Abstract
Ectodermal dysplasia with immune deficiency (EDI) is an immunological and developmental disorder caused by alterations in the gene encoding NF-κB essential modulator (NEMO; also known as IκB kinase γ subunit [IKKγ]). Missense mutations in the gene encoding NEMO are associated with reduced signal-induced nuclear translocation of NF-κB proteins, resulting in defective expression of NF-κB target genes. Here, we report 2 unrelated male patients with EDI, both of whom have normal NEMO coding sequences, but exhibit a marked reduction in expression of full-length NEMO protein. TLR4 stimulation of APCs from these patients induced normal cytoplasmic activation and nuclear translocation of NF-κB. However, cells deficient in full-length NEMO were defective in expression of NF-κB–regulated cytokines, such as IL-12, suggesting a downstream defect in chromatin accessibility for NF-κB transcription factors. TLR4-stimulated APCs from the patients were defective in IKKα-dependent H3 histone phosphorylation at the IL-12 promoter and recruitment of NF-κB heterodimers RelA and cRel to the promoter. Expression of a super-active form of IKKα restored IL-12 production in a NEMO knockdown human monocytic cell line following LPS treatment. Our findings suggest that NEMO regulates the nuclear function of IKKα and offer new insights into the mechanisms underlying diminished NF-κB signaling in patients with EDI.
Introduction
Ectodermal dysplasia with immune deficiency (EDI) is an X-linked immunological and developmental disorder caused by mutations in the gene encoding NF-κB essential modulator (NEMO; also known as IκB kinase γ subunit [IKKγ]). NEMO is a regulatory subunit of the IκB kinase (IKK) complex, which also contains 2 structurally related catalytic subunits, IKKα and IKKβ (1). The IKK complex tightly controls the classical activation pathway of the NF-κB/Rel transcription factor family by promoting phosphorylation and degradation of cytosolic IκBs (2). Although IKKα and IKKβ have a high degree of amino acid homology, these kinases have distinct functional properties (3): IKKβ is primarily cytoplasmic, whereas IKKα is present in both the cytoplasm and the nucleus (4). Gene-targeting studies in mice revealed that IKKβ and NEMO are critical for NF-κB activation in response to proinflammatory stimuli and microbial products. However, IKKα is not essential for cytokine-induced IκB degradation and primarily controls NF-κB–mediated gene expression by 2 mechanisms. The first involves a NEMO-independent NF-κB signaling pathway that is critical for lymphoid organ development and B cell maturation (5, 6). Following activation by NF-κB–inducing kinase (NIK), IKKα homodimers process NF-κB2 to produce the p52 subunit. The NF-κB p52 subunit then associates with another NF-κB family member, RelB, forming a dimer that migrates to the nucleus and stimulates target gene expression (7). This alternative NF-κB signaling pathway is activated by certain members of the TNF family, but not by pattern recognition receptors such as TLR4 (8). A second hypothesis involves a nucleosomal function for IKKα in response to proinflammatory stimuli. Recent reports demonstrate that IKKα is recruited to NF-κB–regulated promoters and phosphorylates specific residues of histone H3 upon activation by cytokines (9–12). However, the precise mechanism by which nuclear IKKα activity is regulated is not known.
The gene encoding NEMO lies on the X chromosome, and depending upon their severity, alterations in the NEMO gene lead to 2 allelic diseases: incontinentia pigmenti (IP) and EDI (13). IP corresponds to a complete loss of NEMO function and is typically lethal in utero in males. Female carriers of NEMO mutations exhibit abnormalities of teeth, hair, skin, nails, and the central nervous system. The severity of such abnormalities in females can be highly variable and depends on lyonization of the X chromosome in individual cells. EDI results from hypomorphic coding mutations of NEMO that partially impair, but do not abolish, IKK complex–dependent phosphorylation and subsequent degradation of cytoplasmic inhibitors of NF-κB (14, 15). Male patients with EDI exhibit abnormal development of ectoderm-derived structures and suffer from recurrent pyogenic infections involving deep-seated organs (16). We have previously demonstrated that B lymphocytes and APCs of individuals with a C417 mutation in the zinc finger domain of NEMO show a profound defect in CD40-mediated IκBα degradation that results in reduced NF-κB nuclear translocation and binding activity (15). In such patients, B cells stimulated with CD40 ligand lack both class-switch recombination and somatic hypermutation in the variable region of immunoglobulin (17). Furthermore, DCs fail to upregulate costimulatory molecules, secrete proinflammatory cytokines such as IL-12, or induce allogeneic T cell proliferation in response to CD40 stimulation (18).
In this study, we report 2 patients with typical features of EDI who fail to express normal levels of full-length NEMO, but do not have mutations in the coding region of the NEMO gene. Interestingly, TLR4-induced cytoplasmic activation and nuclear localization of NF-κB is well conserved in DCs of these patients, whereas expression of NF-κB target genes such as IL-12 is profoundly impaired. The defect in expression of NF-κB target genes is associated with failure to recruit IKKα to certain NF-κB–responsive promoters. These results define a new and unexpected function of NEMO in the regulation of the NF-κB signaling pathway.
Results
Reduced levels of full-length NEMO in patients with EDI.
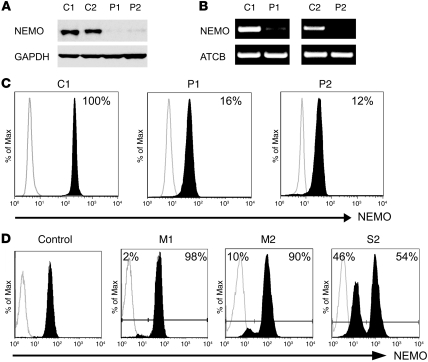
This study describes 2 unrelated male subjects (P1 and P2) who displayed the classic features of EDI, an X-linked recessive disorder associated with mutations in the NEMO gene. We sequenced the coding region of the NEMO gene and 30 other genes, some of which were closely associated with the NF-κB signaling cascade, and did not find any mutations (data not shown). We next measured the expression of NEMO protein in peripheral mononuclear cells (PMCs) from both patients and 2 healthy (control 1 [C1] and C2) individuals by immunoblotting. In contrast with that in healthy subjects, NEMO expression was barely detectable in cellular extracts from the 2 patients (Figure 1A). Multiple cell subsets from the hematopoietic compartment were analyzed, and reduced NEMO expression was always observed (data not shown). To determine whether reduced NEMO protein levels correlate with reduced gene expression, we isolated total mRNA from the affected male patients and controls, and amplified NEMO using specific primers. Semiquantitative analysis with β-actin as a control showed reduced expression of the NEMO transcript in patient cells (Figure 1B), suggesting that the defect lies upstream of NEMO protein synthesis. To further confirm reduced NEMO expression in patient cells, we used flow cytometry (19) to quantitate intracellular NEMO in immune cells. Consistent with the results of immunoblotting, we observed an approximately 5-fold reduction in NEMO expression in patient cells (relative NEMO expression level of 16% and 12% for P1 and P2, respectively, compared with control) (Figure 1C). We also had the opportunity to perform this analysis on cells from the mothers of both patients and P2’s sister. As shown in Figure 1D, the vast majority of cells from both mothers expressed a normal level of NEMO, while a distinct population of cells demonstrated reduced NEMO expression. In contrast, 54% of cells from P2’s sister expressed normal levels of NEMO and 46% of her cells showed reduced expression. Because 1 of the 2 X chromosomes in females is subject to lyonization, these observations in carrier females suggest an X-linked pattern of inheritance and implicate NEMO as the disease susceptibility gene in these 2 kindreds.
Figure 1. EDI patients P1 and P2 exhibit reduced NEMO expression.
