Abstract
The Dinophysis genus is an ecologically and evolutionarily important group of marine dinoflagellates, yet their molecular phylogenetic positions and ecological characteristics such as trophic modes remain poorly understood. Here, a population of Dinophysis miles var. indica was sampled from South China Sea in March 2010. Nuclear ribosomal RNA gene (rDNA) SSU, ITS1-5.8S-ITS2 and LSU, mitochondrial genes encoding cytochrome B (cob) and cytochrome C oxidase subunit I (cox1), and plastid rDNA SSU were PCR amplified and sequenced. Phylogenetic analyses based on cob, cox1, and the nuclear rRNA regions showed that D. miles was closely related to D. tripos and D. caudata while distinct from D. acuminata. Along with morphology the LSU and ITS1-5.8S-ITS2 molecular data confirmed that this population was D. miles var. indica. Furthermore, the result demonstrated that ITS1-5.8S-ITS2 fragment was the most effective region to distinguish D. miles from other Dinophysis species. Three distinct types of plastid rDNA sequences were detected, belonging to plastids of a cryptophyte, a haptophyte, and a cyanobacterium, respectively. This is the first documentation of three photosynthetic entities associated with a Dinophysis species. While the cyanobacterial sequence likely represented an ectosymbiont of the D. miles cells, the detection of the cryptophyte and haptophyte plastid sequences indicates that the natural assemblage of D. miles likely retain more than one type of plastids from its prey algae for temporary use in photosynthesis. The result, together with recent findings of plastid types in other Dinophysis species, suggests that more systematic research is required to understand the complex nutritional physiology of this genus of dinoflagellates.
Introduction
The Dinophysis genus is an ecologically important group of dinoflagellates. Dinophysis spp. play dual roles in the marine ecosystems: as primary (photosynthetic) and secondary (heterotrophic) producers. Furthermore, many Dinophysis species are known to produce potent polyether toxins. For instance, D. caudata and D. miles have formed blooms and caused diarrhetic shellfish poisoning through accumulation of toxins in the green mussel [1]. Therefore, the genus Dinophysis is important in microbial food webs and for its potential influence on public health [2]. In addition, Dinophysis spp. have peculiar and unique morphologies that are not shared by any organisms outside the class of Dinophysiales, making this genus an interesting subject of evolutionary studies. However, until recently their phylogenetic position among dinoflagellates and their ecology such as trophic modes have remained poorly understood in most species due to the paucity of cultures or tools to study wild populations.
The genus Dinophysis has an obscure phylogenetic position among dinoflagellates. Using rRNA gene (rDNA) small subunit (SSU) and mitochondrial genes encoding cytochrome B (cob) and cytochrome C oxidase subunit I (cox1) and its mRNA editing patterns, a natural population of D. acuminata was placed phylogenetically between Gonyaulacales and Prorocentrales [3]. Recently, a sister kinship to Phalacoma was established for the genus Dinophysis [4], [5]. Dinophysioids have diverse trophic modes; some species are heterotrophic feeding on other algae [6], [7], whilst others have intracellular and extracellular cyanobionts and probably acquire carbon fixed by these symbionts. In Histioneis and Ornithocercus, the cyanobionts resides on the cingular lists [4], [8]–[11], whereas Amphisolenia [12], [13] and Sinophysis canaliculata cells [14] host the cyanobionts intracellularly. Typical Dinophysis spp. have been found to contain a plastid of cryptophyte origin [7], [15]–[17], in most cases Teleaulax-derived [2], although whether such uniformity in plastid acquisition is likely in other species and whether the plastids are kleptoplasts or permanent plastids have been debated [17]–[21]. Hackett et al. (2003) detected plastid rDNA sequences of a cryptophyte and a rhodophyte in D. acuminata and attributed the former to plastid and the latter to prey [22]. Meanwhile, D. mitra was found to harbor plastids of haptophyte origin [23].
The recent success in culturing D. acuminata [24] has greatly facilitated physiological, phylogenetic and molecular studies of the genus [25]–[27]. However, because the number of Dinophysis cultures is currently limited, work on many species still relies on natural populations. Work on natural populations not only broadens the range of species to be studied, but also can reveal in situ status of physiology and gene expression (e.g., [28]). A population of D. acuminata was isolated via flow cytometer from Narragansett Bay that enabled both the detection of mitochondrial mRNA editing in this species and its phylogenetic position based on nuclear rDNA SSU [3]. More phylogenetic studies have been conducted for natural populations from Florida embayments [4] and Indian Ocean [5]. rDNA LSU and SSU have been used to determine the relationship between the genera Phalacroma and Dinophysis [4]–[5], although their resolving power has yet to be demonstrated in some species in the Dinophysis genus. For instance, a study showed that rDNA LSU failed to distinguish D. miles from D. tripos, and D. odiosa [5]. To date, hardly any studies have been dedicated to D. miles, and the plastid type of this species remains undocumented. D. miles is recognized as variant D. miles var. schroeteri in Southeast Asia and D. miles var. indica in Indo-West Pacific [29], the latter widely distributed in the northeast area of South China Sea, such as Hainan island and Nansha islands waters [30]. In this study, we have investigated the phylogenetic position and plastid types of D. miles var. indica from South China Sea.
