Abstract
Oncogenic Ras and p53 loss-of-function mutations are common in many advanced sporadic malignancies and together predict a limited responsiveness to conventional chemotherapy. Notably, studies in cultured cells have indicated that each of these genetic alterations creates a selective sensitivity to ataxia telangiectasia and Rad3-related (ATR) pathway inhibition. Here, we describe a genetic system to conditionally reduce ATR expression to 10% of normal levels in adult mice to compare the impact of this suppression on normal tissues and cancers in vivo. Hypomorphic suppression of ATR minimally affected normal bone marrow and intestinal homeostasis, indicating that this level of ATR expression was sufficient for highly proliferative adult tissues. In contrast, hypomorphic ATR reduction potently inhibited the growth of both p53-deficient fibrosarcomas expressing H-rasG12V and acute myeloid leukemias (AMLs) driven by MLL-ENL and N-rasG12D. Notably, DNA damage increased in a greater-than-additive fashion upon combining ATR suppression with oncogenic stress (H-rasG12V, K-rasG12D, or c-Myc overexpression), indicating that this cooperative genome-destabilizing interaction may contribute to tumor selectivity in vivo. This toxic interaction between ATR suppression and oncogenic stress occurred without regard to p53 status. These studies define a level of ATR pathway inhibition in which the growth of malignancies harboring oncogenic mutations can be suppressed with minimal impact on normal tissue homeostasis, highlighting ATR inhibition as a promising therapeutic strategy.
Introduction
The ataxia telangiectasia and Rad3-related (ATR) checkpoint kinase maintains genome integrity during DNA replication through the phosphorylation and activation of CHK1 and other DNA damage response proteins (1). Interference with the ATR pathway in the absence of p53 sensitizes cultured cells to DNA-damaging agents and lethally disrupts tissue homeostasis in adult mice through the accumulation of highly damaged cells (2–12). Notably, recent reports have shown that oncogene expression can induce DNA replication stress (13–22) and, similarly to p53 deficiency, creates an increased reliance on the ATR pathway for genome maintenance during DNA synthesis and cell survival (23). These studies suggest that p53 deficiency and oncogene expression each synergize with ATR pathway suppression to induce DNA damage in cells harboring these transformation-associated genetic alterations. This selective induction of genomic instability elicited by ATR suppression could produce a useful therapeutic index and effectively target malignant cells while sparing normal tissues (24).
Several drugs that suppress one ATR target, CHK1, are in early stage clinical trials. However, the most advanced compounds have been proven to inhibit additional kinases and have shown appreciable toxicity (6), which may be associated either with the essential functions of ATR and CHK1 in genome duplication or with the lack of specificity of these compounds (6, 25–31). However, although CHK1 is a principal target of ATR, several functions of ATR are independent of CHK1, and it is possible that inhibition of these kinases in advanced experimental systems will produce distinct outcomes. Currently, ATR inhibitors are in the earliest stages of development and little is known about their dosage-dependent therapeutic potential in vivo. Thus, the effects of highly specific hypomorphic inhibition of the ATR pathway on normal tissues and advanced malignancies have not been tested.
Recently, a mouse model of Seckel syndrome, a rare human genetic disorder associated with reduced ATR pathway function, has been developed (12, 32). The ATRseckel mutation leads to aberrant transcript splicing and decreased ATR protein levels, thus functioning as a hypomorphic allele of ATR (12, 32). Herein, we have used this allele together with a Cre-lox–conditional allele of ATR (ATRfl) and a drug-activated form of Cre-recombinase (Cre-ERT2) to rapidly reduce ATR expression to 10% of normal levels in adult animals, providing a means to compare the effects of this suppression on the homeostatic maintenance of normal tissues and the growth of oncogenic Ras-driven tumors that are deficient in p53. As described herein, this genetic system has defined a level of ATR expression that can potently stem tumor growth while minimally affecting normal proliferative tissues.
Results
Genetic system to achieve conditional hypomorphic suppression of ATR in mice.
The murine ATRseckel allele was previously generated by replacing murine exons 8 through 10 with an orthologous region from the human Seckel allele (12). To further characterize this allele, the region between murine exon 7 and human exon 8 was isolated and sequenced. This region was found to contain an intact PGK-neomycin resistance gene cassette that was presumably introduced during original targeting of the allele and was not subsequently removed (Figure 1A and Supplemental Figure 1; supplemental material available online with this article; doi: 10.1172/JCI58928DS1). The continued presence of this cassette in the ATRseckel allele suggests several mechanisms by which ATR transcript levels might be reduced, including inhibition of transcriptional elongation, premature termination at the inserted poly A site, promoter competition, and alternative splicing (Figure 1A and Supplemental Figure 2). Although insertion of this expression cassette alone does not dramatically reduce ATR protein levels (12), these mechanisms, in combination with the human Seckel mutation in exon 9, appear to cooperate to decrease the levels of productively spliced ATR mRNA containing human exons 8–10 (Supplemental Figure 2). These diverse and numerous obstructions to ATR expression from this allele imply that the level of ATR suppression should be relatively stable and consistent throughout fully developed adult tissues.
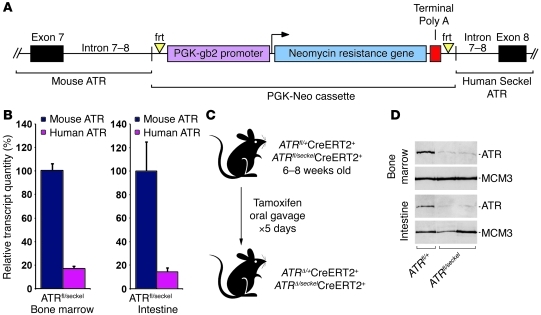
Figure 1. A genetic system to conditionally reduce ATR expression to hypomorphic levels in adult mice.
(A) Illustration of the 5′ region of the humanized ATRseckel allele. (B) Quantification of mouse (ATRfl) and human (ATRseckel) ATR transcript containing exons 8–9 in adult (6 to 8 week old) bone marrow and intestinal epithelium. Relative transcript quantity was measured through qRT-PCR analysis of RNA isolated from the indicated tissues as described in Supplemental Figure 2 (n = 5 mice per genotype, per tissue). Data show mean ± SEM. (C) Treatment used to achieve conditional ATR suppression in adult tissues. (D) Western blot analysis of ATR expression in either adult bone marrow (top panel) or intestinal epithelium (bottom panel) isolated 10 days following initial tamoxifen treatment/ATR suppression. MCM3 is shown as a loading control.
To determine the abundance of functional ATR transcript produced by the ATRseckel allele relative to wild-type ATR and the unrecombined ATRfl allele, PCR primer and probe sets that selectively discriminate mRNA containing murine exons 8–9 from human exons 8–9 were designed and validated (Supplemental Figure 2). Quantitative RT-PCR (qRT-PCR) determination of mRNA abundance from the adult bone marrow and intestinal epithelium indicated that the ATRseckel allele expressed normally spliced ATR transcript at a level that was 82% lower than that of transcript originating from either the unrecombined ATRfl allele or wild-type ATR (Figure 1B and data not shown). This decrease was consistent with the reported level of ATR expression from the ATRseckel mutation in embryonic and early postembryonic tissues (12).
To conditionally reduce ATR expression to hypomorphic levels in adult mice, ATRfl/seckel mice were crossed with mice expressing a drug-inducible form of Cre-recombinase (Cre-ERT2), generating ATRfl/seckelCre-ERT2+ mice (Figure 1C and ref. 31). Consistent with mRNA quantifications (Figure 1B), conditional deletion of the ATRfl allele in ATRfl/seckelCre-ERT2+ mice reduced total ATR expression from approximately 60% to 10% of wild-type levels both in the bone marrow and in the intestinal epithelium (Figure 1, C and D, and data not shown).
Hypomorphic suppression of ATR minimally affects normal tissue homeostasis.
Complete deletion of ATR is cell lethal and toxic to tissues with high cellular turnover rates (31). Remarkably, suppression of ATR to approximately 10% of wild-type levels in adult mice did not grossly affect bone marrow cellularity, nor did it cause an appreciable decline in several proliferative marrow subsets for up to 6 weeks after ATR reduction (Figure 2, A and B, Supplemental Figure 3, and data not shown). ATR suppression also had no discernible impact on the integrity of the intestinal epithelium and did not affect body weight (Figure 2, C and D).
