Abstract
Defining the genetic contribution of rare variants to common diseases is a major basic and clinical science challenge that could offer new insights into disease etiology and provide potential for directed gene- and pathway-based prevention and treatment. Common and rare nonsynonymous variants in the GCKR gene are associated with alterations in metabolic traits, most notably serum triglyceride levels. GCKR encodes glucokinase regulatory protein (GKRP), a predominantly nuclear protein that inhibits hepatic glucokinase (GCK) and plays a critical role in glucose homeostasis. The mode of action of rare GCKR variants remains unexplored. We identified 19 nonsynonymous GCKR variants among 800 individuals from the ClinSeq medical sequencing project. Excluding the previously described common missense variant p.Pro446Leu, all variants were rare in the cohort. Accordingly, we functionally characterized all variants to evaluate their potential phenotypic effects. Defects were observed for the majority of the rare variants after assessment of cellular localization, ability to interact with GCK, and kinetic activity of the encoded proteins. Comparing the individuals with functional rare variants to those without such variants showed associations with lipid phenotypes. Our findings suggest that, while nonsynonymous GCKR variants, excluding p.Pro446Leu, are rare in individuals of mixed European descent, the majority do affect protein function. In sum, this study utilizes computational, cell biological, and biochemical methods to present a model for interpreting the clinical significance of rare genetic variants in common disease.
Introduction
Common human diseases result from the combined effects of genetic susceptibility and environmental factors. Understanding the genetic contribution to disease may offer new insights into disease etiology and provide potential for directed gene- and pathway-based prevention and treatment. Genome-wide association studies (GWAS) have been successful in identifying common genetic variants associated with complex disease heritability by their individual statistical associations (1, 2). Individual rare variants conferring low to moderate risk are not as tractable by this approach (2–4). However, emerging evidence suggests rare variants are also important contributors to complex disease susceptibility (5–10). Rapid advances in exome and whole-genome sequencing, as well as collective efforts such as the 1000 Genomes Project, will uncover many novel variants. However, the fundamental challenges in using sequencing for individual diagnostics lie in developing methodologies for distinguishing variants that have demonstrable functional effect from those that are neutral and in relating this information back to clinical phenotypes. The GCKR gene, encoding glucokinase regulatory protein (protein name, GKRP), is a logical candidate for exploration of these issues, as both common and rare variation in GCKR have been suggested as being clinically important and the biology of GCKR has been extensively studied (11–15).
GKRP posttranslationally regulates hepatic glucokinase (GCK) (16). GCK is the predominant hexokinase in pancreatic β cells and hepatocytes, but GCKR is not appreciably expressed in β cells (13, 17–20). In the liver, GKRP binding inhibits GCK competitively with respect to glucose (15, 21). This inhibition is also associated with nuclear sequestration of GCK at low glucose concentrations (14). Glucose-mediated dissociation of GCK from GKRP activates GCK and exposes its nuclear export signal (22). Additionally, GKRP is regulated by binding of fructose 6-phosphate (F6P) or fructose 1-phosphate (F1P). F6P binding to GKRP promotes GKRP-GCK association, while F1P disrupts this interaction (23).
Studies in model systems have suggested GKRP not only serves as a GCK inhibitor, but also plays a somewhat paradoxical role in enhancing GCK protein stability. Gckr-knockout mice exhibit normal fasting glucose levels, but suffer from postprandial hyperglycemia due to lower hepatic Gck protein levels and activity (24, 25). Similarly, cats lack endogenous hepatic GKRP expression and display low hepatic GCK activity (26). Additionally, adenoviral overexpression of GCK and GCKR in HepG2 cells results in elevated GCK protein levels and activity compared with overexpression of GCK alone (27). GKRP-bound GCK may therefore serve as a functional nuclear reserve that can be rapidly activated and mobilized to the cytoplasm following a glucose challenge.
Given its central role in hepatic glucose metabolism, it is probably not surprising that GCKR has emerged as an important locus for susceptibility to diabetes and related traits (12, 28–40). GWAS initially identified a 400-kb region on chromosome 2 significantly associated with serum triglycerides (28). Comprehensive fine mapping identified rs1260326 (c.1337 C>T, p.Pro446Leu) as the likely causative variant associated with an inverse modulation of fasting glucose and triglyceride levels (12). This association has been widely replicated in additional studies and populations (40–42). Further genome-wide analyses have documented associations of this region with risk of type 2 diabetes (T2D), total cholesterol, fasting insulin, C peptide, homeostasis model assessment of insulin resistance (HOMA-IR), and concentrations of a number of other metabolites, including C-reactive protein (CRP), mannose, the liver enzyme γ-glutamyl transferase, and urate (29–40). Functionally, p.Pro446Leu-GKRP has been shown to decrease F6P-mediated inhibition of GCK and reduce nuclear sequestration of GCK (13, 43). Decreased inhibition and sequestration of GCK may lead to increased GCK activity in the liver, resulting in increased de novo triglyceride and cholesterol synthesis and export, as suggested by recently observed associations of GCKR with VLDL particle concentrations (44). Increased hepatic glucose disposal could act to decrease plasma glucose concentrations, consistent with the observed inverse effects on glucose and lipid levels.
A recent study highlighted the clinical relevance of the collective burden of rare alleles in GCKR, reporting that nonsynonymous variants of minor allele frequency (MAF) of less than 0.01 in GCKR are enriched in cases of extreme hypertriglyceridemia (11). This suggests that many nonsynonymous GCKR variants will have significant effects on protein structure, function, and/or expression and that these changes will lead to demonstrable metabolic effects. However, this study was not able to explore the effects of individual alleles with respect to biochemical function or to assess the impact of GCKR variants on less extreme phenotypes in subjects with trait values more representative of the general population. Understanding such relationships on an individual level will be essential for interpretation of medical sequencing data, especially in the context of genetically complex traits in which rare alleles may be contributory rather than completely penetrant. Such challenges will only become more pronounced due to increasing availability of whole-exome and whole-genome sequence information for large numbers of individuals. Therefore, we aimed to identify and clinically evaluate subjects with nonsynonymous GCKR variants and to couple this work with comprehensive functional analysis.
Results
Identification and clinical characteristics of individuals with GCKR variants.
We sequenced the exons of the GCKR gene in 800 members of the ClinSeq cohort ascertained by April 2010. The majority (88.5%) of ClinSeq participants are non-Hispanic individuals of mixed European descent, reflecting the ethnic and sociodemographic characteristics of Bethesda, Maryland, where the NIH Clinical Center is located (45, 46). Cohort selection was implemented to enrich for coronary atherosclerosis and has been described previously (46). Individuals were between the ages of 45 and 65 years, with a mean age of 56 years. Nineteen (10 novel) nonsynonymous (16 missense, 1 nonsense, and 2 frameshift) GCKR variants were identified by exonic Sanger sequencing in ClinSeq participants (Table 1). All variants apart from p.Pro446Leu had a MAF of less than 0.02 in the cohort.
Table 1 .
GCKR variants discovered by exonic sequencing in the ClinSeq cohort
p.Pro446Leu had significant effects on fasting triglyceride levels in the expected direction (Supplemental Figure 1 and Supplemental Table 1; supplemental material available online with this article; doi: 10.1172/JCI46425DS1) (28). The effect of p.Pro446Leu on fasting glucose did not reach significance (Supplemental Figure 1), most likely due to limited statistical power, as the previously reported effect of this allele on fasting glucose levels is small (0.5 mg/dl per allele in ref. 34) and unlikely to be detected in a cohort of this size. Individuals with either T2D (n = 51) or potential familial hyperlipidemia (family history of dyslipidemia and triglyceride levels > 500 mg/dl; n = 6) were excluded from phenotype comparisons because of direct effects on glucose and lipid phenotypes. Exclusion of individuals on statins or niacin did not alter significance of triglyceride or glucose levels. p.Pro446Leu was in Hardy-Weinberg equilibrium (HWE) in the whole sample and the subset of non-Hispanic individuals of mixed European descent. Allele frequencies were in accordance with data from the International HapMap project ( http://www.hapmap.org).
