
Figure 2.
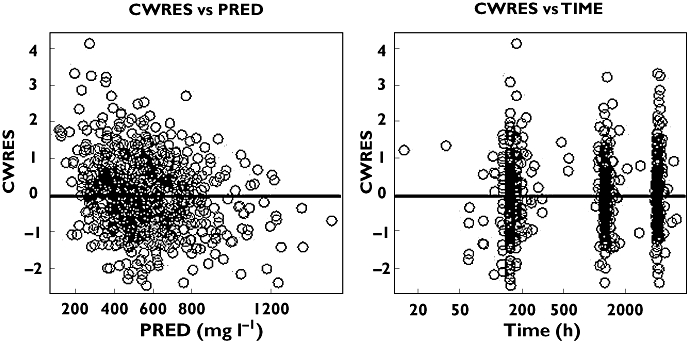
S-Warfarin conditional weighted residual (CWRES) against population predicted concentration (PRED) and time
Figure 3.
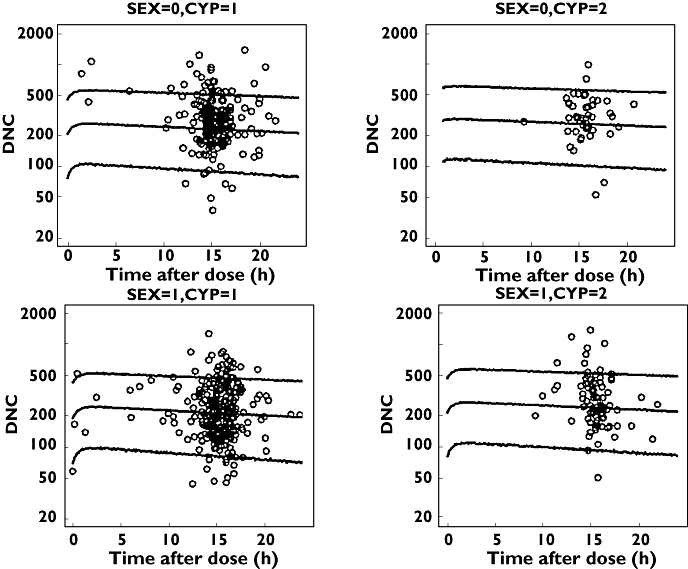
S-Warfarin visual predictive check for some covariate groups using dose-normalized observed and predicted concentrations against time. The lines are the 5th, 50th and 95th percentiles and the open circles are observed plasma concentration
R-Warfarin models
The base model for R-warfarin (Table 2) had a CL of 0.132 l h−1 (95% confidence interval 0.125, 0.139) and a volume of distribution of 9.11 l (95% confidence interval 7.28, 10.9). Unexplained intersubject variability on CL and V was 47.3 and 56.5%, respectively. As with the S-warfarin model, different values for the absorption rate constant did not significantly affect the fit.
The univariate covariate model identified six potential covariates for inclusion in the multivariate model (Table 3); however, only four of these [bodyweight, age, SNP rs3814637 in the CYP2C19 gene (hereafter referred to as CYP2C19 genotype) and SNP rs2242480 in the CYP3A4 genotype (hereafter referred to as CYP3A4 genotype)] were shown to have a significant impact on reducing the objective function of the multivariate model. The genetic variants of CYP1A2 had no effect on the CL of R-warfarin. When these covariates were included as covariates on CL, the objective function was reduced from 9425 to 9335 (P < 0.01), a statistically significant reduction. With the covariates added, the proportion of unexplained variability in CL was reduced from 47.3 to 43.0%. The ETA shrinkage values for clearance and volume were 9.41 and 59.2%, respectively. As with the S-warfarin model, these are relatively high values; consequently, post hoc plots were only used for guidance and all covariate effects were tested formally using the likelihood ratio test. Also, like S-warfarin, the influence of interoccasion variability was found to be nonsignificant, but it was possible to estimate a covariance between the random intersubject effects on CL and V. The additive term in the residual variability model dropped out of the model and was set to a low fixed value (1). The proportional residual variability was 31.9%.
