Abstract
DNA methyltransferase 3B (Dnmt3b) belongs to a family of enzymes responsible for methylation of cytosine residues in mammals. DNA methylation contributes to the epigenetic control of gene transcription and is deregulated in virtually all human tumors. To better understand the generation of cancer-specific methylation patterns, we genetically inactivated Dnmt3b in a mouse model of MYC-induced lymphomagenesis. Ablation of Dnmt3b function using a conditional knockout in T cells accelerated lymphomagenesis by increasing cellular proliferation, which suggests that Dnmt3b functions as a tumor suppressor. Global methylation profiling revealed numerous gene promoters as potential targets of Dnmt3b activity, the majority of which were demethylated in Dnmt3b–/– lymphomas, but not in Dnmt3b–/– pretumor thymocytes, implicating Dnmt3b in maintenance of cytosine methylation in cancer. Functional analysis identified the gene Gm128 (which we termed herein methylated in normal thymocytes [Ment]) as a target of Dnmt3b activity. We found that Ment was gradually demethylated and overexpressed during tumor progression in Dnmt3b–/– lymphomas. Similarly, MENT was overexpressed in 67% of human lymphomas, and its transcription inversely correlated with methylation and levels of DNMT3B. Importantly, knockdown of Ment inhibited growth of mouse and human cells, whereas overexpression of Ment provided Dnmt3b+/+ cells with a proliferative advantage. Our findings identify Ment as an enhancer of lymphomagenesis that contributes to the tumor suppressor function of Dnmt3b and suggest it could be a potential target for anticancer therapies.
Introduction
Cytosine methylation represents a heritable epigenetic modification affecting gene transcription and the integrity of the genome. The DNA methyltransferases (DNMTs) DNMT1, DNMT3A, and DNMT3B are the enzymes primarily responsible for methylation of CpG dinucleotides in mammalian DNA. Basic methylation patterns are established by catalytic activity of de novo methyltransferases Dnmt3a and Dnmt3b on a demethylated genome during early embryogenesis (1). DNMT1 recognizes methylation patterns in parental DNA and replicates them to nascent DNA strands during cell division; thus, DNMT1 is considered a maintenance methyltransferase. Although most CpGs in the genome are methylated, promoters containing clusters of CpG dinucleotides (also referred to as CpG islands) remain unmethylated and are typically transcriptionally active. Methylation of these promoters is associated with transcriptional repression, which plays a role in a variety of normal physiologic processes, including development, X-chromosome inactivation, genomic imprinting, and suppression of unwanted transcription (2).
Previous studies in mice have revealed the importance of DNA methylation in normal hematopoiesis: loss of Dnmt1 severely impairs the development of thymocytes and myeloid lineages (3, 4); Dnmt3a is involved in regulation of the helper T cells Th1 and Th2 (5); and combined loss of Dnmt3a and Dnmt3b in hematopoietic stem cells has no effect on their differentiation into myeloid or lymphoid lineages, but impairs their ability to self renew (6). However, to our knowledge, the individual loss of Dnmt3b in hematopoiesis in vivo has not been analyzed to date.
In humans, deregulated DNA methylation is involved in the pathogenesis of systemic lupus erythematosus, asthma, and other inflammatory processes (7–9). In addition, methylation is deregulated in a variety of hematologic malignancies, including leukemias, lymphomas, and myelodysplastic syndromes (MDS) (10). This deregulation is manifested by global genome-wide hypomethylation and regional promoter hypermethylation (11), both of which have the potential to promote tumorigenesis. For example, hypomethylation of the genome induced by a hypomorphic allele of Dnmt1 resulted in development of T cell lymphomas in mice (12). Additionally, genes upregulated by hypomethylation, such as IGF2 and S100A4, have been implicated as promoters of tumor development (13, 14). In contrast, promoter regions of tumor suppressor genes, such as CDKN2A and CDKN2B, are hypermethylated and silenced in hematologic malignancies (15). The reactivation of such tumor suppressors is thought to be responsible for the success of DNMT inhibitors in the management of MDS, but further studies are required to assess the effects of drugs on DNA methylation patterns in tumors cells (16).
Despite extensive efforts in the DNA methylation field over the last 2 decades, the functions of individual Dnmts in the generation of locus-specific DNA methylation patterns and their maintenance remain poorly understood. This is partially due to the fact that tumor cells grown in culture exhibit methylation patterns different from those of the primary tumors from which they were derived, suggestive of a greater extent of functional overlap between Dnmts in vitro that may skew methylation readouts in certain settings (17). Recent in vivo studies have demonstrated that Dnmt3b, but not Dnmt3a, plays a critical role in de novo promoter DNA methylation in developing mouse embryos. Furthermore, transgenic expression of Dnmt3b in normal mouse colonic mucosa resulted in hypermethylation of 1,500 genes, which suggests that a subset of genes in the genome is intrinsically methylation sensitive (18). Other developmental studies showed that DNMTs may have overlapping functions in de novo and maintenance methylation (2, 19–21), but the extent of the overlap, particularly in tumor development, is not clear.
To determine the contribution of de novo methyltransferase Dnmt3b to the cancer-specific DNA methylation landscape, we used a mouse model of MYC-induced T cell lymphomagenesis. Loss of Dnmt3b did not affect T cell development, but accelerated MYC-induced T cell lymphomagenesis. This process was accompanied by global changes in the methylation landscape, gene transcription, and increased genomic instability. Surprisingly, Dnmt3b played a role in the maintenance of promoter methylation of a large number of genes, including the predicted gene Gm128 (C1ORF56 in humans). Because Gm128 is methylated and silenced in normal thymocytes, we have termed this gene methylated in normal thymocytes (Ment). Further studies identified Ment as a potential proto-oncogene in mouse lymphomagenesis whose continuous methylation and repression depended on the maintenance activity of Dnmt3b. Thus, our results not only provide a mechanism for accelerated lymphomagenesis in Dnmt3b-deficient mice, but also identify MENT as a potential target for anticancer therapies.
Results
Loss of Dnmt3b activity does not affect T cell development.
To evaluate the effects of Dnmt3b on T cell development, we conditionally inactivated Dnmt3b in mouse T cells using a EμSR-tTA;Teto-Cre;Rosa26LOXPEGFP system. In this configuration (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI57292DS1), the tetracycline transactivator protein (tTA) is under the control of the Ig heavy chain enhancer and the SRα promoter (EμSR-tTA; ref. 22). In the absence of doxycycline, tTA is expressed in a subset of hematopoietic stem/progenitor cells, and its expression persists in 40%–60% of T cells (23). Expression of tTA leads to activation of the Teto-Cre transgene, resulting in the excision of the stop cassette located upstream of the Rosa26LOXPEGFP reporter locus and, therefore, coexpression of EGFP and Cre within the same subpopulation of cells. When combined with the conditional knockout allele of Dnmt3b (Dnmt3b2LoxP; ref. 19), Cre-mediated excision of the Dnmt3b conditional allele occurs only in EGFP+ T cells, not EGFP– T cells. To examine the effects of Dnmt3b loss on T cell development, we generated cohorts of EμSR-tTA;Teto-Cre;Rosa26LOXPEGFP;Dnmt3bfl/fl and EμSR-tTA;Teto-Cre;Rosa26LOXPEGFP;Dnmt3b+/+ mice. Although Dnmt3b was efficiently deleted in EGFP+ cells isolated from thymi of Dnmt3b mutant mice (Supplemental Figure 1, B and C), no substantial differences were found in survival over 1 year, nor in the organ size, cellularity, or EGFP or marker expression in thymi, spleens, and lymph nodes of mice at 6–8 weeks or 8–12 months of age (Supplemental Figure 1, D–H). Collectively, these results indicated that Dnmt3b is dispensable for T cell development and that its loss is insufficient to induce lymphomagenesis.
A tumor suppressor function for Dnmt3b in mouse lymphomagenesis.
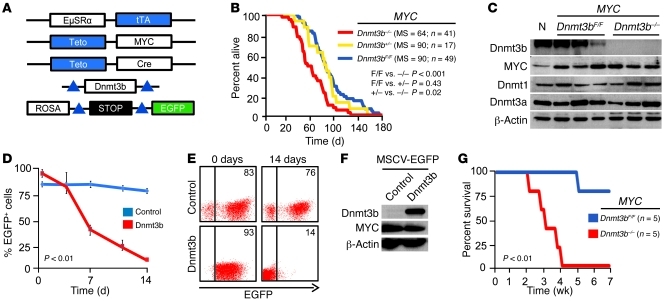
To determine the role of Dnmt3b in MYC-induced lymphomagenesis, we compared tumor development in cohorts of EμSR-tTA;Teto-MYC;Teto-Cre;Rosa26LOXPEGFP;Dnmt3bfl/fl and EμSR-tTA;Teto-MYC;Rosa26LOXPEGFP;Dnmt3bfl/fl mice (referred to herein as MYC;Dnmt3b–/– and MYC;Dnmt3bfl/fl, respectively). Expression of MYC is driven by the tTA protein (Figure 1A) and results in development of T cell lymphomas (22). Using these animal cohorts, we found that although loss of 1 allele of Dnmt3b had no effect (median survival [MS], 90 days), the biallelic inactivation of Dnmt3b significantly accelerated lymphomagenesis (MYC;Dnmt3b–/–, MS 64 days; MYC;Dnmt3bfl/fl, MS 90 days; P < 0.001, Kaplan-Meier; Figure 1B). FACS analysis using antibodies against CD4, CD8, CD44, CD25, B220, and CD11b revealed that MYC;Dnmt3b–/– lymphomas were immunophenotypically indistinguishable from MYC;Dnmt3bfl/fl tumors (Supplemental Figure 2, A and B). Analysis of protein and DNA confirmed the complete ablation of Dnmt3b in tumors arising in MYC;Dnmt3b–/– mice (Figure 1C and Supplemental Figure 3, A and B). Loss of Dnmt3b had no effect on MYC expression (Figure 1C and Supplemental Figure 4), which suggests that the accelerated tumorigenesis in Dnmt3b-deficient mice was not caused by upregulation of transgenic MYC. By virtue of the genetic setting, MYC;Dnmt3b–/– mice, unlike controls, expressed Cre and EGFP. We confirmed that there were no measureable differences in survival between EμSR-tTA;Teto-MYC;Teto-Cre;Rosa26LOXPEGFP (referred to herein as MYC;Dnmt3b+/+) and EμSR-tTA;Teto-MYC mice (ref. 23 and data not shown), which suggests that there are no adverse effects of Cre and EGFP on the course of lymphomagenesis. Importantly, MYC overexpression also did not alter T cell development, since no significant changes in expression of cell surface markers were observed between MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– thymocytes early in tumor development (Supplemental Figure 5). Thus, we concluded that differences in survival can be attributed to loss of Dnmt3b.