Materials and Methods
Sample collection
A phytoplankton sample was collected at 18°11.5′N, 119°27′E (latitude, longitude) near Sanya in the South China Sea with a 55-µm mesh plankton net in March, 2010. The towed sample was transferred into a 500-mL plastic container and preserved with neutral Lugol's solution [31]. The sample was stored in the laboratory in the dark until analysis (within 3 months).
Microscopic observations and cell sorting
Microscopic examination of the preserved phytoplankton sample revealed an abundant population of D. miles. The abundance of this species and other phytoplankton in the sample was determined using Sedgwick-Rafter chamber. Identification of the species was carried-out according to Steidinger (1997) and Wood (1963) [9] [32]. The abundance of this species in the natural environment was estimated by adjusting the cell concentration in the retrieved sample to the volume of water filtered in the net tow. Morphocytological features were examined both under Lugol's staining and after Lugol's stain was removed. To remove Lugol's stain, a subsample was centrifuged and supernatant discarded. The cell pellet was rinsed with 0.45-µm filtered seawater, followed by treatment with 10% (weight/volumn) sodium thiosulfate [33]. DNA was stained using SYBR Green I (35149A, Molecular probes, Invitrogen Corporation, Carlsbad, CA, USA) at 1∶10000 dilution at room temperature for 30 min [34]. DNA and pigment fluorescence was observed under an Olympus BX51 epifluorescence microscope. From the original Lugol's-preserved samples, colonies consisting of eight D. miles cells were isolated under the inverted microscope. The isolated cells were rinsed carefully with 0.45-µm filtered seawater for subsequent DNA extraction.
DNA extraction, PCR, and gene sequencing
Four eight-cell D. miles colonies were resuspended in 0.5 mL DNA lysis buffer (0.1 M EDTA pH 8.0, 1% SDS, 200 µg mL−1 proteinase K) and incubated for 48 hours at 55°C. DNA extraction followed a previously reported protocol [35]. Briefly, after incubation, NaCl was added to achieve 0.7 M, and CTAB was added to the final concentration of 1.7%. The lysate was then extracted in chloroform. After centrifugation, the supernatant was removed and DNA further purified using Zymo DNA Clean and Concentrator kit (Zymo Research Corp., Orange, CA). At last, DNA was eluted in 32 µl Tris-HCl solution so that each µl contained DNA from about 1 cell of D. miles.
Using 1 µl of the extracted DNA as the template, PCR reactions were carried out using a pair of dinoflagellate-specific rDNA SSU primers [31], a pair of rDNA primers extended from internal transcribed spacer (ITS) to LSU regions [4], [36], [37], a pair of cob primers [3], a pair of cox1 primers [3], and a pair of plastid rDNA SSU primers [38]. The sequences of the primers were as shown in Table 1. PCR cycles consisted of one initial cycle of denaturation at 94°C for 3 min followed by 35 cycles of at 94°C for 30 sec, 56°C for 30 sec, and 72°C for 45 sec, followed by 10 min at 72°C for the final extension. PCR products were resolved on an agarose gel electrophoretically and the specific DNA band was excised. DNA was recovered and purified using a Zymo DNA column and sequenced directly using BigDye sequencing kit. For the plastid rDNA SSU, direct sequencing of the PCR product indicated the presence of different sequences. Therefore, the purified PCR product was ligated, cloned, and multiple clones were sequenced on both strands of the DNA.