Figure 2. ATR hypomorphic suppression has a minimal impact on normal tissue homeostasis.
(A) H&E-stained sections of humeral bones from tamoxifen-treated mice at the indicated time points after initial treatment. Original magnification, ×200. (B) Absolute number of myeloid cells (Mac1+Gr1+) obtained from 4 hind limb bones of mice at the indicated time points (n = 5–15 mice per genotype, per time point). (C) H&E-stained sections of intestines from tamoxifen-treated mice at the indicated time points after initial treatment. Original magnification, ×200. (D) Weight of mice at the indicated time points. (E and F) Abundance of the ATRfl-recombined allele (ATRΔ) in the bone marrow (E) and intestines (F) of tamoxifen-treated mice. Time points represent the number of days after tamoxifen treatment. The frequency of ATRfl recombination was determined by qPCR amplification of the ATRfl allele from genomic DNA isolated from each tissue (n = 5–15 mice per genotype, per time point). Data show mean ± SEM.
Previous studies have shown that loss of ATR expression in ATRΔ/– cells causes the rapid death and clearance of these cells from tissues with high proliferation rates. These events are followed by their immediate replacement with residual ATR-heterozygous cells (ATRfl/–) that escaped deletion (31). This cellular turnover leads to a rapid decrease in the representation of the ATRΔ allele in genomic DNA isolates (within 1–2 weeks) from proliferative tissues as measured by qPCR (8). Monitoring the loss of the ATRΔ allele over time in a given tissue provides a means to competitively assess the relative proliferative advantage or disadvantage conferred by ATR reduction. Notably, hypomorphic suppression of ATR in ATRΔ/seckel cells did not induce a rapid decline in their representation, as these cells persisted at high levels (>60%) in the bone marrow, intestinal epithelium, and skin, as determined by ATRΔ allele frequency over time (Figure 2, E and F, and Supplemental Figures 4 and 5). Remarkably, no significant decline in ATRΔ/seckel cells was observed in the intestinal epithelium and skin for up to 16 months after ATRfl deletion (Supplemental Figure 5). Among the tissues analyzed, only the bone marrow exhibited some demonstrable level of decrease in ATRΔ/seckel representation, which began to occur approximately 6 weeks after ATR suppression. However, this rate of decline is in stark contrast to the rapid replacement (1–2 weeks) that occurs in the bone marrow and intestine following mosaic deletion of ATR in ATRmKO mice (31). Consistent with the limited effects of ATR hypomorphic suppression, the bone marrow and intestines of ATRΔ/seckel mice demonstrated only subtle increases in a chromatin marker of DNA damage, H2AX phosphorylation (γH2AX), in the absence of p53, a background that allows such damage to accumulate (Supplemental Figure 6). Given the high cellular turnover rate in these tissues, these results indicate that hypomorphic suppression of ATR to 10% of normal levels is not acutely toxic in the context of cell proliferation that is controlled by normal homeostatic stimuli.
Hypomorphic suppression of ATR inhibits the growth of acute myeloid leukemias driven by MLL-ENL and N-rasG12D.
Oncogenic stress has been shown to cause an increased reliance on the ATR pathway for genome stability and cellular viability in culture (23). Therefore, we sought to determine how oncogene-mediated transformation of bone marrow progenitors would affect sensitivity to hypomorphic ATR suppression. Acute myeloid leukemia (AML) driven by coexpression of the oncogenic fusion protein MLL-ENL with N-rasG12D was chosen as a model system due to its clinical relevance and reported resistance to conventional chemotherapy (33).
Murine fetal liver cells (FLCs), which are enriched for hematopoietic progenitors, were isolated from ATRfl/seckelCreERT2+ embryos, transduced to express both MLL-ENL and N-rasG12D, and transplanted into recipient mice as previously described (33). These transformed marrow progenitors continued to express functional ATR transcript from the ATRseckel allele at levels that were similar to those of normal bone marrow (Figure 2A and Figure 3A), confirming that ATR expression is suppressed similarly by this allele before and after transformation. Tumor-bearing secondary-recipient mice, those in which AML burden reached at least 20% of peripheral white blood cells, were treated with tamoxifen to recombine the ATRfl allele and suppress ATR expression to hypomorphic levels (Figure 3B). These mice were then quantified for the representation of ATRfl-deleted ATRΔ/seckel leukemia cells at several time points after ATR reduction.
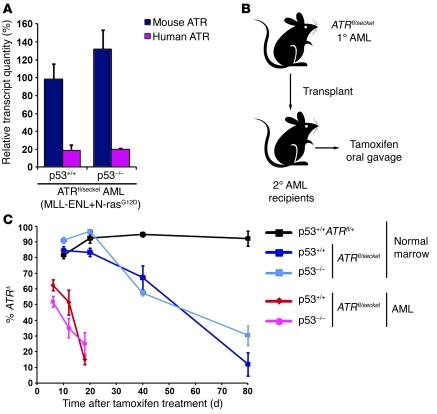
Figure 3. ATR hypomorphic suppression is more limiting to MLL-ENL– and N-rasG12D–transformed AMLs than to normal bone marrow cells.
(A) The amount of correctly spliced ATR transcript from the ATRseckel allele. Transcript levels relative to the murine ATRfl allele are shown. These levels were quantified through qRT-PCR as described in Figure 1 and Methods. Data represent mean ± SEM. (B) Experimental approach to suppressing ATR expression in murine AML. AML cells were transplanted from a primary recipient to secondary recipients, and tamoxifen was given upon detection of substantial representation (>20%) of GFP+ AML cells in peripheral white blood cells. (C) ATR hypomorphic suppression is competitively disadvantageous to AMLs to a greater degree than to normal bone marrow. tamoxifen-treated secondary (AML) recipient mice were sacrificed at various days following initiation of treatment. Genomic DNA was isolated from leukemic bone marrow, and Cre-mediated ATRfl recombination (ATRΔ) was quantified through qPCR. For comparison, bone marrow from systemically treated ATRfl/seckel mice was quantified similarly for the persistence of ATRΔ/seckel cells. No significant differences were observed between p53+/+ and p53–/– backgrounds. Data represent mean ± SEM.
A striking selection against ATRΔ/seckel AML cells was observed in comparison with the relative persistence of nontransformed ATRΔ/seckel normal bone marrow cells. Within 20 days of ATR suppression, the abundance of ATRΔ/seckel AML cells was rapidly reduced to only a minor fraction of total AML cells in treated animals (Figure 3C). This potent selection against ATRΔ/seckel AML cells was observed both in p53+/+ AMLs, in which p53 function is partially inhibited through MLL-ENL expression (34), and in p53–/– AMLs. Due to the robust expansion of residual AML cells that retained ATR expression through an unrecombined ATRfl allele, no survival benefit was readily evident, even with continuous tamoxifen treatment (data not shown). However, the rapid loss of ATRΔ/seckel AML cells and the competitive expansion of ATR-expressing counterparts argues that ATR reduction strongly impedes the growth of this chemoresistant form of AML. Importantly, this loss of ATRΔ/seckel cells was far more rapid than that observed in normal bone marrow (Figure 3C). The distinct effects of hypomorphic ATR suppression on transformed versus normal bone marrow cells suggest that the additional stress produced by hyperproliferative signaling in oncogene-expressing cells confers a selective sensitivity to ATR suppression in vivo.
The subtle disadvantage caused by hypomorphic ATR suppression in tissues is not amplified by p53 deficiency.
The long-term competitive disadvantage of ATRΔ/seckel cells in normal bone marrow provided a model to directly assess the effect of p53 deficiency on cell proliferation following partial reduction of ATR expression. As previously noted (8, 11), p53 loss typically rescues both cellular function and organismal viability in the context of low levels of DNA damage. In contrast, the appreciable genomic instability induced by complete deletion of ATR causes significant delays in tissue regeneration and organism-level synthetic lethality when it is combined with p53 deletion (8). These effects were associated with the accumulation of highly damaged, ATR-deleted cells in several proliferative tissues, including the bone marrow; thus, p53 absence did not accelerate the elimination of ATR-deleted cells.
