Abstract
Glioblastoma multiforme (GBM) is the most common and lethal of all gliomas. The current standard of care includes surgery followed by concomitant radiation and chemotherapy with the DNA alkylating agent temozolomide (TMZ). O6-methylguanine–DNA methyltransferase (MGMT) repairs the most cytotoxic of lesions generated by TMZ, O6-methylguanine. Methylation of the MGMT promoter in GBM correlates with increased therapeutic sensitivity to alkylating agent therapy. However, several aspects of TMZ sensitivity are not explained by MGMT promoter methylation. Here, we investigated our hypothesis that the base excision repair enzyme alkylpurine–DNA–N-glycosylase (APNG), which repairs the cytotoxic lesions N3-methyladenine and N7-methylguanine, may contribute to TMZ resistance. Silencing of APNG in established and primary TMZ-resistant GBM cell lines endogenously expressing MGMT and APNG attenuated repair of TMZ-induced DNA damage and enhanced apoptosis. Reintroducing expression of APNG in TMZ-sensitive GBM lines conferred resistance to TMZ in vitro and in orthotopic xenograft mouse models. In addition, resistance was enhanced with coexpression of MGMT. Evaluation of APNG protein levels in several clinical datasets demonstrated that in patients, high nuclear APNG expression correlated with poorer overall survival compared with patients lacking APNG expression. Loss of APNG expression in a subset of patients was also associated with increased APNG promoter methylation. Collectively, our data demonstrate that APNG contributes to TMZ resistance in GBM and may be useful in the diagnosis and treatment of the disease.
Introduction
The most common primary adult human brain tumors are gliomas, with grade IV glioblastoma multiforme (GBM) astrocytoma being most common and malignant (1–3). With the current standard of treatment (surgery with concomitant radiation therapy [RT] and chemotherapy), the median survival of GBM patients is only approximately 12–16 months (1). There are at least 2 recognized molecular subtypes of GBM (4, 5): primary GBMs occur de novo in older patients, whereas secondary GBMs occur in younger patients, harbor mutations in isocitrate dehydrogenase 1 and 2 (IDH1/IDH2), and develop from malignant progression of preexisting low-grade astrocytomas. Common to all GBMs are aberrations in cell-cycle regulation, mediated by mutations in TP53 and RB pathways, and hyperactivation of PI3K and Ras signaling (6–8). Current knowledge of the molecular pathogenesis of GBM has generated clinical trials with biologically targeted agents, although none to date has conclusively shown efficacy (1). Hence, there are many gaps in our understanding of glioma biology. In addition, recent genome-wide studies by the Cancer Genome Atlas Network and others have shown that remarkable molecular heterogeneity exists among GBMs and that identification of novel cancer-associated genes may be of diagnostic, prognostic, and therapeutic use (9, 10).
Using a retrovirally mediated gene trap strategy on our characterized transgenic mouse models of gliomas, we previously identified GATA4 and GATA6, a subfamily of transcription factors, as novel tumor suppressor genes in human GBM, involved in suppressing tumor formation in vivo and in vitro (11, 12). Furthermore, GATA4 expression sensitized GBM cells to temozolomide (TMZ) (13). TMZ is a DNA alkylating agent and the only current chemotherapeutic agent demonstrated to have some efficacy in the treatment of GBM along with surgery and RT (14). Resistance to TMZ in vitro and in animal models can be mediated by O6-methylguanine–DNA methyltransferase (MGMT), a DNA repair protein that removes TMZ-generated methyl groups from the O6 position of guanine that represents the most cytotoxic lesion (15). GBM patients with a methylated MGMT promoter have increased overall survival and better response to combined TMZ and RT compared with RT alone, with 2-year survival rates of 48.9% (95% CI, 33.7%–62.4%) and 23.9% (95% CI, 12.9%–36.9%), respectively (16, 17). Conversely, patients with MGMT-unmethylated promoters have reduced overall survival, with 2-year survival rates of 14.8% (95% CI, 7.2%–25.0%) with the combination therapy and 1.8% (95% CI, 0.1%–8.6%) with RT only (18). However, 50% of GBM patients with a MGMT-methylated promoter do not survive 2 years, and hence only moderately benefit from TMZ treatment, suggestive of additional factors of resistance (19). Similarly, there are MGMT-unmethylated GBM patients that show some response to TMZ, strongly suggesting that MGMT promoter methylation is not the only predictor of response to TMZ (20, 21).
Our investigation into other DNA repair modulating systems, including GATA4-mediated TMZ sensitivity, showed reduced levels of the DNA base excision repair (BER) enzyme alkylpurine–DNA–N-glycosylase (APNG; also known as DNA methylpurine–N-glycosylase [MPG]). Downregulation of APNG using siRNA also increased TMZ sensitivity in several established and primary GBM cell lines (13), independent of — and additive to — the effect of MGMT downregulation. APNG is the BER enzyme that removes the alkylated DNA bases N3-methyladenine (N3-meA) and N7-methylguanine (N7-meG) from DNA, creating apurinic/apyrimidinic (AP) sites. Subsequent steps involve the AP endonuclease/redox effector factor-1 (APE1/REF-1), which cleaves 5′ to the AP site (22); removal of the resulting deoxyribose; gap filling by DNA polymerase; and, finally, sealing the break by ligation. TMZ also generates N7-meG and N3-meA in DNA, and the latter is well established to be potentially lethal by blocking the progress of the replicative DNA polymerase (23). The resulting AP site is removed and then filled in with a new base by DNA h-polymerase. O6-guanine, N7-guanine, and N3-adenine are the 3 most abundant sites of alkylation by chemotherapeutic agents such as TMZ and carmustine (24, 25). Therefore, we hypothesized APNG may provide TMZ resistance in an additive manner with MGMT, as they repair mutually exclusive alkylated bases.
Results
Characterization of APNG and MGMT in GBM cell lines.
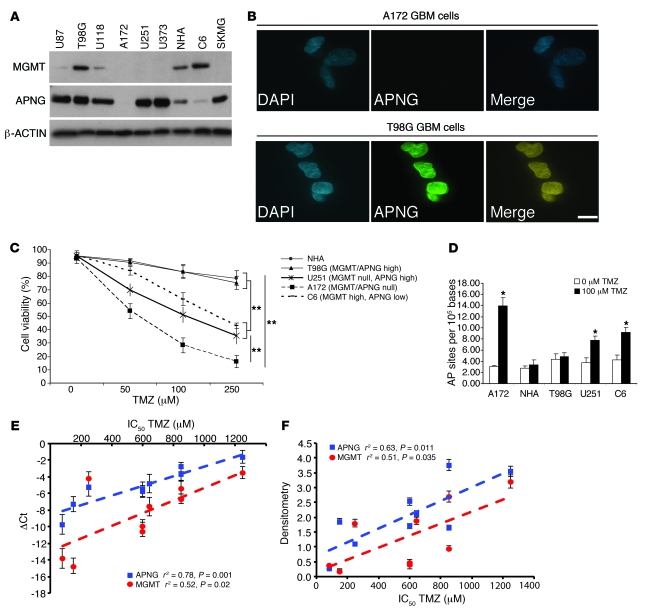
To generate an expression profile of APNG and MGMT, we screened several GBM cell lines by Western blotting and observed a variety of expression patterns for APNG and MGMT (Figure 1A). APNG immunofluorescence (IF) analysis corroborated the immunoblot results, demonstrating nuclear localization in T98G cells and negative staining of A172 cells (Figure 1B). Treating several GBM cell lines with varying concentrations of TMZ revealed that MGMT- and APNG-expressing normal human astrocytes and T98G cells were relatively resistant to TMZ, whereas A172 cells lacking both MGMT and APNG were highly sensitive (P = 0.001). C6 rat glioma cells (which predominantly express MGMT) and U251 human GBM cells (which express APNG, but not MGMT) were more sensitive than T98G cells, but less sensitive than A172 cells, at 100 μM TMZ (P < 0.01; Figure 1C). In addition to reduced cell viability, A172 cells had marked increase of DNA damage, as measured by an increased abundance of AP sites compared with control cells not treated with TMZ (P < 0.05; Figure 1D). Normal human astrocytes and T98G cells did not have any significant increase in AP sites when treated with 100 μM TMZ (Figure 1D). APNG and MGMT RNA levels correlated positively with IC50 levels of TMZ-treated GBM cells, as shown by Pearson correlation (APNG, r2 = 0.78, P = 0.001; MGMT, r2 = 0.52, P = 0.02; Figure 1E). Chemiluminescent densitometric analysis showed that IC50 for TMZ-treated GBM cells also correlated at the protein level for APNG and MGMT (APNG, r2 = 0.63, P = 0.011; MGMT, r2 = 0.51, P = 0.035; Figure 1F). However, these experiments are suggestive (albeit not conclusive) as to APNG and MGMT levels contributing to TMZ resistance, since these cells had additional alterations varying between them (see below), thus requiring further determination of their respective contributions to TMZ-mediated resistance.