Figure 1. Accelerated lymphomagenesis and increased relapse rate in Dnmt3b-deficient mice.
(A) EμSRα-tTA, Teto-MYC, Teto-Cre, and ROSA26LOXPEGFP transgenes and the Dnmt3b conditional knockout allele (Dnmt3b2LoxP) used. Blue indicates that tTA directly regulates transcription from Teto promoters and indirectly recombines loxP sites (triangles). (B) Kaplan-Meier survival curves of MYC;Dnmt3bfl/fl (F/F), MYC;Dnmt3b+/– (+/–), and MYC;Dnmt3b–/– (–/–) mice. MS, n, and P values (log-rank test) are indicated. (C) Expression of Dnmt3b, MYC, Dnmt1, and Dnmt3a assessed by Western blot in normal thymocytes (N), control lymphomas (MYC;Dnmt3bfl/fl), and Dnmt3b-deficient (MYC;Dnmt3b–/–) lymphomas. β-Actin served as a loading control. (D) In vitro growth of unselected MYC;Dnmt3b–/– cells infected with the indicated retroviral constructs. Cells were plated at a concentration of 0.2 × 106 per ml (day 0) and replated every 2 days. Percent EGFP+ cells was determined by FACS. Data are mean ± SEM of 3 independent experiments. P < 0.01, 2-way ANOVA/Tukey test, for the last time point. (E) Representative examples of FACS in D. Percent EGFP+ cells is indicated. (F) Western blot analysis of Dnmt3b levels in MSCV-IRES-EGFP (control) and MSCV-Dnmt3b-IRES-EGFP (Dnmt3b) infected MYC;Dnmt3b–/– cells from D and E. (G) Kaplan-Meier relapse-free survival of MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– cohorts. Terminally ill mice were placed on doxycycline and monitored for tumor relapse. P < 0.01, log-rank test.
To support this conclusion, we infected cells derived from MYC;Dnmt3b–/– tumors with control MSCV-IRES-GFP or Dnmt3b-expressing retroviruses and monitored in vitro growth of EGFP+ cells by FACS. While the percentage of EGFP+ control cells remained similar over 14 days, the percentage of Dnmt3b-overexpressing cells gradually decreased (Figure 1, D–F). These results suggest that expression of Dnmt3b exerts an inhibitory effect on tumor cell proliferation and support a tumor-suppressive function of Dnmt3b in mouse lymphomagenesis.
Inactivation of MYC by doxycycline treatment in terminally ill EμSR-tTA;Teto-MYC mice results in approximately 100% of tumor regression and low frequency of tumor relapse (~20%) over the course of 8 weeks (22). To determine whether primary Dnmt3b-deficient lymphomas relapse upon MYC inactivation, we treated terminally ill MYC;Dnmt3b–/– and MYC;Dnmt3bfl/fl mice with doxycycline. All treated mice revived a few days after treatment and were virtually identical to healthy mice. Whereas 1 of 5 MYC;Dnmt3bfl/fl mice relapsed, all MYC;Dnmt3b–/– mice relapsed, with a mean latency of 22 days (Figure 1G), which suggests that MYC;Dnmt3b–/– cells acquired molecular changes that bypassed inhibition of MYC.
Loss of Dnmt3b increases cellular proliferation during disease progression.
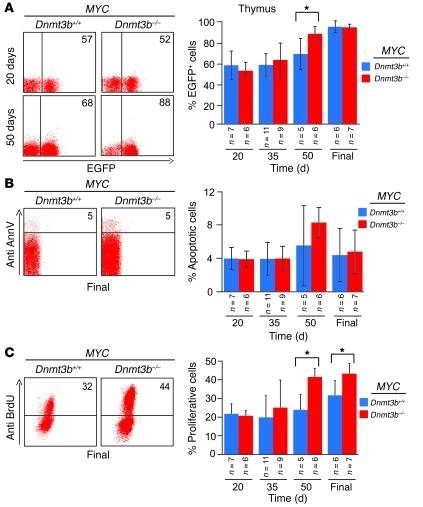
MYC;Dnmt3b+/+ mice are asymptomatic until the age of 42 days, when some start to show early signs of tumor progression (23). The presence of the Rosa26LOXPEGFP reporter transgene allows for evaluation of biological and molecular events occurring in precancerous cells by FACS. To investigate whether loss of Dnmt3b affects initiation or progression of tumorigenesis, we measured levels of EGFP in thymi, spleens, and lymph nodes of MYC;Dnmt3b+/+ and MYC;Dnmt3b–/– mice during tumor development. Whereas no substantial differences in EGFP levels were observed at early stages of lymphomagenesis, MYC;Dnmt3b–/– mice showed increased levels of EGFP at 50 days of age (Figure 2A), which suggests that loss of Dnmt3b affects tumor progression, but not initiation. Similarly, no differences in apoptotic indices were observed between MYC;Dnmt3b+/+ and MYC;Dnmt3b–/– mice at any stage of tumor development (Figure 2B and Supplemental Figure 6, A and B). Whereas BrdU incorporation was similar at early stages of tumor development (age 20 and 35 days), a substantial increase in cells incorporating BrdU was observed at later stages of tumor development (age 50 days and terminally ill mice; Figure 2C and Supplemental Figure 6, A and B). These data suggest that accelerated lymphomagenesis in Dnmt3b-deficient mice is caused by increased proliferation during disease progression.
Figure 2. Loss of Dnmt3b results in enhanced proliferation of tumors in vivo.
EGFP expression (A), apoptosis (B), and BrdU incorporation assay (C), as determined by FACS analysis of thymocytes isolated from MYC;Dnmt3b+/+ and MYC;Dnmt3b–/– mice at 20, 35, and 50 days of age as well as in terminally ill mice (Final). Anti–annexin V and anti-BrdU antibodies were used to evaluate apoptosis and BrdU incorporation, respectively. Representative examples of FACS analysis at the indicated ages are shown; percent positive cells in the FACS profile is shown within each plot. Error bars represent ± SEM; n as indicated. *P < 0.05, pairwise comparisons, Student’s t test with Bonferroni correction.
Dnmt3b deficiency leads to increased genomic instability in mouse lymphomas.
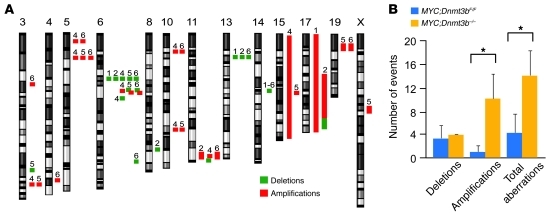
Hypomethylation of the cancer genome is known to be associated with increased genomic instability (12). To determine whether Dnmt3b-deficient lymphomas show higher levels of genomic instability, we performed array comparative genome hybridization (aCGH) on DNA isolated from MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– tumors (Figure 3A). While both tumor groups showed genomic instability, MYC;Dnmt3b–/– tumors had significant increases in the number of amplifications (Figure 3B). Certain aberrations, such as deletions in qC2 of chromosome 14, were consistently detected in both tumor settings, which suggests that these changes are intrinsic to the tumorigenic process and do not depend on Dnmt3b. Only 1 genetic aberration, an amplification in the qA3 locus of chromosome 5, was consistently present in MYC;Dnmt3b–/– lymphomas (Supplemental Table 1). This area is associated with the Lhfpl3 gene, whose expression did not seem to be affected. Thus, the phenomenon of increased chromosomal aberrations is an unlikely explanation for the accelerated lymphomagenesis observed in MYC;Dnmt3b–/– mice.
Figure 3. Ablation of Dnmt3b leads to increased genome instability.
(A) Cytogenetic aberrations found using aCGH for 3 MYC;Dnmt3bfl/fl tumors (tumors 1–3) and 3 MYC;Dnmt3b–/– tumors (tumors 4–6). Positions of amplifications and deletions are indicated next to each chromosome. Only chromosomes with copy number variations are shown. (B) Number of deletions, amplifications, and total aberrations found in MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– tumors (n = 3 per group). *P < 0.05, pairwise comparisons, Tukey test.
Tumor DNA methylation landscape in the absence of Dnmt3b.
To determine the effects of Dnmt3b on global DNA methylation, we first measured the content of 5-methylcytosine in DNA from primary mouse lymphomas using HPLC (24). We observed a 12% decrease in 5-methylcytosine levels in MYC;Dnmt3bfl/fl tumors compared with aged-matched normal thymocytes (Figure 4A). In MYC;Dnmt3b–/– tumors, total 5-methylcytosine decreased 23%, suggesting a nonredundant contribution of Dnmt3b to overall genome methylation in lymphomas. The Dnmt3b-dependent fraction may be partially attributable to methylation of short interspersed nuclear elements (SINEs), as we observed a significant decrease in SINE methylation in MYC;Dnmt3b–/– compared with MYC;Dnmt3bfl/fl tumors (Figure 4B and Supplemental Figure 7).
Figure 4. Dnmt3b plays a role in cancer-specific maintenance methylation.
(A) Total 5′methyl-cytosine content in normal thymocytes (N), MYC;Dnmt3bfl/fl tumors, and MYC;Dnmt3b–/– tumors, determined by HPLC (n = 2 per group) and expressed relative to normal thymocytes. (B) Relative methylation levels of SINE-B1 repeats, determined by pyrosequencing. (C) Analysis of SINE-B1 and LINE repeats using MSCC data. (D) Methylation changes in HpaII sites, determined by negative binomial analysis of MSCC counts obtained from MYC;Dnmt3bfl/fl (n = 5) and MYC;Dnmt3b–/– (n = 5) versus normal thymocytes (n = 3). Yellow wedge indicates number of HpaII sites with statistically significant changes in methylation counts relative to normal. Sites changed in a genotype-specific manner are shown by colored boxes; arrow direction denotes significant increase or decrease in methylation. (E) Gene-body methylation of HpaII sites in the indicated tumors. (F) Number of promoters with significant changes in methylation of the indicated lymphomas, determined by MSCC. Number of HpaII sites within CpG islands is indicated. (G) Analysis of promoters hypomethylated in MYC;Dnmt3b–/– lymphomas. MSCC counts obtained from MYC;Dnmt3b–/– pretumor (24–/–) and MYC;Dnmt3bfl/fl lymphomas were compared with normal thymocytes (single negative binomial analysis). Dashes indicate no significant change in methylation; arrow direction denotes significant increase or decrease in methylation. (A–C) *P < 0.05, 2 sample t test with Bonferroni adjustment. (D–F) Negative binomial analysis was used; only changes with FDRs less than 5% are shown.