The final equations for CL and V in the R-warfarin model were as follows:
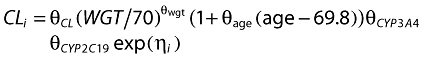 |
(5) |
| (6) |
where θwgt is the exponent on weight (WGT), and θage, θCYP3A4 and θCYP2C19 are the coefficients for age, CYP3A4 genotype (rs2242480) and CYP2C19 genotype (rs3814637), respectively.
The final model implies that the CL of R-warfarin, in a similar manner to S-warfarin, increased with bodyweight and decreased with age. The CL of R-warfarin was reduced according to CYP2C19 genotype. For a patient with a bodyweight of 70 kg and median age, those with the heterozygote genotype for CYP2C19 had a reduction in clearance of 24%, from 0.125 to 0.0951 l h−1. Those with the variant homozygote genotype experienced a much larger reduction in CL, by approximately 51%, from 0.125 to 0.0618 l h−1. Those patients with the heterozygote genotype for CYP3A4 had a 32% increased clearance for R-warfarin.
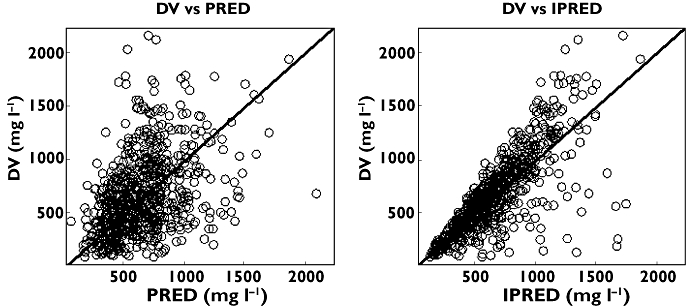
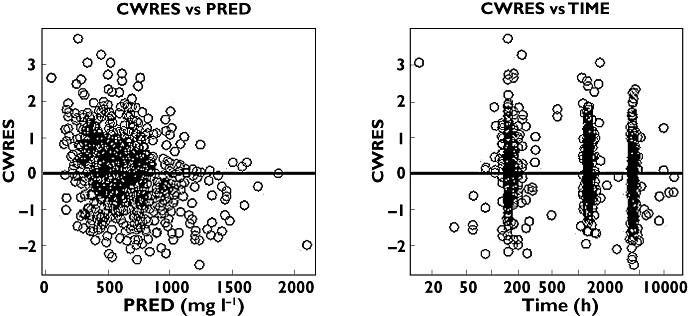
In a similar manner to S-warfarin, Figures 4 and 5 show some diagnostic plots for the fitted model for R-warfarin, also indicating that the model gives an adequate description of the data. Figure 6 shows some plots of the data, stratified by covariate grouping, together with a visual predictive check.
Figure 4.
R-Warfarin observed plasma concentration (DV) against population predicted (PRED) and individual predicted concentrations (IPRED)
Figure 5.
R-Warfarin conditional weighted residual (CWRES) against population predicted concentration (PRED) and time
Figure 6.
R-Warfarin visual predictive check for some covariate groups using dose-normalized observed and predicted concentrations against time. The lines are the 5th, 50th and 95th percentiles and the open circles are observed plasma concentration
Volume of distribution
No covariates influenced the volume of distribution, and the proportion of unexplained variability remained moderately high at 35.8% in the S-warfarin model and 38.3% in the R-warfarin model. These results are consistent with those of Hamberg et al. [5]. The experimental design used in our study was not conducive to the estimation of the volume of distribution. Further studies with different designs are required to identify the covariates that influence the volume of distribution and reduce the proportion of unexplained variability in this PK parameter.
Discussion
The analysis demonstrated that both R- and S-warfarin can be modelled by a one-compartment model with first-order absorption. The following four covariates were identified as influencing the plasma clearance of S-warfarin: bodyweight, age, sex and CYP2C9 genotype. Of these, CYP2C9 genotype was the most influential covariate in determining S-warfarin clearance. Although amiodarone is thought to potentiate the anticoagulant effect of warfarin through the inhibition of CYP2C9 [25], it was not identified as having a significant impact on CL in the present study.