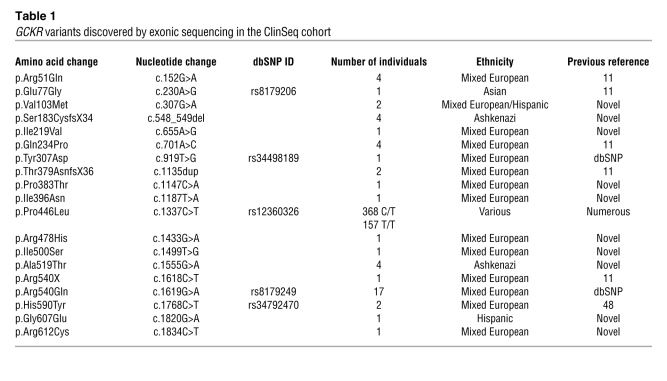
There were 42 ClinSeq individuals with GCKR variants other than p.Pro446Leu. Consistent with the findings of Johansen et al. (11), collective analysis of the group of individuals with rare GCKR variants revealed a significant association with plasma triglycerides compared with the WT reference group (individuals homozygous for Pro at position 446 but with no rare GCKR variants; Figure 1; P = 0.03). Of these 42 individuals, 28 were also heterozygous and 4 were homozygous for the Leu allele at position 446 (Supplemental Table 2). We were able to determine the likely phase for 24 of 28 of the compound heterozygotes through family studies and/or inference from other individuals harboring the same rare allele (Supplemental Table 2). Six individuals carried multiple rare alleles: 1 individual carried p.[Gln234Pro;Arg540X] in cis, 1 individual carried p.[Tyr307Asp(;)Arg540Gln] (phase unknown), and 4 individuals carried p.[Ser183CysfsX34;Ala519Thr] in cis, suggesting p.Ala519Thr may not be relevant at the protein level in these individuals because it occurs downstream of a frameshift variant. One of these 4 individuals also carried p.Arg540Gln in trans. No rare variant produced extreme outlier phenotypes greater than 4 SD from the mean for the ClinSeq cohort (data not shown). Efforts to expand pedigrees did not provide sufficient data to establish segregation.
Figure 1. Triglyceride levels of ClinSeq participants separated by GCKR genotype.
Red bars indicate unadjusted means ± SD. Two-tailed P values are compared with WT (see also Table 3). WT, individuals homozygous for Pro at position 446; P446L, heterozygous individuals at position 446; L446, individuals homozygous for Leu at position 446; rare, individuals heterozygous for 1 or more rare GCKR nonsynonymous variants.
While preliminary comparisons of the collective group of individuals with rare variants with the WT reference group suggested a relationship to triglycerides, we were concerned that the merging of data from certain nonsynonymous variants of potentially no consequence with that of those that cause significant loss or gain of function could considerably diminish the power of the analysis. Finding large numbers of individuals with specific GCKR variants for detailed phenotyping was impractical, given the rarity of these particular variants in 1000 Genomes (47) and previously published studies (11, 48). It thus became imperative to explore evolutionary, cell biological, and biochemical characterization of each variant to evaluate their potential contribution to phenotypes.
Evolutionary characteristics and bioinformatic predictions for GCKR variants.
To aid in the interpretation of genetic and clinical data, we aimed to characterize nonsynonymous variants at the protein level. There is no crystal structure available for GKRP. However, GKRP bears homology to bacterial proteins of the sugar isomerase (SIS) family and contains 2 separate SIS domains that combine to form a single site capable of binding F1P or F6P (23, 49). The location of critical sugar-binding residues appears highly conserved (23). Mapping of variants on these predicted GKRP subdomains is presented in Supplemental Figure 2A. Variant p.Val103Met is located directly within a predicted sugar-binding motif, while p.Ile500Ser immediately precedes a binding motif (Supplemental Figure 2B). p.His590Tyr, p.Gly607Glu, and p.Arg612Cys are C-terminal of SIS domains, as predicted by NCBI Conserved Domains search ( http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). p.Ile219Val and p.Gln234Pro are predicted to localize to a eukaryote-specific α-helix (Supplemental Figure 2, B and C). All variants are in highly conserved residues apart from p.Arg51Gln, p.Arg478His, p.Arg540Gln, and p.His590Tyr (Supplemental Table 3). The effect of amino acid substitution was predicted by SIFT ( http://sift.jcvi.org/) and PolyPhen ( http://genetics.bwh.harvard.edu/pph/) (Supplemental Table 4). These algorithms predict mutation severity based on a number of parameters, including sequence conservation and known functional motifs (50, 51), and their use has been proposed as a method for classifying variants identified by resequencing (52). Together, SIFT and PolyPhen predicted 7 of 15 missense variants as damaging and 5 of 15 as benign, disagreeing on 3 variants. However, the reliability of these prediction algorithms has not been extensively tested. For example, Johansen et al. (11) found a significant excess in the proportion of rare variants predicted by PolyPhen as benign in hypertriglyceridemia cases compared with controls, and SIFT predicted all 8 GCKR missense variants identified by Johansen et al. as benign (data not shown).
The majority of GCKR variants alter cellular localization of the regulatory protein.
As we had concerns about the reliability of prediction algorithms and wanted to compare evolutionary features to protein function, we biochemically characterized GCKR variants to determine their molecular and cellular effects. As GKRP function is intimately linked with nuclear localization, we generated N-terminal yellow fluorescent protein (YFP) fusions to WT or variant GCKR-coding sequences and determined their subcellular location in HeLa cells. The interaction of GCK and GKRP has been previously extensively characterized in this cell type (22, 43). WT YFP-GKRP localized primarily to the nucleus 24 hours after transient transfection into HeLa cells (Figure 2A). Predominantly nuclear localization of WT YFP-GKRP was also observed in COS-1 and HepG2 cells (data not shown).
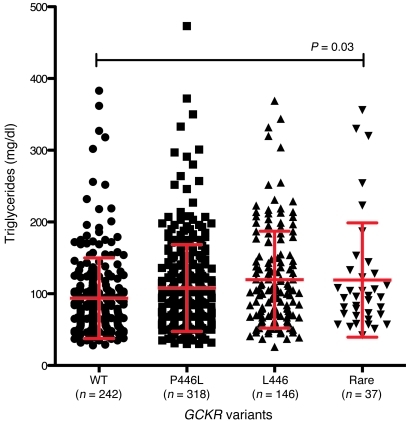
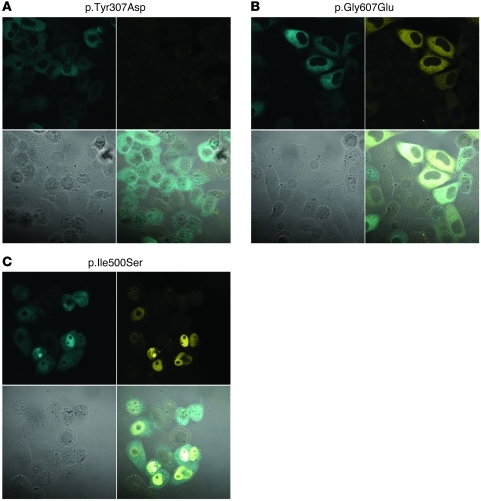
Figure 2. Cellular localization of representative variant YFP-GKRPs on transient transfection into HeLa cells.
(A) WT GKRP localized primarily to the nucleus, as did p.Arg51Gln, p.Glu77Gly, p.Pro383Thr, p.Arg478His, p.Arg540Gln, and p.His590Tyr. (B) p.Ile396Asn, p.Pro446Leu, and p.Ile500Ser localized to both the cytoplasm and nucleus. (C) p.Val103Met, p.Ile219Val, p.Gln234Pro, and p.Tyr307Asp localized primarily to the cytoplasm and showed comparatively lower YFP fluorescence intensity. (D) p.Gly607Glu and p.Arg612Cys were also cytoplasmic, but showed higher levels of YFP expression. Original magnification, ×63. Images use the same laser settings and intensity and are representative of at least 2 transfections of 2 independent plasmid preparations for each variant. A representative image of each localization pattern is shown.
Eighteen nonsynonymous human variants from the ClinSeq project were introduced into the YFP-GKRP plasmid and transiently transfected into HeLa cells (p.Arg51Gln, p.Glu77Gly, p.Val103Met, p.Ser183CysfsX34, p.Ile219Val, p.Gln234Pro, p.Tyr307Asp, p.Thr379AsnfsX36, p.Pro383Thr, p.Ile396Asn, p.Pro446Leu, p.Arg478His, p.Ile500Ser, p.Arg540X, p.Arg540Gln, p.His590Tyr, p.Gly607Glu, and p.Arg612Cys). Only truncating variants p.Ser183CysfsX34, p.Thr379AsnfsX36, and p.Arg540X failed to generate YFP fluorescence, except in a minority of cells where punctate fluorescent aggregates were observed. Both of these phenomena are associated with protein misfolding (53). These results suggest that the p.Gln234Pro variant will not have any additional effect on p.Arg540X in cis for the individual in which these variants were discovered.