To investigate the DNA methylation landscape in lymphomas, we used methyl-sensitive cut counting (MSCC), a technique that profiles the methylation of approximately 1 × 106 CG dinucleotides across the genome using HpaII restriction digests and next-generation sequencing (25). The number of sequence tags (termed counts) inversely correlates with the degree of methylation in HpaII sites. Using MSCC, we profiled methylation patterns of MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– lymphomas along with EμSR-tTA;Teto-Cre;Rosa26LOXPEGFP normal thymocytes and MYC;Dnmt3b–/– pretumor thymocytes (FACS-sorted EGFP+ thymocytes isolated from 24-day-old mice). Reliability of MSCC data was assessed by real-time PCR of HpaII-digested genomic DNA using the strategy shown in Supplemental Figure 8A. Methylation status of HpaII sites correlated with MSCC data in 20 of 26 samples tested (77%; Supplemental Figure 8, B and C), which suggests that MSCC readouts faithfully reflect the degree of DNA methylation. We then used combined bisulfite restriction analysis (COBRA) to detect locus-specific methylation close to HpaII sites. In 16 of 21 loci analyzed (76%), MSCC and COBRA assays yielded identical results (Supplemental Figure 9, A and B). Thus, MSCC readouts are likely to reflect the methylation status of broader areas surrounding HpaII sites. We next analyzed MSCC data to determine effects of Dnmt3b on methylation of long interspersed nuclear elements (LINEs) and SINEs. Consistent with our functional analysis, SINE repeats were hypomethylated in MYC;Dnmt3bfl/fl tumors, and the difference was further increased in MYC;Dnmt3b–/– tumors (Figure 4C). Unlike SINEs, LINEs were hypomethylated in MYC;Dnmt3b–/– tumors compared with control thymocytes, but not compared with MYC;Dnmt3bfl/fl lymphomas (Figure 4C). Together, these data suggest that Dnmt3b contributes to methylation of SINE repeats, but not LINE repeats.
Further analysis of MSCC data revealed that the methylation status of 42,422 of 1,291,893 HpaII sites changed between tumors and normal thymocytes (Figure 4D). Of these, 3,715 methylation changes were specific to MYC;Dnmt3bfl/fl, with 714 HpaII sites hypermethylated and 3,001 HpaII sites demethylated. Similarly, 32,146 changes occurred only in MYC;Dnmt3b–/– lymphomas, with 6,250 sites hypermethylated and 25,896 sites hypomethylated. The methylation of the remaining 6,561 HpaII sites was changed in both tumor settings, which suggests that Dnmt3b does not contribute to their methylation.
We next performed 2-dimensional clustering analysis using MSCC counts significantly changed (7,753 tags) between MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– tumors. This analysis resulted in almost-perfect segregation of tumors in a Dnmt3b-specific manner, with normal thymocytes clustering closer to MYC;Dnmt3bfl/fl tumors than MYC;Dnmt3b–/– tumors (Supplemental Figure 10). Thus, Dnmt3b plays an important role in shaping DNA methylation landscape during tumor development.
In accord with these data, COBRA analysis showed that methylation for some loci (e.g., Clic4 and Isyna1) was comparable between the different genetic settings (Supplemental Figure 9, A and B). In contrast, methylation of Pnldc1 and Arhgef18 was virtually eliminated in all MYC;Dnmt3b–/– tumors (Supplemental Figure 11, A and B), which suggests that Dnmt3b plays a role promoter-specific methylation. Interestingly, while demethylation of the Pnldc1 promoter resulted in increased transcription, Arhgef18 levels remained unchanged (Supplemental Figure 11, C and D), suggesting involvement of other factors in gene regulation. Collectively, the data are consistent with global hypomethylation and regional hypermethylation of the cancer genome, a ubiquitous feature observed in tumors (2).
We next analyzed the contribution of Dnmt3b to intragenic methylation, as increased gene-body methylation has recently been linked to increased transcription (25). Loss of Dnmt3b resulted in hypomethylation of at least 1 HpaII site in 2,297 genes and hypermethylation of 887 genes in MYC;Dnmt3b–/– tumors compared with MYC;Dnmt3bfl/fl tumors (Figure 4E). Thus, Dnmt3b seems to play a role in both hyper- and hypomethylation of intragenic regions, but more detailed analysis is required to understand the molecular basis of this effect. We next compared methylation of promoter-specific HpaII sites (–1,500 to +500 relative to the transcription start site) between MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– tumors. This analysis showed that 260 HpaII sites (51 located within CpG islands) were hypomethylated in MYC;Dnmt3b–/– tumors, and 60 HpaII sites (17 located within CpG islands) were hypermethylated in MYC;Dnmt3b–/– tumors, with 22,016 HpaII sites unaffected between these 2 tumor groups (Figure 4F and Supplemental Table 2). Thus, 260 promoters represent potential targets of Dnmt3b-specific methylase activity. We next analyzed methylation status of these promoters in normal thymocytes and MYC;Dnmt3b–/– pretumor thymocytes. Compared with normal thymocytes, 201 promoters were hypomethylated in MYC;Dnmt3b–/– lymphomas, but not in MYC;Dnmt3b–/– pretumor thymocytes or MYC;Dnmt3bfl/fl tumors (Figure 4G), which suggests that Dnmt3b is dispensable for their de novo methylation during normal development, but irreplaceable for their tumor-specific maintenance. Conversely, methylation of 5 promoters increased in MYC;Dnmt3bfl/fl tumors compared with normal thymocytes, but was lost in MYC;Dnmt3b–/– tumors. These promoters represent potential targets of cancer-specific Dnmt3b de novo methylation. In addition, 69 promoters that were hypomethylated in MYC;Dnmt3b–/– lymphomas were also hypomethylated in MYC;Dnmt3b–/– pretumor thymocytes, which suggests that they might be targets of Dnmt3b de novo activity during either normal or tumor development. The de novo and maintenance groups had an overlap of 15 genes, as each promoter contained more than 1 HpaII site with dissimilar methylation readouts that met the requirements for different groups. Collectively, these data indicate that the contribution of Dnmt3b to the methylation landscape appears to consist not only of de novo activity, but mainly of tumor-specific maintenance of methylation patterns.
Dnmt3b deficiency results in deregulated transcription in lymphomas.
To further elucidate the mechanism of accelerated lymphomagenesis in MYC;Dnmt3b–/– mice, we screened for genes whose transcription was changed between control and Dnmt3b-deficient tumors. Microarray-based gene expression profiling identified 2,402 genes whose expression was significantly different between MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– lymphomas (Supplemental Table 3; P < 0.05). Of these, transcript levels of 40 genes increased more than 3-fold, whereas transcript levels of 25 genes decreased more than 3-fold in Dnmt3b-deficient tumors (Supplemental Table 4). Real-time quantitative RT-PCR (qRT-PCR) confirmed the altered expression of a subset of selected test genes (Supplemental Figure 12). We reasoned that the most likely candidates to explain Dnmt3b tumor suppressor function would be genes that were demethylated and overexpressed specifically in MYC;Dnmt3b–/– tumors. Analysis of MSCC data and gene expression yielded 4 putative Dnmt3b target genes potentially responsible for the tumor-suppressive function of Dnmt3b (Figure 5A). qRT-PCR and COBRA confirmed that Rpl39l, Psma8, Pnldc1, and Ment were overexpressed and demethylated in MYC;Dnmt3b–/– tumors (Figure 5, B and C). We next sought to determine whether siRNA-mediated inhibition of these genes could affect proliferation of Dnmt3b–/– lymphoma cells in vitro. Whereas a decrease in Rpl39l, Psma8, and Pnldc1 transcripts had no appreciable effect, knockdown of Ment resulted in an approximately 30% decrease in cellular proliferation caused by an increased number of cells in the G0/G1 phase of the cell cycle and a decreased number in the S and G2/M phases (Figure 5, D and E, and Supplemental Figure 13, A and B). Taken together, these data suggest that Ment is involved in control of cellular proliferation.
Figure 5. Gene expression, DNA methylation, and functional analysis of genes overexpressed in Dnmt3b-deficient tumors.
(A) Heat maps for genes with at least 3-fold induction of expression and at least 4-fold decrease in promoter methylation in MYC;Dnmt3b–/– lymphomas, determined by microarray (left) and MSCC (right). Average values from MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– lymphomas, normal thymocytes, and MYC;Dnmt3b–/– pretumor thymocytes are shown. Red and green indicate induction and reduction, respectively, in gene expression; blue and yellow indicate increase and decrease, respectively, in methylation levels. P values for microarray (Bayesian t test), and FDR for MSCC (negative binomial), and n for each group are shown. (B) qRT-PCR analysis of selected Dnmt3b target genes in normal thymocytes (n = 2) and in MYC;Dnmt3bfl/fl (n = 3) and MYC;Dnmt3b–/– (n = 3) lymphomas. Mean ± SEM fold difference is shown. (C) COBRA of selected Dnmt3b target genes in normal thymocytes, Dnmt3b–/– thymocytes (N3b), MYC;Dnmt3bfl/fl and MYC;Dnmt3b–/– tumors, and a fully methylated control (CpG). Ment was digested with TaiI; all other PCR fragments were digested with BstUI. Undigested (U) and digested (D) fragments correspond to unmethylated and methylated DNA, respectively. (D) siRNA-mediated knockdown in MYC;Dnmt3b–/– cells. Cells were transfected with control or SmartPool siRNA against the indicated genes and counted 48 hours later. Data derived from 3 experiments were normalized to control. (E) FACS-based cell cycle analysis of MYC;Dnmt3b–/– cells transfected with control siRNA or siRNA against Ment, determined 48 hours after transfection. (B and D) Error bars represent ± SEM. *P < 0.05, Tukey test.