Studies have suggested that age may play a role in determining interindividual variability in S-warfarin metabolism, with older patients requiring a lower dose than younger patients, with the suggestion that weekly maintenance dose requirement fall by 0.4 mg for each year of age [26, 27]. This was supported by the results from the present study, because age was shown to be related to clearance. It has also been suggested that women require smaller doses of warfarin than men, and this was again identified by the S-warfarin model, with men having higher clearance rates than women.
Our data are consistent with the accumulating evidence on the role of CYP2C9 polymorphism in the clearance of S-warfarin [5, 15, 28–30] and in daily dose requirements [18, 21, 31]. There is an association between CYP2C9 genotype and the risk of bleeding, with patients having one or more CYP2C9 variant alleles being at higher risk [18]. Our data show that the *3/*3 genotype resulted in a reduction in clearance by up to 71%, which is consistent with the much lower daily dose requirement in these patients. This is, however, limited by the fact that it is based on only two patients, but is nevertheless consistent with the estimate (85.2%) of Hamberg et al. [5], the only other assessment of CYP2C9 genotypes on the population pharmacokinetics of warfarin. The models derived using plasma concentrations taken over a relatively long period of time demonstrate that bodyweight influences the clearance of both R- and S-warfarin.
R-Warfarin is less potent than S-warfarin and undergoes metabolism via CYP1A2, CYP3A4 and CYP2C19[1]. Noncompartmental analyses have indicated a role for CYP2C19 in determining the clearance of R-warfarin [10, 30]; consistent with this, our data showed that SNP rs3814637 in CYP2C19 significantly influenced CL. This SNP is not in linkage disequilibrium with the CYP2C19*2 allele in this patient population (data not shown), and whether it is functional or acts as a marker for a functional SNP that is in linkage disequilibrium is unknown. CYP1A2 polymorphisms had no effect on R-warfarin clearance. Interestingly, a novel finding in our population was the effect of the CYP3A4*1G polymorphism (rs2242480), which showed a significant influence, with heterozygotes having a higher clearance of R-warfarin. The functional effect of this polymorphism has been subject to controversy, with some studies suggesting that this is a gain-in-function polymorphism [32, 33], while others suggesting that it leads to a reduction in CYP3A4 activity [34, 35]. Interestingly, more recently, use of a luciferase reporter system showed that the A allele had significantly higher transcriptional activity than the G allele [36]. The contradictory reports on the functional effect of this polymorphism are also not helped by the inconsistency of the nomenclature used in the literature. Given the discordant findings regarding the functional effect of this polymorphism, our finding with R-warfarin needs to be confirmed in further specifically designed studies. The major influence on the R-warfarin model was bodyweight, which had a significant impact on CL. Age was also shown to be associated with CL, with CL decreasing with age, after accounting for bodyweight.
The final models for both R- and S-warfarin suggest that there is still a significant proportion of the interindividual variability in the PK parameters that is unexplained. This is likely to be due to the numerous other factors, including diet, the presence of other diseases and non-adherence to warfarin.
In conclusion, the study provides a PK model for patients on long-term warfarin therapy, as opposed to other studies that concentrate on the short-term pharmacokinetics following a single dose of warfarin. The analysis suggests that bodyweight, age, sex and particularly CYP2C9 genotype have a profound effect on S-warfarin clearance, while bodyweight, age and CYP2C19 and CYP3A4 genotypes had a significant impact on R-warfarin clearance. No covariates were significantly related to the volume of distribution and, owing to the lack of early data after dosing, the absorption rate could not be estimated. The data presented have related clinical and genetic factors to exposure rather than dose, which is more prone to error because of adherence. This has allowed us to provide quantitative estimates of how different clinical and genetic factors affect the clearance of not only S-warfarin, but also of R-warfarin, which will be important for future attempts at developing individualized dosing algorithms. The relationship between warfarin concentration and anticoagulant response is being considered separately using alternative approaches, such as artificial neural networks.
Acknowledgments
The authors acknowledge the support of the Department of Health (NHS Chair of Pharmacogenetics) and the Wellcome Trust Sanger Institute. MP is an NIHR Senior Investigator.
Competing Interests
There are no competing interests to declare.