Figure 1. APNG expression in GBM.
(A) Western blot of APNG and MGMT in several GBM cell lines demonstrating variable expression levels. NHA, normal human astrocytes. (B) IF of A172 and T98G GBM cells (APNG-negative and -positive, respectively) stained with DAPI (blue) for nuclei. APNG expression (green), which was seen only in T98G cells, colocalized to the nucleus (yellow in merged image). (C) Cell viability of GBM cell lines in response to TMZ. (D) DNA damage, measured as the number of AP sites in response to TMZ. (E) Real-time qRT-PCR of APNG and MGMT. ΔCt values are reported by subtracting the Ct value for β-ACTIN housekeeping gene from that for APNG or MGMT. Pearson r2 values and P values are shown. (F) Chemiluminescent densitometric analysis of APNG and MGMT protein levels, normalized to housekeeping gene β-ACTIN. Pearson r2 values and P values are shown. *P < 0.05, **P < 0.01.
APNG confers resistance to TMZ.
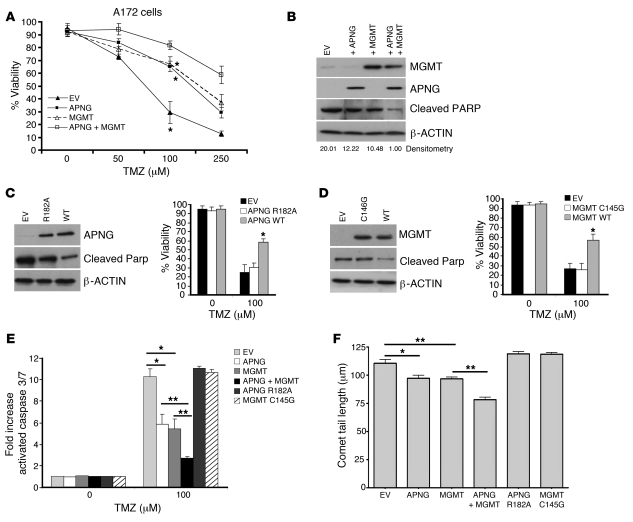
To determine whether APNG directly confers resistance to TMZ, A172 cells (lacking both APNG and MGMT expression; Figure 1A) were transfected with pcDNA3.1 constructs containing APNG or MGMT alone under the control of a CMV promoter, both APNG and MGMT combined, or empty vector (EV). Stable expression of APNG or MGMT did not alter cell viability compared with EV controls under non-TMZ conditions (P < 0.05, ANOVA; Figure 2A). A172 cells expressing APNG had increased cell viability in response to 100 μM TMZ (P = 0.0001; Figure 2, A and B), with similar increased viability of A172 cells expressing MGMT. There was a synergistic increase in viability in A172 cells expressing both APNG and MGMT (P < 0.05; Figure 2A). The increased viability due to APNG or MGMT expression was caused by decreased apoptosis, as measured by decreases in cleaved PARP (Figure 2B) and in activated caspases (i.e., caspase 3/7; Figure 2E).This was also supported by increased numbers of cells in the sub-G1 phase (DNA content less than 2N) of the cell cycle (Supplemental Figure 1A; supplemental material available online with this article; doi: 10.1172/JCI59334DS1).
Figure 2. APNG confers resistance to GBM cells in vitro.
(A) Cell viability of A172 cells expressing EV, MGMT, APNG, or both MGMT and APNG in response to varying amounts of TMZ. (B) Representative Western blot of A172 cells stably expressing APNG, MGMT, or both under 100 μM TMZ. Cleaved PARP was used as a measure of apoptosis. Densitometry values (cleaved PARP relative to β-ACTIN) are shown below. (C) Western blot and cell viability in response to TMZ of site-directed APNG mutant R182A compared with WT APNG. (D) Western blot and cell viability in response to TMZ of site-directed MGMT mutant C145G compared with WT MGMT. (E) Activated cleaved caspase 3/7 levels of A172 cells expressing APNG, MGMT, or both. (F) Comet tail assay measuring DNA damage of A172 cells exposed to 100 μM TMZ. Comet tail length, indicative of less DNA damage, was reduced by expression of APNG and/or MGMT, but not their corresponding catalytic dead mutants. *P < 0.05, **P < 0.01.
To confirm that TMZ resistance is the result of APNG and MGMT functional activity, catalytically dead mutants were created using site-directed mutagenesis. Transient expression of the APNG R182A mutant was unable to confer resistance to TMZ compared with WT APNG (P < 0.05), with viability comparable to that of EV controls (Figure 2C). Similar results were obtained with the MGMT C145G mutant (Figure 2D). Cells expressing catalytically dead APNG and MGMT mutants displayed increased apoptosis and cell death compared with their WT counterparts, as measured by activated caspases and increased levels of cleaved PARP (Figure 2, C–E). Furthermore, the comet assay, indicative of DNA damage, demonstrated that APNG- or MGMT-transfected A172 cells had reduced comet tail lengths compared with EV controls (P < 0.05; Figure 2F), or cells transfected with the catalytically dead mutants. In addition to A172 cells, transient expression of APNG in C6 and SKMG GBM cells (which express moderate or low levels of APNG) increased their resistance to TMZ, as evaluated by cell viability and activated caspase 3/7 levels (Supplemental Figure 1, B and C). In summary, APNG expression conferred TMZ resistance that was comparable to that conferred by MGMT expression, and there was additive resistance when both DNA repair proteins were expressed.
APNG loss sensitizes cells to TMZ.
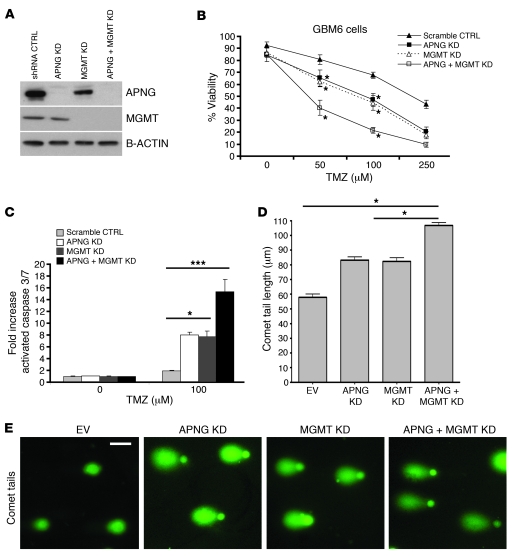
If gain of APNG function could confer resistance to TMZ in GBM cells, we argued that loss of APNG should cause sensitization. Primary GBM6 cells from serially passaged orthotopic xenografts retain the heterogeneity of GBMs and are known to be MGMT- and APNG-positive (26). shRNA gene-mediated silencing was used to generate pools of stable GBM6 cells with knockdown of APNG, MGMT, or both (Figure 3A). In untreated cells, there was no difference in viability between GBM6 cells with loss of APNG, MGMT, or both compared with GBM6 cells with control shRNA (Figure 3B). In comparison, TMZ-treated GBM6 cells expressing scrambled shRNA control were relatively resistant to TMZ, whereas single knockdown of APNG or MGMT conferred sensitivity to TMZ (P < 0.01; Figure 3B). Consistent with reduced cell viability, these cells had increased levels of activated caspase 3/7 as well as increased levels of DNA damage, as measured by the comet assay (P < 0.05; Figure 3, C–E). This and the significant increase of the number of cells in the sub-G1 population (Supplemental Figure 1D) collectively demonstrated increased cell death and apoptosis. Simultaneous downregulation of both APNG and MGMT led to further reduction in cell viability and increased levels of DNA damage and activated caspases (P < 0.05; Figure 3, C–E, and Supplemental Figure 1D). Furthermore, results similar to those in primary GBM6 cells were found in established T98G GBM cells after transient siRNA-mediated knockdown of APNG, MGMT, or both (Supplemental Figure 2, A and B).