The remaining nonsynonymous YFP-GKRP variants showed 4 distinct patterns of localization. First, variants p.Arg51Gln, p.Glu77Gly, p.Pro383Thr, p.Arg478His, p.Arg540Gln, and p.His590Tyr localized primarily to the nucleus like WT GKRP (see Supplemental Figure 3 for all variants). Second, consistent with our previous findings, common variant p.Pro446Leu localized to both the cytoplasm and nucleus (Figure 2B and ref. 43). Rare variants p.Ile396Asn and p.Ile500Ser also displayed this behavior. Third, p.Val103Met, p.Ile219Val, p.Gln234Pro, and p.Tyr307Asp showed almost exclusive cytoplasmic localization and comparatively low YFP fluorescence (Figure 2C). Finally, variants p.Gly607Glu and p.Arg612Cys localized to the cytoplasm and exhibited greater fluorescence intensity than other cytoplasmic variants (Figure 2D). Western blot analysis showed that expression levels of p.Gly607Glu and p.Arg612Cys were similar to those of WT YFP-GKRP, while other cytoplasmic variants showed reduced expression (data not shown).
For 3 rare variants (p.Glu77Gly, p.Pro383Thr, and p.Arg540Gln), the individuals harboring them were confirmed as also having the Leu allele at position 446 in cis. We therefore assessed the combined effect of these variants. While p.[Pro446Leu;Arg540Gln] showed diffuse localization similar to that of p.Pro446Leu, p.[Glu77Gly;Pro446Leu] and p.[Pro383Thr;Pro446Leu] showed almost exclusive cytoplasmic localization, suggesting the p.Pro446Leu variant may amplify otherwise modest cellular effects for these 2 variants (Supplemental Figure 4).
Cellular interaction of GKRP variant proteins with GCK emphasizes distinct mislocalization subtypes.
To understand the consequences of GKRP mislocalization further, we assessed the effect of WT and variant GKRPs on the cellular localization of GCK. Human liver GCK was cloned as an N-terminal cyan fluorescent protein (CFP) fusion protein. Without cotransfected GKRP, CFP-GCK localized exclusively to the cytoplasm in HeLa cells (Figure 3A). The same result was seen in HepG2 and COS-1 cells (data not shown). To assess interaction between GCK and GKRP, a single plasmid containing separate promoters, coding sequences, and polyadenylation signals for CFP-GCK and YFP-GKRP was utilized to ensure consistent coexpression of GKRP and GCK in individual cells across experiments. This was particularly important because a number of variant GKRPs showed very low levels of expression.
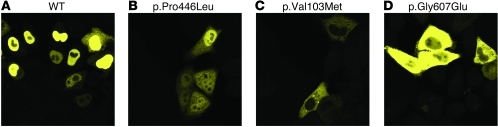
Figure 3. WT YFP-GKRP is necessary and sufficient to sequester CFP-GCK to the HeLa cell nucleus.
(A) CFP-GCK localized to the cytoplasm upon transient transfection into HeLa cells. (B) Cotransfection of YFP-GKRP expressed from the same plasmid sequestered CFP-GCK to the nucleus in HeLa cells. All YFP-GKRP variants that localized primarily to the nucleus were capable of sequestering CFP-GCK. Top left, CFP channel; top right, YFP channel; bottom left, phase channel; bottom right, combined channels. Original magnification, ×63. Images were taken using the same laser settings and intensity and are representative of at least 2 transfections of 2 independent plasmid preparations for each variant.
Transient transfection of the CFP-GCK and YFP-GKRP dual-expression plasmid resulted in nuclear localization of WT GKRP and nuclear sequestration of CFP-GCK (Figure 3B). In accordance with previous observations, GCK exhibited similar subcellular localization patterns for multiple glucose concentrations tested (data not shown) (43). Therefore, images from 1 glucose concentration (25 mM) are presented and representative of other concentrations. The 6 variants exhibiting WT-like localization as YFP fusion proteins (p.Arg51Gln, p.Glu77Gly, p.Pro383Thr, p.Arg478His, p.Arg540Gln, and p.His590Tyr) remained localized to the nucleus and sequestered GCK. Evolutionary analysis of these variants revealed that 4 of these variants (p.Arg51Gln, p.Arg478His, p.Arg540Gln, and p.His590Tyr) were in amino acid residues showing the lowest conservation in mammals among the GCKR variants tested. The observed variant amino acid at residues 51, 478, and 540 is the reference amino acid in 3 or more nonhuman mammalian species (Supplemental Table 3).
Diffuse and cytoplasmic GKRP variants showed distinct responses to coexpression of GCK. Variants p.Gly607Glu and p.Arg612Cys did not sequester GCK to the nucleus, but showed significant cytoplasmic YFP fluorescence (Figure 4A). Five variants (p.Val103Met, p.Ser183CysfsX34, p.Tyr307Asp, p.Thr379AsnfsX36, and p.Arg540X) did not traffic GCK to the nucleus and showed low or undetectable intensity of fluorescent GKRP (Figure 4B). In the presence of GCK, GKRP variant proteins p.Ile219Val, p.Gln234Pro, p.Ile396Asn, p.Pro446Leu, and p.Ile500Ser relocalized to the nucleus and sequestered GCK (Figure 4C). Previous studies have suggested GCK promotes nuclear localization of GKRP (22, 54), which may in part explain this observation. However, we have shown by quantification of nuclear and whole-cell fluorescence intensity that the degree of GKRP nuclear relocalization and GCK sequestration is significantly reduced for p.Pro446Leu compared with WT GKRP (43). p.[Glu77Gly;Pro446Leu], p.[Pro383Thr;Pro446Leu], and p.[Pro446Leu;Arg540Gln] also relocalized to the nucleus and sequestered GCK.
Figure 4. Diffuse and cytoplasmic YFP-GKRP variants show distinct classifications of behavior in the presence of CFP-GCK on transient transfection into HeLa cells.
(A) Variants p.Val103Met, p.Ser183CysfsX34, p.Tyr307Asp, p.Thr379AsnfsX36, and p.Arg540X did not sequester CFP-GCK to the nucleus and did not show appreciable YFP fluorescence when intensity matched to WT YFP-GKRP. (B) Variants p.Gly607Glu and p.Arg612Cys did not sequester CFP-GCK, but showed appreciable YFP fluorescence intensity. (C) Variants p.Ile219Val, p.Gln234Pro, p.Ile396Asn, p.Pro446Leu, and p.Ile500Ser interacted with GCK and were relocalized to the nucleus. Original magnification, ×63. Images were made using the same laser settings and intensity and are representative of at least 2 transfections of 2 independent plasmid preparations for each variant. Top left, CFP channel; top right, YFP channel; bottom left, phase channel; bottom right, combined channels. A representative image of each localization pattern is shown.
Residues near the C terminus of the regulatory protein affect nuclear localization and GCK sequestration.
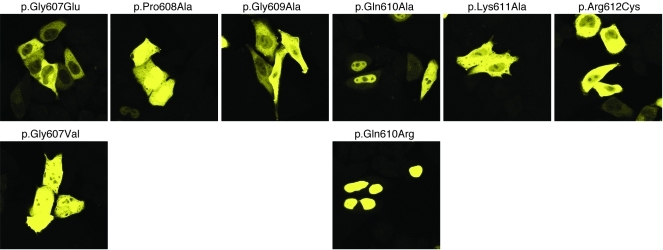
Two variants near the C terminus of the protein, p.Gly607Glu and p.Arg612Cys, were exclusively cytoplasmic, demonstrated high fluorescence intensity, and did not sequester GCK. Alignment of C-terminal GKRP residues 595–625 with 3 other GKRP orthologs known to localize to the nucleus showed decreased amino acid conservation, particularly in the case of Xenopus laevis GKRP (Supplemental Table 5 and Supplemental Figure 5). The region containing ClinSeq variants p.Gly607Glu and p.Arg612Cys was more highly conserved. To determine whether mutation of intervening residues had similar effects, residues 607–612 were mutated to Ala or to residues present in X. laevis GKRP. Mutation to X. laevis residues p.Gly607Val or p.Gln610Arg maintained partial and complete nuclear localization, respectively. Variants p.Gly609Ala and p.Lys611Ala localized to the cytoplasm and did not sequester GCK, while variants p.Pro608Ala and p.Gln610Ala showed a mixture of nuclear and cytoplasmic localization. All variants showed fluorescence intensity similar to that of p.Gly607Glu and p.Arg612Cys (Figure 5).
Figure 5. YFP-GKRP C-terminal variants have an important role in nuclear localization.
Mutations of YFP-GKRP conserved residues to ClinSeq variants p.Gly607Glu and p.Arg612Cys, residues present in other GKRP orthologs (p.Gly607Val, p.Gln610Arg) or Ala (p.Pro608Ala, p.Gly609Ala, p.Gln610Ala, p.Lys611Ala). Original magnification, ×63. Images were taken the same laser settings and intensity and are representative of at least 2 transfections of 2 independent plasmid preparations for each variant.
Kinetic analysis of selected variant regulatory proteins supports distinct functional variant classes.