Ment transcription is regulated by Dnmt3b-dependent methylation in lymphomas.
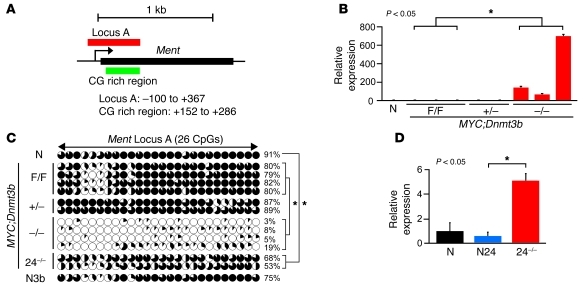
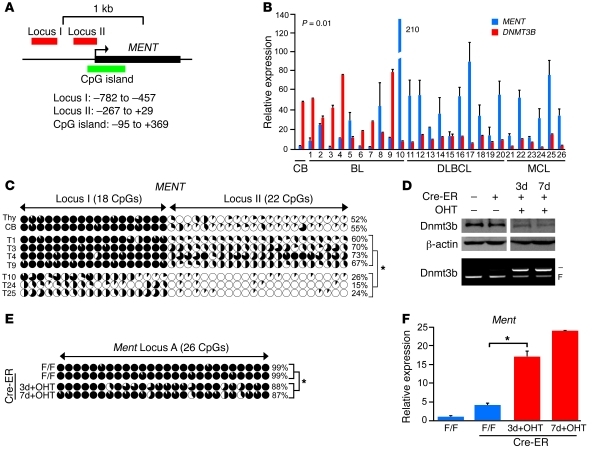
Ment was first identified in a functional screening for secreted proteins as a predicted protein encoding sequence (Gm128 in mouse, C1ORF56 in humans; ref. 26). The mouse Ment gene is located on chromosome 3 and encodes a protein composed of 350 amino acids with a predicted molecular weight of 39 kDa (Figure 6A). Ment protein has 73% homology to its human counterpart localized on chromosome 1 and no significant homology to other known proteins. In normal mouse tissues, Ment expression was high in testis, but low in other tissues (Supplemental Figure 14A). Consistent with its expression pattern, the promoter of Ment was not methylated in testis, but was methylated in all other mouse tissues tested, including thymus, kidney, heart, and lung (Supplemental Figure 14, B and C). Similarly, Ment was methylated and repressed in normal thymocytes as well as in MYC;Dnmt3bfl/fl and EμSRα-tTA;Teto-MYC;Teto-Cre;ROSA26LOXPEGFP;Dnmt3b+/fl (referred to herein as MYC;Dnmt3b+/–) tumors (Figure 6, B and C). In contrast, almost no methylation was observed in MYC;Dnmt3b–/– lymphomas, and this was accompanied by an approximately 100- to 600-fold increase in transcript levels (Figure 6, B and C). Furthermore, in MYC;Dnmt3b–/– pretumor thymocytes, Ment was partially derepressed, and mild promoter demethylation was observed relative to both Dnmt3b+/+ and Dnmt3b–/– normal thymocytes (Figure 6, C and D). These results suggest that Dnmt3b is dispensable for de novo methylation of Ment in normal thymocytes and that the contribution of Dnmt3b to methylation of this area is limited to its maintenance function in a tumor setting.
Figure 6. Ment is a target of Dnmt3b-mediated methylation in mouse lymphomas.
(A) Mouse genomic region of Ment. (B) qRT-PCR analysis of Ment expression in normal thymocytes and in MYC;Dnmt3bfl/fl, MYC;Dnmt3b+/–, and MYC;Dnmt3b–/– lymphomas. *P < 0.05, ANOVA. (C) Bisulfite sequencing of Ment locus A in DNA from thymocytes, MYC;Dnmt3bfl/fl lymphomas (n = 4), MYC;Dnmt3b+/– lymphomas (n = 2), MYC;Dnmt3b–/– lymphomas (n = 4), MYC;Dnmt3b–/– pretumor thymocytes (n = 2), and EGFP+ EμSR-tTA;Teto-Cre;Rosa26LOXPEGFP;Dnmt3bfl/fl thymocytes (N3b). Graphic denotes individual CpG dinucleotides within the locus; black shading denotes the percentage of methylated alleles. Shown is methylation analysis of 10 clones, excepting MYC;Dnmt3b+/– lymphomas (8 clones). Average methylation levels were calculated for the entire locus and are shown as percentage for each sample. Total average methylation of the Ment locus was calculated by averaging all samples within the group. *P < 0.05, Student’s t test. (D) qRT-PCR analysis of Ment expression in normal thymocytes, normal pretumor thymocytes (N24), and MYC;Dnmt3b–/– pretumor thymocytes (n = 2 per group). Error bars represent ± SEM. *P < 0.05, Student’s t test.
We next sought to determine whether methylation is important for regulation of human MENT (Figure 7A). Transcript levels of DNMT3B and MENT inversely correlated in human lymphoma cell lines (P < 0.05, Spearman rank correlation; Supplemental Figure 15A). The promoter locus I of MENT almost completely lacked methylated cytosines in cell lines expressing high levels of MENT (JVM-2, JY, JeKo-1, and RAJI), whereas high levels of methylation were found in low expressors of MENT (Akata, REH, and Daudi; Supplemental Figure 15, B and C). We further observed a statistically significant inverse correlation between levels of MENT and DNMT3B in 26 human tumors that consisted of Burkitt, diffuse large B cell, and mantle cell lymphomas (P < 0.05, Spearman rank correlation; Figure 7B). Methylation analysis of loci I and II revealed increased methylation in primary tumors with low levels of MENT (Supplemental Figure 16). Bisulfite sequencing performed on selected tumors with high levels of MENT and low levels of DNMT3B (tumors 10, 24, and 25) showed very little methylation in loci I and II in the MENT promoter (average methylation, 21.7%; Figure 7C). In contrast, tumors with high levels of DNMT3B and low levels of MENT (tumors 1, 3, 4, and 9) showed high levels of methylation in both areas (average methylation, 67.5%; Figure 7C). These data strongly suggest that methylation regulates human MENT transcription and that it is carried out by DNMT3B. To determine whether loss of Dnmt3b affects Ment methylation in vitro, we generated an inducible cell line, MYC;Dnmt3bfl/fl;Cre-ER, in which Cre-mediated excision of Dnmt3b conditional alleles was activated by addition of 4-hydroxytamoxifen (4-OHT; ref. 27). Activation of Cre-ER in vitro resulted in approximately 30%–50% cell death in culture (data not shown). Cells harvested 3 and 7 days after initiation of 4-OHT treatment showed approximately 50%–60% reduction in the Dnmt3b conditional allele and in Dnmt3b protein levels (Figure 7D). Importantly, despite upregulation of Dnmt1 and Dnmt3a in response to decreased Dnmt3b levels, a moderate but significant approximately 12% decrease in cytosine methylation was detected in cells at 2 independent time points (Figure 7E and Supplemental Figure 17). This demethylation led to subsequent upregulation of Ment (Figure 7F), further supporting the idea that Dnmt3b plays a role in maintenance methylation of Ment. Thus, in both mouse and human lymphoid malignancies, DNA methylation is likely to play a role in transcriptional regulation of MENT and its extent positively correlates with DNMT3B levels.
Figure 7. Dnmt3b-dependent methylation regulates transcript levels of human and mouse Ment.
(A) Human genomic region of MENT. (B) Inverse correlation between DNMT3B and MENT transcript in human primary Burkitt (BL), diffuse large B cell (DLBCL), and mantle cell (MCL) lymphomas, determined by qRT-PCR and Spearman rank correlation (rs = –0.5; P = 0.01). Normal centroblasts (CB) served as a control. (C) Bisulfite sequencing of MENT locus I and II in controls (thymocytes [Thy], and centroblasts [CB]) and primary human tumors (T1,3,4,9,10,24,25). Methylation data are presented as in Figure 6C. (D) Western blot analysis and PCR-based genotyping of Dnmt3b in MYC;Dnmt3bfl/fl cells and MYC;Dnmt3bfl/fl cells expressing Cre-ER without 4-OHT and after 3 or 7 days of incubation with 4-OHT. β-Actin served as a loading control. (E) Bisulfite sequencing of Ment locus A in cells as in D. (F) qRT-PCR analysis of Ment in cells as in D. Average methylation levels by bisulfite sequencing are shown for each sample, and tumor groups were pooled and averaged. 10 clones are shown unless noted. (C, E, and F) *P < 0.05, Student’s t test. Error bars represent ± SEM.
Ment plays a role in proliferation in mouse and human lymphomas.
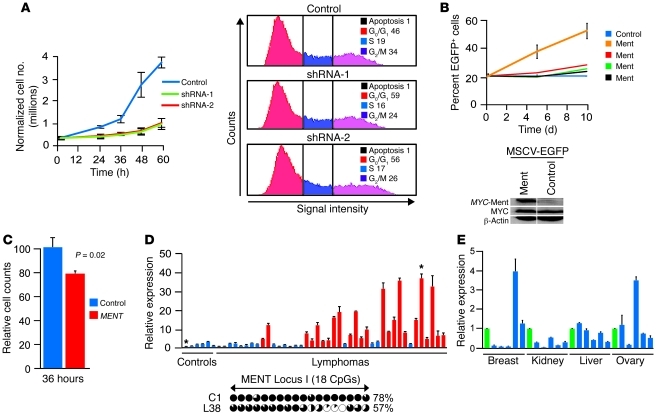
To further examine the role of Ment in lymphomagenesis, we used MYC;Dnmt3b–/– cells to derive stable cell lines expressing shRNA constructs targeting Ment. As shown in Figure 8A and Supplemental Figure 18, 40%–60% knockdown of Ment resulted in substantial inhibition of cellular proliferation, with a 10%–13% increase in G0/G1 phase relative to control cells. Minimal changes in cell death were observed. On the other hand, overexpression of Ment conferred a growth advantage in vitro to 3 of 4 MYC;Dnmt3bfl/fl cell lines (Figure 8B and Supplemental Figure 19). These results further confirm the proproliferative function of Ment and suggest that increased Ment transcription substantially contributes to the accelerated tumor progression observed in MYC;Dnmt3b–/– mice.
Figure 8. Upregulation of Ment contributes to mouse and human lymphomagenesis.