(A) NEMO protein levels in whole-cell extracts prepared from PMCs were determined by immunoblot analysis with anti-NEMO antibodies. (B) NEMO mRNA levels in PMCs were measured by semiquantitative RT-PCR. Primer pairs were designed to amplify NEMO exon 2. (C) NEMO protein expression was measured by flow cytometry. Cells were fixed, permeabilized, and stained with anti-NEMO antibody (black histograms) or an isotype-matched antibody (white histograms). Percentages represent NEMO expression relative to reference control. (D) NEMO protein levels were measured in PMCs from P1’s mother (M1), P2’s mother, and P2’s sister (S2) after immunostaining, as described above.
A genomic rearrangement leads to reduced NEMO expression in patient P2.
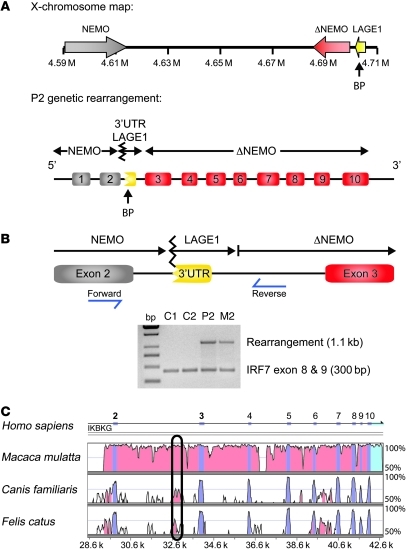
A NEMO pseudogene (ΔNEMO) is located telomeric to the coding NEMO gene on the X chromosome (20). The start codon for NEMO is located in exon 2, whereas ΔNEMO contains only exons 3–10 and is therefore not expressed. Because segmental duplication of genes in the mammalian genome provides substrates for homologous recombination, we looked for a rearrangement between NEMO and ΔNEMO as a cause of reduced NEMO levels. Southern blot analysis and direct sequencing of genomic regions that span NEMO introns did not reveal a genomic abnormality in patient P1; however, in patient P2, we identified a nucleotide sequence in intron 2 of NEMO that corresponded to the 3′ UTR region of the CTAG2/LAGE1 gene that normally neighbors ΔNEMO. A schematic representation of the genomic rearrangement is depicted in Figure 2A. To confirm the presence of this sequence modification in P2, we designed primers complementary to NEMO exon 2 and the ΔNEMO 5′ domain, as shown in Figure 2B. A set of primers for IRF7 was simultaneously used in the PCR reaction as an internal control. As expected, an amplicon of the predicted size of 1.1 kb was detected for the patient (P2), but not for the healthy individuals (Figure 2B). The experiment was repeated with cells from the patient’s mother (M2), and an identical band was detected, indicating that she also carries the genomic rearrangement. To determine how this sequence alteration in the second intron might affect NEMO expression, we performed a computational analysis of genomic NEMO sequences from different species. This analysis identified a highly conserved region in intron 2 that may function as a noncoding regulatory element for NEMO gene transcription (Figure 2C).
Figure 2. Intronic rearrangement in P2 might prevent normal NEMO expression.
(A) Schematic representation of the locus containing NEMO and ΔNEMO on the X chromosome. DNA sequencing in the patient indicated a genomic rearrangement involving the NEMO and ΔNEMO CTAG2/LAGE1. BP, breaking point. (B) Validation of this genomic rearrangement in the patient and his mother. Primer pairs were designed to generate a PCR amplicon of 1.1 kb in the presence of the genomic rearrangement. Products of the predicted size were detected in P2 and M2, but not in the healthy controls. Primers amplifying exons 8 and 9 of IRF7 were used as a PCR internal control (300 bp). (C) NEMO sequences from various species were compared using a computational analysis tool (mVISTA software) to identify conserved regions that might regulate NEMO expression. A conserved domain was identified in intron 2 of all species.
Impaired expression of NF-κB–regulated cytokines in patient cells.
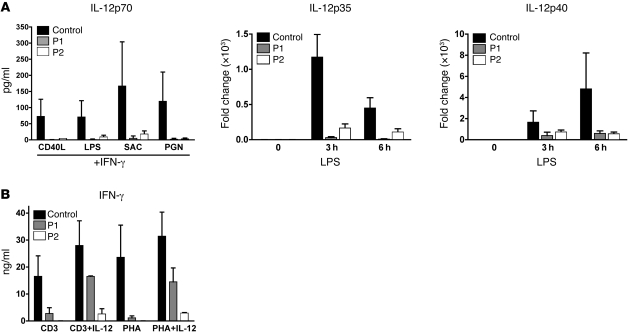
To evaluate the impact of reduced NEMO expression on immune function, we first measured the secretion of proinflammatory cytokines after stimulation of patient PMCs with various stimuli. Both P1 and P2 failed to secrete IL-12p70 in response to CD40L, LPS, Staphylococcus aureus Cowan strain I (SAC), and peptidoglycan (PGN) in the presence of IFN-γ (Figure 3A). We next stimulated DCs generated from patient’s monocytes (monocyte-derived DCs [MoDCs]) with the TLR4 ligand (LPS) for 3 and 6 hours and measured IL-12p35 and IL-12p40 transcripts by quantitative real time RT-PCR. In contrast with normal MoDCs, IL-12p35 expression was not detectable and IL-12p40 mRNA production was markedly reduced in MoDCs from both patients (Figure 3A). The secretion of TNF-α in response to CD40L plus IFN-γ or LPS plus IFN-γ was also diminished in patient PMCs (data not shown). T cell cytokine production was also tested using stimulation with anti-CD3 or phytohemagglutinin (PHA) in the presence or absence of IL-12. In contrast to that in normal individuals, IFN-γ production by patient cells was greatly reduced after both anti-CD3 and PHA stimulation (Figure 3B). Interestingly, the addition of IL-12 partially rescued the expression of IFN-γ. Together, these results suggest that low NEMO levels have an impact on the signaling cascade, leading to reduced secretion of NF-κB–regulated cytokines.
Figure 3. Reduced NEMO expression is associated with impaired production of proinflammatory cytokines.
(A, left panel) IL-12p70 secretion by monocytes in PMCs was measured by a microbead-based assay after 36 hours stimulation with CD40L (2.5 μg/ml), LPS (2.5 μg/ml), SAC (1/10,000 w/v), or PGN (10 μg/ml) in the presence of 2,000 U/ml recombinant IFN-γ. (A, right panels) Measurement of IL-12 subunits (p35 and p40) by quantitative real-time RT-PCR in MoDCs following stimulation with LPS for 3 and 6 hours. Results were normalized to GAPDH expression and represented as a fold change compared with unstimulated conditions. (B) IFN-γ secretion by T cells in PBMCs following stimulation with anti-CD3 (10 μg/ml), anti-CD3 plus IL-12 (10 ng/ml), PHA, or PHA plus IL-12. Data shown are mean ± SD.
NF-κB and MAPK activity are preserved in patient MoDCs.