This study indicated that the degenerative effects of ATR and p53 loss were not due to a cell-autonomous lethal effect, but rather to the accumulation of functionally compromised cells that inhibit effective tissue renewal (8, 11). These studies also implied that the previously demonstrated cell-lethal effects of combining p53 deficiency with ATR pathway inhibition may be manifested only in conjunction with additional exogenous DNA-damaging agents (2–6) because such cell synthetic–lethal effects were not observed in vivo upon combining these deletions alone (8, 11). These findings indicate that any genome-destabilizing effect of this interaction in the absence of exogenous DNA damage agents is not sufficiently caustic to overwhelm the survival benefit conferred by p53 loss.
With these prior results in mind, we wished to determine whether partial suppression of ATR expression, which caused only subtle deleterious effects over time (Figure 3C), would be altered by p53 deficiency. ATRfl/seckelCre-ERT2+ mice were crossed onto a p53-null background, and ATR expression was repressed through tamoxifen-induced Cre activation in adult mice (6–8 weeks postnatum). Subsequently, the representation of ATRΔ/seckel cells in the bone marrow was monitored over the course of 3 months. Notably, the absence of p53 expression appeared to have little effect on the competitive disadvantage of ATRΔ/seckel cells in the bone marrow (Figure 3C). The status of p53, null or wild type, also did not affect the representation of ATRΔ/seckel cells in the intestine (Supplemental Figure 6). Given the sensitivity of this competitive assay, these results demonstrate that the long-term effects of ATR hypomorphic suppression on cell proliferation are neither accentuated nor suppressed by p53 deficiency during normal homeostatic renewal. Therefore, although complete loss of ATR in the absence of p53 leads to the accumulation of functionally compromised cells that obstruct normal tissue homeostasis, the subtle disadvantageous effects of hypomorphic ATR suppression are unaffected by p53 loss in tissues.
ATR reduction inhibits the growth of H-rasG12V–transformed fibrosarcomas.
The apparent indifference of ATR-suppressed bone marrow cells to p53 loss (Figure 3) indicated that p53 deficiency neither specifically sensitizes cells to ATR reduction alone, nor does it confer resistance to ATR suppression. In other words, p53-deficient cells remained sensitive to ATR inhibition and, in all circumstances, displayed a toxicity profile similar to that of cells with wild-type p53 function. These results argue that the survival advantage and resistance to conventional chemotherapy typically conferred by p53 deficiency fail to operate in response to ATR suppression.
To further investigate the effects of oncogenic stress and p53 deficiency on cell-autonomous sensitivity to ATR suppression, p53–/–ATRfl/seckelCre-ERT2+ murine embryonic fibroblasts (MEFs) were infected with retrovirus-expressing oncogenic H-rasG12V (Supplemental Figure 7). Once again, the ATRseckel allele in p53–/–ATRfl/seckelCre-ERT2+ MEF lines expressed functional ATR transcript at levels comparable to those observed in the bone marrow and intestinal epithelium (Figure 2A and Figure 4A). H-rasG12V–expressing p53–/–ATRfl/seckelCreERT2+test and H-rasG12V–expressing p53–/–ATRfl/+CreERT2+ control lines were injected into the flanks of recipient mice, and tumors were allowed to grow to 100–200 mm3 before deleting the ATRfl allele through tamoxifen-induced Cre activation.
Figure 4. ATR reduction potently suppresses the growth of p53-null fibrosarcomas driven by H-rasG12V and increases genomic instability.
(A) Quantification of correctly spliced mouse (ATRfl) and human (ATRseckel) ATR transcript in p53–/–ATRfl/seckelCreERT2+ cell lines expressing H-rasG12V. Relative transcript quantity was measured through qRT-PCR analysis of RNA isolated from the indicated cultures (n = 4 independent cell lines). Data represent mean ± SEM. (B) Representative images of tamoxifen-treated ATRfl/+CreERT2+ (top panel) or ATRfl/seckelCreERT2+ (bottom panel) tumors 10 days following the initiation of treatment. TAM, tamoxifen. (C) Measurement of fibrosarcoma growth following ATRfl deletion. Tumors were allowed to grow to 100–200 mm3 before initiation of tamoxifen treatment to recombine the ATRfl allele. Tamoxifen treatment days are indicated by arrows. *P < 0.01 (n = 4–13 tumors per genotype, per measurement). Percentages listed above at specific time points indicate the abundance of the ATRfl-recombined allele (ATRΔ) in tumors isolated from that time point (n = 1–3 tumors per genotype/measurement). Data represent mean ± SEM. (D) H&E-stained sections (left panels) or γH2AX/DAPI–stained sections (right panels; γH2AX, green; DAPI, blue) of H-rasG12V–expressing tumors (A–C) isolated 10 days after initial ATRfl deletion. Enlarged cells with aberrant nuclei are indicated with arrows. Original magnification, ×200. (E) Quantification of γH2AX-positive cells in H-rasG12V–expressing tumors (A–C) isolated 9–10 days after initial ATRfl deletion. The abundance of γH2AX-positive cells was determined from 10 high power field images (HPF, ×200) from each tumor type (n = 4–6 tumors per genotype). Only nucleated (DAPI-positive) cells were scored in this analysis. Data represent mean ± SEM.
As expected, p53–/–ATRΔ/+Cre-ERT2+ control tumors grew rapidly during the 5 consecutive days of tamoxifen treatment and beyond, reaching sizes greater than 1,000 mm3 by 10 days after the initiation of treatment (Figure 4, B and C). In contrast, p53–/–ATRΔ/seckel Cre-ERT2+ tumors appeared to stop growing shortly after the completion of tamoxifen treatment (Figure 4, B and C). Histological examination of p53–/–ATRΔ/seckelCre-ERT2+ tumors at early time points (days 8–10) revealed a less uniform architecture in comparison with control tumors (Figure 4D). Moreover, hypomorphic ATR suppression led to a significant increase in DNA damage, as indicated by detection of γH2AX (Figure 4, D and E).
Because the selective disadvantage caused by ATR hypomorphic suppression in the bone marrow is unaffected by p53 status, we asked whether full or partial inhibition of p53 function in fibrosarcomas would alter the therapeutic effect of ATR hypomorphic suppression. Despite the suppression of p16INK4A and p19ARF, H-rasG12V–transformed fibrosarcomas that expressed p53 at wild-type levels grew at an inadequate rate for accurate comparison to p53-null tumors (Supplemental Figure 8). To overcome this obstacle, a cancer-associated p53 point mutant, p53R175H, was expressed at near endogenous levels in a p53 wild-type background (Supplemental Figure 8). This level of expression permitted tumor growth that was comparable to p53-null tumors (Figure 5A and Supplemental Figure 8); however, these cell lines largely retained DNA damage–induced p53 transcriptional activity (Supplemental Figure 8), as expected from previous studies (35).
Figure 5. H-rasG12V-transformed p53+/+ fibrosarcomas that express low levels of p53R175H are also sensitive to hypomorphic ATR reduction.
(A) Quantification of correctly-spliced mouse (ATRfl) and human (ATRseckel) ATR transcript in p53+/+ATRfl/seckelCreERT2+ cell lines ectopically expressing p53R175H and H-rasG12V. Data represent mean ± SEM. (B) Measurement of p53R175H-expressing tumor growth following ATRfl deletion. Tamoxifen treatment was performed on days 1 through 5, as shown in Figure 4. Data represent mean ± SEM. (C) Quantification of γH2AX-positive cells in H-rasG12V–expressing tumors (B) following ATRfl deletion. Tumors were isolated 9–10 days after initial tamoxifen treatment and quantified for γH2AX-positive cells as described in Figure 4E. Data represent mean ± SEM.