REFERENCES
- 1.Wittkowsky AK. Warfarin and other coumarin derivatives: pharmacokinetics, pharmacodymanics and drug interactions. Semin Vasc Med. 2003;3:221–30. doi: 10.1055/s-2003-44457. [DOI] [PubMed] [Google Scholar]
- 2.Cannegieter SC, Rosendaal FR, Wintzen AR, van der Meer FJ, Vandenbroucke JP, Briët E. Optimal oral anticoagulant therapy in patients with mechanical heart valves. N Engl J Med. 1995;333:11–7. doi: 10.1056/NEJM199507063330103. [DOI] [PubMed] [Google Scholar]
- 3.Hylek EM, Skates SJ, Sheehan MA, Singer DE. An analysis of the lowest effective intensity of prophylactic anticoagulation for patients with non-rheumatic atrial fibrillation. N Engl J Med. 1996;335:540–6. doi: 10.1056/NEJM199608223350802. [DOI] [PubMed] [Google Scholar]
- 4.Kearon C, Ginsberg JS, Kovacs MJ, Anderson DR, Wells P, Julian JA, MacKinnon B, Weitz JI, Crowther MA, Dolan S, Turpie AG, Geerts W, Solymoss S, van Nguyen P, Demers C, Kahn SR, Kassis J, Rodger M, Hambleton J, Gent M. Extended Low-Intensity Anticoagulation for Thrombo-Embolism Investigators. Comparison of low intensity warfarin therapy with conventional-intensity warfarin therapy for long-term prevention of recurrent venous thromboembolism. N Engl J Med. 2003;349:631–9. doi: 10.1056/NEJMoa035422. [DOI] [PubMed] [Google Scholar]
- 5.Hamberg A, Dahl M, Barban M, Scordo M, Wadelius M, Pengo V, Padrini R, Jonsson E. A PK-PD model for predicting impact of age, CYP2C9 and VKORC1 genotype on individualization of Warfarin therapy. Clin Pharmacol Ther. 2007;81:529–38. doi: 10.1038/sj.clpt.6100084. [DOI] [PubMed] [Google Scholar]
- 6.Siguret V, Pautas E, Gouin-Thibault I. Warfarin therapy: influence of pharmacogenetic and environmental factors on the anticoagulant response to wearfarin. Vitam Horm. 2008;78:247–64. doi: 10.1016/S0083-6729(07)00012-X. [DOI] [PubMed] [Google Scholar]
- 7.Oake N, Fergusson DA, Forster AJ, van Walraven C. Frequency of adverse events in patients with poor anticoagulation: a meta-analysis. CMAJ. 2007;176:1589–94. doi: 10.1503/cmaj.061523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pirmohamed M, James S, Meakin S, Green C, Scott A, Walley T, Farrar K, Park B, Breckenridge A. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18,820 patients. BMJ. 2004;329:15–9. doi: 10.1136/bmj.329.7456.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wienkers LC, Worden CJ, Storch E, Kunge KL, Rettie AE, Trager WF. Formulation of (R) -8-hydroxywarfarin in human liver microsomers. A new metabolic marker for the (S) mephenytoin hydroxlose P4502C19. Drug Metab Dispos. 1996;24:610–4. [PubMed] [Google Scholar]
- 10.Kaminsky LS, Zhang ZY. Human P450 metabolism of warfarin. Pharmacol Ther. 1997;73:67–74. doi: 10.1016/s0163-7258(96)00140-4. [DOI] [PubMed] [Google Scholar]
- 11.Daly AK, King BP. Pharmacogenetics of oral anticoagulants. Pharmacogenetics. 2003;13:247–52. doi: 10.1097/00008571-200305000-00002. [DOI] [PubMed] [Google Scholar]
- 12.Loebstein R, Yonath H, Peleg D, Almog S, Rotenberg M, Lubetski A, Roitelman J, Harats D, Halkin H, Ezra D. Interindividual variability in sensitivity to warfarin-Nature or nurture? Clin Pharmacol Ther. 2001;70:159–64. doi: 10.1067/mcp.2001.117444. [DOI] [PubMed] [Google Scholar]