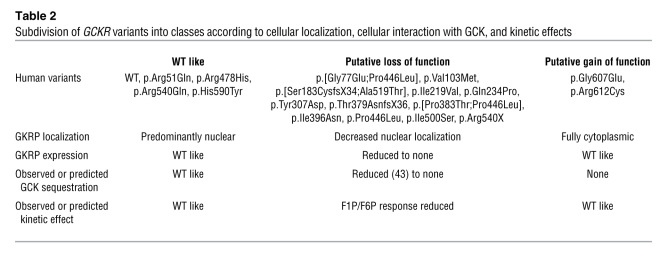
The combination of these cellular results assessing human GCKR nonsynonymous variants suggested GKRP variant proteins have a spectrum of effects on localization and interaction with GCK (Table 2). To further explore these observations, we selected a subset of variants for recombinant expression in E. coli, purification, and kinetic comparison with WT GKRP. We selected 4 variants that each showed reduced protein expression, reduced nuclear localization, and potential reduction in GCK sequestration (p.Val103Met, p.Ile219Val, p.Gln234Pro, and p.Ile500Ser). We also selected p.Arg612Cys to represent C-terminal variants displaying high cytoplasmic YFP expression. Finally, we selected variant p.Pro383Thr, as this variant was strongly sensitive to p.Pro446Leu genotype in cis, was in a highly conserved residue, was predicted to be deleterious by both SIFT and PolyPhen, was observed only once in the ClinSeq cohort, and had not been previously reported.
Table 2 .
Subdivision of GCKR variants into classes according to cellular localization, cellular interaction with GCK, and kinetic effects
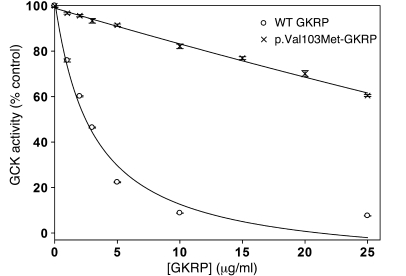
Variant regulatory proteins appreciably inhibited recombinant human GCK, with the exception of p.Val103Met-GKRP, a variant that showed very low expression in HeLa cells. p.Val103Met-GKRP had significantly reduced ability to inhibit GCK over a 0 to 25 μg/ml concentration range, with 60.5% ± 0.4% GCK activity remaining at 25 μg/ml (mean ± SEM; P < 0.001 for all GKRP concentrations). In contrast, GCK activity was only 7.6% ± 0.1% that of uninhibited GCK in the presence of 25 μg/ml WT GKRP (Figure 6). As protein misfolding is dependent on temperature, the assay temperature was reduced to 30°C. This led to only a slight improvement in p.Val103Met-GKRP function, with 50% inhibition of GCK predicted at 22.5 μg/ml protein (data not shown).
Figure 6. p.Val103Met-GKRP shows significantly reduced inhibition of GCK.
Inhibition of 10 mU/ml recombinant GCK by increasing concentrations of recombinant WT GKRP (circles) and p.Val103Met-GKRP (X’s). GCK activity (mean ± SEM; n = 8; P < 0.001 for all GKRP concentrations tested) is plotted as a percentage of that obtained in the absence of regulatory protein at 5 mM glucose.
We calculated 50% inhibition of 10 mU/ml GCK at 5 mM glucose and 37°C (defined as one GKRP unit) for WT GKRP and all variants excluding p.Val103Met (Supplemental Table 6). p.Ile219Val-GKRP and p.Gln234Pro-GKRP showed a reduction in activity, while p.Pro383Thr-GKRP, p.Ile500Ser-GKRP, and p.Arg612Cys-GKRP were nearly indistinguishable from WT GKRP. GCK inhibition by 1 unit of WT GKRP was compared with inhibition by 1 unit p.Ile219Val-GKRP, p.Gln234Pro-GKRP, p.Pro383Thr-GKRP, p.Ile500Ser-GKRP, or p.Arg612Cys-GKRP over a 0 to 100 mM glucose concentration range. No significant differences were observed between WT and variant proteins (Supplemental Figure 6). One unit of each of these variant proteins was then tested for ability to interact with the phosphate esters F6P and F1P. C-terminal variant p.Arg612Cys showed no difference in response to either phosphate ester (Figure 7A). Accordingly, this variant appears to result in a GKRP protein that is a fully active, exclusively cytoplasmic inhibitor of GCK and hence may be considered as a potential gain-of-function mutation. p.Pro383Thr, a variant that localizes to the nucleus, showed decreased response to F1P (Figure 7B), but showed no difference in response to F6P. p.Ile219Val, a variant relocalized to the nucleus in the presence of GCK, also showed no difference in response to F6P and had a decreased response to F1P (Figure 7C). Variants p.Gln234Pro and p.Ile500Ser, which were also relocalized to the nucleus by GCK, had significantly diminished response to both F1P and F6P (Figure 7, D and E). The relative amplitudes of response to F1P and F6P are listed in Supplemental Table 6.
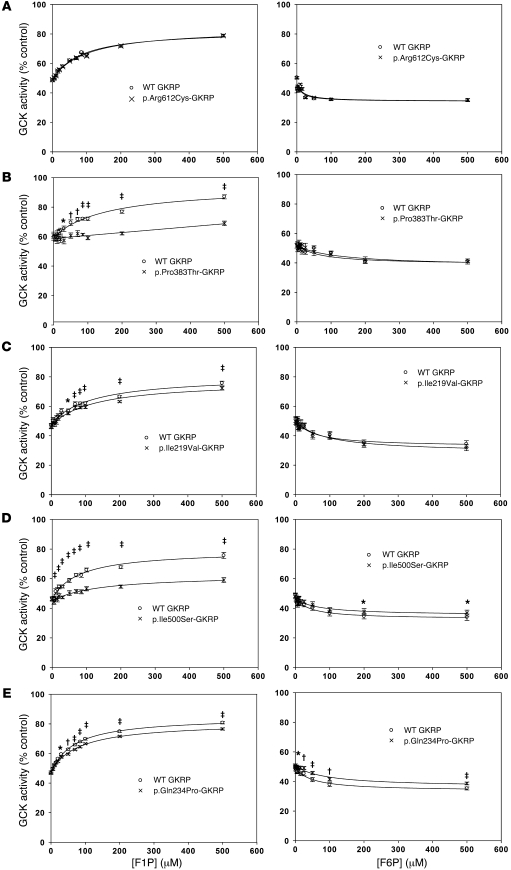
Figure 7. Effect of 0–500 μM F1P or F6P on inhibition of 10 mU/ml recombinant GCK by 1 unit of selected GKRP variants.
Comparisons of recombinant proteins with WT GKRP. (A) p.Arg612Cys showed no significant difference (P > 0.1) in response to F1P (n = 12) or F6P (n = 12). (B and C) p.Pro383Thr (n = 12; P < 0.02 for 30–500 μM F1P< 0.04 for 5–500 μM F1P) showed a significantly reduced response to F1P but not F6P (P > 0.1). (D and E) p.Ile500Ser and p.Gln234Pro showed a significantly reduced response to F1P (n = 12 and P < 0.03 for 15–500 μM for p.Ile500Ser; n = 38 and P < 0.02 for 20–500 μM for p.Gln234Pro) and F6P (n = 19 and P < 0.05 for 200–500 μM for p.Ile500Ser; n = 41 and P < 0.02 for 10–500 μM for p.Gln234Pro). GCK activity is plotted as a percentage of that obtained in the absence of regulatory protein at 5 mM glucose. Data points are mean ± SEM. *P < 0.05; †P < 0.01; ‡P < 0.001.
To assess the kinetic effect of Leu at position 446 in cis, p.[Pro383Thr;Pro446Leu]-GKRP and p.Pro446Leu-GKRP were generated. Consistent with previous observations, p.Pro446Leu-GKRP showed reduction in activity compared with WT GKRP (13). p.[Pro383Thr;Pro446Leu]-GKRP showed reduced activity compared with WT GKRP, p.Pro446Leu-GKRP, and p.Pro383Thr-GKRP (Supplemental Figure 7A). However, comparison of activity-matched p.Pro446Leu-GKRP with p.[Pro383Thr;Pro446Leu]-GKRP showed no additional defects in response to F1P or F6P beyond those observed when comparing p.Pro383Thr-GKRP and WT GKRP (Supplemental Figure 7B). This suggests there is no combined kinetic effect of these 2 variants on F6P and F1P affinity. As p.Pro446Leu-GKRP showed no significant difference in response to F1P and a very modest decrease in response to F6P (Supplemental Table 6), it is probable that Leu446 in cis would contribute little to the F1P- and F6P-binding characteristics of rare variants. However, our results suggest cis effects on protein activity and cellular localization may be more prominent.