(A) Growth curves of MYC;Dnmt3b–/– cell lines infected with retroviruses encoding either control shRNA or independent shRNAs against Ment, and their corresponding cell cycle profiles 36 hours after plating. (B) Percent EGFP+ cells at different time points during in vitro growth of 1 control and 4 Ment-infected cell lines. Western blot of control and Ment-infected MYC;Dnmt3bfl/fl cells using anti-MYC antibodies is also shown. (C) siRNA-mediated knockdown of human MENT in JURKAT cells. Cells transfected with control siRNA or with SmartPool siRNA against MENT were counted 36 hours later. Data from 3 experiments were normalized to control; error bars represent ± SEM. P = 0.02, Student’s t test. (D) Expression of MENT, determined by qRT-PCR analysis of 42 human primary lymphomas present in TissueScan Lymphoma panel I (Origene). Data from 2 independent experiments are presented as fold difference relative to the average of 6 control samples. Red bars represent lymphomas with statistically significant fold increases in MENT expression. Below, methylation status of MENT locus I in 1 control and 1 lymphoma sample (corresponding to the bars marked with asterisks above), as determined by bisulfite sequencing. (E) Relative MENT transcript levels, as determined by qRT-PCR, in human breast, kidney, liver, and ovary tumors from the Origene TissueScan starter kit. Green bars represent control samples within each tissue. (A–E) Error bars represent ± SEM. *P < 0.05, Student’s t test.
We next determined whether inhibition of MENT affects growth of JURKAT (human T cell lymphoma) cells. Consistent with data obtained in the mouse model, siRNA-mediated knockdown of MENT resulted in decreased cell growth (Figure 8C and Supplemental Figure 20). Analysis of MENT levels in primary human B cell and T cell tumors (Lymphoma Panel I; Origene) revealed that 24 of 42 samples (~57%) had 2.5-fold or higher levels of human MENT compared with an average of 6 control samples (Figure 8D). Promoter methylation of the high MENT expressor lymphoma 38 was decreased relative to normal control (Figure 8D). In addition, MENT expression increased at least 2.5-fold in 18 of 26 BL, DLBCL, and MCL lymphomas compared with control CD77+CD38hi centroblasts isolated from a healthy donor (Figure 7B). On the contrary, only 1 of 5 breast and ovarian cancer samples obtained from Origene had elevated levels of MENT (Figure 8E). These data suggest that MENT is important for the progression and/or maintenance of the tumor phenotype in lymphoid malignancies, but a possible role in the pathogenesis of solid malignancies might be limited to subset of tumors or tumor types. A more comprehensive analysis is required to address the role of MENT in pathogenesis of human malignancies.
Discussion
The DNA methylation landscape is substantially altered in cancer, resulting in profound effects on gene transcription and genomic stability. Since Dnmts are enzymes mediating methylation, dissecting their contribution to the process of tumorigenesis represents an important step toward understanding the mechanism(s) by which aberrant DNA methylation in cancer is generated. In the tumor prevention study presented here, we identified Dnmt3b as a gene in a mouse model of MYC-induced T cell lymphomagenesis that functioned as a tumor suppressor, at least in part through its ability to maintain promoter methylation of the previously uncharacterized gene Ment during MYC-induced lymphomagenesis. In the absence of Dnmt3b, stepwise demethylation and derepression of Ment during tumor progression resulted in accelerated lymphomagenesis, thus identifying Ment as a potential tumor modifier.
Our data are consistent with a recent report in which overexpression of a truncated version of DNMT3B frequently expressed in human cancer, DNMT3B7, accelerated MYC-induced B cell lymphomagenesis (28, 29). These data, together with the recent discovery of mutations in the DNMT3B relative DNMT3A in patients with acute myeloid leukemia and MDS, suggest that DNMT3A and DNMT3B may have tumor suppressor functions in the hematopoietic compartment in humans (10, 30, 31). This notion is further supported by large-scale profiling of human lymphomas and leukemias that show DNMT3B expression to be downregulated in a vast majority of tested samples (32–34). Thus, decreased activity of DNMT3B in hematopoietic cells, through naturally occurring polymorphisms, lower transcript levels, posttranscriptional or posttranslational modifications, and/or overexpression of the dominant-negative isoform DNMT3B7, may expose certain people to an increased risk for tumor development. Identification of a large set of potential Dnmt3b targets in the present study may help to develop DNA methylation–based assays to test intracellular DNMT3B activity in easily obtainable peripheral blood leukocytes. In addition, our data indicate that treatment of patients with MDS may result not only in reexpression of epigenetically silenced tumor suppressor genes, but also in reactivation of genes that might contribute to the transformation of MDS to AML. Thus, inhibitors of specific Dnmts and/or concomitant inhibition of Ment and other as-yet unidentified oncogenes may be needed for more effective treatment.
Ment is an oncogenic modifier contributing to the tumor suppressor function of Dnmt3b.
Loss of DNA methylase activity can result in hypomethylation of the genome with subsequent induction of genomic instability and/or deregulation of proto-oncogenes. For example, genome-wide hypomethylation induced T cell lymphomas in Dnmt1chip/– mice characterized by increased genomic instability (12). Our present studies showed that loss of Dnmt3b had no effect on T cell development and did not induce lymphomas, but accelerated MYC-induced lymphomagenesis. Similar to Dnmt1chip/– and mediastinal lymphomas derived from Myc/DNMT3B7 mice (12, 28), MYC;Dnmt3b–/– lymphomas showed increased genomic instability. Whether acquired genetic alterations contribute to accelerated lymphomagenesis is less clear. Unlike Dnmt1chip/– lymphomas, which acquire trisomy of chromosome 15, genetic alterations in MYC;Dnmt3b–/– lymphomas are heterogeneous. The only consistent aberration is amplification in qA3 of chromosome 5, which is associated with the gene Lhfpl3. Since, based on our microarray data, Lhfpl3 expression was not affected in Dnmt3b–/– lymphomas, there is no apparent evidence for a connection to the tumor suppressor function of Dnmt3b. Interestingly, reintroduction of Dnmt3b into lymphoma cell lines established from MYC;Dnmt3b–/– tumors inhibited cellular proliferation in vitro, which suggests that the Dnmt3b tumor suppressor function operates even in cells that have already acquired a full repertoire of tumor-related genetic aberrations. Thus, while we cannot rule out that genetic alterations contribute to enhanced tumorigenesis, Dnmt3b-specific methylation events are more likely to be responsible for the tumor suppressor function of Dnmt3b.
We have also provided data to show that Dnmt3b affects tumor progression. Loss of Dnmt3b was insufficient to result in the immediate initiation of lymphomagenesis, which implicates additional players in the accelerated lymphomagenesis in MYC;Dnmt3b–/– mice. Also consistent with this, proliferation of MYC;Dnmt3b–/– T cells increased in late, but not early, stages of tumorigenesis. Functional analysis of several genes hypomethylated and overexpressed in Dnmt3b–/– lymphomas revealed that Ment was responsible, at least in part, for the Dnmt3b tumor suppressor function. Whereas Ment was silenced in normal thymocytes and control tumors, its expression increased more than 100-fold in MYC;Dnmt3b–/– tumors. Ment derepression occurs gradually, as MYC;Dnmt3b–/– pretumor thymocytes showed a moderate, approximately 5-fold increase in transcript levels. This would be expected if the absence of Dnmt3b leads to successive demethylation of Ment. Moreover, knockdown of Ment inhibited cellular proliferation of Dnmt3b–/– lymphomas, which would also be a predicted outcome if Dnmt3b activity normally suppresses the proliferation of tumor cells, at least in part through methylation of the Ment promoter. Finally, by mimicking a single molecular event consistently found in Dnmt3b–/– lymphomas (overexpression of Ment), MYC;Dnmt3bfl/fl lymphoma cells obtained a growth advantage in vitro. Thus, Ment is likely an oncogenic modifier whose overexpression contributes to accelerated lymphomagenesis in MYC;Dnmt3b–/– mice. By virtue of its high expression in testis and lymphomas and low expression in nongermline normal tissues, Ment exhibited the features of cancer germline (CG) antigens, a group of epigenetically regulated genes characterized by increased expression during tumor progression whose protein products induce spontaneous humoral and immune responses in cancer patients (35). Some of these genes, such as melanoma antigen genes (MAGEs), form multigene families that are present on the X chromosome (CG-X antigens), whereas other CG antigens are located on autosomal chromosomes as single-copy genes that show less restriction of expression to the testis (non–CG-X antigens). The function of more than 130 known CG antigens in cancer remains largely unknown, but MAGEA11 was previously shown to stimulate the activity of the androgen receptor and thereby contribute to the development of castration-resistant prostate tumor growth (35, 36). Unlike solid tumors, leukemia and lymphoma rarely express CG antigens. Thus, Ment is an unusual example of a CG antigen expressed in lymphomas that appears to contribute to tumor progression. Due to their cancer-specific expression patterns and high immunogenicity, CG antigens are considered good targets for development of tumor vaccines. The immunogenicity and overall feasibility of Ment targeting remain to be tested.
Contribution of Dnmt3b to the tumor-specific landscape by maintenance and de novo methylation.
By comparing methylation patterns in normal mouse thymocytes, MYC;Dnmt3b–/– pretumor thymocytes, Dnmt3b+/+ lymphomas, and Dnmt3b–/– lymphomas, we identified 260 promoters whose methylation appears to depend on Dnmt3b. Of these, 201 promoters are likely targets of tumor-specific Dnmt3b maintenance activity, as these were exclusively hypomethylated in Dnmt3b–/– lymphomas. The requirement of Dnmt3b for methylation of some promoters ranged from critical (Ment, Arhgef18, and Pnldc1) to partial (Rpl39l and S100a6), presumably due to overlapping functions with Dnmt3a and/or Dnmt1. The irreplaceable function of Dnmt3b in maintenance methylation of specific loci can be best illustrated in the case of the Ment promoter (Figure 9). First, the Ment promoter is almost completely methylated in normal mouse thymocytes. Ablation of Dnmt3b does not eliminate methylation, which suggests that Dnmt3b is dispensable for both de novo and maintenance methylation of this locus during normal thymocyte development. Second, the Ment promoter shows evidence of demethylation in MYC;Dnmt3b–/– pretumor thymocytes, but remains largely methylated. Third, in control MYC;Dnmt3bfl/fl lymphomas, methylation of the Ment promoter remains similar to normal thymocytes. However, loss of Dnmt3b results in near-complete elimination of cytosine methylation. Finally, the degree of DNA methylation in the MENT promoter directly correlates with levels of DNMT3B in human lymphomas. Fluctuation in Dnmt3b levels observed in tumors is more likely to affect maintenance rather than de novo methylation due to its immediate and persistent impact on methylation patterns. Thus, we propose that Dnmt3b is involved in the maintenance methylation of Ment and that this maintenance activity is likely limited to highly proliferative tumor cells. Our interpretation is consistent with recent speculations that while Dnmt1 is primarily responsible for maintenance methylation, Dnmt3a and Dnmt3b contribute to this process by localizing to CpG islands and mediating methylation of cytosines that were omitted by Dnmt1 during replication (21).