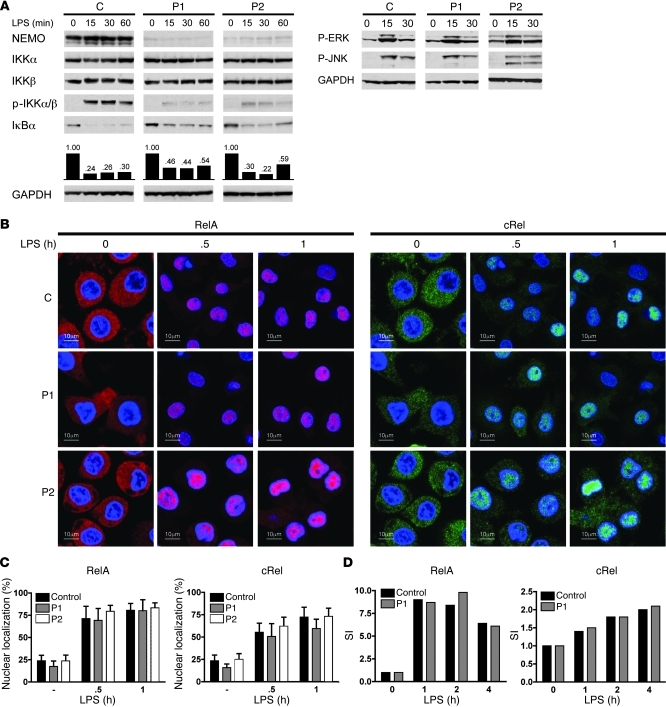
The cellular responses to immune stimuli in EDI patients with coding NEMO mutations are usually associated with NF-κB and MAPK signaling defects (15, 17, 21). We thus subjected MoDCs from controls and patients with reduced full-length NEMO to LPS stimulation and then analyzed the extracts of stimulated cells by immunoblotting with antibodies specific for proximal NF-κB and MAPK intermediates. Whereas control MoDCs displayed easily detectable phospho-IKKα/IKKβ, patient MoDCs displayed greatly reduced phosphorylation of these 2 kinases. Interestingly, this reduced phosphorylation of IKK kinases was not associated with the expected impairment of IκBα degradation (Figure 4A) Indeed, protein quantification by densitometry revealed that IκBα degradation, while reduced in patient MoDCs, was still relatively preserved when compared with that in the control (Figure 4A). We next characterized MAPK signaling events in patient MoDCs (Figure 4A). In response to LPS, phosphorylation of ERK and JNK in patient MoDCs occurred normally despite reduced NEMO levels (Figure 4B). Hence, these results indicate that reduced expression of full-length NEMO is not associated with marked defects in IκBα and MAPK processing following LPS stimulation (Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI42534DS1).
Figure 4. NF-κB and MAPK activity are preserved in patient MoDCs.
(A) Reduced levels of NEMO expression did not alter LPS-stimulated NF-κB and MAPK activity. Control and patient MoDCs were stimulated with LPS (2.5 μg/ml) for the indicated times; then proteins were extracted and subjected to Western blot analysis. Blots were probed with antibodies to NEMO, IKKα, IKKβ, phospho-IKKα/β (p-IKKα /β), IκBα, or GAPDH. Representative data from 2 independent experiments are shown. (B) Reduced NEMO expression does not affect NF-κB nuclear migration and DNA binding upon LPS stimulation. After LPS activation, MoDCs were fixed, permeabilized, and immunostained with anti-RelA antibody (left panels, red) or anti-cRel antibody (right panels, green) together with a nuclear-specific marker (blue). Original magnification, ×40. (C) Quantification of nuclear RelA and cRel staining. 15 images were selected for quantification. Data shown are mean ± SD. SI, stimulation index. (D) In cellular extracts prepared from LPS-activated (2.5 μg/ml) MoDCs, NF-κB DNA–binding activity was measured by ELISA (TransAM). One experiment of 2 is shown.
Following IκBα degradation, released NF-κB molecules translocate from the cytoplasm to the nucleus where they activate target gene expression through association with NF-κB promoters. Using immunostaining and confocal microscopy, we next investigated the subcellular distribution of RelA and cRel in MoDCs following LPS stimulation. As shown in Figure 4B, migration of RelA and cRel from the cytoplasm to the nuclei proceeded normally in control MoDCs, and maximal nuclear accumulation of both transcription factors was observed after 1 hour of LPS stimulation (Figure 4B). Interestingly, nuclear translocation of RelA and cRel was also noted in LPS-stimulated patient MoDCs (Figure 4B). Using computational analysis of acquired images, we found that the relative distribution of RelA and cRel between the nucleus and cytoplasm of LPS-stimulated MoDCs from patients with reduced expression of full-length NEMO was comparable to that in control MoDCs (Figure 4C). Next, we determined the DNA binding activity of both transcription factors using an NF-κB DNA–binding ELISA assay. Binding of both RelA and cRel to NF-κB–specific oligonucleotides was readily detectable in cellular extracts prepared from LPS-stimulated patient MoDCs at levels equivalent to those observed in LPS-stimulated control MoDCs (Figure 4D). Taken together, these results indicate that very low levels of full-length NEMO in MoDCs are sufficient for cytoplasmic activation, nuclear translocation, and DNA-binding of NF-κB molecules upon TLR4 stimulation.
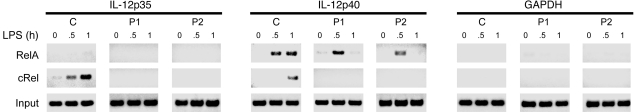
Impaired recruitment of RelA and cRel to IL-12 promoters in patient MoDCs.
Although reduced NEMO expression does not prevent nuclear transport or DNA binding of NF-κB molecules, stimulation-induced expression of NF-κB–regulated cytokines was impaired in patient cells. We therefore used chromatin immunoprecipitation assays to determine whether RelA and cRel were recruited to the promoters of NF-κB target genes. In agreement with previously published results, LPS treatment of normal MoDCs induced recruitment of RelA to the IL-12p40 promoter, but no binding was detectable at the IL-12p35 promoter (Figure 5). Recruitment of cRel to the IL-12p35 promoter was evident in normal MoDCs after 30 minutes of stimulation with LPS and reached a maximum at 60 minutes (Figure 5). Binding was also observed for cRel on the IL-12p40 promoter at 60 minutes in LPS-stimulated normal MoDCs. The GAPDH promoter does not have known consensus NF-κB–binding sites, and as expected, neither RelA nor cRel bound to the GAPDH promoter (Figure 5). In sharp contrast to the binding of cRel to the IL-12 promoter in normal MoDCs, there was no detectable association of cRel at the IL-12p35 and –p40 promoters in LPS-treated patient MoDCs (Figure 5). Moreover, although RelA bound to the IL-12p40 promoter in LPS-treated patient MoDCs, binding was transient and was not detectable at 60 minutes of LPS stimulation. These results indicate that the defective production of IL-12 in cells expressing low levels of NEMO protein may be due to impaired recruitment of NF-κB molecules at the IL-12 promoter.
Figure 5. Reduced NEMO expression prevents normal recruitment of RelA and cRel to IL-12 subunit promoters.
ChIP assays were carried out on MoDCs stimulated with LPS (2.5 μg/ml) for the indicated times. After immunoprecipitation with antibodies against RelA or cRel, chromatin was amplified using primer pairs that selectively surround the κB site of IL-12p35 and IL-12p40 promoters. Primers in the GAPDH promoter were used as a negative control. The input lane corresponds to 10% of total isolated chromatin amplified with indicated primers.
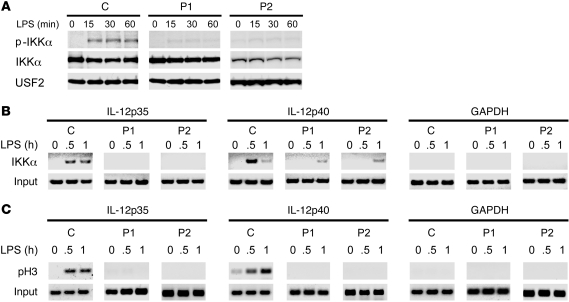
Reduced NEMO expression is associated with reduced nuclear IKKα phosphorylation.
It has recently been shown that IKKα functions in the nucleus to regulate gene expression through its association with NF-κB–responsive promoters and subsequent phosphorylation of Ser10 on the N-terminal tail of histone H3 (10, 12). To address whether reduced levels of full-length NEMO affect the activation of nuclear IKKα, we stimulated patient and control MoDCs with LPS and measured the phosphorylation of IKKα in nuclear lysates by Western blot with antibody specific for phospho-IKKα. Endogenous IKKα can be detected in nucleus and cytoplasm of MoDCs, whereas NEMO and IKKβ are primarily in the cytoplasm. Following LPS stimulation of MoDCs from controls, phosphorylation of nuclear IKKα was detectable at 15 minutes and maintained for at least 60 minutes (Figure 6A). In contrast, there was no increase in nuclear IKKα phosphorylation in MoDCs from patients following LPS stimulation. These results suggest that NEMO can affect nuclear IKKα activation, which is distinct from its previously described role in regulating the phosphorylation of IκB proteins by the IKK complex.