- 13.Linder MW, Looney S, Adams J, Johnson N, Antonito-Green D, Lacefield N, Bukaveckas B, Valdes R. Warfarin dose adjustment based on CYP2C9 genetic polymorphisms. J Thromb Thrombolysis. 2002;14:227–32. doi: 10.1023/a:1025052827305. [DOI] [PubMed] [Google Scholar]
- 14.Kamali F, Khan TI, King BD, Frearson R, Kesteven P, Wood P, Daly AK, Wynne H. Contribution of age, body size, and CYP2C9 genotype to anticoagulation response to Warfarin. Clin Pharmacol Ther. 2004;75:204–12. doi: 10.1016/j.clpt.2003.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Herman D, Locatelli I, Grabnar I, Peternel P, Stegnar M, Mrhar A, Breskvar K, Dolzan V. nfluence of CYP2C9 polymorphisms, demographic factors and concomitant drug therapy on warfarin metabolism and maintenance dose. Pharmacogenomics J. 2005;5:193–202. doi: 10.1038/sj.tpj.6500308. [DOI] [PubMed] [Google Scholar]
- 16.Rettie AE, Tai G. The pharmacogenomics of Warfarin: closing in on personalized medicine. Mol Interv. 2006;6:223–7. doi: 10.1124/mi.6.4.8. [DOI] [PubMed] [Google Scholar]
- 17.AH W. Use of genetic and nongenetic factors in Warfarin dose algorithms. Pharmacogenomics. 2007;8:851–61. doi: 10.2217/14622416.8.7.851. [DOI] [PubMed] [Google Scholar]
- 18.Lindh JD, Holm L, Andersson ML, Rane A. Influence of CYP2C9 genotype on warfarin dose requirements – a systematic review and meta-analysis. Eur J Clin Pharmacol. 2009;65:365–75. doi: 10.1007/s00228-008-0584-5. [DOI] [PubMed] [Google Scholar]
- 19.Ufer M. Comparative pharmacokinetics of vitamin K antagonists: warfarin, phenprocoumon and acenocoumarol. Clin Pharmacokinet. 2005;44:1227–46. doi: 10.2165/00003088-200544120-00003. [DOI] [PubMed] [Google Scholar]
- 20.Hamberg A, Wadelius M, Lindh J, Dahl M, Pengo V, Padrini R, Deloukas P, Rane A, Jonsson E. A pharmacometric model describing the relationship between Warfarin dose and INR response with respect to variations in CYP2C9, VKORC1 and age. Clin Pharmacol Ther. 2010;87:727–34. doi: 10.1038/clpt.2010.37. [DOI] [PubMed] [Google Scholar]
- 21.Jorgensen A, Al-Zubiedi S, Zhang E, Keniry A, Hanson A, Hughes D, Van Eker D, Stevens L, Hawkins K, Toh C, Kamali F, Daly A, Fitzmaurice D, Coffey A, Williamson P, Deloukas P, Pirmohamed M. Genetic and environmental factors determining clinical outcomes and cost of warfarin therapy: a prospective study, accepted. Pharmacogenet Genomics. 2009;19:800–12. doi: 10.1097/FPC.0b013e3283317ab5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Naidong W, Lee JW. Development and validation of a high-performance liquid chromatographic method for the quantification of Warfarin emantiomers in human plasma. J Pharm Biomed Anal. 1993;11:785–92. doi: 10.1016/0731-7085(93)80070-h. [DOI] [PubMed] [Google Scholar]
- 23.Matthews I, Aarons L. A population pharmacokinetic model for S-Warfarin application of a mixture model to determine genotype/phenotype. Proceedings of the 15th annual meeting of the population approach group in Europe, Bruges, Belgium 14–6 June 2006.