Figure 9. Critical role of Dnmt3b in cancer-specific methylation maintenance of Ment.
Dnmt3b is dispensable for de novo methylation of the Ment promoter during normal thymocyte development. De novo methylation of Ment is likely performed by either Dnmt1 or Dnmt3a and results in gene silencing. In MYC;Dnmt3b–/– pretumor thymocytes, Ment is partially demethylated, with a concomitant increase in gene expression. Failure of methylation machinery to maintain Ment promoter methylation in the absence of Dnmt3b during tumor progression results in complete demethylation of Ment, its overexpression, and accelerated lymphomagenesis.
In addition to the proposed function of Dnmt3b in maintenance methylation, we have also considered alternative explanations for the demethylation of Ment or — in a broader sense — the 201 promoters specifically demethylated in Dnmt3b–/– lymphomas. One possibility is that in MYC;Dnmt3b–/– lymphomas, activity of other DNA methylases could be decreased. However, this is unlikely, since levels of Dnmts were unaffected by the loss of Dnmt3b (Figure 1C). While the activity of Dnmts could be compromised by other means, such a change would likely have a greater effect on the global methylation landscape, rather than the relatively modest 5% decrease in methylation we observed in MYC;Dnmt3b–/– lymphomas. Another possibility is that the loss of Dnmt3b results in the transformation of an alternative cellular subtype in which a set of 201 promoters is not methylated, and therefore no maintenance activity is involved in their demethylation. However, such a fundamental switch in tumor biology would be expected to have an immunophenotypic manifestation. In reality, MYC;Dnmt3b–/– lymphomas were immunophenotypically indistinguishable from controls (Supplemental Figure 2, A and B).
Finally, a lack of methylation of 201 promoters is unlikely to result indirectly from upregulation of potential demethylases such as Apobec1, Apobec2, Apobec3, Mbd4, Gadd45b, and Tet1, as our microarray data failed to reveal substantial differences in levels of transcript for a majority of these genes. Moreover, we found that only 18 of the greater than 5,000 proposed target genes of demethylase activity overlapped with our set of 201 potential Dnmt3b maintenance methylation targets (Supplemental Table 5 and ref. 37). Thus, the simplest explanation for hypomethylation of 201 promoters in MYC;Dnmt3b–/– lymphomas is loss of maintenance activity in the absence of Dnmt3b. Since we did not address whether Dnmt3b is physically present on any of these promoters, we cannot rule out the possibility that Dnmt3b may have indirect effects on the maintenance methylation of a subset of these promoters. Nonetheless, lymphoid malignancies appear to represent the first in vivo mouse and human tumor settings in which Dnmt3b maintenance activity was detected.
Of 260 Dnmt3b-dependent promoters, 5 appeared to be cancer-specific targets of Dnmt3b de novo activity, as they were specifically hypermethylated in Dnmt3b+/+ tumors. This number is likely underestimated, as statistical approaches tend to select for consistent observations, which would therefore diminish cases in which biological frequency is not 100% penetrant. We have previously shown that the frequency of de novo methylation varies considerably in this T cell lymphoma mouse model (23). As a result, the variation between biological replicates can lead to elimination of de novo methylation observations due to a lack of statistical significance. Thus, more detailed studies need to be performed to carefully evaluate the contribution of Dnmt3b to tumor-specific de novo methylation. Our present findings underscore the importance of identification of downstream effectors of DNA methyltransferases, not only as potential therapeutic targets, but also to better understand how aberrant methylation patterns evolve during tumorigenesis.
Methods
Generation and maintenance of mice.
Dnmt3b2LoxP (Dnmt3bfl) and EμSR-tTA;Teto-MYC mice were obtained from E. Li (Novartis Institutes for Biomedical Research, Cambridge, Massachusetts, USA) and D.W. Felsher (Stanford University, Stanford, California, USA), respectively. Rosa26LOXPEGFP (38) and Teto-Cre (39) mice were obtained from The Jackson Laboratory. All studies were performed on mice backcrossed 6 times into FVB/N strain. Standard genetic crosses were used to generate cohorts of mice used in these studies. Mice of appropriate genotypes were monitored and harvested at the terminal stage, as determined by overall health. Genomic DNA isolated from mouse tails was used in PCR-based genotyping. For developmental studies, organ weight and cellularity of thymus, spleen, and lymph nodes were compared between EμSR-tTA;Teto-Cre;Rosa26LOXPEGFP;Dnmt3bfl/fl and EμSR-tTA;Teto-Cre;Rosa26LOXPEGFP;Dnmt3b+/+ mice at either 6–8 weeks or 8–12 months of age. Expression of cell surface markers was compared in both EGFP+ and EGFP– populations by FACS. For survival studies, tumor development was compared among MYC;Dnmt3b–/– (EμSR-tTA;Teto-MYC;Teto-Cre;Rosa26LOXPEGFP;Dnmt3bfl/fl), MYC;Dnmt3bfl/fl (EμSR-tTA;Teto-MYC;Rosa26LOXPEGFP;Dnmt3bfl/fl), and MYC;Dnmt3b+/fl (EμSR-tTA;Teto-MYC;Teto-Cre;Rosa26LOXPEGFP;Dnmt3b+/fl) littermates. The Kaplan-Meier method was used to estimate overall survival distributions, and the log-rank test was used to compare survival distributions between genetic cohorts. Tumor initiation studies and BrdU incorporation and apoptosis assays were performed by comparing EGFP+ populations between MYC;Dnmt3b+/+ (EμSR-tTA;Teto-MYC;Teto-Cre;Rosa26LOXPEGFP) and nonlittermate MYC;Dnmt3b–/– mice at days 20, 35, and 50. Normal thymocytes for all studies were obtained from MYC;Dnmt3b+/+ mice. For tumor regression and relapse studies, terminally ill mice were given drinking water containing doxycycline at a concentration of 100 μg/ml and monitored over a 7-week period for signs of relapse.
Construction of the MSCC library, data collection, and analysis.
High-throughput global methylation analysis was performed using the MSCC assay previously developed by Ball et al. (25). This procedure uses the methyl-sensitive restriction endonuclease HpaII, which cuts genomic DNA at all unmethylated CCGG sites. In the mouse genome (mm9), there are a total of 820,040 unique HpaII sites. Of these, 638,216 sites have duplicate (forward and reverse) tags and 181,824 sites have a single unique tag. The unique character of each HpaII site is based on the requirements that it must be greater than 40 bases from another adjacent HpaII site and that the tag sequence is present only once in the genome. Briefly, 2 μg genomic DNA was digested with 20 U of the restriction enzyme HpaII (NEB), and an adapter containing a recognition site for the restriction enzyme MmeI was ligated to digested DNA fragments using T4 DNA ligase (NEB). DNA was ethanol precipitated and nick repaired with 8 U Bst DNA polymerase (NEB). The DNA was digested with 2 U MmeI to capture the 18 bases adjacent to HpaII sites, and the fragments were subsequently ligated to a second adapter to allow PCR amplification and final high-throughput sequencing. After appropriate tag size purification using a 10% PAGE gel, DNA was PCR amplified using BioRad iProof high-fidelity polymerase and quantitative PCR procedures to avoid overamplification and skewing the production of final tags. Final tags were checked for proper size and concentration using a Bioanalyzer High Sensitivity DNA chip (Agilent) and Qubit Fluorometer (Invitrogen). High-throughput sequencing of library tags was performed by Tufts University Core Facility and UNMC using an Illumina Genome Analyzer II. The resulting 18-bp sequence tags were aligned with the mouse genome (mm9) using the short read sequence aligner Bowtie (40). The library was matched to all possible unique tags (courtesy of M. Ball, Harvard University, Boston, Massachusetts, USA), and the appropriate gene using perl scripts was developed by the UNO/UNMC Genetic Sequence Analysis Facility using Microsoft Access and Excel software. Counts of each sequence tag representing a unique unmethylated HpaII site were determined. A change in tag count between control and sample group indicated a corresponding change in the methylation status of each HpaII CpG. All HpaII sites for all samples were batch analyzed by edgeR (a bioconductor package for R programming language), which uses Bayesian estimation and exact tests based on the negative binomial distribution to make pairwise comparisons between groups (41–43). The false discovery rate (FDR) was estimated based on the Benjamini Hochberg method, and an FDR of up to 5% was considered significant. For Figure 4E, the gene body represented +1 bp relative to the transcription start site through the end of the transcript. For Figure 4, F and G, the promoter was defined as –1,500 to +500 bps relative to the transcription start site, and the prerequisite for group assignment was significantly changed (FDR < 0.05) methylation of at least 1 HpaII site in promoters. If the methylation readout for 2 or more HpaII sites located within the same promoter differed, the promoter was categorized to more than 1 group. For Figure 5A, genes were selected as follows: (a) 3-fold and greater expression in MYC;Dnmt3b–/– relative to MYC;Dnmt3bfl/fl lymphomas, as determined by microarray expression profiling with subsequent confirmation by qRT-PCR; and (b) 4-fold and greater decrease in methylation in MYC;Dnmt3b–/– relative to MYC;Dnmt3bfl/fl lymphomas, as determined by MSCC (FDR < 0.05) with subsequent locus-specific confirmation of DNA hypomethylation by COBRA. If available, methylation readout for both forward and reverse tags at the HpaII site had to fulfill these criteria.
To confirm MSCC data at selected loci, genomic DNA was digested with HpaII enzyme for 8 hours and subsequently used as a template in real-time PCR analysis using primers specific to individual HpaII sites along with SYBR Green Supermix (BioRad) in a final volume of 20 μl (Supplemental Table 6). Real-time PCR was performed as described above (Supplemental Figure 8, A–C). MSCC data have been deposited in NCBI’s Gene Expression Omnibus (accession no. GSE33849; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE33849; ref. 44).