Fibrosarcomas derived from these p53R175H-expressing cell lines exhibited a response to ATR suppression that was nearly identical to that seen with the p53-null lines, with inhibition of growth occurring soon after tamoxifen treatment (Figure 5B). Once again, this growth suppression was accompanied by prominent γH2AX staining (Figure 5C). Notably, growth of ATR-suppressed tumors, either p53-null or p53R175H-expressing, at later time points was consistently derived from residual tumor cells that failed to undergo ATRfl recombination (data not shown). Collectively, these findings demonstrate that genetic suppression of ATR to 10% of normal levels can potently inhibit the growth of H-rasG12V-driven tumors, regardless of the type and degree of p53 mutation, and indicate that this therapeutic effect correlates with increased genomic instability.
ATR suppression cooperates principally with H-rasG12V to induce DNA damage.
ATR hypomorphic suppression had only a modest effect on bone marrow homeostasis, but potently suppressed the growth of malignancies driven by oncogenic Ras. The selective sensitivity of oncogene-driven cancers to ATR suppression correlated with a striking increase in DNA damage in tumor tissue (Figure 4, D and E, and Figure 5C). Low levels of p53R175H expression would not be expected to inhibit the DNA damage–induced function of p53 (35); however, these tumors also exhibited a strong increase in γH2AX following hypomorphic ATR suppression (Figure 5C). These data suggest that p53 deficiency may not play a dominant role in sensitizing these tumors to ATR inhibition. Nevertheless, the elevated level of DNA damage in ATRΔ/seckel fibrosarcomas could still be influenced additively by the effect of H-rasG12V expression when combined with any level of p53 suppression.
To distinguish the influences of H-rasG12V expression and p53 deficiency, ATR expression was reduced to hypomorphic levels in cultured fibroblasts, and the impact of oncogenic Ras expression and p53 deficiency on genome stability was compared. As quantified through H2AX phosphorylation and chromatid breaks, ATR suppression modestly increased genomic instability in cells expressing wild-type H-ras and p53 (Figure 6, A and B). This finding is consistent with the minimal effect of ATR suppression on normal, nontransformed tissues (Figure 2). ATR suppression in combination with p53 deficiency did elevate H2AX phosphorylation upon ATR suppression; however, this enhancement was closely associated with increased S-phase DNA content and additional cell doublings (Figure 6A). This result agrees well with the reported effects of chemical inhibition of ATR in p53-deficient cells, as quantified by γH2AX staining alone (36, 37). However, this increase in H2AX phosphorylation was not reflected in an increase in chromatid breaks in p53-deficient cells following ATR suppression (Figure 6B). Therefore, although an intermediate marker of a DNA damage response was elevated, p53 deficiency alone ultimately failed to significantly exacerbate the long-term genomic instability caused by ATR suppression, at least within a limited number of cell cycles.
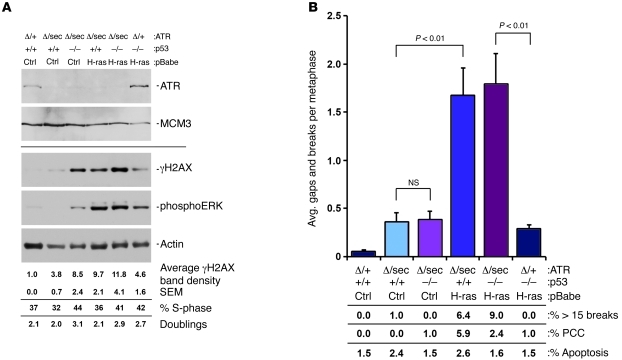
Figure 6. Genomic instability caused by ATR suppression is significantly enhanced by H-rasG12V expression.
(A) Western blot analysis of immortalized MEF cultures 48 hours following initial 4-OHT treatment. Separate blots from similarly prepared protein lysates were detected for ATR and MCM3 (top panels) or γH2AX, phospho-ERK, and actin (bottom panels). S-phase content was quantified through EdU incorporation/detection in cultures harvested 48 hours after initial 4-OHT treatment. Cumulative doublings were obtained through cell counting at 48 hours after initial 4-OHT treatment. (B) Chromosomal breakage analysis of MEF cultures from A. Cultures were collected for metaphase analysis 48 hours after initial 4-OHT treatment. Gaps/breaks per metaphase were scored in readily assessable metaphase spreads, while metaphase spreads with more than 15 breaks were scored separately and weighted into this analysis. The percentage of metaphase spreads with more than 15 breaks is listed in the lower panel. PCC was also scored, and the percentage of metaphase spreads marked by PCC is listed in the lower panel. Data represent mean ± SEM.
In contrast to the effect of p53 deficiency, H-rasG12V expression caused a synergistic increase in both H2AX phosphorylation and chromatid breaks in the context of ATR suppression (Figure 6, A and B). These increases were not strongly associated with constitutively elevated rates of proliferation (Figure 6A), suggesting that H-rasG12V signaling produces an increased reliance on the ATR pathway within individual cell cycles. Notably, the enhanced genomic instability produced by suppressing ATR in H-rasG12V–expressing cells was greater than that caused by combining ATR suppression with p53 deficiency alone and was not further elevated when H-rasG12V expression was combined with p53 deficiency (Figure 6B). These findings, along with previous reports (23), indicate that ATR suppression potently cooperates with oncogenic H-rasG12V to induce DNA damage. Furthermore, these data are consistent with the model that oncogenic Ras expression strongly contributes to the tumor-selective effects of ATR hypomorphic reduction.
H-rasG12V, K-rasG12D, and Myc overexpression create an increased reliance on ATR for genomic stability.
The ability of oncogenic H-ras mutants to sensitize cells to ATR pathway inhibition suggested the possibility that other forms of oncogenic stress may also lead to an increased reliance on the ATR pathway for genome stability. Indeed, expression of other oncogenes, in particular c-Myc, has been shown to be sufficient to promote ATR pathway activation (15, 18). To assess the generality of this interaction, H-rasG12V, K-rasG12D, a stabilized form of cyclin D1 (38, 39), and c-Myc were overexpressed in NIH3T3 cells (clone 7). These cells maintain functional p53 expression, but harbor a deletion in the INK locus. Following expression of these oncogenes, all lines exhibited phenotypic changes characteristic of transformation (data not shown). A recently described highly specific inhibitor of ATR, ATR-45 (40), was then used to suppress the ATR pathway in these cell lines. This inhibitor permitted quantification of the short-term effects of ATR pathway suppression on genomic instability within the normal transition of S to M phase in 1 cell cycle, avoiding the potential additional effects of multiple rounds of replication. Use of this inhibitor also permitted the independent validation of results obtained using the ATRfl/seckel system (Figure 6) and shRNAs targeting ATR (23).
Asynchronous oncogene-expressing and control cell lines were treated with the ATR-45 inhibitor for 7 hours and harvested for quantification of H2AX phosphorylation and chromatid breaks. In accord with results utilizing the ATRfl/seckel suppression system and previous findings (23), ATR inhibition led to greater-than-additive increases in H2AX phosphorylation and chromatid breaks in H-rasG12V–expressing lines in comparison with the levels seen following treatment of control lines (Figure 7, A–C). H2AX phosphorylation was predominantly observed in S phase (Figure 7B), consistent with the view that oncogenic stress causes an increased reliance on ATR signaling to maintain genome stability during DNA replication. Notably, cooperative increases in chromatid breaks and H2AX phosphorylation were also observed upon ATR inhibition in K-rasG12D and c-Myc–overexpressing cell lines (Figure 7, A–C). Increased H2AX phosphorylation following ATR inhibition in these lines was again observed in cells exhibiting S phase DNA content (Figure 7B and data not shown). These results indicate that several sources of oncogenic stress, including H-rasG12V, K-rasG12D, and c-Myc overexpression, produce an increased reliance on the ATR kinase activity for genome stabilization during S phase.
Figure 7. ATR inhibition synergizes with H-rasG12V, K-rasG12D, and c-Myc overexpression to cause increased genomic instability and cell synthetic lethality.