Figure 3. APNG loss confers sensitivity to primary GBM6 cells.
(A) Western blot showing protein expression of pooled shRNA stables. KD, knockdown. (B) Cell viability of GBM6 cells expressing pooled stable shRNA of APNG, MGMT, or both. (C) Cleaved caspase assay of GBM6 cells with knockdown of APNG, MGMT, or both when exposed to TMZ. (D) Comet tail assay measuring DNA damage of GBM6 cells with knockdown of APNG, MGMT, or both when exposed to 100 μm TMZ. Tail lengths are denoted. Knockdown of either APNG or MGMT increased tail length; knockdown of both enzymes produced a synergistic effect. (E) Representative images of comet tails quantified in D. *P < 0.05, ***P < 0.001.
APNG modulation in vitro affects sensitivity to TMZ.
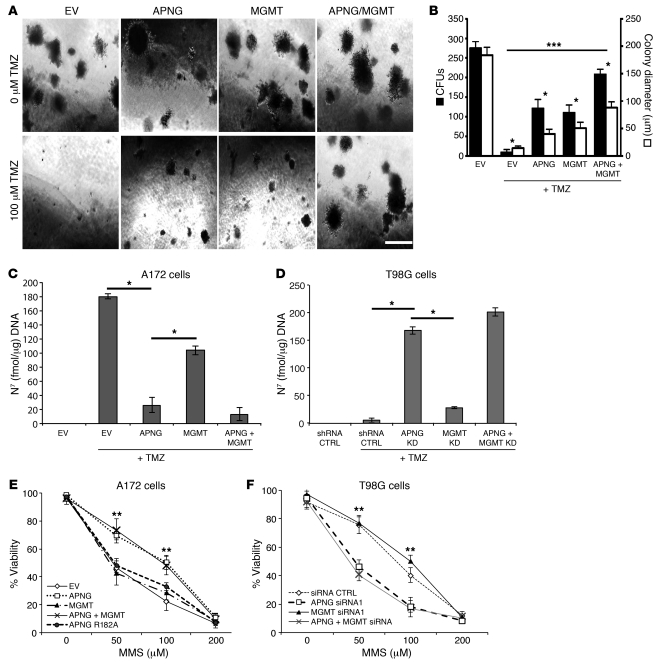
We next assessed the anchorage-independent growth of GBM cells using soft agar assays. A172 cells transfected with EV, APNG, MGMT, or APNG and MGMT combined grew robustly in the absence of TMZ, with no marked differences in colony number or size (P < 0.05, ANOVA; Figure 4, A and B). However, after TMZ treatment, A172 cells expressing EV had a significant reduction in colony formation compared with untreated A172 cells, whereas A172 cells expressing APNG had a partial rescue of colony number and colony size compared with treated A172 cells expressing EV (P < 0.05; Figure 4, A and B). A similar effect was seen with A172 cells expressing MGMT, whereas A172 cells expressing both APNG and MGMT had an additive increase in colony formation and colony size (P < 0.05; Figure 4, A and B).
Figure 4. APNG affects in vitro transformation.
(A) Anchorage-independent growth assay of A172 cells grown in soft agar with 100 μM TMZ. Scale bar: 100 μm. (B) Summary of anchorage-independent growth assay. Black bars, colony number; white bars, colony size. (C and D) Direct measurement of alkylated (methylated) N7-guanine DNA adducts in A172 and T98G cells. (E and F) Cell viability assay of A172 cells expression and T98G cells with knockdown of APNG, MGMT, or both, treated with varying concentrations of MMS. MMS generated N7-guanine and N3-adenine, but not O6-guanine, alkylated bases. *P < 0.05, **P < 0.01, ***P < 0.001.
To complement these A172 rescue experiments, we used the soft agar assay to assess the effect of shRNA-mediated knockdown of APNG, MGMT, or both in GBM6 lines (Figure 3B). In this case, in the absence of TMZ treatment, attenuation of APNG or MGMT expression again had little or no effect on colony formation, but simultaneous attenuation reduced colony formation and colony size. However, after TMZ treatment, colony formation and size were significantly decreased relative to the shRNA control by knockdown of APNG or MGMT and enhanced by concomitant knockdown of both DNA repair proteins (P < 0.01; Supplemental Figure 3, A and B).
APNG expression modulates repair of TMZ-induced DNA damage.
APNG removes the TMZ methylation products N7-meG and N3-meA from DNA (27–29). To confirm the functional activity of APNG, we directly quantified N7-meG in DNA extracted from TMZ-treated cells using an immunoblot method. A172 cells expressing APNG had a significant reduction in N7-meG levels compared with A172 cells expressing EV or MGMT (P < 0.01; Figure 4C). Simultaneous expression of APNG and MGMT did not reduce N7-meG levels more so than APNG expression alone (P < 0.05; Figure 4C). To conversely evaluate whether APNG loss increases N7-meG levels, siRNA was used to knock down APNG, MGMT, or both in T98G cells (which expressed both enzymes; Figure 1A). siRNA-mediated downregulation of APNG, but not MGMT, in T98G cells significantly increased N7-meG levels compared with T98G cells with shRNA control (P < 0.001), and knockdown of both APNG and MGMT had no additional effect (Figure 4D).
The cytoprotective effect of APNG was further examined using the methylating agent methylmethanesulfonate (MMS), which generates predominantly N7-meG and N3-meA, but very little O6-meG, in DNA (24, 30). A172 cells transfected with APNG or with both APNG and MGMT, but not with MGMT alone, showed increased resistance to MMS compared with EV cells (P < 0.05; Figure 4E). Expression of a catalytically dead APNG R182A mutant did not increase cell viability. Furthermore, in T98G and GBM6 cells, knockdown of APNG, but not MGMT, increased sensitivity to MMS (P < 0.05; Figure 4F and Supplemental Figure 2C). Treatment with MMS resulted in elevated levels of caspase 3/7 in A172 and T98G cells (Supplemental Figure 2, D and E), suggestive of increased apoptosis.
APNG confers resistance to TMZ in in vivo xenograft mouse models.