Figure 6. Reduced NEMO expression prevents phosphorylation of nuclear IKKα, recruitment of IKKα to IL-12 promoters, and phosphorylation of histone H3 following LPS stimulation.
(A) Nuclear proteins from LPS-activated MoDCs were analyzed by Western blotting using antibodies against IKKα, anti–phospho-IKKα, and USF2. (B and C) ChIP assays were performed on MoDCs stimulated with LPS. IKKα (B) and phospho-Ser10 histone H3 (C) were immunoprecipitated using specific antibodies, and the κB site of IL-12p35 and IL-12p40 promoters was amplified. GAPDH was used as a negative control, and the input lane corresponds to 10% of total isolated chromatin amplified with corresponding primers.
Defective IKKα binding at the IL-12 promoter in patient DCs.
To address whether nuclear IKKα is involved in IL-12 expression, we measured association of IKKα with the IL-12 promoter in LPS-stimulated MoDCs. As shown in Figure 6B, we found that IKKα is recruited to IL-12p35 and IL-12p40 promoters in control MoDCs treated with LPS for 30 and 60 minutes. In contrast, patient MoDCs completely lacked IKKα binding to the IL-12p35 promoter, and IKKα recruitment to the IL-12p40 promoter was clearly delayed compared with that of normal cells stimulated with LPS (Figure 6B). These findings indicate that recruitment of IKKα to the IL-12 promoter is defective in LPS-stimulated MoDCs expressing reduced levels of NEMO. Because IKKα has been shown to phosphorylate histone H3 on Ser10, we measured the kinetics of LPS-mediated association of phosphorylated histone H3 with the IL-12p35 and IL-12p40 promoters. ChIP assays of control MoDCs demonstrated that LPS treatment led to increased levels of phosphorylated H3 at the IL-12p35 and IL-12p40 promoters at 30 and 60 minutes (Figure 6C). In contrast, no modified histone H3 was detected at either the IL-12p35 or IL-12p40 promoter following LPS treatment of patient MoDCs (Figure 6C). Neither IKKα binding nor H3 phosphorylation was detected at the GAPDH promoter in patient or control MoDCs. Taken together, these data suggest that NEMO is important for binding of IKKα and enhanced association of Ser10-phosphorylated histone H3 on the IL-12p35 and –p40 promoters in LPS-stimulated MoDCs.
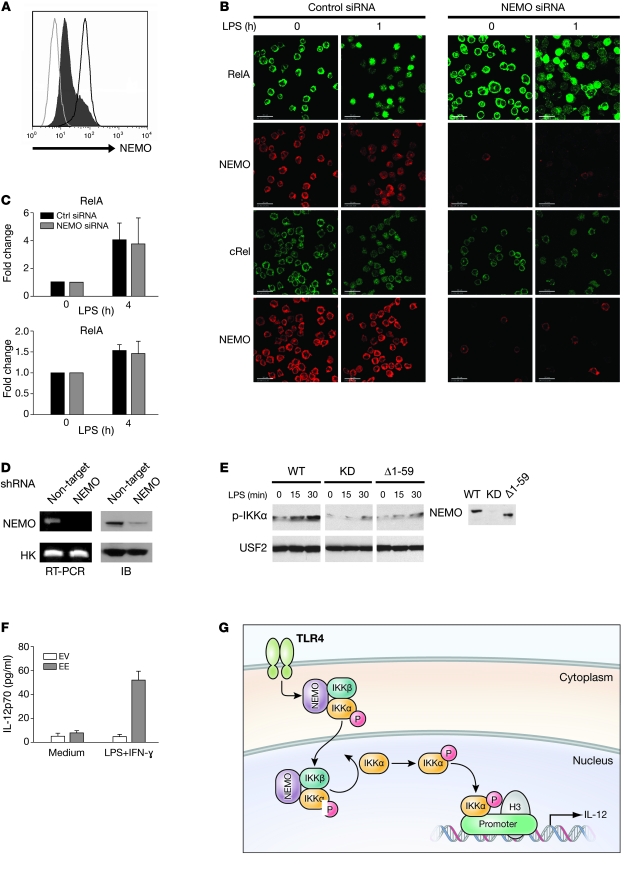
IKKα (EE) expression in NEMOlo THP1 cells restores IL-12 production.
It was important to confirm that a reduced level of full-length NEMO limits IKKα activity and IL-12 synthesis with an in vitro functional assay. We utilized both siRNA and lentivirus-based shRNA approaches to knock down NEMO expression in the monocytic cell line THP1. For siRNA NEMO knockdown, THP1 cells were transfected twice with a pool of 4 siRNAs targeting NEMO, and residual NEMO expression was determined by flow cytometry. As shown in Figure 7A, greater than 5-fold reduction in NEMO expression was observed in THP1 NEMO knockdown (NEMOlo) cells, which closely corresponded to NEMO protein levels in patient cells. We next determined whether low NEMO expression in THP1 affected NF-κB nuclear translocation following LPS activation. NEMOlo THP1 cells were stimulated with LPS, and nuclear NF-κB subunits RelA and cRel were detected by confocal microscopy. NEMO knockdown did not alter the nuclear translocation of RelA and cRel at 60 minutes after LPS treatment (Figure 7B). We also measured the formation of NF-κB DNA–binding complexes in NEMOlo THP1 after 4 hours stimulation with LPS. As expected, reduced levels of full-length NEMO in THP1 cells did not change the DNA-binding activity of RelA and cRel following LPS stimulation (Figure 7C).
Figure 7. Complementation experiments in THP1 cells.
(A) THP1 cells were transfected with either control siRNA (dark gray line) or NEMO siRNA (black histogram), and residual NEMO expression was determined by flow cytometry. Isotype-matched control antibody staining is represented by the light gray line. (B) Nuclear translocation of NF-κB in siRNA-transfected THP1 was assayed by confocal microscopy. THP1 cells transfected either with control siRNA (left panels) or NEMO siRNA (right panels) were stimulated with LPS (2.5 μg/ml) for 60 minutes and then stained with either anti-RelA (green) or anti-cRel (green) antibody. Anti-NEMO antibody (red) was also used to assess NEMO expression in visualized cells. Scale bars: 30 μm. (C) RelA and cRel DNA–binding activity was measured in cellular extracts prepared from LPS-stimulated NEMO knockdown THP1 cells by ELISA. Mean and SD of 3 independent experiments are displayed. Ctrl, control. (D) NEMO expression determined by either immunoblot and semiquantitative PCR in THP1 cells transduced with either nontargeting or NEMO-targeting shRNA-expressing lentivirus. (E) A direct interaction between NEMO and IKKα is necessary for nuclear IKKα phosphorylation. THP1 NEMO knockdown (KD) cells were reconstituted with WT, empty vector, or a truncated form of NEMO that lacks IKKα binding. Following LPS stimulation for the indicated time, nuclear extracts were prepared and subjected to Western blot analysis with phospho-IKKα/β or USF2 antibodies. A Western blot of cellular NEMO levels is shown in the right panel. (F) IL-12p70 measurements in supernatants from THP1 NEMO knockdown cells transfected with either IKKα (EE) or empty vector (EV) following stimulation with LPS (2.5 μg/ml) plus IFN-γ (2,000 U/ml). (G) Proposed model for the dual function of NEMO in the NF-κB signaling pathway. Upon LPS stimulation, the pool of phosphorylated IKK complexes constitutively enter the nucleus, where they can phosphorylate IKKα. Activated nuclear IKKα phosphorylates histone H3 on the promoters of several NF-κB–responsive genes to mediate gene transcription.