(A) Phosphorylation of H2AX in response to ATR inhibition in the context of oncogene expression. NIH3T3 cells were transduced with retroviruses expressing the indicated oncogenes or empty control vector (pBabe-puro). Following drug resistance marker selection, cell lines were expanded and treated with 1 μM ATR-45 inhibitor for 7 hours. Cells were then harvested for Western blot detection of the indicated proteins. (B) Cell-cycle distribution of γH2AX following ATR inhibition. Oncogene-expressing and control cell lines were treated with 4 μM ATR-45 inhibitor for 24 hours and detected for phospho-S139 H2AX and DNA content (propidium iodide staining). Aphidicolin (0.5 μM) was added to control cells to induce exogenous replication stress and serve as a positive control for its effects. (C) Chromatid breaks following short-term ATR inhibition in oncogene-expressing and control cell lines. Cell lines were treated with 2 μM ATR-45 inhibitor for 7 hours, as described in A, were harvested for mitotic spreads, and chromatid breaks were quantified. Nocodazole (0.5 μM) was added 4 hours prior to harvest. Data represent mean ± SEM.
Consistent with the model that increased genomic instability in oncogene-expressing cells causes tumor-selective toxicity in response to ATR suppression, low doses of ATR inhibitor suppressed the proliferation of H-rasG12V and c-Myc–overexpressing cells more so than untransformed counterparts (Figure 8A). This growth suppression was accompanied by an increase in sub-G1 DNA content, consistent with elevated cell death (Figure 8B). Although higher levels of ATR inhibition (>10 μM) were capable of causing significant chromatid breaks, proliferation defects, and cell death in control cells, the lower doses of inhibitor used in the experiments above (1–4 μM) did not. Together, these data indicate that oncogenic stress is sufficient to selectively sensitize cells to partial ATR inhibition.
Figure 8. ATR inhibition selectively suppresses the expansion of H-rasG12V– and c-Myc–transformed fibroblasts and increases cell death.
(A) Effects of ATR inhibition on the proliferation of oncogene-expressing and control cell lines. Asynchronously growing cell lines were treated with ATR inhibitor (2 μM); medium and inhibitor were replaced at replating every 2 days. Cells were counted at replating, and cumulative doublings were quantified. Data represent mean ± SEM. (B) Cell death following ATR inhibition. Sub-G1 DNA content was quantified by propidium iodide staining and flow cytometry (left panels) both 2 days (right bar graph) and 4 days (left bar graph) after continuous ATR inhibitor treatment as described in A. Peak levels of cell death were observed in the respective oncogene-transformed cell lines at these time points. SEM bars are indicated from 3–5 independent experiments.
Interestingly, partial inhibition of ATR did not cause a linear reduction in Chk1 phosphorylation. Indeed, some low levels of ATR inhibition produced an increase in this signaling event (Supplemental Figure 9). This counterintuitive outcome is likely the product of increased levels of resected double-strand breaks, which can robustly stimulate Chk1 phosphorylation through residual ATR function. Similar signaling relationships have been reported using Chk1 inhibitors and following suppression of ATR-signaling mediators (41, 42). In this light, direct detection of double-strand break formation provides a superior readout for the potential therapeutic effect of partial ATR inhibition, at least in comparison with monitoring changes in the phosphorylation of Chk1 or other ATR substrates.
In addition, it is noteworthy that not all forms of oncogenic stress produced a similar ultimate reliance on ATR for genome stability and cell viability. Although ATR inhibition in cyclin D1T286A–expressing cell lines caused an increase in H2AX phosphorylation (Figure 7, A and B), it neither led to elevated chromatid breaks (Figure 7C) nor selectively suppressed growth in culture at the same low concentrations of ATR inhibitor (1–2 μM) that produced significant effects in conjunction with other forms of oncogenic stress (Figure 7 and data not shown). Nevertheless, higher doses of ATR inhibitor for long incubation times (4–10 μM for 24 hours) were capable of causing an increase in chromatid breaks in cyclin D1T286A–expressing cell lines (data not shown), indicating that these cells are refractory but not completely resistant to the effects of ATR inhibition. In addition, while K-rasG12D–expressing lines exhibited mild increases in γH2AX and synergistic increases in chromatid breaks following ATR inhibition (Figure 7, A and B), these increases did not produce a significant proliferative defect (data not shown).
Together, these findings indicate that the type and degree of genomic instability and proliferation disadvantage produced by ATR inhibition may be strongly influenced by additional distinguishing effects of different oncogenes on cellular functions. These distinguishing features may include specific replication-associated activities (39, 43) or the additional independent regulation of cell survival and death mechanisms (44). In addition, it is certainly conceivable that different oncogenes may render distinct sensitivities to ATR inhibition in manners that depend on the cell type in which they are expressed (e.g. epithelial versus mesenchymal). In any case, these small molecule studies independently support findings obtained in animals using the ATRfl/seckel-suppression system, which collectively indicate that certain levels of ATR inhibition can be tolerated in nontransformed cells and, at the same time, elicit antiproliferative and toxic effects in oncogene-expressing cells.
Discussion
Herein, we describe the characterization and use of a genetic system to achieve conditional hypomorphic suppression of ATR in adult mice. Use of this system has demonstrated that ATR suppression to 10% of normal levels is tolerable in normal tissues, wherein cellular proliferation is under the control of normal homeostatic stimuli. However, a similar reduction of ATR expression in cells exposed to oncogenic stress leads to a high degree of genomic instability, which correlates with increased cell toxicity. This genetic interaction is observed even in the context of p53 deficiency (Figures 3–6). In cultured cells, the cooperative genome-destabilizing interaction between oncogenic stress and ATR reduction was observed using both the ATRfl/seckel system and a specific ATR inhibitor (Figures 6 and 7). Furthermore, this relationship was readily apparent in a variety of oncogenic environments, including H- and K-ras mutations and c-Myc overexpression (Figure 7). In addition, given the ability of ATR hypomorphic reduction to potently suppress the growth of MLL-ENL– and N-rasG12D–driven AML (Figure 3), the genome-destabilizing effect of this interaction may translate to a variety of additional oncogenic stresses that hyperactivate cell growth signaling.
The mechanism by which oncogenic stress leads to an increased reliance on the ATR pathway is largely unknown. According to our studies, this interaction can take place within an individual cell cycle in S phase (Figures 6 and 7 and ref. 23). ATR is known to stabilize uncoupled replication forks, which exhibit an increased accumulation of single-strand DNA (1). Therefore, it is plausible that oncogenic stress may generate circumstances that increase the frequency of these replication fork uncoupling events and thereby generate an increased reliance on ATR function. The direct cause of such replication fork uncoupling events under oncogenic stress includes Cdk2/cyclin E hyperactivation and premature origin firing, replisome component deficiency, dNTP deficiency, altered origin density and placement, and increased oxidative DNA damage (13–22). In addition, it is interesting to note that MLL fusions have recently been shown to be defective in regulating late origin firing in response to DNA damage (45). Thus, it is conceivable that distinct oncogene-specific mechanisms may operate in driving an increased reliance on ATR, a central regulator of responses to the numerous and diverse means by which DNA replication can be perturbed (1).
Notably, oncogenic Ras expression appeared to enhance sensitivity to ATR reduction more than p53 deficiency did, at least in the absence of exogenous DNA-damaging agents. This distinction was most evident in cultured cells (Figures 6–8), but was also observed in the bone marrow, where p53 deficiency led to no selective disadvantage to ATRΔ/seckel cells. Consistent with this data, the tissue renewal defects and synthetic lethality induced by combined loss of ATR and p53 in adult mice appear not to be the consequence of an enhanced elimination of ATR-deficient cells, but rather from the accumulation of highly damaged cells and the non–cell autonomous obstruction of normal tissue renewal (8, 11). Such a relationship was not observed after ATR hypomorphic suppression in p53-deficient mice, most likely because this level of ATR reduction caused only the slow accrual of low levels of damage, which over time could be readily compensated for.
Nevertheless, it is also important to recognize that p53 deficiency also did not preserve ATRΔ/seckel cells in the bone marrow and other tissues. The lack of this well-characterized effect of p53 loss on DNA repair mutants is, at the very least, an indication that the cell-autonomous deleterious effect of hypomorphic ATR suppression cannot be rescued by p53 deficiency. The genome-destabilizing interaction between p53 loss and ATR or CHK1 suppression has been proposed to be associated with the combined loss of G1 and S-G2 checkpoints (2, 4–6, 46). The deleterious effect of this is an interaction that becomes more readily apparent in the presence of exogenous DNA-damaging agents (2, 4–6, 46). Interestingly, because oncogenic stress also appears to rely on the ATR pathway for genome stabilization, it is conceivable that the previously observed synthetic-lethal relationships between p53 deficiency and ATR/CHK1 pathway suppression may be partly attributable to the ability of p53 loss to permit further amplification of oncogenic signaling pathways in cancer cells (47–49).