The p.Pro383Thr-GKRP and p.[Pro383Thr;Pro446Leu]-GKRP proteins were also used to model the potential kinetic effect of the p.Pro446Leu variant in trans. Protein activities were calculated for WT GKRP, p.Pro383Thr-GKRP, p.Pro446Leu-GKRP, and p.[Pro383Thr;Pro446Leu]-GKRP in conjunction with 1:1 mixtures (by concentration) of these proteins (Supplemental Table 7). The maximal responses of these proteins and mixtures of proteins to F1P and F6P were also calculated. All mixtures resulted in intermediate activity values and responses to F1P and F6P when compared with variant proteins assessed in isolation, suggesting none of these proteins had dominant kinetic effects.
Classification of phenotypes based on variant functional effects.
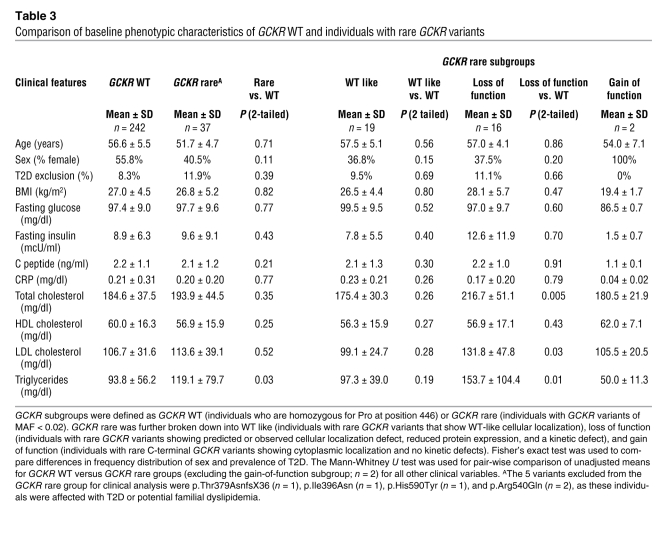
The combination of cellular and kinetic assays suggested 3 broad classifications of variant GKRPs (Table 2). Based on our biochemical analysis, we subdivided GCKR rare variant heterozygotes (n = 42) into subgroups with variants that were functionally similar to WT (Rare, WT-like; n = 22), C-terminal potential gain-of-function variants (Rare, GOF; n = 2), and putative loss-of-function variants that were functionally similar to p.Pro446Leu in showing reduced to complete loss of protein expression, reduced nuclear localization, and reduced interaction with F1P and/or F6P (Rare, LOF; n = 18) (Table 3). Four individuals were excluded because of T2D (1 individual with the p.Thr379AsnfsX36 variant, p.Ile396Asn, 1 individual with the p.Arg540Gln variant, and 1 individual with the p.His590Tyr variant). A second individual with p.Arg540Gln was excluded on the basis of potential familial hyperlipidemia, leaving 37 individuals for analysis. Compared with the WT reference group, the Rare, WT-like group showed no significant differences for all phenotypes tested (P > 0.1). However, the Rare, LOF group had significantly higher levels of total cholesterol (P = 0.005), LDL cholesterol (P = 0.03), and triglycerides (P = 0.01) (Table 3 and Supplemental Figure 8). The difference in total cholesterol (P = 0.002), LDL cholesterol (P = 0.02), and triglycerides (P = 0.001) for the Rare, LOF group remained significant after adjustment for covariates including genotype at p.Pro44Leu (Supplemental Table 8 and Supplemental Methods). An effect estimate for the Rare, LOF group (using P trend, see Supplemental Table 9) was 50.1 ± 14.7 mg/dl (mean ± SEM; P < 0.001) for triglycerides. This compares with an estimate of 11.3 ± 3.0 mg/dl (mean ± SEM; P < 0.001) for the Leu allele at position 446 alone. The p.Pro446Leu variant was not overrepresented in either the overall GCKR rare group (MAF = 0.37) or the Rare, LOF group (MAF = 0.37) compared with the entire cohort (MAF = 0.42).
Table 3 .
Comparison of baseline phenotypic characteristics of GCKR WT and individuals with rare GCKR variants
We also sought to determine whether rare GCKR variants with more severe effects on protein function such as null alleles would have an even more significant effect on phenotype. Loss-of-function variant GKRPs show distinctive kinetic parameters, cellular localization, and interactions with GCK. However, data from Gckr-knockout mice suggest more severe LOF variants may predispose to development of diabetes-associated phenotypes, particularly in the context of additional factors such as high-fat and high-sugar diets (24). The variants with most severe cellular and kinetic defects (severe LOF, n = 11) appeared to be associated with even greater elevations of triglycerides, cholesterol, BMI, and fasting insulin in the ClinSeq cohort compared with the WT reference group (all P < 0.05; Supplemental Figure 9), although numbers of individuals were too small to make definitive conclusions.
Replication genotyping efforts.
Four variants shown to affect protein function were observed multiple times in the ClinSeq cohort (p.Val103Met, p.[Ser183CysfsX34;Ala519Thr], p.Gln234Pro, and p.Thr379AsnfsX36). For these variants, the prospects for identifying additional carriers through population-based genotyping were greatest, and we genotyped them in T2D case-control cohorts. Variants p.Gln234Pro and p.Thr379AsnfsX36 were present in ClinSeq samples of non-Hispanic mixed European ancestry and were therefore genotyped in well-characterized sample sets of Finnish and German origin (n = 1800–11000; Supplemental Table 10 and refs. 31, 55–57). Both variants were more common in samples of German origin compared with samples of Finnish origin (p.Gln234Pro, MAF = 0.001 and 0.0001, respectively; p.Thr379AsnfsX36, MAF = 0.002 and 0.0003, respectively). Variant p.[Ser183CysfsX34;Ala519Thr] was observed in 4 ClinSeq samples of Ashkenazi descent but was not detected in 1000 Genomes or Finnish or German samples. This variant had a MAF of 0.006 in 1206 individuals of Ashkenazi descent (Supplemental Methods). p.Val103Met was detected in 2 ClinSeq individuals and in 2 1000 Genomes samples of reported Mexican-American descent. This variant had a MAF of 0.009 in 1,528 samples of Mexican-American ancestry (Supplemental Methods and ref. 58). Accordingly, all of these variants were rare (MAF < 0.01) even when screened in populations matched for ethnicity. Consistent with our phenotype analyses, severe LOF variants p.Val103Met, p.[Ser183CysfsX34;Ala519Thr], and p.Thr379AsnfsX36 appeared enriched in individuals with impaired glycemia (Supplemental Table 11).
Discussion
Many observers are predicting that whole-genome sequencing will become part of standard medical care within the next decade (59). That potential has heightened interest in interpretation of genetic variants that might provide insight into assessing future risks of illness, refining present diagnoses, or predicting drug response. Hundreds of common variants shown to be associated with disease risk or quantitative traits have emerged in the last 5 years as a result of the GWAS approach. However, most of these have been shown to have a relatively modest effect on risk, and there appears to be “missing heritability” for many diseases and traits that may at least in part be attributable to rare variants, some of which may have larger effects (2).
While successful discovery of such rare variants has been achieved in some studies by focusing on individuals at the extremes of a quantitative trait (6–8, 10), a major challenge for the future is how to interpret such variants when they occur in a less selected population. For that purpose, we studied a clinical population modestly enriched for cardiovascular disease risk and looked for rare variants in GCKR.
Variants in GCKR have been previously shown to affect a range of metabolic processes. For common variant p.Pro446Leu, it has been relatively easy to detect such associations because of the availability of phenotype information on large numbers of individuals. As an alternative approach to finding rare alleles, sequencing GCKR in individuals with extremely high triglycerides has shown utility in relating such variants collectively to phenotypes (11). However, for GCKR or indeed any gene, determining whether a specific mutation is functionally important in an individualized clinical setting remains a major challenge. Bioinformatic methods such as mutation prediction algorithms and assessment of evolutionary conservation are useful preliminary tools in analysis of whether a particular variant might have functional consequences, but these approaches showed limitations in accuracy and consistency between prediction programs both in this study and in the study by Johansen et al. (11).
Accordingly, we utilized existing information about GKRP function and undertook individual molecular characterization of all 18 GCKR variants from the ClinSeq project. We observed defects in cellular localization for the majority of these variants (12/18) as YFP-tagged GKRP constructs in HeLa cells and refined our analysis by GCK coexpression and kinetic characterization. Functional rare variants could be broadly subdivided into putative LOF and GOF subtypes (Table 2).
Potential GOF mutations in conserved C-terminal residues of GKRP abolished GKRP nuclear localization and GCK sequestration, while p.Arg612Cys-GKRP showed no significant differences in kinetic properties compared with WT GKRP (Figure 2D and Figure 5). This suggests that the region surrounding residues 607–612 could be part of the unknown mechanism by which GKRP is localized to the nucleus. As only 2 individuals carried potential GOF mutations (Table 3 and Supplemental Figure 9), further studies will be needed to determine the phenotypic effects of such variants. However, physiologically, these mutations increase cytoplasmic GKRP and thus may serve to decrease GCK activity by decreasing both the pool of sequestered nuclear GCK and of active, cytoplasmic GCK. This would be predicted to decrease hepatic glycogen, triglyceride, and cholesterol synthesis.