To address whether IKKα is important for IL-12 synthesis, we used a lentiviral vector expressing shRNAs to specifically and stably knock down the expression of NEMO in THP1 cells. Semiquantitative PCR and Western blot were used to demonstrate shRNA knockdown for NEMO (Figure 7D). To determine whether direct complex between NEMO and IKKα is required for nuclear IKKα phosphorylation, THP1 cells with stable NEMO knockdown were transfected with either WT NEMO, empty vector, or a truncated form of NEMO lacking the aminoterminal α-helical region that has previously been shown to associate with a carboxyterminal segment of IKKα (22). Phosphorylation of IKKα was readily detected at 15 minutes and peaked at 30 minutes in nuclear lysates prepared from LPS-stimulated NEMO knockdown THP1 cells reconstituted with WT NEMO (Figure 7E). In contrast, minimal IKKα phosphorylation was noted in nuclear lysates prepared from NEMO knockdown THP1 cells following LPS stimulation or those reconstituted with the truncated form of NEMO that lacks IKKα binding. These results suggest that a stable complex consisting of IKKα and NEMO is required for nuclear IKKα activity. We next transfected the shRNA NEMO knockdown THP1 cell line with a constitutively active kinase mutant IKKα (EE) plasmid construct where the phospho-acceptor sites are converted to glutamic acids. THP1 NEMO knockdown cells transfected with IKKα (EE) or empty vector were then treated with LPS, and IL-12 was measured in supernatants with antigen-specific ELISA. IL-12 production was markedly increased following LPS stimulation in NEMO knockdown THP1 cells transfected with IKKα (EE), whereas IL-12 production was unchanged in control cells (Figure 7F). These results indicate that IKKα (EE) overexpression rescues the defect in LPS-mediated IL-12 production in cells with reduced expression of full-length NEMO.
Discussion
The transcription factor NF-κB regulates expression of genes involved in multiple cellular processes, including immune and inflammatory responses, cell adhesion, and cell survival. Mutations in the genes encoding NF-κB essential modulator NEMO and the NF-κB inhibitor IκBα cause EDI, a clinical disorder characterized by severe immune deficiency, with reduced or absent sweat glands and hair follicles. The study of such individuals has provided insight into the role of the IKK signalosome in NF-κB activation; however, much less is known about the function of NEMO in downstream signaling events. In this study, we describe 2 unrelated individuals with reduced expressed of full-length NEMO who have no mutations in the NEMO coding sequence. These patients exhibit clinical features of ectodermal dysplasia with a history of recurrent pyogenic infections indicative of immune deficiency. Our finding that NEMO is weakly expressed in these individuals shows for what we believe is the first time that EDI can be caused by noncoding mutations that affect NEMO expression. Moreover, the identification of a genomic rearrangement involving the second intron in 1 patient suggests that the UTR region of NEMO may contain a regulatory element that is important for transcript stability and therefore protein synthesis. The fact that this second intronic region is highly conserved among several species is consistent with this hypothesis and warrants further investigation.
As is the case for EDI patients with mutations in the coding region of NEMO, PMCs from both P1 and P2 were defective in the expression of NF-κB–regulated proinflammatory cytokines. Reduced IL-12 production was also observed in LPS-stimulated NEMO knockdown THP1 cells, indicating that normal expression levels of full-length NEMO are required for induction of cytokines following stimulation. Coding mutations in NEMO inhibit activation and nuclear translocation of NF-κB following stimulation, resulting in defective expression of NF-κB target genes. Unexpectedly, APCs from both patients expressing low levels of full-length NEMO had preserved cytoplasmic activation and nuclear translocation of NF-κB following stimulation, but were defective in binding of NF-κB to the cytokine IL-12 promoter. This failure to recruit NF-κB at the IL-12 promoter was associated with impaired nuclear phosphorylation of IKKα as well as a failure of IKKα to bind to the IL-12 promoter and phosphorylate histone H3. Combined with similar findings in THP1 cells with RNAi-mediated NEMO knockdown, our data define a mechanism for nuclear IKKα activation that requires expression of full-length NEMO. Thus, these findings suggest that NEMO plays a role in the induction of NF-κB–responsive genes through a mechanism that is distinct from IKK complex–mediated degradation of the inhibitors of NF-κB.
Coding mutations in NEMO occur throughout the protein and generally affect NF-κB signaling function through impaired degradation of IκBα (13–15). Integration of these NEMO mutations with biochemical analysis has revealed an important role for K63-linked polyubiquitin chains in this degradation process (18, 23, 24). The majority of NEMO mutations in EDI patients are located in the zinc finger domain, which is a site for lysine 63–linked (K63-linked) polyubiquitination (25). We observed that, in contrast with EDI patients with coding NEMO mutations, the proximal signaling cascade was conserved in patients P1 and P2 with reduced NEMO expression. Indeed, proteosomal degradation of the cytosolic inhibitor IκBα after LPS stimulation was not significantly impaired in patient MoDCs. The simplest explanation is that these IKK complexes containing reduced levels of full-length NEMO still have sufficient kinase activity to stimulate degradation of the IκBα proteins.
DNA damage leads to the phosphorylation, ubiquitination, and sumoylation of NEMO (26–28). This posttranslational modification of the protein in response to genotoxic stress allows NEMO to shuttle between the cytoplasm and nucleus to activate NF-κB. However, NEMO does not exhibit increased nuclear translocation in response to LPS treatment (Supplemental Figure 2). IKKα has a nuclear localization signal, and NEMO-IKKα complexes may constitutively shuttle between the nucleus and cytoplasm as do IκBα and RelA complexes in cells in the absence of exogenous stimuli (26, 29–31). A report of NEMO shuttling in resting HeLa cells in a CRM1-dependent manner supports this hypothesis (32). Since the IKK complex is activated in the cytoplasm in response to LPS stimulation, it is conceivable that the pool of phosphorylated IKK complexes constitutively entering the nucleus, where it can phosphorylate IKKα and regulate nuclear IKKα activity, also increases. Thus, patients with reduced cellular NEMO levels would be expected to also have a reduced pool of activated nuclear IKK complexes available to phosphorylate free IKKα. While we were able to establish that a complex between NEMO and IKKα is necessary for nuclear IKKα phosphorylation, we were not successful in pulling down NEMO from the IL-12 promoter with chromatin immunoprecipitation (our unpublished observations). While this may relate to the limited suitability of the anti-NEMO antibody for chromatin immunoprecipitation, our inability to detect NEMO at the IL-12 promoter is supportive of a hypothesis in which an activated IKK complex phosphorylates free nuclear IKKα. Interestingly, the nuclear IKKα level in patient P2’s MoDCs was reduced in comparison with those of the other patient and reference control, thus suggesting that intact levels of NEMO are necessary to stabilize this kinase in the nucleus. Taken together, these mechanisms may explain why IKKα does not efficiently target histone H3 in our patients.
In a recently published paper, a patient was found to have a mutation in the 5′ UTR region of the NEMO gene that was associated with reduced NEMO protein expression (33). In vitro studies revealed an impairment of TLR7-mediated RelA activation in EBV-transformed B cells and reduced NF-κB reporter activity in IL-1–stimulated transformed fibroblasts. While this patient experienced infections with pyogenic organisms, he did not have dysmorphic features associated with ectodermal dysplasia and was able to generate antigen-specific antibody responses. These clinical findings suggest that, despite a mutation in the 5′ UTR region of NEMO, the NF-κB signaling function is sufficient for the development of ectoderm-derived tissues and certain adaptive immune responses. Interestingly, this patient developed Mycobacterium avium infection. IL-12 has been shown to have an important role in the host defense against mycobacteria, and the synthesis of IL-12 by activated macrophages is often severely suppressed in patients with a variety of NEMO hypomorphic mutations. Similarly, mice that harbor an L153P mutation in the first coiled-coil domain of NEMO also lack the typical feature of ectodermal dysplasia (34). Macrophages isolated from these animals have preserved degradation of IκBα following stimulation by a variety of immune stimuli, yet the synthesis of IL-12 was virtually absent. It is interesting to speculate on whether the impairments of NF-κB–directed gene expression that are associated with other NEMO mutations also result from defective nuclear function of IKKα.