- 24.Mosteller R. Simplified Calculation of Body Surface Area. N Engl J Med. 1987;317:1098. doi: 10.1056/NEJM198710223171717. [DOI] [PubMed] [Google Scholar]
- 25.Heimark I, Wienkers L, Kunze K, Gibaldi M, Eddy A, Trager W, O'Reilly R, Goulart D. The mechanism of the interaction between Amiodarone and Warfarin in humans. Clin Pharmacol Ther. 1992;51:398–407. doi: 10.1038/clpt.1992.39. [DOI] [PubMed] [Google Scholar]
- 26.Wynne H, Kamali F, Edwards C, Long A, Kelly P. Effects of aging upon Warfarin dose requirements: a longitudinal study. Age Ageing. 1996;26:429–31. doi: 10.1093/ageing/25.6.429. [DOI] [PubMed] [Google Scholar]
- 27.Garcia D, Regan S, Crowther M, Hughes R, Hylek E. Warfarin maintenance dosing patterns in clinical practice: implications for safer anticoagulation in the elderly population. Chest. 2005;127:2049–56. doi: 10.1378/chest.127.6.2049. [DOI] [PubMed] [Google Scholar]
- 28.Ouellet D, Bramson C, Carvajal-Gonzalez S, Roman D, Randinitis E, Remmers A, Gardner MJ. Effects of lasofoxifene on the pharmacokinetics and pharmacodynamics of single-dose warfarin. Br J Clin Pharmacol. 2006;61:741–5. doi: 10.1111/j.1365-2125.2006.02589.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim JS, Nafziger AN, Gaedigk A, Dickmann LJ, Rettie AE, Bertino JS., Jr Effects of oral vitamin K on S- and R-warfarin pharmacokinetics and pharmacodynamics: enhanced safety of warfarin as a CYP2C9 probe. J Clin Pharmacol. 2001;41:715–22. doi: 10.1177/00912700122010618. [DOI] [PubMed] [Google Scholar]
- 30.Uno T, Sugimoto K, Sugawara K, Tateishi T. The effect of CYP2C19 genotypes on the pharmacokinetics of warfarin enantiomers. J Clin Pharm Ther. 2008;33:67–73. doi: 10.1111/j.1365-2710.2008.00887.x. [DOI] [PubMed] [Google Scholar]
- 31.Warfarin I. Pharmacogenetics Consortium. Estimation of the warfarin dose with clinical and pharmacogenetic data. N Engl J Med. 2009;360:753–64. doi: 10.1056/NEJMoa0809329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu XY, Jiao Z, Zhang M, Zhong LJ, Liang HQ, Ma CL, Zhang L, Zhong MK. Association of MDR1, CYP3A4*18B, and CYP3A5*3 polymorphisms with cyclosporine pharmacokinetics in Chinese renal transplant recipients. Eur J Clin Pharmacol. 2008;64:1069–84. doi: 10.1007/s00228-008-0520-8. [DOI] [PubMed] [Google Scholar]
- 33.Hu YF, Tu JH, Tan ZR, Liu ZQ, Zhou G, He J, Wang D, Zhou HH. Association of CYP3A4*18B polymorphisms with the pharmacokinetics of cyclosporine in healthy subjects. Xenobiotica. 2007;37:315–27. doi: 10.1080/00498250601149206. [DOI] [PubMed] [Google Scholar]
- 34.Zhang W, Chang Y-Z, Kan Q-C, Zhang L-R, Li Z-S, Lu H, Wang Z-Y, Chu Q-J, Zhang J. CYP3A4*1G genetic polymorphism influences CYP3A activity and response to fentanyl in Chinese gynecologic patients. Eur J Clin Pharmacol. 2010;66:61–6. doi: 10.1007/s00228-009-0726-4. [DOI] [PubMed] [Google Scholar]
- 35.Gao Y, Zhang LR, Fu Q. CYP3A4*1G polymorphism is associated with lipid-lowering efficacy of atorvastatin but not of simvastatin. Eur J Clin Pharmacol. 2008;64:877–82. doi: 10.1007/s00228-008-0502-x. [DOI] [PubMed] [Google Scholar]
- 36.He BX, Shi L, Qiu J, Tao L, Li R, Yang L, Zhao SJ. A functional polymorphism in the CYP3A4 gene is associated with increased risk of coronary heart disease in the Chinese Han population. Basic Clin Pharmacol Toxicol. 2011;108:208–13. doi: 10.1111/j.1742-7843.2010.00657.x. [DOI] [PubMed] [Google Scholar]