Analysis of the subset of ClinSeq individuals heterozygous for rare GCKR LOF variants collectively showed a significant increase in total cholesterol, LDL cholesterol, and triglyceride levels (Table 3). These variants were both functionally and phenotypically similar to p.Pro446Leu, showing reduced expression, reduced nuclear localization, potential reduction in GCK sequestration, and reduced interaction with F6P and/or F1P. As has been proposed for p.Pro446Leu (13, 43), reduced nuclear localization and GCK sequestration would likely increase fasting hepatic glucose uptake and disposal through synthetic pathways including de novo lipogenesis. For p.Pro446Leu, the phenotypic effect was much stronger on triglycerides than on fasting glucose; presumably the same phenomenon is present in our collection of rare LOF variants, as even with small numbers, we were able to detect the effect on lipids, but not on fasting glucose. However, while variants within this group are qualitatively similar, they displayed a range in the magnitude of cellular and kinetic effects.
The most severe loss-of-function variants, such as p.Val103Met, appear to form very little, if any, functional protein, characterized by low cellular fluorescence and dramatically reduced ability to inhibit GCK. Physiologically, these variants may be indistinguishable from null mutations and might therefore be compared with heterozygous Gckr-knockout mice. Gckr+/– mice show reduced liver GCK levels and activity (24, 25) as well as trends toward lower hepatic glycogen content and higher blood glucose levels 30 and 60 minutes after an oral glucose tolerance test (24). The reduction in glycemic control is likely attributable to a decrease in the Gkrp-bound Gck nuclear pool that is normally mobilized in response to a glucose challenge. This loss of nuclear stabilization and/or glucose-dependent translocation has been shown to be associated with impaired glucose tolerance in rodent models (24, 60). Phenotype comparisons and results of genotyping for the severe LOF subgroup were consistent with these findings.
As the common p.Pro446Leu variant has been shown to have both cellular and kinetic effects (13, 43), it is useful to consider the effect of this variant in cis or in trans with rare variants. Experiments assessing Leu446 in cis suggested this variant may amplify defects in protein expression, localization, and activity, but will have fairly mild effects on F1P and F6P interaction. Kinetic results modeling the effect of Leu446 in trans suggested intermediate effects on activity and phosphate ester response (Supplemental Table 7). Using our cellular model system, it is difficult to assess the effect of Leu446 in trans on GCK sequestration. However, as loss-of-function variants are similar to p.Pro446Leu (Table 2), cellular effects of Leu446 in trans with a LOF rare variant are likely to include reduced GCK sequestration (43), GKRP activity, and inhibition from both chromosomes. Phenotypic effects may range from those of Leu446 homozygotes (as suggested by GWAS) to those approaching Gckr-knockout mice for more severe rare variants.
ClinSeq sequencing, combined with previous studies and emerging 1000 Genomes data, suggests it is unlikely there are additional common nonsynonymous GCKR variants in the general western European population (11, 47, 48). Follow-up genotyping confirmed the rarity of individual variants, but highlighted the importance of considering ethnicity for replication of rare variants. Accordingly, our findings supported a collective analysis of rare variants to explore relationships with phenotypes. However, functional characterization revealed that not every variant is likely to have the same biochemical characteristics and therefore the same phenotypic consequences (Table 2). This is an important limitation of in silico predictions, as bioinformatic methods often cannot distinguish between different types of functional variants. The separation of phenotypes based on functional classification in the ClinSeq cohort suggests the mutational load of low-frequency GCKR variants may still play a significant role in heritability of human glucose and lipid traits.
Some of the challenges highlighted by this study are likely to be amplified as more sequencing data becomes available. Resequencing studies will identify a large number of rare variants per individual, many of which may be novel. Phenotyping a large number of individuals for each rare variant will often not be practical. Unless there are reliable computational, cell biological, or biochemical methods for determining the functional consequences of a variant (as we have been able to do here with GCKR), it will generally be difficult to interpret the significance of rare sequence changes. While the era of complete genome sequencing holds much promise for identifying heritable risk factors and ushering in the era of personalized medicine, the leap from sequence discovery to functional inference and medical consequence will often not be trivial.
Methods
Subject recruitment.
ClinSeq participants were evaluated at the NIH Clinical Center or Suburban Hospital (Bethesda, Maryland, USA). The target enrollment was 1,000 nonsmokers between the ages of 45 and 65. Participants were selected to represent the full spectrum of risk for developing coronary artery disease (CAD) using the Framingham score (61). Participants were divided into groups of 250 participants with a Framingham 10-year CAD risk of less than 5%, 5%–10%, or more than 10% as well as an additional group with known CAD.
Clinical and laboratory analysis.
Participants were evaluated by an abbreviated medical history focused on cardiovascular disease. Clinical assessment included measurement of height, weight, blood pressure, head and abdominal circumferences, and electrocardiogram, echocardiogram, and measurement of coronary artery calcification (by multidetector computed tomography). Blood and urine were collected to assess baseline lipids, glucose metabolism, renal function, and specific markers for CAD risk. Genomic DNA was isolated by standard techniques for PCR-based Sanger sequencing of GCKR. Bidirectional sequence generation and analysis using PolyPhred followed by manual review were performed as previously described (46). Primer sequences are presented in Supplemental Methods.
The cohort consisted of 800 participants enrolled in the ClinSeq study between January 5, 2007, and April 21, 2010. Fifty-seven individuals were excluded from clinical analysis on the basis of T2D or extremely high triglycerides, leaving 743 individuals for analysis. ClinSeq participants were subdivided into genotypic subgroups for comparison of baseline clinical parameters. The mutational status of rs1260326 (c.1337C>T, p.Pro446Leu) was determined by Sanger sequencing or TaqMan assay (for participants with low-quality Sanger sequencing at this nucleotide) for all subjects.
Cloning of fluorescent plasmids.
The pcDNA-DEST53 vector (Invitrogen) was modified to express the YFP variant ZsYellow1 or CFP variant AmCyan1 (Clontech) (Supplemental Methods). Plasmids were verified by sequencing. The Mammalian Gene Collection full-length cDNA of human GCKR and Xenopus Gene Collection full-length cDNA of X. laevis gckr (ATCC) were amplified with Phusion DNA polymerase (New England Biolabs) according to the manufacturer’s instructions using primers containing attB1 and attB2 recombination sequences (Supplemental Methods). PCR products were recombined in frame for N-terminal fusions into the Gateway entry vector pDONR-201 using Gateway cloning technology (Invitrogen). The Mammalian Gene Collection full-length cDNA of human β cell GCK (ATCC) was also PCR amplified and cloned in frame for N-terminal fusions into pDONR-201. The liver-specific exon 1 and shared exon 2 of GCK were synthesized (Integrated DNA Technologies) and cloned into this N-terminal vector, replacing the β cell–specific exon 1 using a SacII restriction site. Mutations in GCKR were introduced into the Gateway entry vector using PCR-based site-directed mutagenesis according to the manufacturer’s instructions (Agilent Biotechnologies; Supplemental Methods). WT or mutant GCKR were recombined into the YFP-expressing Gateway vector. Two independent preparations were generated for each variant; all mutations were verified by sequencing. GCK was recombined into the CFP gateway expression vector.
The CFP-GCK promoter, ORF, and poly(A) signal were cloned into the YFP Gateway destination vector so that the CMV promoters upstream of CFP and YFP were in opposite orientations (Supplemental Methods). The GCKR coding sequence (either WT or mutant) was recombined into this vector to generate a plasmid expressing both YFP-GKRP and CFP-GCK. Two independent preparations were generated for each variant and verified by sequencing.
Cell culture and transfection.
HeLa cells were cultured in DMEM with 25 mM glucose (Invitrogen) supplemented with 10% FBS and penicillin/streptomycin. 4 × 104 cells were seeded into 4-chamber slides (BD Biosciences) and cultured for 24 hours. Transfection was carried out with a set amount of plasmid DNA (200 ng for YFP-GKRP plasmids; 1 μg for dual-expression plasmids) and 3 μl of lipofectamine 2000 (Invitrogen) according to the manufacturer’s instructions. Medium was replaced 5 hours after transfection. Slides were fixed with formaldehyde 24 hours after transfection and mounted with VectaShield Mounting Medium (Vector Laboratories). Confocal images were acquired using a Zeiss LSM 510 NLO Meta System mounted on a Zeiss Axiovert 200M microscope with an oil immersion Plan-Apochromat 63×/1.4 DIC objective lens. Each preparation was transfected at least 2 times.