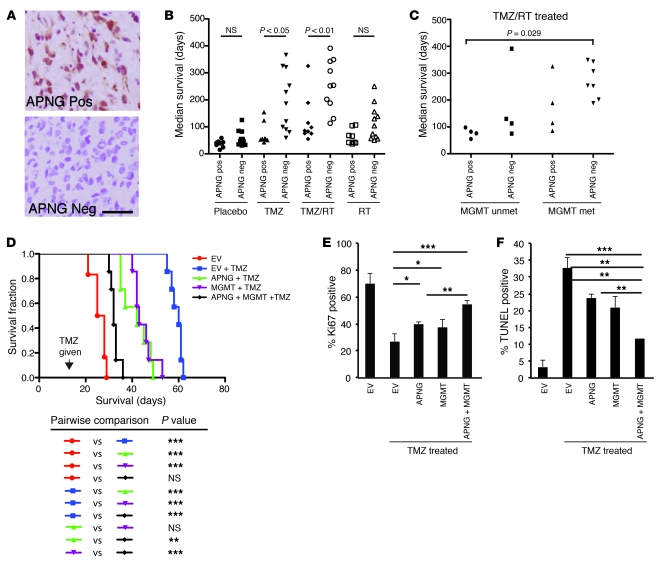
Having established modulation of TMZ sensitivity by either knockdown or reexpression of APNG, our next hypothesis was that APNG modulates the response to TMZ in vivo. Xenograft models of human GBM explants recapitulate several hallmarks of GBM, including proliferation and necrosis. We first analyzed the status of APNG in 19 such GBM explant xenograft models that were given human-equivalent treatments for GBM or none (placebo) (26, 31). A summary of treatment, overall survival, and number of mice per group is given in Supplemental Table 1. GBM xenograft models were identified as APNG-positive or -negative by immunohistochemistry (IHC; Figure 5A). When treated with TMZ alone or with concomitant TMZ and 20 Gy RT, median survival was significantly better in APNG-negative than APNG-positive GBM xenografts (P < 0.05; Figure 5B). Median survival of GBM xenografts did not vary with respect to APNG expression in the nontreated placebo group or the group receiving RT alone (Figure 5B). We next sought to determine whether APNG expression combined with MGMT promoter methylation status has an additive effect. When treated with TMZ and RT or with TMZ alone, APNG-negative and MGMT promoter–methylated (inferred as MGMT-negative) GBM xenografts had better median survival than did those that were APNG-positive and MGMT promoter–unmethylated (inferred as MGMT-positive) (Figure 5C and Supplemental Figure 4A). Median survival of GBM xenografts did not vary with respect to APNG and MGMT expression in the nontreated placebo group or in the group receiving RT alone (Supplemental Figure 4B).
Figure 5. Role of APNG and MGMT in GBM xenograft models.
(A) Representative IHC images of APNG-positive and -negative GBM explants. Scale bar: 50 μm. (B) Xenograft GBM explants were subjected to various modalities of treatment and compared within each treatment with respect to median overall survival, based on APNG IHC expression. (C) Xenograft GBM explants were subjected to TMZ and RT and compared within each treatment with respect to median overall survival, based on combined APNG IHC expression and MGMT promoter methylation. (D) Overall survival of A172 cells expressing EV, APNG, MGMT, or both APNG and MGMT injected into orthotopic (intracranial) xenograft models treated with TMZ. n = 7 (TMZ); 5 (untreated). (E and F) IHC quantification of Ki67 (E) and TUNEL (F) staining. 10 low-magnification fields per mouse were used, and 4 mice total per group assessed. Mean and SEM are shown. Data are mean and SEM. *P < 0.05, **P < 0.01, ***P < 0.001.
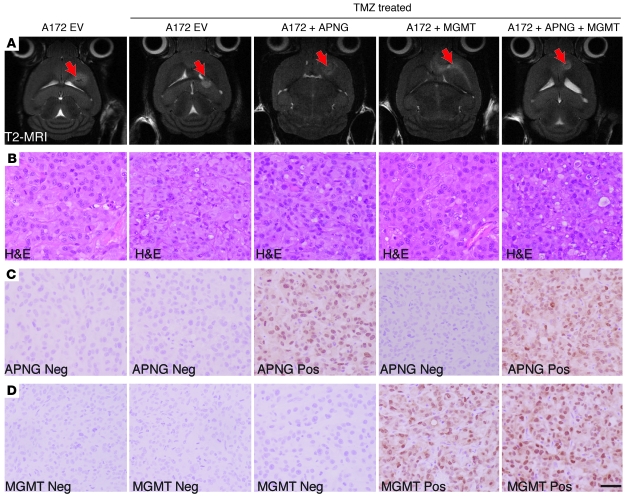
To control for cell line heterogeneity and directly determine the role of APNG in vivo, we performed isogenic experiments using our A172 cells from Figure 2. A172 cells expressing EV, APNG, MGMT, or both MGMT and APNG were injected intracranially into immunocompromised mice. Mice were given TMZ at 2 weeks at a human-equivalent single-day dose of 66 mg/kg for 2 days. Mice were confirmed to have tumors at 2 weeks by T2-weighted MRI imaging before being given TMZ (Figure 6A). Mice injected with A172 cells expressing control EV had the best overall survival when treated with TMZ. TMZ-treated xenograft models expressing both APNG and MGMT had significantly shorter overall survival than did TMZ-treated A172 EV xenograft models (P < 0.001; Figure 5D). Overall survival of TMZ-treated xenograft models expressing either APNG or MGMT was better than those expressing both APNG and MGMT, but significantly shorter than TMZ-treated A172 EV xenograft models (Figure 5D). In APNG- or MGMT-expressing A172 xenograft models treated with TMZ, overall survival was comparable (Figure 5D). We also performed IHC analysis to verify APNG and MGMT expression and representative H&E stain for tumor cytostructure (Figure 6, B–D). IHC analysis of Ki67, a marker for proliferation, demonstrated that compared with TMZ-injected A172 cells expressing EV, TMZ-injected A172 cells expressing either MGMT or APNG had a significant increase in proliferation, with an additive effect in cells expressing both APNG and MGMT (Figure 5E). Finally, TUNEL staining was undertaken to measure differences in apoptosis among several groups. Compared with TMZ-injected A172 cells expressing EV, TMZ-injected A172 cells expressing either MGMT or APNG had a significant decrease in TUNEL staining, with an additive effect in cells expressing both APNG and MGMT (Figure 5F). In summary, APNG and MGMT conferred resistance to TMZ-mediated death in vivo.
Figure 6. IHC analysis of APNG and other markers in xenograft models.
(A) T2 MRI images of mice 2 weeks after injection of cells. Arrow denotes location of tumor. (B–D) Representative staining of H&E (B), APNG (C), and MGMT (D). Scale bar: 50 μm.
APNG is epigenetically regulated.
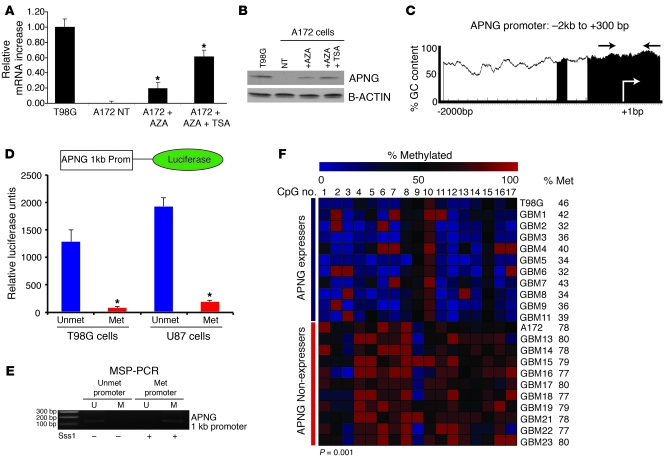
The levels of APNG protein expression varied among GBM cells and tumor biopsy specimens (Figure 1A and see below). Because MGMT can be epigenetically silenced in GBM, we hypothesized that loss of APNG expression may also be caused by promoter methylation. In support of this, treatment of A172 cells with 5-azacytidine (AZA) significantly upregulated APNG transcription, as measured by quantitative RT-PCR (qRT-PCR; Figure 7A). APNG mRNA expression was further enhanced approximately 3-fold using AZA combined with trichostatin A (TSA; Figure 7A), a histone deacetylase (HDAC) inhibitor also involved in epigenetic regulation. The restoration of APNG expression was also demonstrated at the protein level as measured by immunoblotting, with T98G cells used as a positive control (Figure 7B).
Figure 7. APNG is epigenetically regulated.
(A) qRT-PCR of A172 cells treated with AZA or with both AZA and TSA. (B) Western blot of experiments in A. (C) APNG promoter CpG island plot. Shaded regions indicate CpG islands of GC content greater than 50%. (D) The APNG promoter-luciferase construct (top) drove luciferase expression when unmethylated. (E) MSP-PCR of in vitro methylated APNG promoter showed complete methylation (only methylated [M] product detected) compared with nonmethylated APNG-luciferase vector (only unmethylated [U] product detected). (F) Bisulfite sequencing summary of 17 CpG sites of the promoter/intron 1 of the APNG gene locus. APNG-expressers had lower methylation at these sites (37% ± 5%) than did APNG-nonexpressers (77% ± 6%). *P < 0.05, **P < 0.01, ***P < 0.001.