In summary, in this study, we describe 2 EDI patients with reduced expression of full-length NEMO protein caused by noncoding mutations. Impaired expression of NF-κB–induced cytokines in cells from these patients indicates that failure to express full-length NEMO results in the immune deficiency that is characteristic of EDI. Detailed analysis of the signaling abnormalities in these patients revealed a previously unappreciated role for NEMO in regulating IKKα function in the nucleus (see proposed model in Figure 7G).
Methods
Case report.
We studied 2 kindreds, each containing 1 male patient with classical features of anhidrotic EDI at the Clinical Center (CRC) of the NIH (protocol 06-1-0049). At the time of the study, P1 was a 15-year-old male of mixed European descent and had a history of recurrent pneumonia and cellulitis in infancy that were successfully treated with antibiotics. At the age of 9 months, he was noted to have a marked reduction in serum γ globulin levels and absent responses to protein and lipopolysaccharide antigens. Since then, he has been maintained on i.v. immunoglobulin therapy. Cells of various blood lineages, including lymphoid subsets (CD3, CD4, CD8, CD19, CD14, CD56), were present in normal numbers. The B cells were naive with invariable coexpression of IgD and IgM on the surface. His brother died at the age of 16 months due to pneumococcal meningitis.
At the time of the study, P2 was a 4-year-old African-American male who had multiple hospitalizations early in infancy for cellulitis, Gram-positive Bacillus bacteremia, and Pneumocystis carinii pneumonia. A skin biopsy at 11 months demonstrated the absence of adnexal tissue, with eczematous dermatitis and pigmentary incontinence. At 1 year of age, his serum immunoglobulin levels were as follows: IgG 172 (range 345–1213), IgA 9 (range 43–173), IgM 14 (range 14–106), IgE 25 (range 0.80–15.2). He was placed on i.v. γ globulin substitution therapy and trimethoprim/sulfamethizole, which reduced the frequency of infectious complications. By the age of 3, the patient was noted to have atopic symptoms consisting of asthma, atopic dermatitis, and adverse reactions to multiple foods, resulting in severe nutritional deficiency. His gastrointestinal symptoms initially improved with enteral feeding with an elemental formula. However, he subsequently developed eosinophilic colitis and failure to thrive. Treatment with immunosuppressive therapy and total parenteral nutrition was not successful, and his clinical course was notable by frequent line infections and sepsis. He has since undergone allogeneic bone marrow transplantation. There was no family history of immunodeficiency or early pediatric death. Blood lineage cells, including lymphoid subsets (CD3, CD4, CD8, CD19, CD14, CD56), were present in normal numbers before transplantation.
Cell culture and reagents.
PMCs were obtained by gradient density centrifugation of venous blood. MoDCs were prepared by culturing elutriated monocytes in RPMI 1640 (Invitrogen) supplemented with 10% heat-inactivated FBS, GM-CSF (Berlex Laboratories), and IL-4 (R&D Systems) for 6 days. The following reagents were used for stimulation: LPS (2.5 μg/ml; Sigma-Aldrich), CD40L (2.5 μg/ml; Amgen), IFN-γ (2,000 U/ml; Genzyme), SAC (1/10,000 w/v; Calbiochem), PGN (10 μg/ml; Invivogen), anti-CD3 (OKT3; 10 μg/ml; gift of Ortho Biotech), PHA (Invitrogen), and human recombinant IL-12p70 (10 ng/ml; R&D Systems).
Western blotting.
Whole-cell extracts were prepared with a lysis buffer containing 50 mM Tris-HCl, 150 mM NaCl, 10% glycerol, 0.5% NP-40, 40 mM β-glycerophosphate, 1 mM Na3VO4, 30 mM NaF, 5 mM EDTA, 1 mM DTT, 0.1 mM PMSF, and complete protease inhibitor mixture (Roche Diagnostics). Nuclear extracts were prepared using Nuclear Extract Kit from Active Motif. Briefly, cells were incubated in a hypotonic buffer for 15 minutes before addition of detergent. After brief centrifugation, the nuclei were resuspended in a complete lysis buffer according to the manufacturer’s instructions for 30 minutes. Supernatants containing soluble materials were heat denatured at 70°C for 10 minutes in a 4-fold dilution of NuPAGE LDS Sample Buffer (Invitrogen) supplemented with 2.5% 2-ME. Samples were resolved on a 4%–12% polyacrylamide gel (Invitrogen), and proteins were transferred onto nitrocellulose membrane and probed with specific antibodies against proteins of interest. Antibodies against NEMO (clone FL-419), GAPDH (0411), USF2 (C20), and RelA (C20) were from Santa Cruz Biotechnology Inc. Antibodies against IκBα, cRel, IKKα, IKKβ, and phospho-IKKα/β (Ser176/180) were from Cell Signaling Technologies. Detection was performed using HRP-conjugated donkey anti-rabbit or mouse IgG (GE Healthcare) and developed by the chemiluminescence method (GE Healthcare).
Measurement of cytokine expression.
After 36 hours of stimulation, culture supernatants were harvested and cytokine secretion was measured by the Luminex method. For quantitative real-time PCR evaluation, cells were stimulated for 3 and 6 hours with LPS and total RNA was extracted using an RNeasy Kit (QIAGEN) following the manufacturer’s protocol. Purified RNA was reverse transcribed into cDNA using a high-capacity cDNA reverse transcription kit (Applied Biosystems). Real-time PCR was then carried out on a 7500 Real-Time PCR System following the manufacturer’s instructions (Applied Biosystems). Primers and probes for the following human genes were used: IL12A (Hs00168405_m1; Applied Biosystems), IL12B (Hs00233688_m1), and GAPDH (4326317E).
Confocal microscopy.
MoDCs (1 × 105/slide) were directly stimulated on poly-l-lysine–coated slides (Sigma-Aldrich) for the indicated time. Cells were then fixed with a 4% paraformaldehyde solution for 15 minutes at room temperature. Membrane permeabilization was achieved by incubation with a 0.1% Triton X-100 solution for 15 minutes at room temperature. After washing with PBS, cells were incubated with different combinations of specific antibodies in the presence of 1% BSA for 1 hour at room temperature. The following primary antibodies were used: mouse anti-RelA and rabbit anti-RelA (clones F-6 and C-20, respectively; Santa Cruz Biotechnology Inc.), rabbit anti-cRel (Cell Signaling Technologies), and mouse anti-NEMO (clone 54; BD Biosciences). Slides were washed and incubated with Alexa Fluor 488 goat anti-rabbit antibodies, Alexa Fluor 594 goat anti-mouse antibodies (Invitrogen), and DAPI (1:500 dilution) for 30 minutes at room temperature. After extensive washing, slides were mounted with the SlowFade Light Antifade Kit (Invitrogen), and images were collected on a Leica TCS-NT/SP1 confocal microscope (Leica Microsystems) using a ×40 oil immersion objective (NA 1.32, zoom ×4). Images were processed using Leica TCS-NT/SP software (version 1.6.587), Imaris 3.3.2 (Bitplane AG), and Adobe Photoshop 7.0.
Flow cytometry.
Cells were fixed and permeabilized using BD Cytofix/Cytoperm kit (BD Biosciences) according to the manufacturer’s instructions. Cells were incubated with either unconjugated mouse anti-NEMO (clone 54; BD Biosciences) at a final concentration of 1 μg/ml or an isotype control for 30 minutes at 4°C. After washing, PE-coupled antibody was added (1:1000 dilution) for 30 minutes at 4°C in the dark. After a final wash, cells were resuspended in Perm/Wash Buffer and images were collected on a FACSCalibur (BD Biosciences) flow cytometer. Acquired events were analyzed using FlowJo software (Treestar).