Table 1. Primers used in the present study.
| Primer name | Sequence (5′–3′) | References |
| Dino18SF1 | AAGGGTTGTGTTYATTAGNTACARAAC | Lin et al., 2006 |
| 18ScomR1 | CACCTACGGAAACCTTGTTACGAC | Zhang et al.,2005 |
| Dino1662 F | CCGATTGAGTGWTCCGGTGAATAA | Handy et al., 2008 |
| 25R | CTTGGTCCGTGTTTCAAGAC | Yamaguchi et al., 2005 |
| Dinocob1F | ATGAAATCTCATTTACAWWCATATCCTTGTCC | Zhang et al., 2008 |
| Dinocob2R | CGAGCATAAGATAKAAACWTCTCTTGAGG | Zhang et al., 2008 |
| DinocoxF | AAAAATTGTAATCATAAACGCTTAGG | Zhang et al., 2008 |
| DinocoxR | TGTTGAGCCACCTATAGTAAACATTA | Zhang et al., 2008 |
| CYA361f | GGAATTTTCCGCAATGGG | Martin et al., 2008 |
| CYA785r | GACTACWGGGGTATCTAATCC | Martin et al., 2008 |
Phylogenetic analyses
DNA sequences were trimmed of primers and the two strands were merged. The assembled sequences were analyzed using Basic Local Search Tool (BLAST) against databases in GenBank to determine what organisms these rDNA sequences represented. Sequences showing significant similarity in BLAST to the sequences obtained in this study were retrieved from the databases. Phylogenies based on partial SSU, ITS1-5.8S-ITS2, partial LSU (D1-D2, 700-bp; [4]), cob (334-bp), and cox1 (840-bp) regions were used to investigate the phylogenetic position of D. miles. Phylogenetic trees were also inferred from plastid rDNA SSU to analyze the plastid type in D. miles. These datasets were separately aligned using ClustalX. The alignments were run through ModelTest to select the most appropriate evolutionary model. The selected General Time Reversible (GTR) model with gamma distribution was employed for Maximum Likelihood analysis using PhyML3.0 aLRT [39]. Categories of substitution rates were set at 4, and other parameters were estimated based on the datasets. The proportion of invariable sites and gamma shape parameter were 0.464 and 0.583, respectively for the SSU dataset, 0.127 and 1.296 for ITS, 0.185 and 0.689 for LSU, 0.098 and 1.130 for cob, 0.000 and 0.725 for cox1, and 0.214 and 0.360 for plastid SSU.
Nucleotide sequence accession numbers
The sequences obtained in this study were deposited in GenBank under accession numbers JN982970-JN982975.
Results
Microscopic observations
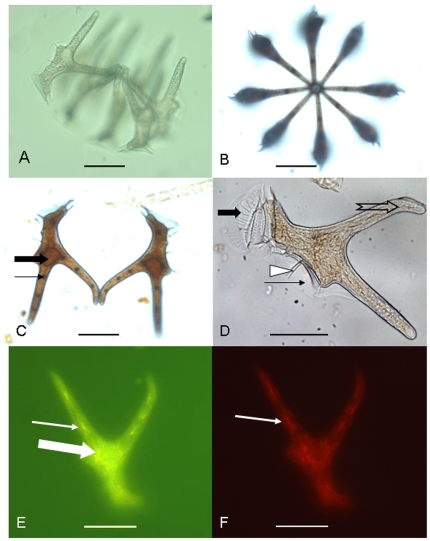
Microscopic examination confirmed that the isolated cells (Fig. 1) were morphologically identical to D. miles var. indica. The cells had two posterior projections that extended from the end of the hypotheca, which are characteristic of D. miles and D. tripos. In contrast to D. tripos, our sorted cells had slim cell bodies and the dorsal process was longer than that of D. tripos, plus the ends of the processes were smooth, which is typical of D. miles. The angle between the two projections was about 70°, matching that of D. miles var. indica [32]. The cell concentration ranged from 28 to 34 cells L−1. The size of D. miles cell was about 16–21 µm in width and 140–165 µm in length. Most of the cells were found in eight-cell colonies (Fig. 1A, B) except two-cell pairs in some cases (Fig. 1C). The eight cells formed a ring by attaching to each other at the end of the dorsal process of the cell (Fig. 1C), i.e. the process opposite to the sulcal list (Fig. 1D). In the cells of D. miles that were examined under the microscope, 5–10 plastids-like entities (n = 10) were observed, which showed dark staining of starch deposit by Lugol's solution (Fig. 1D), indicating plastids likely of cryptophyte origin. After removal of Lugol's stain followed by DNA staining using SYBR Green I, DNA fluorescence (Fig. 1E) and pigment autofluorescence (Fig. 1F) were apparent under the epifluorescence microscope.
Figure 1. Micrographs of Dinophysis miles collected in this study.
a) Side view of a 8-cell colony. b) Apical view of the 8-cell colony. c) Close-up view of two cells to show their attachment to each other at the end of the dorsal process, the visible nucleus (thick arrow), and the dark-stained plastid by Lugol's indicative of starch storage (thin arrow). d) A cell after Lugo's stain was removed, showing the anterior list (thick arrow), the sulcal list (thin arrow), and ribs (dashed arrow). e) Green fluorescence under blue light excitation of DNA stained with SYBR Green I in the nucleus (thick arrow) and plastid (thin arrow). f) Orange fluorescence from phycoerythrin in the plastids (arrow) under green excitation light. Scale bar = 50 µm in Fig. 1 A–F.