Analyzing the proximal promoter of APNG (–2,000 to +300 bp) revealed CpG-dense islands close to the transcriptional start site (+1 bp; Figure 7C). To confirm that APNG promoter methylation silences gene expression, we cloned an approximately 1-kb fragment of the APNG promoter with the transcriptional start site into a luciferase reporter vector. Transfection of this construct into T98G and U87 cells resulted in substantial luciferase expression (Figure 7D). To silence the APNG promoter, it was treated in vitro with Sss1 (an enzyme that methylates cytosines at CpG dinucleotide repeats; Figure 7E), which ablated luciferase expression (P < 0.001; Figure 7D). To determine whether APNG promoter methylation is associated with APNG protein expression in human GBM biopsy samples, we undertook bisulfite sequencing of the 17 CpG sites in the promoter/intron 1 region of the APNG gene. Among 22 samples, APNG expressers (as evaluated by IHC) had significantly lower levels of methylation than did APNG nonexpressers (37% ± 5% vs. 77% ± 6%; P = 0.001; Figure 7F). These 17 CpG sites were also contained in our promoter luciferase construct (Figure 7F).
APNG confers poorer overall survival in GBM patients.
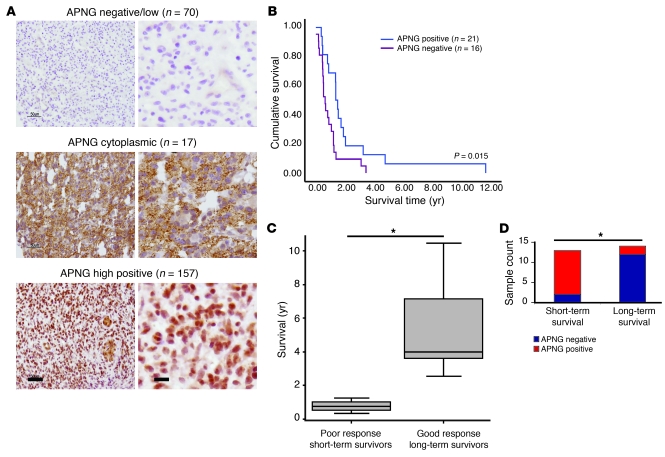
The results of the in vitro experiments raised the important question of whether APNG expression correlates with GBM patient survival. As demonstrated above, IHC analysis was an accurate method of determining APNG expression, with good correlation to APNG promoter methylation. Hence, we used IHC to determined expression of APNG in 244 GBM specimens in total over several sample sets. There was differential staining of APNG, with 29% (n = 70) of GBMs staining negative, 7% (n = 17) with cytoplasmic nonnuclear expression, and 64% (n = 157) with strong nuclear staining with or without cytoplasmic expression (Figure 8A). Given that APNG repairs DNA, only samples showing nuclear staining were scored as positive.
Figure 8. APNG predicts poorer overall survival in GBM patients.
(A) Representative APNG staining patterns of APNG-negative, APNG-cytoplasmic, and APNG-positive GBM patients. (B) Kaplan Meier survival curves of GBM patients from A (n = 37). 21 patients had strong nuclear expression and 16 were nonnuclear (14 negative and 2 cytoplasmic). (C) Difference between patients with good (>1 year; n = 14) and poor (<1 year; n = 13) overall survival. Both cohorts were MGMT-methylated. (D) APNG staining summary of good and poor overall survival groups. *P < 0.05.
Of the various clinical sample sets evaluated, one was a tissue microarray (TMA) of 37 GBM from patients who received the current standard of care (i.e., maximal safe surgery, then RT with TMZ) (32). A log-rank survival test showed that GBM patients scoring positive for APNG expression had poorer overall survival than did APNG-negative tumors (P = 0.015; Figure 8B). These data were also corroborated by RNA expression analysis from the Repository of Molecular Brain Neoplasia Data (REMBRANDT; http://rembrandt-db.nci.nih.gov/), in which high RNA expression of APNG conferred poor overall survival in glioma patients (Supplemental Figure 5A). Although MGMT promoter methylation is a well-established marker for predicting overall survival and response to TMZ, there is a significant proportion of GBM patients with MGMT promoter methylation that have short-term survival (<12 months) with the current standard of treatment (18). A second clinical group of 27 GBMs with methylated MGMT promoter, as determined by quantitative methylation-specific PCR (MSP-PCR) (33), was stratified into 2 groups: favorable response to treatment (i.e., good overall survival, >12 months) and poor response to treatment (i.e., poor overall survival, <12 months) (P = 0.001; Figure 8C). Analysis of APNG by IHC revealed significantly higher levels of APNG-positive specimens in the short-term survival group (P = 0.0004, Fisher exact test; Figure 8D). Age was not significantly different between the 2 groups (P = 0.15; Supplemental Figure 5B).
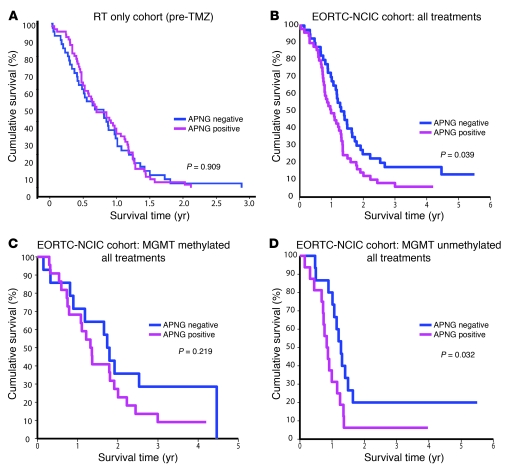
APNG expression was also evaluated on TMAs of 2 large independent GBM patient cohorts. The patients of the first were treated only with surgery and RT (pre-TMZ time) (19, 34). In this set, APNG expression bore no significant relationship to survival (P = 0.909; Figure 9A). The second TMA consisted of patient samples from the pivotal trial carried out by the European Organization for Research and Treatment of Cancer (EORTC) and National Cancer Institute of Canada (NCIC), which led to the current standard of treatment that includes TMZ (14) and also identified the predictive and prognostic value of MGMT promoter methylation (17). We first examined the expression of APNG in all patients with respect to all treatments (RT alone and TMZ plus RT). In this patient cohort, APNG positivity correlated with significantly poorer overall survival compared with APNG negativity (median survival, 12 vs. 16 months; P = 0.039, log-rank test; Figure 9B). In the MGMT-methylated subgroup with respect to all treatments, patients with negative APNG expression had better overall survival, but this did not reach statistical significance (Figure 9C). Stratification of MGMT-methylated patients into RT only or RT plus TMZ groups also yielded no significant effect of APNG expression (data not shown). However, in the MGMT-unmethylated group (associated with worse overall survival), APNG-negative patients had significantly better overall survival than did APNG-positive patients with respect to all treatments (P = 0.032; Figure 9D). Further stratification of the MGMT-unmethylated group revealed no significant difference with respect to APNG expression in those receiving RT only (Supplemental Figure 6B). Finally, in the MGMT-unmethylated group that received RT and TMZ, there was a trend toward increased overall survival in APNG-negative patient GBMs (P = 0.068; Supplemental Figure 6A). We next evaluated whether a MSP-PCR–based assay can also predict survival. Using MSP-PCR on a small subset of GBM samples for which we could compare MSP-PCR results with IHC in terms of survival (n = 24), we observed that IHC, but not MSP-PCR status of APNG, was able to predict overall survival (Supplemental Figure 7, A–C).
Figure 9. In large clinical trial datasets, APNG expression predicts poorer overall survival.
Patient samples were analyzed on TMAs by IHC for APNG expression. (A) There was no statistical difference in overall survival with respect to APNG expression in a GBM patient cohort treated with surgery and RT only (pre-TMZ era). (B) In patients treated within the EORTC-NCIC clinical trial, there was a statistically significant decrease in overall survival in patients whose tumor exhibited nuclear APNG. (C) In the EORTC-NCIC subgroup containing patients with a methylated MGMT promoter, APNG-positive patients had poorer survival than did their APNG-negative counterparts, but the difference was not significant. (D) In the EORTC-NCIC subgroup with an unmethylated MGMT, there was a significant decrease in overall survival in APNG-positive patients.