Determination of NF-κB DNA–binding activity.
An ELISA-based method, TransAM (Active Motif), was used to quantify RelA and cRel DNA–binding activity according to the manufacturer’s protocol. Cellular extracts (5 μg) from LPS-activated (2.5 μg/ml) MoDCs were incubated in wells that were precoated with κB consensus oligonucleotide. After washing, wells were incubated sequentially with anti-RelA or anti-cRel antibody and secondary HRP-conjugated antibody for 1 hour. Detection was performed by addition of a developing solution, and absorbance was measured at 450 nm.
ChIP.
ChIP assays were performed using ChIP-IT Express from Active Motif according to the manufacturer’s protocol. The primers and probes used for ChIP assays were as follows: IL-12p35 forward primer, 5′-CGAACATTTCGCTTTCATTTTGG-3′; IL-12p35 reverse primer, 5′-GCTGGATGCAGACCTGTGGC-3′; IL-12p40 forward primer, 5′-GCATACAGTTGTTCCATCC-3′; IL-12p40 reverse primer, 5′-CTCTACTCCTTTCTGATGGA-3′; GAPDH forward primer, 5′-TACTAGCGGTTTTACGGGCG-3′; and GAPDH reverse primer, 5′-TCGAACAGGAGGAGCAGAGAGCGA-3′.
NEMO siRNA.
Nontargeting or NEMO siRNA ON-TARGETplus SMARTpool oligonucleotides (Dharmacon) were constructed as follows: nontargeting, 5′-AUGAACGUGAAUUGCUCAA-3′, 5′-UAAGGVUAUGAAGAGAUAC-3′, 5′-AUGUAUUGGCCUGUAUUAG-3′, 5′-UAGCGACUAAACACAUCAA-3′; NEMO, 5′-AACAGGAGGUGAUCGAUAAUU-3′, 5′-GAAGCGGCAUGUCGAGGUCUU-3′, 5′-GAAUGCAGCUGGAAGAUCUUU-3′, and 5′-GGAAGAGCCAACUGUGUGAUU-3′. THP1 cells were plated in 24-well plates (1 × 105 cells/well) in antibiotic-free medium and transfected with 100 nM siRNA using the siRNA-optimized transfection reagent Dharmafect II (Dharmacon) according to the manufacturer’s protocol. After 3 days, cell suspensions were split and a second transfection was carried out. Protein knockdown efficiency was measured after another 2 days by flow cytometry.
shRNA-mediated NEMO silencing and plasmid transfection.
To establish stable NEMO-knockdown THP1 cells, a set of SMARTvector shRNA Lentiviral Particles containing shRNAs targeted to human NEMO was purchased from Dharmacon. After transduction, cells were selected in the presence of puromycin (10 μg/ml) for 2 weeks. Cells that stably expressed the NEMO-silencing constructs were tested for GFP expression and NEMO expression, and the most efficiently knocked down cells were chosen for subsequent plasmid transfection. NEMO knockdown THP1 cells were transfected with Amaxa Nucleofector according to the manufacturer’s protocol. A total of 8 × 106 cells were transfected with 3.2 μg pEF1-mIKKα (EE) or vector control together with 0.8 μg pDsRed-Monomer-C1 (Clontech) expressing RFP to monitor transfection efficiency by FACS. After 24 hours, FACS analysis indicated 50%–60% transfection efficiency. Dead cells were removed by the Dead Cell Removal Kit (Miltenyi Biotech), and live cells were transferred to medium containing LPS plus IFN-γ for an additional 24 hours. The amount of IL-12p70 in the medium was quantitated by ELISA (eBioscience). For reconstitution in NEMO KD THP1 cells, lentivirus vector pLVX-IRES-Neo (Clontech) was used to clone δ 1–59 NEMO and WT NEMO. The QuikChange Site-Directed Mutagenesis Kit (Stratagene) was used to introduce mutations in the wobble positions of the codons in the siRNA target region.
Mutation detection and sequence analysis.
For genotyping analysis, DNA and total RNA were extracted from lymphocytes using standard methods. PCR of genomic DNA was performed with the GC Rich Kit (Roche Diagnostics) using the following PCR cycling conditions: 10 cycles with 1 minute extension followed by 22 cycles with an increment of 1 second per cycle. To detect the patient’s rearrangement, the following sets of primers were used: 5′-CCCTTGCCCTGTTGGATGAAT-3′ (located in the exon 2 of NEMO gene) and 5′-TTACTTTAGAGTCAGGGTCTCG-3′ (located between NEMO pseudogene and CTAG2/LAGE1 gene). As an internal control for PCR amplification, we used primers that flanked exons 8 and 9 from the IRF7 gene: 5′-TGTCACCCTCCCCAAGCG-3′ and 5′-GCATCAGGCGTCTGTCAGTGG-3′. cDNA was obtained from RNA by RT-PCR using NEMO-specific primers. PCR products were purified and cycle sequenced at both the 5′ and 3′ ends using dye terminator dideoxynucleotides. Analysis of nucleotide sequences was performed using annotation available in NCBI. The genomic region of the NEMO gene in different species was analyzed using the software mVISTA (http://genome.lbl.gov/vista/mvista/submit.shtml).
Study approval.
The 2 affected patients and healthy controls without immunological abnormalities were studied at the Clinical Center, NIAID, NIH, under an IRB-approved protocol (NIAID/NIH protocol 00-I-006) and with informed consent.
Statistics.
Statistical analyses were done using GraphPad Prism 4.0. Data shown in Figures 3, 4, and 7 are from 1 experiment that is representative of multiple independent experiments and are given as mean ± SD. Statistical analysis of significance (P values) was based on the 2-tailed Student’s t test for all the figures. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank the families studied for their invaluable contribution to this project; W. Strober and J. Ashwell for helpful discussions; O.M. Schwartz and J. Kabat for microscopy assistance; and Phil Murphy for careful review of the manuscript.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(1):315–326. doi:10.1172/JCI42534.
Stephane T. Temmerman’s present address is: Institute for Medical Immunology (IMI), Université Libre de Bruxelles (ULB), Gosselies, Belgium.