Collectively, these findings suggest that ATR and CHK1 inhibitors may be useful for a range of human malignancies, including those that harbor p53 mutations. Recent reports agree with this conclusion, which appears to apply to a variety of malignancies (50–52). Although the cancer-selective attributes of ATR inhibition are highly encouraging, caution is certainly warranted when the therapeutic window is narrowed both by the general toxicity produced from high levels of inhibition and the risk of therapy-related cancers that may result from lower levels of inhibition (Figure 6). Indeed, ATR and CHK1 haploinsufficiency have been shown to be tumor promoting (23, 25, 26, 30, 53–61), although the risk of therapy-related cancers may be no greater than that caused by DNA-damaging conventional chemotherapeutics. With these selectivity issues in mind, the most promising therapeutic use of ATR inhibition may be in specifically targeting certain combinatorial genetic changes that make a given malignancy distinctly reliant on ATR. Such a strategy will prominently distinguish the cancer from normal tissues. Nevertheless, these findings have defined a specific level of ATR pathway inhibition that permits homeostatic renewal of essential tissues that are typically dose limiting for conventional chemotherapeutic treatment and simultaneously suppresses the growth of oncogene-transformed tumors, even those that are deficient in p53.
Methods
Further information can be found in Supplemental Methods.
Mice.
The ATRfl, ATR+/–, and Cre-ERT2+ mouse lines (129SvEv/C57BL/6 mixed) were generated as previously described (25, 31) and crossed to ATRseckel mice (12). p53–/– (Trp53tm1Tyj) mice were also used where indicated. NCr nude recipient mice (NCr-Foxn1nu) for tumor experiments were obtained from Taconic. For systemic ATR deletion, 4–6 mg of tamoxifen (Sigma-Aldrich) was administered to adult (6 to 8 week old) mice through 5 consecutive daily oral gavage treatments. Each treatment consisted of 200 μl of a 20–30 mg/ml tamoxifen solution solubilized in corn oil (Sigma-Aldrich) with 2% ethanol.
qRT-PCR.
RNA was isolated from tissues and processed as described in Supplemental Methods. The relative amount of ATR transcript was quantified through qRT-PCR using discriminating primer/probe sets as described in Supplemental Figure 2.
Histological analysis and bone marrow (FACS) analysis.
Tissues were collected and processed as described in Supplemental Methods. γH2AX was detected by immunofluorescence with mouse anti–phospho-H2AX (Ser139) FITC (1:50 dilution, 16-202A; Millipore).
qPCR.
Genomic DNA was isolated from tissues, and the abundance of the ATRfl-recombined allele (ATRΔ) was quantified as detailed in Supplemental Methods.
FLC isolation, infections, and transplant.
FLCs were collected, infected, and transplanted as previously described (33, 62). Briefly, FLCs were harvested from day 13.5–14.5 postcoitus embryos, manually dissociated through a 70-μM cell strainer, and cultured overnight in Iscove’s modified Dulbecco medium (IMDM) supplemented with IL-3, IL-6, and SCF (PeproTech) along with 20% FBS (StemCell Technologies). The following day, FLCs were infected with MSCV-(MLL-ENL)-GFP and MSCV-luciferase-(N-rasG12D) retroviral supernatant (1:1) during a 1.5-hour spin infection. They were infected again 1 day later, cultured overnight, and transplanted into the retroorbital sinus of irradiated (9.5 Gy) primary recipient mice. Following the development of significant leukemia burden in primary recipients (>70% AML cells in peripheral blood), leukemic bone marrow was harvested and transplanted into irradiated (5.5 Gy) secondary recipient mice. Secondary recipients were allowed to develop a readily identifiable leukemia (>20% AML cells in peripheral blood) and treated with tamoxifen as detailed above (see Mice).
MEF isolation, infections, and treatments.
MEFs were isolated from day-14.5 postcoitus embryos through mincing and digestion in 0.25% trypsin. MEFs were cultured in a 3% oxygen incubator in DMEM (Cellgro) supplemented with 10% FBS (Hyclone), 1% l-glutamine, and 1% penicillin/streptomycin. Passage 1 and 2 MEFs were infected with lentivirus expressing an shRNA targeting both p16INK4A and p19ARF (TRCN0000077813; Open Biosystems) and selected in 40 μg/ml puromycin (Calbiochem). MEFs were then infected with either control retrovirus (pBabe-empty-GFP or pWZL-empty-hygro) or retrovirus expressing H-rasG12V (pBabe-H-rasG12V-GFP) or p53R175H (pWZL-p53R175H-hygro). GFP+ cells were sorted on a FACSVantage SE. Hygromycin-resistant cells were selected in 5 μg/ml hygromycin (Mediatech). To delete ATR in culture, MEFs were treated with 500 nM 4-hydroxy-tamoxifen (Calbiochem) for 24 hours.
Oncogene expression and ATR-45 treatment of NIH3T3 murine fibroblasts.
NIH3T3 (clone 7, p19ARF mutant) was transduced with murine retroviruses expressing various oncogenes as described (23). Oncogene-expressing vectors utilized were as follows: pBabe-H-rasG12V-puro, pBabe-murine-K-rasG12D-puro (63), pBabe-murine-c-Myc-puro, pBabe–murine–cyclin D1T286A–puro and pBabe-puro control vector. Transduced and selected lines were treated with ATR-45 inhibitor, and proliferation assays were performed as described (Figure 6 and ref. 23, respectively). Codetection of phospho-S139 H2AX and propidium iodide staining by flow cytometry was performed as described (64).
Tumor transplant experiments.
Recipient mice (NCr nude) received subcutaneous injections of 2–4 × 106 GFP+ cells in each flank. When tumors reached a size of 100–200 mm3, with clear evidence of growth, mice were treated with tamoxifen as detailed above (see Mice).
Western blots.
Cells were resuspended in PBS and lysed with 1× SDS sample buffer (2× = 20% glycerol, 0.15M SDS, 125 mM Tris, pH 6.8). Protein concentration was determined with a BCA Protein Assay Kit (Thermo Scientific), and whole-cell lysates were separated by SDS-PAGE and transferred to 0.45 μm PVDF membranes. Blots were detected for ATR (N-19; Santa Cruz Biotechnology Inc.), MCM3 (124A; Bethyl), phospho-H2AX (Ser139, JBW301; Millipore), phospho-ERK (4370; Cell Signaling), and actin (I-19; Santa Cruz Biotechnology Inc.).
Cell-cycle and apoptosis analysis.
Cells were processed as described in Supplemental Methods.
Metaphase spread analysis.
Cells were treated with 0.5 μM Nocodazole (Calbiochem) for 2.5 hours, collected, incubated in 75 mM KCl at 37°C for 18 minutes, and fixed in 3:1 methanol/glacial acetic acid. Cells were dropped on glass slides (Fisher) and stained with Giemsa; metaphase spreads were assessed for gaps/breaks and evidence of premature chromosome condensation (PCC).
Statistics.
Microsoft Excel was used for all statistical analysis. Student’s unpaired, 2-tailed t test was used to calculate P values for all data sets. Single SEMs are shown as bars on each graph. P ≤ 0.05 was considered significant.
Study approval.
Animal studies were performed with the approval of, and according to guidelines provided by, the University of Pennsylvania IACUC.
Supplementary Material
Acknowledgments
We are indebted to Scott Lowe, Anil Rustgi, David Tuveson, Kevin Smith, Yaroslava Ruzankina, Gabrielle Wong, Austin Thiel, Dan Zlotoff, and Laura Pontano for reagents, protocols, and advice and are grateful to the AFCRI Histology Core for tissue processing. These studies were supported by the National Institute on Aging (R01AG027376 and F30AG034027), an NIH training grant (R25CA101871), a Department of Defense Breast Cancer Research Program Idea award (W81XWH-09-1-0669), the Abramson Family Cancer Research Institute, and the Mary Kay Ash Charitable Trust.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(1):241–252. doi:10.1172/JCI58928.