Kinetic characterization.
Recombinant glutathione S-transferase–tagged (GST-tagged) human pancreatic GCK and WT and variant FLAG-tagged GKRP were prepared as described previously (13, 62). Purity and concentration were measured by the Agilent 230 Protein Kit (Agilent Technologies) and Bio-Rad Bradford reagent assay (Bio-Rad Laboratories), respectively. GKRP preparations ranged in yield between 54 and 100 μg/l broth, and GCK yields averaged 1.5 mg/l broth.
GKRP inhibition of GCK activity was determined spectrophotometrically using glucose 6-phosphate dehydrogenase–linked (G6PDH-linked) assays (Sigma-Aldrich) as described previously (13). Independent WT GKRP preparations were generated for each variant preparation. Three preparations were generated for p.Gln234Pro, 2 for p.[Pro383Thr;Pro446Leu], and 1 each for p.Arg612Cys, p.Val103Met, p.Ile219Val, p.Ile500Ser, p.Pro446Leu, and p.Pro383Thr. Except for p.Val103Met, assays were standardized by matching WT and variant GKRP activity (1 unit; corresponding to the amount required for half-inhibition of 10 mU/ml GCK at 5 mM glucose). On average, one GKRP unit resulted from 3 μg/ml of WT GKRP. Relative activities are listed in Supplemental Table 6.
Glucose-dependence assays were carried out over a glucose concentration range of 0 to 100 mM. F1P and F6P assays (0–500 μM) were performed as described previously (13). Both phosphate esters were purchased from Sigma-Aldrich.
Genotyping.
GCKR variants were genotyped using the iPLEX Sequenom MassARRAY platform on samples of German and/or Finnish origin. The study design and phenotypic characteristics of these samples have been previously described (31, 55–57, 63). All genotyped SNPs had a genotyping call rate of more than 95%, and polymorphic variants had an HWE P value of more than 0.001. p.[Ser183CysfsX34;Ala519Thr] and p.Val103Met follow-up genotyping were performed using TaqMan technology (Applied Biosystems) in samples of Ashkenazi and Mexican-American origin, respectively (Supplemental Methods and ref. 58). c.1555G>A (p.Ala519Thr) was queried to determine p.[Ser183CysfsX34;Ala519Thr] genotype. Two-step PCR was performed on an ABI7900HT machine, before post-read allelic discrimination using SDS2.3 software.
Statistics.
HWE was confirmed using a χ2 test with 1 degree of freedom. HWE calculation was made with the Online Encyclopedia for Genetic Epidemiology Studies HWE calculator ( http://www.oege.org/software/hwe-mr-calc.shtml). Nonparametric, unpaired (2-tailed) tests were performed to compare baseline clinical measurements in GCKR subgroups. Mann-Whitney U test was used for continuous variables and Fisher’s exact test for categorical variables using GraphPad InStat v3.1a. Results are presented as mean ± SD. Effect estimates of the GCKR Leu446 allele and rare variant subgroups on baseline laboratory measurements were derived from P trend linear regression. Results are shown as estimate ± SEM and P value for trend. Stepwise multivariate (race, sex, age, BMI, genotype status) logistic regression was used to identify covariates that have a significant effect on baseline clinical laboratory values. Least-square means were derived to adjust for the effects of confounding covariates (Supplemental Methods). Results are shown as mean ± SEM. P trend tests, stepwise logistic regressions, and calculations of least-square means were performed with SAS v.9.2. For all tests, 2-tailed P values of less than 0.05 were considered significant. Statistical analysis for kinetic experiments utilized paired 2-tailed t tests, with a cutoff for significance of P < 0.05.
Study approval.
The ClinSeq protocol was approved by the Institutional Review Boards of the National Human Genome Research Institute, NIH, and Suburban Hospital. All study participants gave informed consent upon enrollment.
Supplementary Material
Acknowledgments
We thank Stephen Wincovitch at the National Human Genome Research Institute (NHGRI) Microscopy Core for assistance gathering fluorescence images; Pedro Cruz for variant detection in GCKR exons across the ClinSeq sample cohort; Lori Bonnycastle, Mike Erdos, Narisu Narisu, Peter Chines, Heather Stringham, Anne Jackson, and Caitlin Krause for assistance with variant genotyping; Chris Groves, Mira Korner, and Ben Glaser for assistance with p.Ala519Thr genotyping; Nancy J. Cox, Graeme I. Bell, Veronica P. Paz, and Craig L. Hanis for assistance with p.Val103Met genotyping; Neal Oden for statistical discussions; Tanya Teslovich for discussions regarding SIFT and PolyPhen predictions; and Nicole Stone and Michael Stitzel for advice. This study was supported by the NIH Division of Intramural Research and NHGRI project number Z01-HG000024 (to F.S. Collins), in Oxford by the Medical Research Council (MRC) (81696) and the Wellcome Trust (095101/Z/10/Z), and by US Public Health Service grants DK020595 (to Graeme I. Bell.) and DK073451 and DK085501 (to Craig L. Hanis ). A.L. Gloyn is a Wellcome Trust Senior Fellow in Basic Biomedical Research.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(1):205–217. doi:10.1172/JCI46425.
Contributor Information
Collaborators: NISC Comparative Sequencing Program
References
- 1.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360(17):1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Manolio TA, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. doi: 10.1038/nature08494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McCarthy MI, Hirschhorn JN. Genome-wide association studies: potential next steps on a genetic journey. Hum Mol Genet. 2008;17(R2):R156–165. doi: 10.1093/hmg/ddn289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McCarthy MI, et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat Rev Genet. 2008;9(5):356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 5.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324(5925):387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305(5685):869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 7.Cohen JC, et al. Multiple rare variants in NPC1L1 associated with reduced sterol absorption and plasma low-density lipoprotein levels. Proc Natl Acad Sci U S A. 2006;103(6):1810–1815. doi: 10.1073/pnas.0508483103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kotowski IK, et al. A spectrum of PCSK9 alleles contributes to plasma levels of low-density lipoprotein cholesterol. Am J Hum Genet. 2006;78(3):410–422. doi: 10.1086/500615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schork NJ, Murray SS, Frazer KA, Topol EJ. Common vs. rare allele hypotheses for complex diseases. Curr Opin Genet Dev. 2009;19(3):212–219. doi: 10.1016/j.gde.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Romeo S, et al. Rare loss-of-function mutations in ANGPTL family members contribute to plasma triglyceride levels in humans. J Clin Invest. 2009;119(1):70–79. doi: 10.1172/JCI37118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johansen CT, et al. Excess of rare variants in genes identified by genome-wide association study of hypertriglyceridemia. Nat Genet. 2010;42(8):684–687. doi: 10.1038/ng.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orho-Melander M, et al. Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes. 2008;57(11):3112–3121. doi: 10.2337/db08-0516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beer NL, et al. The P446L variant in GCKR associated with fasting plasma glucose and triglyceride levels exerts its effect through increased glucokinase activity in liver. Hum Mol Genet. 2009;18(21):4081–4088. doi: 10.1093/hmg/ddp357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Toyoda Y, Miwa I, Satake S, Anai M, Oka Y. Nuclear location of the regulatory protein of glucokinase in rat liver and translocation of the regulator to the cytoplasm in response to high glucose. Biochem Biophys Res Commun. 1995;215(2):467–473. doi: 10.1006/bbrc.1995.2488. [DOI] [PubMed] [Google Scholar]
- 15.Vandercammen A, Van Schaftingen E. The mechanism by which rat liver glucokinase is inhibited by the regulatory protein. Eur J Biochem. 1990;191(2):483–489. doi: 10.1111/j.1432-1033.1990.tb19147.x. [DOI] [PubMed] [Google Scholar]
- 16.Van Schaftingen E. A protein from rat liver confers to glucokinase the property of being antagonistically regulated by fructose 6-phosphate and fructose 1-phosphate. Eur J Biochem. 1989;179(1):179–184. doi: 10.1111/j.1432-1033.1989.tb14538.x. [DOI] [PubMed] [Google Scholar]
- 17.Dipietro DL, Sharma C, Weinhouse S. Studies on glucose phosphorylation in rat liver. Biochemistry. 1962;1:455–462. doi: 10.1021/bi00909a014. [DOI] [PubMed] [Google Scholar]
- 18.Salas M, Vinuela E, Sols A. Insulin-dependent synthesis of liver glucokinase in the rat. J Biol Chem. 1963;238:3535–3538. [PubMed] [Google Scholar]
- 19.Walker DG, Rao S. The role of glucokinase in the phosphorylation of glucose by rat liver. Biochem J. 1964;90(2):360–368. doi: 10.1042/bj0900360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Matschinsky FM, Ellerman JE. Metabolism of glucose in the islets of Langerhans. J Biol Chem. 1968;243(10):2730–2736. [PubMed] [Google Scholar]