Discussion
The identification and characterization of novel genes and pathways involved in chemotherapeutic resistance in GBM has been of long-standing interest. Currently, TMZ is the only chemotherapy demonstrated to have some clinical efficacy in the treatment of GBM along with surgery and RT (14). Therefore, identifying novel resistance mechanisms to TMZ in GBM could provide attractive novel molecular targets for inhibitors. Here, our detailed mechanistic in vitro studies experiments, complemented by analysis of clinical datasets, demonstrated for the first time to our knowledge that APNG may be an important modulator of TMZ resistance in human GBMs.
TMZ methylates DNA bases at 12 different sites, of which O6-meG is known to be the most toxic lesion. MGMT repairs O6-meG by a suicidal reaction, thereby providing resistance to TMZ. However, TMZ to a greater extent yields N3-meA, also known to be toxic to cells, and this site is repaired exclusively by APNG (35). We found that APNG expression varied among human GBM cell lines, with most TMZ-resistant cell lines expressing both APNG and MGMT. We showed that siRNA-mediated downregulation of APNG or MGMT in established (T98G) or serially passaged primary GBM cells led to TMZ sensitivity, with increased apoptosis, reduced anchorage-independent growth, and increased markers of DNA damage (i.e., DNA fragmentation in the comet assay and increased numbers of AP sites). We further showed that in TMZ-sensitive GBM cells lacking APNG and MGMT expression, transfection of APNG conferred TMZ resistance. Expression of both APNG and MGMT led to an additive increase in survival. In contrast, site-directed mutants encoding catalytically inactive APNG and MGMT did not confer TMZ resistance.
The biochemical effects of APNG downregulation and transfection were corroborated by quantification of N7-meG, a substrate exclusively repaired by APNG, in DNA extracted from TMZ-treated cells. Similar modulations of APNG expression had comparable effects on cell sensitivity to MMS, a methylating agent that generates mainly N7-meG and N3-meA in DNA (24, 30). The prior literature, although scant, is consistent with our observations on APNG and GBM cells. Suppression of APNG expression in HeLa cells led to increased sensitization to TMZ, MMS, and BCNU (3). Furthermore, loss of APNG in GBM cells led to sensitization to methyl-lexitropsin, which predominantly produces N3-meA in DNA (2).
As is well established for MGMT, we found that methylation of the APNG promoter attenuated APNG expression. APNG-nonexpressing A172 cells treated with the demethylating agents AZA and TSA showed upregulation of APNG protein. Furthermore, APNG promoter luciferase reporters were silenced by in vitro CpG methylation. A point of interest is that, in contrast to APNG, MGMT promoter evaluation by MSP-PCR and IHC expression often do not correlate (36, 37). This has been speculated to be the result of several factors, including normal infiltrating cells in the tumor that stain positive for MGMT (38).
The magnitude of the effects we observed in the various GBM cell lines suggested that MGMT and APNG substrates contribute approximately equally to the overall toxicity of TMZ. This encouraged us to examine the extent to which APNG expression is of clinical relevance in GBM response to therapy that included TMZ. Analysis of APNG expression in GBM biopsies from several datasets demonstrated that patients with nuclear APNG staining had significantly poorer overall survival. In addition, in a subset of specimens available for analysis in the large randomized EORTC-NCIC clinical trial, overall survival was significantly higher when the GBM was negative for APNG. As anticipated, APNG was not predictive in a RT-only cohort, in line with the fact that RT leads primarily to DNA double strand breaks, which are not the target of MGMT or APNG repair. These data are further supported by our findings of in vivo mouse model xenograft studies of human GBM explants, in which explants expressing APNG and MGMT had the worst overall survival when treated with TMZ compared with explants expressing none.
Given that MGMT promoter methylation is currently the best predictor of overall survival in response to TMZ, we investigated APNG expression in relation to MGMT promoter methylation. The group of GBM patients with a methylated MGMT promoter and good survival also had a greater portion of APNG-negative tumors, which suggests that concomitant downregulation of both MGMT and APNG leads to better TMZ response. However, patients from the same group that were MGMT-methylated but who actually had a short survival (median <12 months) harbored a substantially higher number of APNG expressers (Figure 8, C and D), which indicates that APNG expression might be used to predict survival in this group. Our data lead us to propose that evaluation of APNG expression by IHC, which can be routinely undertaken by pathology labs, will be beneficial in allowing selection of those patients with a methylated MGMT promoter that more likely will benefit from TMZ therapy. However, it must be noted that this was a retrospective evaluation of APNG; future predictive and prospective studies would shed full light on APNG’s role in TMZ resistance and applicability as a prognostic marker.
Although APNG expression may be an attractive target for inhibition, overexpression of APNG — although promoting chemotherapeutic resistance — may also be used as the Achilles heel of GBM and other cancer cells. In studies involving breast, glioma, and ovarian cancer cell lines, APNG overexpression conferred sensitivity to alkylating agents, including TMZ. These results demonstrate that overexpression of DNA glycosylases can impair proper BER. The production of abasic sites from overexpression of APNG was then exploited using methoxyamine, which binds AP sites caused by APNG activity and makes them recalcitrant to subsequent repair with downstream BER enzymes (39). This stalling of the BER pathway by APNG overexpression and targeting downstream BER genes leads to accumulating single-strand DNA breaks, DNA damage, and apoptosis of these cancer cell lines when subjected to chemotherapeutics including TMZ (40–42). It would be of great interest to determine whether some GBM patients with extremely high levels of APNG have impaired BER and might respond better to TMZ with methoxyamine. In summary, we have demonstrated that APNG is a DNA repair enzyme, which, along with MGMT, is critical in modulating resistance to TMZ in GBM.
Methods
Further information can be found in Supplemental Methods; some protocols were as described previously (43–45).
Cell culture and treatments
A172 and T98G cell lines were obtained from ATCC. Cells were grown in DMEM containing 10% FBS at 37°C in a 95% air, 5% CO2 atmosphere. GBM6 xenograft lines were established from serially passaged mouse xenograft models as previously described (26). Cells were exposed to 0–250 μM TMZ (provided by Merck/Schering-Plough) or MMS (0–250 μM; catalog no. M4016, Sigma-Aldrich) for the times indicated.
Western blots
Cells were lysed with standard PLC lysis buffer containing protease and phosphatase inhibitors (Sigma-Aldrich). Protein concentration was determined using the bicinechoninic acid (BCA) assay (Pierce Chemical Co.). Lysates containing 30 μg total protein were loaded onto 10% or 12% SDS-PAGE gels and electrophoresed. Proteins were then transferred onto PVDF membranes (NEN Research Products) using a semidry transfer apparatus (Bio-Rad). Membranes were probed for varying proteins overnight in 5% nonfat milk. Antibodies were as follows: β-actin (1:10,000 dilution; catalog no. A2228, Sigma-Aldrich), MGMT (1:1,000 dilution; catalog no. 2739, Cell Signaling), APNG (1:500 dilution; catalog no. SC-101237, Santa Cruz), cleaved PARP (ASN214) (1:1,000 dilution; catalog no. 9546, Cell Signaling). After incubation, membranes were washed 3 times for 10 minutes with PBS with 0.1% Tween 20 and incubated with horseradish peroxidase–conjugated antibodies specific for the primary antibody (BioRad Laboratories). Binding was detected using Chemiluminescence Reagent Plus (PerkinElmer).
IHC and IF
Paraffin-embedded blocks were cut into 5-μm sections and were dewaxed in xylene followed by rehydration in a standard alcohol series. Antigen retrieval was achieved by pressure cooking for 20 minutes in citrate buffer (pH 6.0), followed by blocking of endogenous peroxidase in 0.3% H2O2.