References
- 1.Paulsen RD, Cimprich KA. The ATR pathway: fine-tuning the fork. DNA Repair (Amst). 2007;6(7):953–966. doi: 10.1016/j.dnarep.2007.02.015. [DOI] [PubMed] [Google Scholar]
- 2.Nghiem P, Park PK, Kim Y, Vaziri C, Schreiber SL. ATR inhibition selectively sensitizes G1 checkpoint-deficient cells to lethal premature chromatin condensation. Proc Natl Acad Sci U S A. 2001;98(16):9092–9097. doi: 10.1073/pnas.161281798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koniaras K, Cuddihy AR, Christopoulos H, Hogg A, O’Connell MJ. Inhibition of Chk1-dependent G2 DNA damage checkpoint radiosensitizes p53 mutant human cells. Oncogene. 2001;20(51):7453–7463. doi: 10.1038/sj.onc.1204942. [DOI] [PubMed] [Google Scholar]
- 4.Nghiem P, Park PK, Kim YS, Desai BN, Schreiber SL. ATR is not required for p53 activation but synergizes with p53 in the replication checkpoint. J Biol Chem. 2002;277(6):4428–4434. doi: 10.1074/jbc.M106113200. [DOI] [PubMed] [Google Scholar]
- 5.Zhou BB, Bartek J. Targeting the checkpoint kinases: chemosensitization versus chemoprotection. Nat Rev Cancer. 2004;4(3):216–225. doi: 10.1038/nrc1296. [DOI] [PubMed] [Google Scholar]
- 6.Ma CX, Janetka JW, Piwnica-Worms H. Death by releasing the breaks: CHK1 inhibitors as cancer therapeutics. Trends Mol Med. 2011;17(2):88–96. doi: 10.1016/j.molmed.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Toledo LI, Murga M, Fernandez-Capetillo O. Targeting ATR and Chk1 kinases for cancer treatment: a new model for new (and old) drugs. Mol Oncol. 2011;5(4):368–373. doi: 10.1016/j.molonc.2011.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ruzankina Y, Schoppy DW, Asare A, Clark CE, Vonderheide RH, Brown EJ. Tissue regenerative delays and synthetic lethality in adult mice after combined deletion of Atr and Trp53. Nat Genet. 2009;41(10):1144–1149. doi: 10.1038/ng.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Greenow KR, Clarke AR, Jones RH. Chk1 deficiency in the mouse small intestine results in p53-independent crypt death and subsequent intestinal compensation. Oncogene. 2009;28(11):1443–1453. doi: 10.1038/onc.2008.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yazinski SA, Westcott PM, Ong K, Pinkas J, Peters RM, Weiss RS. Dual inactivation of Hus1 and p53 in the mouse mammary gland results in accumulation of damaged cells and impaired tissue regeneration. Proc Natl Acad Sci U S A. 2009;106(50):21282–21287. doi: 10.1073/pnas.0904965106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schoppy DW, Ruzankina Y, Brown EJ. Removing all obstacles: a critical role for p53 in promoting tissue renewal. Cell Cycle. 2010;9(7):1313-1319. doi: 10.4161/cc.9.7.11194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Murga M, et al. A mouse model of ATR-Seckel shows embryonic replicative stress and accelerated aging. Nat Genet. 2009;41(8):891–898. doi: 10.1038/ng.420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tanaka S, Diffley JF. Deregulated G1-cyclin expression induces genomic instability by preventing efficient pre-RC formation. Genes Dev. 2002;16(20):2639–2649. doi: 10.1101/gad.1011002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ekholm-Reed S, Mendez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J Cell Biol. 2004;165(6):789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bartkova J, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434(7035):864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 16.Gorgoulis VG, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434(7035):907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 17.Bartkova J, et al. Oncogene-induced senescence is part of the tumorigenesis barrier imposed by DNA damage checkpoints. Nature. 2006;444(7119):633–637. doi: 10.1038/nature05268. [DOI] [PubMed] [Google Scholar]
- 18.Di Micco R, et al. Oncogene-induced senescence is a DNA damage response triggered by DNA hyper-replication. Nature. 2006;444(7119):638–642. doi: 10.1038/nature05327. [DOI] [PubMed] [Google Scholar]
- 19.Abulaiti A, Fikaris AJ, Tsygankova OM, Meinkoth JL. Ras induces chromosome instability and abrogation of the DNA damage response. Cancer Res. 2006;66(21):10505–10512. doi: 10.1158/0008-5472.CAN-06-2351. [DOI] [PubMed] [Google Scholar]
- 20.Halazonetis TD, Gorgoulis VG, Bartek J. An oncogene-induced DNA damage model for cancer development. Science. 2008;319(5868):1352–1355. doi: 10.1126/science.1140735. [DOI] [PubMed] [Google Scholar]
- 21.Loeb KR, et al. A mouse model for cyclin E-dependent genetic instability and tumorigenesis. Cancer Cell. 2005;8(1):35–47. doi: 10.1016/j.ccr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 22.Mailand N, Diffley JFX. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122(6):915–926. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 23.Gilad O, et al. Combining ATR suppression with oncogenic Ras synergistically increases genomic instability, causing synthetic lethality or tumorigenesis in a dosage-dependent manner. Cancer Res. 2010;70(23):9693–9702. doi: 10.1158/0008-5472.CAN-10-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaelin WG., Jr The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5(9):689–698. doi: 10.1038/nrc1691. [DOI] [PubMed] [Google Scholar]
- 25.Brown EJ, Baltimore D. ATR disruption leads to chromosomal fragmentation and early embryonic lethality. Genes Dev. 2000;14(4):397–402. [PMC free article] [PubMed] [Google Scholar]
- 26.Liu Q, et al. Chk1 is an essential kinase that is regulated by Atr and required for the G(2)/M DNA damage checkpoint. Genes Dev. 2000;14(12):1448–1459. [PMC free article] [PubMed] [Google Scholar]
- 27.Takai H, et al. Aberrant cell cycle checkpoint function and early embryonic death in Chk1(-/-) mice. Genes Dev. 2000;14(12):1439–1447. [PMC free article] [PubMed] [Google Scholar]
- 28.de Klein A, et al. Targeted disruption of the cell-cycle checkpoint gene ATR leads to early embryonic lethality in mice. Curr Biol. 2000;10(8):479–482. doi: 10.1016/S0960-9822(00)00447-4. [DOI] [PubMed] [Google Scholar]
- 29.Brown EJ, Baltimore D. Essential and dispensable roles of ATR in cell cycle arrest and genome maintenance. Genes Dev. 2003;17(5):615–628. doi: 10.1101/gad.1067403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lam MH, Liu Q, Elledge SJ, Rosen JM. Chk1 is haploinsufficient for multiple functions critical to tumor suppression. Cancer Cell. 2004;6(1):45–59. doi: 10.1016/j.ccr.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 31.Ruzankina Y, et al. Deletion of the developmentally essential gene ATR in adult mice leads to age-related phenotypes and stem cell loss. Cell Stem Cell. 2007;1(1):113–126. doi: 10.1016/j.stem.2007.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.O’Driscoll M, Ruiz-Perez VL, Woods CG, Jeggo PA, Goodship JA. A splicing mutation affecting expression of ataxia-telangiectasia and Rad3-related protein (ATR) results in Seckel syndrome. Nat Genet. 2003;33(4):497–501. doi: 10.1038/ng1129. [DOI] [PubMed] [Google Scholar]
- 33.Zuber J, et al. Mouse models of human AML accurately predict chemotherapy response. Genes Dev. 2009;23(7):877–889. doi: 10.1101/gad.1771409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiederschain D, Kawai H, Shilatifard A, Yuan ZM. Multiple mixed lineage leukemia (MLL) fusion proteins suppress p53-mediated response to DNA damage. J Biol Chem. 2005;280(26):24315–24321. doi: 10.1074/jbc.M412237200. [DOI] [PubMed] [Google Scholar]