- 21.van Schaftingen E, Vandercammen A, Detheux M, Davies DR. The regulatory protein of liver glucokinase. Adv Enzyme Regul. 1992;32:133–148. doi: 10.1016/0065-2571(92)90013-P. [DOI] [PubMed] [Google Scholar]
- 22.Shiota C, Coffey J, Grimsby J, Grippo JF, Magnuson MA. Nuclear import of hepatic glucokinase depends upon glucokinase regulatory protein, whereas export is due to a nuclear export signal sequence in glucokinase. J Biol Chem. 1999;274(52):37125–37130. doi: 10.1074/jbc.274.52.37125. [DOI] [PubMed] [Google Scholar]
- 23.Veiga-da-Cunha M, Van Schaftingen E. Identification of fructose 6-phosphate- and fructose 1-phosphate-binding residues in the regulatory protein of glucokinase. J Biol Chem. 2002;277(10):8466–8473. doi: 10.1074/jbc.M105984200. [DOI] [PubMed] [Google Scholar]
- 24.Farrelly D, et al. Mice mutant for glucokinase regulatory protein exhibit decreased liver glucokinase: a sequestration mechanism in metabolic regulation. Proc Natl Acad Sci U S A. 1999;96(25):14511–14516. doi: 10.1073/pnas.96.25.14511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grimsby J, et al. Characterization of glucokinase regulatory protein-deficient mice. J Biol Chem. 2000;275(11):7826–7831. doi: 10.1074/jbc.275.11.7826. [DOI] [PubMed] [Google Scholar]
- 26.Hiskett EK, Suwitheechon OU, Lindbloom-Hawley S, Boyle DL, Schermerhorn T. Lack of glucokinase regulatory protein expression may contribute to low glucokinase activity in feline liver. Vet Res Commun. 2009;33(3):227–240. doi: 10.1007/s11259-008-9171-6. [DOI] [PubMed] [Google Scholar]
- 27.Slosberg ED, et al. Treatment of type 2 diabetes by adenoviral-mediated overexpression of the glucokinase regulatory protein. Diabetes. 2001;50(8):1813–1820. doi: 10.2337/diabetes.50.8.1813. [DOI] [PubMed] [Google Scholar]
- 28.Saxena R, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 29.Ridker PM, et al. Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women’s Genome Health Study. Am J Hum Genet. 2008;82(5):1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolz M, et al. Meta-analysis of 28,141 individuals identifies common variants within five new loci that influence uric acid concentrations. PLoS Genet. 2009;5(6):e1000504. doi: 10.1371/journal.pgen.1000504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saxena R, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42(2):142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Illig T, et al. A genome-wide perspective of genetic variation in human metabolism. Nat Genet. 2010;42(2):137–141. doi: 10.1038/ng.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ingelsson E, et al. Detailed physiologic characterization reveals diverse mechanisms for novel genetic loci regulating glucose and insulin metabolism in humans. Diabetes. 2010;59(5):1266–1275. doi: 10.2337/db09-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dupuis J, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kottgen A, et al. New loci associated with kidney function and chronic kidney disease. Nat Genet. 2010;42(5):376–384. doi: 10.1038/ng.568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamatani Y, et al. Genome-wide association study of hematological and biochemical traits in a Japanese population. Nat Genet. 2010;42(3):210–215. doi: 10.1038/ng.531. [DOI] [PubMed] [Google Scholar]
- 37.Tang W, et al. Genome-wide association study identifies novel loci for plasma levels of protein C: the ARIC study. Blood. 2010;116(23):5032–5036. doi: 10.1182/blood-2010-05-283739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chambers JC, et al. Genome-wide association study identifies loci influencing concentrations of liver enzymes in plasma. Nat Genet. 2011;43(11):1131–1138. doi: 10.1038/ng.970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Suhre K, et al. Human metabolic individuality in biomedical and pharmaceutical research. Nature. 2011;477(7362):54–60. doi: 10.1038/nature10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teslovich TM, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tam CH, et al. Interaction effect of genetic polymorphisms in glucokinase (GCK) and glucokinase regulatory protein (GCKR) on metabolic traits in healthy Chinese adults and adolescents. Diabetes. 2009;58(3):765–769. doi: 10.2337/db08-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chambers JC, et al. Common genetic variation near melatonin receptor MTNR1B contributes to raised plasma glucose and increased risk of type 2 diabetes among Indian Asians and European Caucasians. Diabetes. 2009;58(11):2703–2708. doi: 10.2337/db08-1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Rees MG, et al. Cellular characterisation of theGCKR P446L variant associated with type 2 diabetes risk [published online ahead of print October 25, 2011].Diabetologia . doi:10.1007/s00125–011–2348–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stancakova A, et al. Effects of 34 risk loci for type 2 diabetes or hyperglycemia on lipoprotein subclasses and their composition in 6,580 nondiabetic Finnish men. Diabetes. 2011;60(5):1608–1616. doi: 10.2337/db10-1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Facio FM, Feero WG, Linn A, Oden N, Manickam K, Biesecker LG. Validation of my family health portrait for six common heritable conditions. Genet Med. 2010;12(6):370–375. doi: 10.1097/GIM.0b013e3181e15bd5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Biesecker LG, et al. The ClinSeq Project: piloting large-scale genome sequencing for research in genomic medicine. Genome Res. 2009;19(9):1665–1674. doi: 10.1101/gr.092841.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Veiga-da-Cunha M, et al. Mutations in the glucokinase regulatory protein gene in 2p23 in obese French caucasians. Diabetologia. 2003;46(5):704–711. doi: 10.1007/s00125-003-1083-y. [DOI] [PubMed] [Google Scholar]
- 49.Veiga-da-Cunha M, Sokolova T, Opperdoes F, Van Schaftingen E. Evolution of vertebrate glucokinase regulatory protein from a bacterial N-acetylmuramate 6-phosphate etherase. Biochem J. 2009;423(3):323–332. doi: 10.1042/BJ20090986. [DOI] [PubMed] [Google Scholar]
- 50.Ng PC, Henikoff S. Predicting deleterious amino acid substitutions. Genome Res. 2001;11(5):863–874. doi: 10.1101/gr.176601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ramensky V, Bork P, Sunyaev S. Human non-synonymous SNPs: server and survey. Nucleic Acids Res. 2002;30(17):3894–3900. doi: 10.1093/nar/gkf493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kaganovich D, Kopito R, Frydman J. Misfolded proteins partition between two distinct quality control compartments. Nature. 2008;454(7208):1088–1095. doi: 10.1038/nature07195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bosco D, Meda P, Iynedjian PB. Glucokinase and glucokinase regulatory protein: mutual dependence for nuclear localization. Biochem J. 2000;348 pt 1:215–222. [PMC free article] [PubMed] [Google Scholar]
- 55.Saaristo T, et al. Cross-sectional evaluation of the Finnish Diabetes Risk Score: a tool to identify undetected type 2 diabetes, abnormal glucose tolerance and metabolic syndrome. Diab Vasc Dis Res. 2005;2(2):67–72. doi: 10.3132/dvdr.2005.011. [DOI] [PubMed] [Google Scholar]
- 56.Stancakova A, Javorsky M, Kuulasmaa T, Haffner SM, Kuusisto J, Laakso M. Changes in insulin sensitivity and insulin release in relation to glycemia and glucose tolerance in 6,414 Finnish men. Diabetes. 2009;58(5):1212–1221. doi: 10.2337/db08-1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Below JE, et al. Genome-wide association and meta-analysis in populations from Starr County, Texas, and Mexico City identify type 2 diabetes susceptibility loci and enrichment for expression quantitative trait loci in top signals. Diabetologia. 2011;54(8):2047–2055. doi: 10.1007/s00125-011-2188-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470(7333):187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 60.Shin JS, Torres TP, Catlin RL, Donahue EP, Shiota M. A defect in glucose-induced dissociation of glucokinase from the regulatory protein in Zucker diabetic fatty rats in the early stage of diabetes. Am J Physiol. 2007;292(4):R1381–R1390. doi: 10.1152/ajpregu.00260.2006. [DOI] [PubMed] [Google Scholar]
- 61.Wilson PW, D’Agostino RB, Levy D, Belanger AM, Silbershatz H, Kannel WB. Prediction of coronary heart disease using risk factor categories. Circulation. 1998;97(18):1837–1847. doi: 10.1161/01.cir.97.18.1837. [DOI] [PubMed] [Google Scholar]
- 62.Liang Y, et al. Variable effects of maturity-onset-diabetes-of-youth (MODY)-associated glucokinase mutations on substrate interactions and stability of the enzyme. Biochem J. 1995;309(pt 1):167–173. doi: 10.1042/bj3090167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voight BF, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.