The MGMT and APNG antibodies (each at 1:100 dilution) were added and incubated overnight at 4°C. Detection used biotinylated secondary antibodies for 30 minutes, the ABC reagent kit (Vector Labs), and DAB chromagen (Vector Labs). Sections were counterstained in hematoxylin (Fisher Scientific Inc.) for 30 seconds; dehydrated in 70%, 80%, and 100% ethanol; briefly washed in xylene; and mounted in Permount (Fisher Scientific Inc.). H&E sections were stained using standard protocols (Eosin Yellowish Solution, 1% w/v; Fisher Scientific Inc.). See Supplemental Methods for IF procedures.
Cell viability assay
A172 cells or GBM6 stable cells with varying expression of APNG or MGMT or EV control (1 × 105 cells) were plated into 6-well dishes in 2 ml DMEM containing 10% FBS. After incubation with or without TMZ, cells were collected and analyzed for cell death using trypan blue. Cells were counted using the Beckman Coulter Vi-CELL (12-Sample Carousel) Cell Viability Analyzer (Beckman Coulter).
Stable cell line generation
Full-length myc-tagged APNG and MGMT constructs in PCDNA 3.1 were obtained from SIDNET Resources of Hospital for Sick Children. APNG, MGMT, or EV control (3 μg) was transfected into A172 cells in 10-cm dishes using Fugene HD (3:1 ratio; Roche). After 48 hours, G418 was added to a final concentration of 400 μg/μl. Resistant clones were selected 7–14 days later, pooled, and assayed for APNG and MGMT. For dual expression, APNG-transfected and selected cells were cotransfected with MGMT and a linearized hygromycin marker (Clontech) at a 10:1 ratio. For selection, 40 μg/ml hygromycin was used. shRNA sets were obtained from Open Biosystems (APNG, catalog no. RHS4533-NM-001015052; MGMT, catalog no. RHS4533-NM_002412). Briefly, 3 μg APNG or MGMT (each 1 μg/construct) or 3 μg shRNA control vectors were transfected into GBM6 GBM cultures in 10-cm dishes using Fugene HD. After 48 hours, puromycin was added to a final concentration of 2 μg/ml. Resistant clones were selected 5 days later, pooled, and assayed for knockdown using immunoblotting.
DNA damage assays
Comet (single-cell gel electrophoresis) assays used the Trevigen method (catalog no. 4250-050-01, Trevigen). Abasic sites were quantified using the AP sites Colorimetric Assay Kit (catalog no. K253-25, Biovision Inc.).
Mouse xenograft studies
Isogenic studies.
Stereotactically guided intracranial injections in NOD-SCID mice were performed by injecting 450,000 A172 cells with EV or A172 cells expressing APNG, MGMT, or both. Injections were made into the frontal cortex (coordinates, x = –1.0, y = 1.5, z = 2.4; Bregma serving as the 0 point for x and y). At 2 weeks, mice were given TMZ by oral gavage (66 mg/kg daily for 2 days) and sacrificed upon signs of sickness. 1 mouse per group was subjected to a T2-weighted MRI at 2 weeks to confirm tumor take before delivery of TMZ.
Associative studies.
19 previously established human GBM explants were serially passaged in mice, and each group (n = 5–10) was given placebo treatment, TMZ alone (66 mg/kg daily for 5 days), RT alone (2 Gy twice daily for 5 days; total 20 Gy), or concomitant TMZ and RT (31). IHC analysis of mouse tumor xenografts (samples were on a TMA to allow for multiple tumor samples to be on the same slide) were undertaken to evaluate APNG expression. Groups were dichotomized as APNG-positive or -negative. APNG protein expression analysis with respect to treatment and overall survival was analyzed using the median survival of each xenograft group and compared using ANOVA for statistical significance.
Statistics
All experiments were performed in triplicate, with SEM reported where appropriate. Significance was determined using pairwise 2-tailed Student’s t test, ANOVA (multivariate analysis), Fisher exact test (categorical data), and log-rank/Mantel-Cox test (survival curves); a P value less than 0.05 was considered significant.
Study approval
This study was approved by the Hospital for Sick Children research ethics board under protocol no. REB 1000018702. Animal use protocols were approved by the animal care committee of University Health Network (Toronto, Ontario, Canada) under protocol nos. AUP 884.14 and AUP 1191.13. Normal human brain and GBM biopsy samples were obtained from the Institutional Research Ethics Board–approved Nervous System Tumour Bank at University Health Network. Additional GBM samples were collected from the research ethics board–approved centers of MD Anderson Cancer Center, UCLA, and Centre Hospitalier Universitaire Vaudois. The EORTC-NCIC TMA containing clinical data was obtained from the European Organization for Research and Treatment of Cancer (EORTC) with a material transfer agreement. All GBM samples were reviewed and verified by a minimum of 2 independent pathologists. All patient samples were obtained with informed consent under the various institutional study approvals.
Supplementary Material
Acknowledgments
S. Agnihotri was partially funded by Ontario Cancer Institute Research. This study was partially funded by Canadian Cancer Research Society (CCRS; formerly NCIC-Canada) and Canadian Institute of Health Resources (CIHR) grants to A. Guha. Work at the Paterson Institute for Cancer Research was supported by Cancer Research–UK and CHEMORES. We thank Kelly Burrel, Warren Foltz, and the Spatio-Temporal Targeting and Amplification of Radiation Response (STTARR) program and its affiliated funding agencies for MRI imaging.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J Clin Invest. 2012;122(1):253–266. doi:10.1172/JCI59334.
References
- 1.Reardon DA, Rich JN, Friedman HS, Bigner DD. Recent advances in the treatment of malignant astrocytoma. J Clin Oncol. 2006;24(8):1253–1265. doi: 10.1200/JCO.2005.04.5302. [DOI] [PubMed] [Google Scholar]
- 2.Bobola MS, et al. Human glioma cell sensitivity to the sequence-specific alkylating agent methyl-lexitropsin. Clin Cancer Res. 2007;13(2 pt 1):612–620. doi: 10.1158/1078-0432.CCR-06-1127. [DOI] [PubMed] [Google Scholar]
- 3.Paik J, Duncan T, Lindahl T, Sedgwick B. Sensitization of human carcinoma cells to alkylating agents by small interfering RNA suppression of 3-alkyladenine-DNA glycosylase. Cancer Res. 2005;65(22):10472–10477. doi: 10.1158/0008-5472.CAN-05-1495. [DOI] [PubMed] [Google Scholar]
- 4. Kleihues P, Cavanee WK.World Health Organization Classification of Tumors: Pathology and Genetic: Tumors of the Nervous System . New York, New York, USA; 2000. [Google Scholar]
- 5.Maher EA, et al. Marked genomic differences characterize primary and secondary glioblastoma subtypes and identify two distinct molecular and clinical secondary glioblastoma entities. Cancer Res. 2006;66(23):11502–11513. doi: 10.1158/0008-5472.CAN-06-2072. [DOI] [PubMed] [Google Scholar]
- 6.Chow LM, Baker SJ. PTEN function in normal and neoplastic growth. Cancer Lett. 2006;241(2):184–196. doi: 10.1016/j.canlet.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 7.Guha A, Mukherjee J. Advances in the biology of astrocytomas. Curr Opin Neurol. 2004;17(6):655–662. doi: 10.1097/00019052-200412000-00004. [DOI] [PubMed] [Google Scholar]
- 8.Shih AH, Holland EC. Developmental neurobiology and the origin of brain tumors. J Neurooncol. 2004;70(2):125–136. doi: 10.1007/s11060-004-2746-3. [DOI] [PubMed] [Google Scholar]
- 9.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321(5897):1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Agnihotri S, Wolf A, Picard D, Hawkins C, Guha A. GATA4 is a regulator of astrocyte cell proliferation and apoptosis in the human and murine central nervous system. Oncogene. 2009;28(34):3033–3046. doi: 10.1038/onc.2009.159. [DOI] [PubMed] [Google Scholar]