- 35.Brosh R, Rotter V. When mutants gain new powers: news from the mutant p53 field. Nat Rev Cancer. 2009;9(10):701–713. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 36.Toledo LI, et al. A cell-based screen identifies ATR inhibitors with synthetic lethal properties for cancer-associated mutations. Nat Struct Mol Biol. 2011;18(6):721–727. doi: 10.1038/nsmb.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reaper PM, et al. Selective killing of ATM- or p53-deficient cancer cells through inhibition of ATR. Nat Chem Biol. 2011;7(7):428–430. doi: 10.1038/nchembio.573. [DOI] [PubMed] [Google Scholar]
- 38.Diehl JA, Zindy F, Sherr CJ. Inhibition of cyclin D1 phosphorylation on threonine-286 prevents its rapid degradation via the ubiquitin-proteasome pathway. Genes Dev. 1997;11(8):957–972. doi: 10.1101/gad.11.8.957. [DOI] [PubMed] [Google Scholar]
- 39.Aggarwal P, et al. Nuclear accumulation of cyclin D1 during S phase inhibits Cul4-dependent Cdt1 proteolysis and triggers p53-dependent DNA rereplication. Genes Dev. 2007;21(22):2908–2922. doi: 10.1101/gad.1586007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Charrier JD, et al. Discovery of potent and selective inhibitors of ataxia telangiectasia mutated and Rad3 related (ATR) protein kinase as potential anticancer agents. J Med Chem. 2011;54(7):2320–2330. doi: 10.1021/jm101488z. [DOI] [PubMed] [Google Scholar]
- 41.Syljuasen RG, et al. Inhibition of human Chk1 causes increased initiation of DNA replication, phosphorylation of ATR targets, and DNA breakage. Mol Cell Biol. 2005;25(9):3553–3562. doi: 10.1128/MCB.25.9.3553-3562.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Smith KD, Fu MA, Brown EJ. Tim-Tipin dysfunction creates an indispensible reliance on the ATR-Chk1 pathway for continued DNA synthesis. J Cell Biol. 2009;187(1):15–23. doi: 10.1083/jcb.200905006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Davidson IF, Li A, Blow JJ. Deregulated replication licensing causes DNA fragmentation consistent with head-to-tail fork collision. Mol Cell. 2006;24(3):433–443. doi: 10.1016/j.molcel.2006.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffman B, Liebermann DA. Apoptotic signaling by c-MYC. Oncogene. 2008;27(50):6462–6472. doi: 10.1038/onc.2008.312. [DOI] [PubMed] [Google Scholar]
- 45.Liu H, et al. Phosphorylation of MLL by ATR is required for execution of mammalian S-phase checkpoint. Nature. 2010;467(7313):343–346. doi: 10.1038/nature09350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reinhardt HC, Aslanian AS, Lees JA, Yaffe MB. p53-deficient cells rely on ATM- and ATR-mediated checkpoint signaling through the p38MAPK/MK2 pathway for survival after DNA damage. Cancer Cell. 2007;11(2):175–189. doi: 10.1016/j.ccr.2006.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarkisian CJ, et al. Dose-dependent oncogene-induced senescence in vivo and its evasion during mammary tumorigenesis. Nat Cell Biol. 2007;9(5):493–505. doi: 10.1038/ncb1567. [DOI] [PubMed] [Google Scholar]
- 48.Feldser DM, et al. Stage-specific sensitivity to p53 restoration during lung cancer progression. Nature. 2010;468(7323):572–575. doi: 10.1038/nature09535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Junttila MR, et al. Selective activation of p53-mediated tumour suppression in high-grade tumours. Nature. 2010;468(7323):567–571. doi: 10.1038/nature09526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kawasumi M, et al. Protection from UV-induced skin carcinogenesis by genetic inhibition of the ataxia telangiectasia and Rad3-related (ATR) kinase. Proc Natl Acad Sci U S A. 2011;108(33):13716–13721. doi: 10.1073/pnas.1111378108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tho LM, Libertini S, Rampling R, Sansom O, Gillespie DA. Chk1 is essential for chemical carcinogen-induced mouse skin tumorigenesis [published online ahead of print August 1, 2011]. Oncogene. doi: 10.1038/onc.2011.326. doi:10.1038/onc.2011.326. [DOI] [PubMed] [Google Scholar]
- 52.Ferrao PT, Bukczynska EP, Johnstone RW, McArthur GA. Efficacy of CHK inhibitors as single agents in MYC-driven lymphoma cells [published online ahead of print August 15, 2011]. Oncogene. doi: 10.1038/onc.2011.358. doi:10.1038/onc.2011.358. [DOI] [PubMed] [Google Scholar]
- 53.Menoyo A, Alazzouzi H, Espin E, Armengol M, Yamamoto H, Schwartz S., Jr Somatic mutations in the DNA damage-response genes ATR and CHK1 in sporadic stomach tumors with microsatellite instability. Cancer Res. 2001;61(21):7727–7730. [PubMed] [Google Scholar]
- 54.Vassileva V, Millar A, Briollais L, Chapman W, Bapat B. Genes involved in DNA repair are mutational targets in endometrial cancers with microsatellite instability. Cancer Res. 2002;62(14):4095–4099. [PubMed] [Google Scholar]
- 55.Fang Y, et al. ATR functions as a gene dosage-dependent tumor suppressor on a mismatch repair-deficient background. EMBO J. 2004;23(15):3164–3174. doi: 10.1038/sj.emboj.7600315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tort F, et al. Checkpoint kinase 1 (CHK1) protein and mRNA expression is downregulated in aggressive variants of human lymphoid neoplasms. Leukemia. 2005;19(1):112–117. doi: 10.1038/sj.leu.2403571. [DOI] [PubMed] [Google Scholar]
- 57.Davies H, et al. Somatic mutations of the protein kinase gene family in human lung cancer. Cancer Res. 2005;65(17):7591–7595. doi: 10.1158/0008-5472.CAN-05-1855. [DOI] [PubMed] [Google Scholar]
- 58.Lewis KA, et al. Heterozygous ATR mutations in mismatch repair-deficient cancer cells have functional significance. Cancer Res. 2005;65(16):7091–7095. doi: 10.1158/0008-5472.CAN-05-1019. [DOI] [PubMed] [Google Scholar]
- 59.Maniwa Y, et al. His239Arg SNP of HRAD9 is associated with lung adenocarcinoma. Cancer. 2006;106(5):1117–1122. doi: 10.1002/cncr.21705. [DOI] [PubMed] [Google Scholar]
- 60.Kim CJ, et al. Chk1 frameshift mutation in sporadic and hereditary non-polyposis colorectal cancers with microsatellite instability. Eur J Surg Oncol. 2007;33(5):580–585. doi: 10.1016/j.ejso.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 61.Lewis KA, Bakkum-Gamez J, Loewen R, French AJ, Thibodeau SN, Cliby WA. Mutations in the ataxia telangiectasia and rad3-related-checkpoint kinase 1 DNA damage response axis in colon cancers. Genes Chromosomes Cancer. 2007;46(12):1061–1068. doi: 10.1002/gcc.20486. [DOI] [PubMed] [Google Scholar]
- 62.Schmitt CA, Fridman JS, Yang M, Baranov E, Hoffman RM, Lowe SW. Dissecting p53 tumor suppressor functions in vivo. Cancer Cell. 2002;1(3):289–298. doi: 10.1016/S1535-6108(02)00047-8. [DOI] [PubMed] [Google Scholar]
- 63.Tuveson DA, et al. Endogenous oncogenic K-ras(G12D) stimulates proliferation and widespread neoplastic and developmental defects. Cancer Cell. 2004;5(4):375–387. doi: 10.1016/S1535-6108(04)00085-6. [DOI] [PubMed] [Google Scholar]
- 64.Chanoux RA, Yin B, Urtishak KA, Asare A, Bassing CH, Brown EJ. ATR and H2AX cooperate in maintaining genome stability under replication stress. J Biol Chem. 2009;284(9):5994–6003. doi: 10.1074/jbc.M806739200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.