References
- 1.Zandi E, Rothwarf DM, Delhase M, Hayakawa M, Karin M. The IkappaB kinase complex (IKK) contains two kinase subunits, IKKα and IKKβ, necessary for IκB phosphorylation and NF-κB activation. Cell. 1997;91(2):243–252. doi: 10.1016/S0092-8674(00)80406-7. [DOI] [PubMed] [Google Scholar]
- 2.Hayden MS, Ghosh S. Signaling to NF-κB. Genes Dev. 2004;18(18):2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 3.Hacker H, Karin M. Regulation and function of IKK and IKK-related kinases. Sci STKE. 2006;2006(357):re13. doi: 10.1126/stke.3572006re13. [DOI] [PubMed] [Google Scholar]
- 4.Birbach A, Gold P, Binder BR, Hofer E, de Martin R, Schmid JA. Signaling molecules of the NF-κB pathway shuttle constitutively between cytoplasm and nucleus. J Biol Chem. 2002;277(13):10842–10851. doi: 10.1074/jbc.M112475200. [DOI] [PubMed] [Google Scholar]
- 5.Hu Y, et al. Abnormal morphogenesis but intact IKK activation in mice lacking the IKKα subunit of IκB kinase. Science. 1999;284(5412):316–320. doi: 10.1126/science.284.5412.316. [DOI] [PubMed] [Google Scholar]
- 6.Senftleben U, et al. Activation by IKKα of a second, evolutionary conserved, NF-κB signaling pathway. Science. 2001;293(5534):1495–1499. doi: 10.1126/science.1062677. [DOI] [PubMed] [Google Scholar]
- 7.Bonizzi G, et al. Activation of IKKα target genes depends on recognition of specific κB binding sites by RelB:p52 dimers. EMBO J. 2004;23(21):4202–4210. doi: 10.1038/sj.emboj.7600391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dejardin E, et al. The lymphotoxin-β receptor induces different patterns of gene expression via two NF-κB pathways. Immunity. 2002;17(4):525–535. doi: 10.1016/S1074-7613(02)00423-5. [DOI] [PubMed] [Google Scholar]
- 9.Gloire G, et al. Promoter-dependent effect of IKKα on NF-κB/p65 DNA binding. J Biol Chem. 2007;282(29):21308–21318. doi: 10.1074/jbc.M610728200. [DOI] [PubMed] [Google Scholar]
- 10.Yamamoto Y, Verma UN, Prajapati S, Kwak YT, Gaynor RB. Histone H3 phosphorylation by IKK-α is critical for cytokine-induced gene expression. Nature. 2003;423(6940):655–659. doi: 10.1038/nature01576. [DOI] [PubMed] [Google Scholar]
- 11.Park KJ, Krishnan V, O’Malley BW, Yamamoto Y, Gaynor RB. Formation of an IKKα-dependent transcription complex is required for estrogen receptor-mediated gene activation. Mol Cell. 2005;18(1):71–82. doi: 10.1016/j.molcel.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 12.Anest V, Hanson JL, Cogswell PC, Steinbrecher KA, Strahl BD, Baldwin AS. A nucleosomal function for IkappaB kinase-alpha in NF-κB-dependent gene expression. Nature. 2003;423(6940):659–663. doi: 10.1038/nature01648. [DOI] [PubMed] [Google Scholar]
- 13.Smahi A, et al. Genomic rearrangement in NEMO impairs NF-κB activation and is a cause of incontinentia pigmenti. The International Incontinentia Pigmenti (IP) Consortium. Nature. 2000;405(6785):466–472. doi: 10.1038/35013114. [DOI] [PubMed] [Google Scholar]
- 14.Doffinger R, et al. X-linked anhidrotic ectodermal dysplasia with immunodeficiency is caused by impaired NF-κB signaling. Nat Genet. 2001;27(3):277–285. doi: 10.1038/85837. [DOI] [PubMed] [Google Scholar]
- 15.Jain A, Ma CA, Liu S, Brown M, Cohen J, Strober W. Specific missense mutations in NEMO result in hyper-IgM syndrome with hypohydrotic ectodermal dysplasia. Nat Immunol. 2001;2(3):223–228. doi: 10.1038/85277. [DOI] [PubMed] [Google Scholar]
- 16.Zonana J, et al. A novel X-linked disorder of immune deficiency and hypohidrotic ectodermal dysplasia is allelic to incontinentia pigmenti and due to mutations in IKK-gamma (NEMO). Am J Hum Genet. 2000;67(6):1555–1562. doi: 10.1086/316914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain A, et al. Specific NEMO mutations impair CD40-mediated c-Rel activation and B cell terminal differentiation. J Clin Invest. 2004;114(11):1593–1602. doi: 10.1172/JCI21345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Temmerman ST, et al. Impaired dendritic-cell function in ectodermal dysplasia with immune deficiency is linked to defective NEMO ubiquitination. Blood. 2006;108(7):2324–2331. doi: 10.1182/blood-2006-04-017210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishikomori R, et al. X-linked ectodermal dysplasia and immunodeficiency caused by reversion mosaicism of NEMO reveals a critical role for NEMO in human T-cell development and/or survival. Blood. 2004;103(12):4565–4572. doi: 10.1182/blood-2003-10-3655. [DOI] [PubMed] [Google Scholar]
- 20.Aradhya S, et al. Multiple pathogenic and benign genomic rearrangements occur at a 35 kb duplication involving the NEMO and LAGE2 genes. Hum Mol Genet. 2001;10(22):2557–2567. doi: 10.1093/hmg/10.22.2557. [DOI] [PubMed] [Google Scholar]
- 21. Ma CA, et al. Dendritic cells from humans with hypomorphic mutations in NEMO have impaired mitogen–activated protein kinase activity [published online ahead of print January 25, 2011].Hum Mutat . doi:10.1002/humu.21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marienfeld RB, Palkowitsch L, Ghosh S. Dimerization of the I kappa B kinase-binding domain of NEMO is required for tumor necrosis factor α-induced NF-κB activity. Mol Cell Biol. 2006;26(24):9209–9219. doi: 10.1128/MCB.00478-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ea CK, Deng L, Xia ZP, Pineda G, Chen ZJ. Activation of IKK by TNF-alpha requires site-specific ubiquitination of RIP1 and polyubiquitin binding by NEMO. Mol Cell. 2006;22(2):245–257. doi: 10.1016/j.molcel.2006.03.026. [DOI] [PubMed] [Google Scholar]
- 24.Wu CJ, Conze DB, Li T, Srinivasula SM, Ashwell JD. Sensing of Lys 63-linked polyubiquitination by NEMO is a key event in NF-κB activation [corrected]. Nat Cell Biol. 2006;8(4):398–406. doi: 10.1038/ncb1384. [DOI] [PubMed] [Google Scholar]
- 25.Zhou H, et al. Bcl10 activates the NF-κB pathway through ubiquitination of NEMO. Nature. 2004;427(6970):167–171. doi: 10.1038/nature02273. [DOI] [PubMed] [Google Scholar]
- 26.Huang TT, Wuerzberger-Davis SM, Wu ZH, Miyamoto S. Sequential modification of NEMO/IKKgamma by SUMO-1 and ubiquitin mediates NF-kappaB activation by genotoxic stress. Cell. 2003;115(5):565–576. doi: 10.1016/s0092-8674(03)00895-x. [DOI] [PubMed] [Google Scholar]
- 27.Wu ZH, Shi Y, Tibbetts RS, Miyamoto S. Molecular linkage between the kinase ATM and NF-κB signaling in response to genotoxic stimuli. Science. 2006;311(5764):1141–1146. doi: 10.1126/science.1121513. [DOI] [PubMed] [Google Scholar]
- 28.Niu J, Shi Y, Iwai K, Wu ZH. LUBAC regulates NF–κB activation upon genotoxic stress by promoting linear ubiquitination of NEMO. . EMBO J. 2011;30(18):3741–3753. doi: 10.1038/emboj.2011.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson C, Van Antwerp D, Hope TJ. An N-terminal nuclear export signal is required for the nucleocytoplasmic shuttling of IκBα. EMBO J. 1999;18(23):6682–6693. doi: 10.1093/emboj/18.23.6682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodriguez MS, Thompson J, Hay RT, Dargemont C. Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J Biol Chem. 1999;274(13):9108–9115. doi: 10.1074/jbc.274.13.9108. [DOI] [PubMed] [Google Scholar]
- 31.Tam WF, Lee LH, Davis L, Sen R. Cytoplasmic sequestration of rel proteins by IκBα requires CRM1-dependent nuclear export. Mol Cell Biol. 2000;20(6):2269–2284. doi: 10.1128/MCB.20.6.2269-2284.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verma UN, Yamamoto Y, Prajapati S, Gaynor RB. Nuclear role of I κ B Kinase-gamma/NF-κB essential modulator (IKKgamma/NEMO) in NF-κB-dependent gene expression. J Biol Chem. 2004;279(5):3509–3515. doi: 10.1074/jbc.M309300200. [DOI] [PubMed] [Google Scholar]
- 33.Mooster JL, et al. Immune deficiency caused by impaired expression of nuclear factor-κB essential modifier (NEMO) because of a mutation in the 5' untranslated region of the NEMO gene. . J Allergy Clin Immunol. 2010;126(1):127–132. doi: 10.1016/j.jaci.2010.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Siggs OM, et al. A mutation of Ikbkg causes immune deficiency without impairing degradation of IκBα. Proc Natl Acad Sci U S A. 2010;107(7):3046–3051. doi: 10.1073/pnas.0915098107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.