MSCC for repeat elements.
The alignment and counting of all generated library tags was performed as described above using Bowtie alignment software combined with perl scripts, except that nonunique tags associated with repeat elements were not removed. Sequences associated with SINE and LINE repeats were identified using a perl script developed by the UNMC Center for Bioinformatics and Systems Biology. The perl script identified the portion of the library associated with each repeat type using the Institute for System Biology ( http://www.repeatmasker.org; mm9, July 2007, RepeatMasker, 3.2.8, Repeat Library 20090604). Total repeat tags were calculated and analyzed using Microsoft Excel.
Human cell lines and tumor samples.
The human cell lines — including Burkitt lymphoma cell lines RAJI, Daudi, and Akata, mantle cell lymphoma line JeKo-1, B cell leukemia line JVM-2, T cell lymphoblast–like line JURKAT, acute lymphocytic leukemia line REH, and EBV-immortalized B cell line JY — were obtained from ATCC. Cells were maintained in DMEM or RPMI 1640 (Invitrogen) containing 10% fetal bovine serum (100 U/ml), penicillin, and 100 μg/ml streptomycin. Cell lines were cultured at 37°C in a humidified 5% CO2 atmosphere and were passaged according to ATCC recommendations. 10 samples each of primary Burkittl and diffuse large B cell lymphomas as well as 6 samples of mantle cell lymphomas were obtained from the Nebraska Lymphoma Study Group registry. The TissueScan starter kit, Lymphoma Cancer Panel I, as well as DNA samples from C1 (lymph node) and L38 (lymphoma), were purchased from Origene.
FACS analysis, proliferation, and apoptosis.
Single-cell suspensions were prepared from thymi, spleens, lymph nodes, or tumor masses and stained with the eBioscience antibodies specific for mouse T, B, and myeloid cells, including PE-conjugated anti-CD4, anti-B220, and anti-CD44; PE-Cy5–conjugated anti-CD4, anti-CD8, and anti-CD11b; and allophycocyanin-conjugated anti-CD8 and anti-CD25. Stained cells were detected by a FACSCalibur II flow cytometer and analyzed using BD FACSDiva software (Becton Dickinson). BrdU labeling of T cells in 20-, 35-, and 50-day-old or terminally ill mice was performed via i.p. injection of BrdU (100 mg/g body weight) 2 hours before harvest. Normal thymocytes or thymic tumor cells were stained with CD4-conjugated anti-CD4 and PE-Cy5–conjugated anti-CD8, and BrdU-positive cells were quantified using allophycocyanin-conjugated anti-BrdU (BrdU-Flow Kit; BD Biosciences — Pharmingen), as described by the manufacturer. For analysis of apoptosis, thymocytes or tumor cells were stained with allophycocyanin-conjugated annexin V antibody and analyzed by FACS according to the manufacturer’s recommendations (eBioscience). Cell cycle analysis was performed using Vybrant Violet according to the manufacturer’s instructions (Invitrogen). All samples were analyzed using FACSCalibur II or LSRII flow cytometers and FACSDiva software (Becton Dickinson) at UNMC’s Flow Cytometry facility.
Thymocytes from EμSR-tTA;Teto-MYC;Teto-Cre;Rosa26LOXPEGFP, EμSR-tTA;Teto-MYC;Teto-Cre;Rosa26LOXPEGFP;Dnmt3bfl/fl pretumor, and EμSR-tTA;Teto-Cre;Rosa26LOXPEGFP;Dnmt3bfl/fl (N3b) mice were harvested at 24 days of age and sorted for EGFP+ cells using FACS Aria (Beckton Dickinson) as described previously (23).
Generation of retroviruses and infection of cell lines.
The MSCV-EGFP-Myc-Dnmt3b-Flag construct was generated by PCR amplification using AGGAATTCCACCATGGAACAAAAACTTATTTCTGAAGAAGATCTGAAGGGAGACAGCAG and AGGAATTCCTACTTGTCATCGTCGTCCTTGTAGTCTTCACAGGCAAAGTAGTCCTTC primers, and cDNA was prepared from mouse universal RNA as template. The PCR product was digested with EcoRI and cloned into MSCV-EGFP vector. Similarly, the mouse MSCV-EGFP-Myc-Ment-Flag construct was amplified using primers AGGAATTCCACCATGGAACAAAAACTTATTTCTGAAGAAGATCTGGTCCCCGCCGCCTGC and GAGGAATTCTTACTTGTCATCGTCGTCCTTGTAGTCCCTCTGGATTCTTTCTACTGG, and the PCR product was cloned using an EcoRI site in the MSCV-EGFP vector. Finally, MSCV-EGFP-CreER was generated using GAGAGAGAATTCCACCATGTCCAATTTACTGACCGTAC and GAGAGAGAATTCTCAGATCGTGTTGGGGAAGC primers, and the product was cloned using an EcoRI site in the MSCV-EGFP vector (27). Constructs were verified by sequencing.
Tumor cells were adapted to in vitro growth by placing cell suspension isolated from EμSR-tTA;Teto-MYC;Rosa26LOXPEGFP;Dnmt3bfl/fl or EμSR-tTA;Teto-MYC;Teto-Cre;Rosa26LOXPEGFP lymphomas into RPMI 1640 medium supplemented with 10% FBS and 0.025 mM 2-mercaptoethanol. Retroviruses for infecting tumor T cells were produced by transient transfection of MSCV-EGFP–based retroviral constructs carrying mouse myc-tagged Dnmt3b or Ment cDNA into the Phoenix-Eco packaging cell line. Tumor T cells were infected by incubating for 5 hours with the supernatants containing retrovirus and supplemented with 1 mg/ml Sequabrene (Sigma-Aldrich). After 35 hours, infection efficiency was determined by measuring percent EGFP+ cells by FACS. Cells were plated at a concentration of 0.2 × 106/ml and replated every 48 hours. Aliquots of cells were taken to monitor percentage of EGFP+ cells. MYC;Dnmt3bfl/fl Cre-ER–infected cells expressing EGFP were sorted by FACS. Cre-mediated excision of Dnmt3b2LoxP alleles was initiated by addition of 4-OHT (150 nM final concentration; Sigma-Aldrich). DNA, RNA, and protein extracts were prepared from cells that were harvested at different time points (0, 3, and 7 days) after initiation of 4-OHT treatment.
HPLC.
HPLC for quantification of methylated cytosine was performed at UNMC as described previously (24). Briefly, genomic DNA from normal thymi, MYC;Dnmt3bfl/fl tumors, and MYC;Dnmt3b–/– tumors was purified by the DNeasy Tissue Kit (Qiagen) and digested down to nucleosides by DNA Degradase Plus (Zymo Research). Nucleoside standards 5-methyl-2′-deoxycytidine, 2′-deoxycytidine, 2′-deoxyguanosine, 2′-deoxyadenosine monohydrate, and thymidine were obtained from MP Biomedicals and separated on a standard C18 HPLC column to determine elution times. The area under the curve was used to calculate the relative methylation levels between samples.
Affymetrix chip–based microarray analyses.
Total RNA was isolated using the TRIzol reagent (Invitrogen) and repurified using RNAeasy kit from Qiagen. A total of 200 ng total RNA was reverse transcribed, and cRNA was generated per the manufacturer’s instructions using the Ambion WT Expression Kit (Ambion). Resultant cRNA probes were hybridized to the Affymetrix GeneChip Mouse Gene 1.0 ST Array per the manufacture’s instructions, and chips were scanned using a GeneChip 3,000 6G scanner by the UNMC DNA Microarray Core Facility. The resultant datasets were scaled using GCOS software and evaluated with respect to quality assurance parameters to include background, hybridization kinetics, and reverse transcription efficiency. Intensities were imported into Partek Genomics Suite software using Robust Multi-chip Averaging (RMA) background correction and quantile normalization on a linear scale. Regularized t test analysis of control versus treatment comparisons was performed using a Bayesian approach to estimate the within-treatment variation among replicates using Cyber-T software (45, 46). This principle uses the weighted average of local standard deviations for genes with similar expression levels, resulting in a stabilized within-treatment variance estimate. The data discussed in this article have been deposited in NCBI’s Gene Expression Omnibus (accession no. GSE30126; http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE30126; ref. 44).
Statistics.
For the data in Figure 1B, the Kaplan-Meier method was used to calculate overall survival, and the log-rank test was used to compare survival distributions between groups (P < 0.001). For the data in Figure 6C, Figure 7, C and E, and Figure 8D, bisulfite sequencing data were analyzed by paired t tests (P < 0.05). Continuous variables were compared using 2-sample Student’s t tests; results are shown with error bars representing 1 SEM. Bonferroni corrections were used to adjust for multiple comparisons. Relative gene expression was calculated by adjusting control samples (Figure 5B, Figure 6, B and D, Figure 7F, and Figure 8E) to unity and using this as the baseline for all other comparisons. In the case of multiple control samples, the average was used as a baseline. For primary human tumors in Figure 7B, sample 15 was chosen as a baseline. For microarray data, regularized t test analysis of control versus sample comparisons was performed using a Bayesian approach to estimate the within-group variation among replicates using Cyber-T software (44, 45). This principle uses the weighted average of local SDs for genes with similar expression levels, resulting in a stabilized within-group variance estimate. This allows for multiple testing to occur. Spearman rank correlation was used to calculate coefficients between expression levels of MENT and DNMT3B (Figure 7B). Overall, P values less than 0.05 were considered statistically significant.
Study approval.
All animal experiments were approved by the IACUC at UNMC. The Institutional Review Board of UNMC approved the human studies, and patients provided informed consent prior to the study in accordance with the Declaration of Helsinki.
Western blotting, real-time PCR, pyrosequencing, COBRA, aCGH, and siRNA- and shRNA-mediated knockdown experiments.
See Supplemental Methods.