- 12.Kamnasaran D, Qian B, Hawkins C, Stanford WL, Guha A. GATA6 is an astrocytoma tumor suppressor gene identified by gene trapping of mouse glioma model. Proc Natl Acad Sci U S A. 2007;104(19):8053–8058. doi: 10.1073/pnas.0611669104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agnihotri S, et al. A GATA4-regulated tumor suppressor network represses formation of malignant human astrocytomas. J Exp Med. 2011;208(4):689–702. doi: 10.1084/jem.20102099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stupp R, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 15.Ochs K, Kaina B. Apoptosis induced by DNA damage O6-methylguanine is Bcl-2 and caspase-9/3 regulated and Fas/caspase-8 independent. Cancer Res. 2000;60(20):5815–5824. [PubMed] [Google Scholar]
- 16.Hegi ME, et al. Clinical trial substantiates the predictive value of O-6-methylguanine-DNA methyltransferase promoter methylation in glioblastoma patients treated with temozolomide. Clin Cancer Res. 2004;10(6):1871–1874. doi: 10.1158/1078-0432.CCR-03-0384. [DOI] [PubMed] [Google Scholar]
- 17.Hegi ME, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 18.Stupp R, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised phase III study:5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi: 10.1016/S1470-2045(09)70025-7. [DOI] [PubMed] [Google Scholar]
- 19.Murat A, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 20.Krex D, et al. Long-term survival with glioblastoma multiforme. Brain. 2007;130(pt 10):2596–2606. doi: 10.1093/brain/awm204. [DOI] [PubMed] [Google Scholar]
- 21.Weller M, et al. MGMT promoter methylation in malignant gliomas: ready for personalized medicine? Nat Rev Neurol. 2010;6(1):39–51. doi: 10.1038/nrneurol.2009.197. [DOI] [PubMed] [Google Scholar]
- 22.Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat Res. 2000;461(2):83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- 23.Engelward BP, Dreslin A, Christensen J, Huszar D, Kurahara C, Samson L. Repair-deficient 3-methyladenine DNA glycosylase homozygous mutant mouse cells have increased sensitivity to alkylation-induced chromosome damage and cell killing. EMBO J. 1996;15(4):945–952. [PMC free article] [PubMed] [Google Scholar]
- 24.Denny BJ, Wheelhouse RT, Stevens MF, Tsang LL, Slack JA. NMR and molecular modeling investigation of the mechanism of activation of the antitumor drug temozolomide and its interaction with DNA. Biochemistry. 1994;33(31):9045–9051. doi: 10.1021/bi00197a003. [DOI] [PubMed] [Google Scholar]
- 25.Hang B, Singer B, Margison GP, Elder RH. Targeted deletion of alkylpurine-DNA-N-glycosylase in mice eliminates repair of 1,N6-ethenoadenine and hypoxanthine but not of 3,N4-ethenocytosine or 8-oxoguanine. Proc Natl Acad Sci U S A. 1997;94(24):12869–12874. doi: 10.1073/pnas.94.24.12869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sarkaria JN, et al. Use of an orthotopic xenograft model for assessing the effect of epidermal growth factor receptor amplification on glioblastoma radiation response. Clin Cancer Res. 2006;12(7 pt 1):2264–2271. doi: 10.1158/1078-0432.CCR-05-2510. [DOI] [PubMed] [Google Scholar]
- 27.Smith SA, Engelward BP. In vivo repair of methylation damage in Aag 3-methyladenine DNA glycosylase null mouse cells. Nucleic Acids Res. 2000;28(17):3294–3300. doi: 10.1093/nar/28.17.3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomas L, Yang CH, Goldthwait DA. Two DNA glycosylases in Escherichia coli which release primarily 3-methyladenine. Biochemistry. 1982;21(6):1162–1169. doi: 10.1021/bi00535a009. [DOI] [PubMed] [Google Scholar]
- 29.Ye N, Holmquist GP, O’Connor TR. Heterogeneous repair of N-methylpurines at the nucleotide level in normal human cells. J Mol Biol. 1998;284(2):269–285. doi: 10.1006/jmbi.1998.2138. [DOI] [PubMed] [Google Scholar]
- 30.Newlands ES, Stevens MF, Wedge SR, Wheelhouse RT, Brock C. Temozolomide: a review of its discovery, chemical properties, pre-clinical development and clinical trials. Cancer Treat Rev. 1997;23(1):35–61. doi: 10.1016/S0305-7372(97)90019-0. [DOI] [PubMed] [Google Scholar]
- 31.Carlson BL, et al. Radiosensitizing effects of temozolomide observed in vivo only in a subset of O6-methylguanine-DNA methyltransferase methylated glioblastoma multiforme xenografts. Int J Radiat Oncol Biol Phys. 2009;75(1):212–219. doi: 10.1016/j.ijrobp.2009.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo D, et al. EGFR signaling through an Akt-SREBP-1-dependent, rapamycin-resistant pathway sensitizes glioblastomas to antilipogenic therapy. Sci Signal. 2009;2(68):ra82. doi: 10.1126/scisignal.2000446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rivera AL, et al. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12(2):116–121. doi: 10.1093/neuonc/nop020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Godard S, et al. Classification of human astrocytic gliomas on the basis of gene expression: a correlated group of genes with angiogenic activity emerges as a strong predictor of subtypes. Cancer Res. 2003;63(20):6613–6625. [PubMed] [Google Scholar]
- 35.Fronza G, Gold B. The biological effects of N3-methyladenine. J Cell Biochem. 2004;91(2):250–257. doi: 10.1002/jcb.10698. [DOI] [PubMed] [Google Scholar]
- 36.Brell M, Ibáñez J, Tortosa A. O6–Methylguanine–DNA methyltransferase protein expression by immunohistochemistry in brain and non–brain systemic tumours: systematic review and meta–analysis of correlation with methylation–specific polymerase chain reaction. BMC Cancer. 2011;11:35. doi: 10.1186/1471-2407-11-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karayan-Tapon L, et al. Prognostic value of O6-methylguanine-DNA methyltransferase status in glioblastoma patients, assessed by five different methods. J Neurooncol. 2010;97(3):311–322. doi: 10.1007/s11060-009-0031-1. [DOI] [PubMed] [Google Scholar]
- 38.Sciuscio D, et al. Extent and patterns of MGMT promoter methylation in glioblastoma- and respective glioblastoma-derived spheres. Clin Cancer Res. 2011;17(2):255–266. doi: 10.1158/1078-0432.CCR-10-1931. [DOI] [PubMed] [Google Scholar]
- 39.Liuzzi M, Talpaert-Borle M. A new approach to the study of the base-excision repair pathway using methoxyamine. J Biol Chem. 1985;260(9):5252–5258. [PubMed] [Google Scholar]
- 40.Fishel ML, He Y, Smith ML, Kelley MR. Manipulation of base excision repair to sensitize ovarian cancer cells to alkylating agent temozolomide. Clin Cancer Res. 2007;13(1):260–267. doi: 10.1158/1078-0432.CCR-06-1920. [DOI] [PubMed] [Google Scholar]
- 41.Rinne M, Caldwell D, Kelley MR. Transient adenoviral N-methylpurine DNA glycosylase overexpression imparts chemotherapeutic sensitivity to human breast cancer cells. Mol Cancer Ther. 2004;3(8):955–967. [PubMed] [Google Scholar]
- 42.Tang JB, et al. N-methylpurine DNA glycosylase and DNA polymerase {beta} modulate BER inhibitor potentiation of glioma cells to temozolomide. Neuro Oncol. 2011;13(5):471–486. doi: 10.1093/neuonc/nor011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Eads CA, Danenberg KD, Kawakami K, Saltz LB, Danenberg PV, Laird PW. CpG island hypermethylation in human colorectal tumors is not associated with DNA methyltransferase overexpression. Cancer Res. 1999;59(10):2302–2306. [PubMed] [Google Scholar]
- 44.Kamnasaran D, Guha A. Expression of GATA6 in the human and mouse central nervous system. Brain Res Dev Brain Res. 2005;160(1):90–95. doi: 10.1016/j.devbrainres.2005.07.012. [DOI] [PubMed] [Google Scholar]
- 45.Pegg AE. Repair of O(6)-alkylguanine by alkyltransferases. Mutat Res. 2000;462(2–3):83–100. doi: 10.1016/s1383-5742(00)00017-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.