Supplementary Material
Acknowledgments
We thank Michael Brattain (Eppley Cancer Institute, UNMC), Robert Lewis (Eppley Cancer Institute, UNMC), and Harold Saavedra (Emory University) for critical reading of this manuscript and useful comments. This work was supported by an Eppley Cancer Center pilot project grant (to R. Opavsky) and by Nebraska Center for Cellular Signaling NIH grant P20 RR018759 from the National Center for Research Resources. T.C. Greiner is a grantee of the Lymphoma Research Foundation Mantle Cell Grant program. The UNMC Next Generation Sequencing Core received support from National Center for Research Resources (NCRR) grants, entitled UNMC SIG Supporting Next-generation DNA Sequencing Technology 1S10RR027754-01, The Nebraska Research Network in Functional Genomics NE-INBRE P20 RR16469, the Molecular Biology of Neurosensory Systems CoBRE 2P20RR018788-06, and the Nebraska Research Initiative. The article contents are the sole responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. The authors also acknowledge the University of Nebraska Epigenomics Core Facility for performing the MSCC processing and analysis; Chittibabu Guda and Adam Cornish (Center for Bioinformatics and Systems Biology, UNMC) for developing perl scripts for MSCC repeat sequence analysis; and Jiangtao Luo for contributing to the statistical analysis.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(1):163–177. doi:10.1172/JCI57292.
References
- 1.Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nat Rev Genet. 2009;10(5):295–304. doi: 10.1038/nrg2540. [DOI] [PubMed] [Google Scholar]
- 2.McCabe MT, Brandes JC, Vertino PM. Cancer DNA methylation: molecular mechanisms and clinical implications. Clin Cancer Res. 2009;15(12):3927–3937. doi: 10.1158/1078-0432.CCR-08-2784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee PP, et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity. 2001;15(5):763–774. doi: 10.1016/S1074-7613(01)00227-8. [DOI] [PubMed] [Google Scholar]
- 4.Trowbridge JJ, Snow JW, Kim J, Orkin SH. DNA methyltransferase 1 is essential for and uniquely regulates hematopoietic stem and progenitor cells. Cell Stem Cell. 2009;5(4):442–449. doi: 10.1016/j.stem.2009.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gamper CJ, Agoston AT, Nelson WG, Powell JD. Identification of DNA methyltransferase 3a as a T cell receptor-induced regulator of Th1 and Th2 differentiation. J Immunol. 2009;183(4):2267–2276. doi: 10.4049/jimmunol.0802960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tadokoro Y, Ema H, Okano M, Li E, Nakauchi H. De novo DNA methyltransferase is essential for self-renewal, but not for differentiation, in hematopoietic stem cells. J Exp Med. 2007;204(4):715–722. doi: 10.1084/jem.20060750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patel DR, Richardson BC. Epigenetic mechanisms in lupus. Curr Opin Rheumatol. 2010;22(5):478–482. doi: 10.1097/BOR.0b013e32833ae915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho SM. Environmental epigenetics of asthma: an update. J Allergy Clin Immunol. 2010;126(3):453–465. doi: 10.1016/j.jaci.2010.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wierda RJ, Geutskens SB, Jukema JW, Quax PH, van den Elsen PJ. Epigenetics in atherosclerosis and inflammation. J Cell Mol Med. 2010;14(6A):1225–1240. doi: 10.1111/j.1582-4934.2010.01022.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rice KL, Hormaeche I, Licht JD. Epigenetic regulation of normal and malignant hematopoiesis. Oncogene. 2007;26(47):6697–6714. doi: 10.1038/sj.onc.1210755. [DOI] [PubMed] [Google Scholar]
- 11.Jones PA, Baylin SB. The epigenomics of cancer. Cell. 2007;128(4):683–692. doi: 10.1016/j.cell.2007.01.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gaudet F, et al. Induction of tumors in mice by genomic hypomethylation. Science. 2003;300(5618):489–492. doi: 10.1126/science.1083558. [DOI] [PubMed] [Google Scholar]
- 13.Vu TH, Chuyen NV, Li T, Hoffman AR. Loss of imprinting of IGF2 sense and antisense transcripts in Wilms’ tumor. Cancer Res. 2003;63(8):1900–1905. [PubMed] [Google Scholar]
- 14.Senolt L, et al. S100A4 (Mts1): is there any relation to the pathogenesis of rheumatoid arthritis? Autoimmun Rev. 2006;5(2):129–131. doi: 10.1016/j.autrev.2005.09.010. [DOI] [PubMed] [Google Scholar]
- 15.Galm O, Herman JG, Baylin SB. The fundamental role of epigenetics in hematopoietic malignancies. Blood Rev. 2006;20(1):1–13. doi: 10.1016/j.blre.2005.01.006. [DOI] [PubMed] [Google Scholar]
- 16.Issa JP, Kantarjian HM. Targeting DNA methylation. Clin Cancer Res. 2009;15(12):3938–3946. doi: 10.1158/1078-0432.CCR-08-2783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smiraglia DJ, et al. Excessive CpG island hypermethylation in cancer cell lines versus primary human malignancies. Hum Mol Genet. 2001;10(13):1413–1419. doi: 10.1093/hmg/10.13.1413. [DOI] [PubMed] [Google Scholar]
- 18.Steine EJ, et al. Genes methylated by DNA methyltransferase 3b are similar in mouse intestine and human colon cancer. J Clin Invest. 2011;121(5):1748–1752. doi: 10.1172/JCI43169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okano M, Bell DW, Haber DA, Li E. DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell. 1999;99(3):247–257. doi: 10.1016/S0092-8674(00)81656-6. [DOI] [PubMed] [Google Scholar]
- 20.Chen T, Ueda Y, Dodge JE, Wang Z, Li E. Establishment and maintenance of genomic methylation patterns in mouse embryonic stem cells by Dnmt3a and Dnmt3b. Mol Cell Biol. 2003;23(16):5594–5605. doi: 10.1128/MCB.23.16.5594-5605.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jones PA, Liang G. Rethinking how DNA methylation patterns are maintained. Nature Rev Genet. 2009;10(11):805–811. doi: 10.1038/nrg2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4(2):199–207. doi: 10.1016/S1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- 23.Opavsky R, et al. CpG island methylation in a mouse model of lymphoma is driven by the genetic configuration of tumor cells. PLoS Genet. 2007;3(9):1757–1769. doi: 10.1371/journal.pgen.0030167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jeltsch A, Christ F, Fatemi M, Roth M. On the substrate specificity of DNA methyltransferases. adenine-N6 DNA methyltransferases also modify cytosine residues at position N4. J Biol Chem. 1999;274(28):19538–19544. doi: 10.1074/jbc.274.28.19538. [DOI] [PubMed] [Google Scholar]
- 25.Ball MP, et al. Targeted and genome-scale strategies reveal gene-body methylation signatures in human cells. Nature Biotechnology. 2009;27(4):361–368. doi: 10.1038/nbt.1533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Clark HF, et al. The secreted protein discovery initiative (SPDI), a large-scale effort to identify novel human secreted and transmembrane proteins: a bioinformatics assessment. Genome Res. 2003;13(10):2265–2270. doi: 10.1101/gr.1293003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dor Y, Brown J, Martinez OI, Melton DA. Adult pancreatic beta-cells are formed by self-duplication rather than stem-cell differentiation. Nature. 2004;429(6987):41–46. doi: 10.1038/nature02520. [DOI] [PubMed] [Google Scholar]
- 28.Shah MY, et al. DNMT3B7, a truncated DNMT3B isoform expressed in human tumors, disrupts embryonic development and accelerates lymphomagenesis. Cancer Res. 2010;70(14):5840–5850. doi: 10.1158/0008-5472.CAN-10-0847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ostler KR, et al. Cancer cells express aberrant DNMT3B transcripts encoding truncated proteins. Oncogene. 2007;26(38):5553–5563. doi: 10.1038/sj.onc.1210351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ley TJ, et al. DNMT3A mutations in acute myeloid leukemia. N Engl J Med. 2010;363(25):2424–2433. doi: 10.1056/NEJMoa1005143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walter MJ, et al. Recurrent DNMT3A mutations in patients with myelodysplastic syndromes. Leukemia. 2011;25(7):1153–1158. doi: 10.1038/leu.2011.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hummel M, et al. A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354(23):2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 33.Lossos IS, et al. Transformation of follicular lymphoma to diffuse large-cell lymphoma: alternative patterns with increased or decreased expression of c-myc and its regulated genes. Proc Natl Acad Sci U S A. 2002;99(13):8886–8891. doi: 10.1073/pnas.132253599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi YL, et al. A genomic analysis of adult T-cell leukemia. Oncogene. 2007;26(8):1245–1255. doi: 10.1038/sj.onc.1209898. [DOI] [PubMed] [Google Scholar]
- 35.Akers SN, Odunsi K, Karpf AR. Regulation of cancer germline antigen gene expression: implications for cancer immunotherapy. Future Oncol. 2010;6(5):717–732. doi: 10.2217/fon.10.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karpf AR, Bai S, James SR, Mohler JL, Wilson EM. Increased expression of androgen receptor coregulator MAGE-11 in prostate cancer by DNA hypomethylation and cyclic AMP. Mol Cancer Res. 2009;7(4):523–535. doi: 10.1158/1541-7786.MCR-08-0400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rai K, et al. DNA demethylase activity maintains intestinal cells in an undifferentiated state following loss of APC. Cell. 2010;142(6):930–942. doi: 10.1016/j.cell.2010.08.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mao X, Fujiwara Y, Chapdelaine A, Yang H, Orkin SH. Activation of EGFP expression by cre-mediated excision in a new ROSA26 reporter mouse strain. Blood. 2001;97(1):324–326. doi: 10.1182/blood.V97.1.324. [DOI] [PubMed] [Google Scholar]
- 39.Perl A, Wert SE, Nagy A, Lobe CG, Whitsett JA. Early restriction of peripheral and proximal cell lineages during formation of the lung. Proc Natl Acad Sci U S A. 2002;99(16):10482–10487. doi: 10.1073/pnas.152238499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10(3):R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. R Development Core Team.A Language And Environment For Statistical Computing . Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 42.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robinson MD, Smyth GK. Moderated statistical tests for assessing differences in tag abundance. Bioinformatics. 2007;23(21):2881–2887. doi: 10.1093/bioinformatics/btm453. [DOI] [PubMed] [Google Scholar]
- 44.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baldi P, Long AD. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics. 2001;17(6):509–519. doi: 10.1093/bioinformatics/17.6.509. [DOI] [PubMed] [Google Scholar]
- 46. Baldi P, Hatfield GW.DNA Microarrays And Gene Expression: From Experiments To Data Analysis And Modeling . Cambridge, United Kingdom: Cambridge University Press; 2002. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.