Phylogentic position of D. miles based on nuclear rDNA and mitochondrial cob and cox1
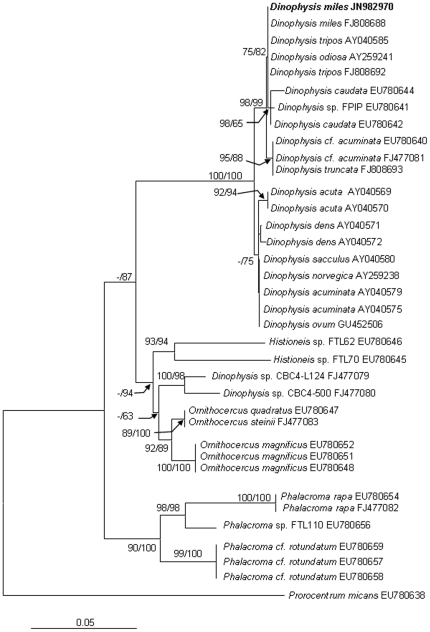
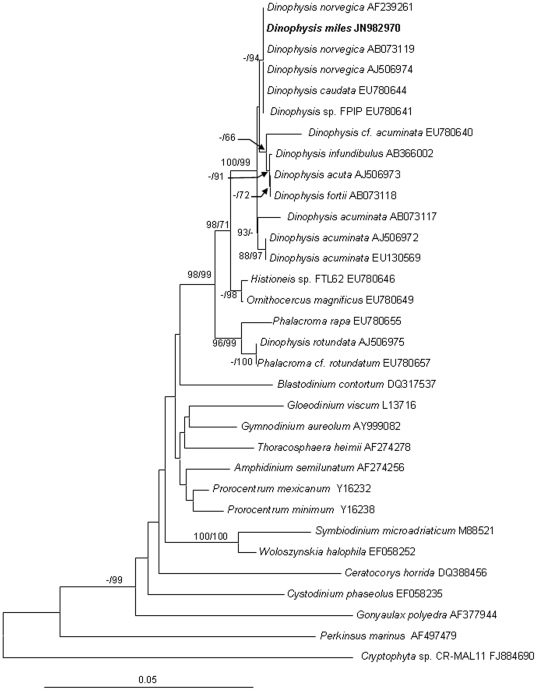
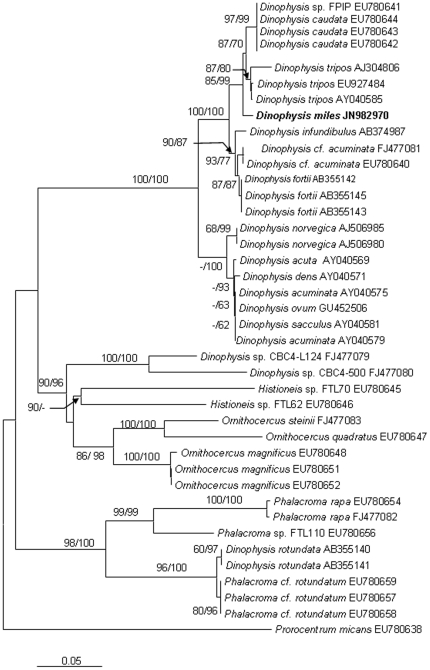
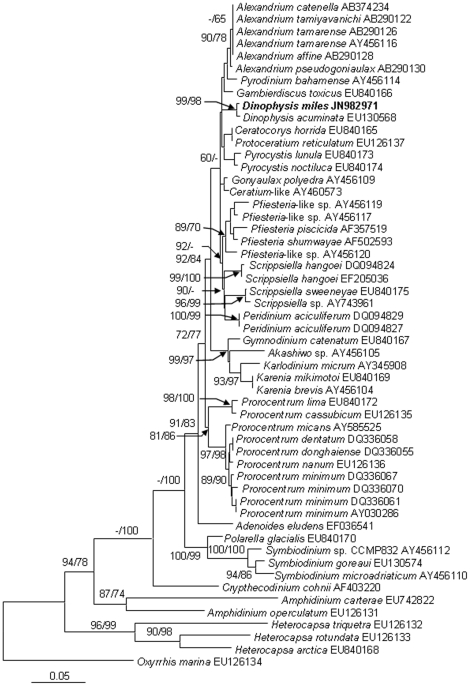
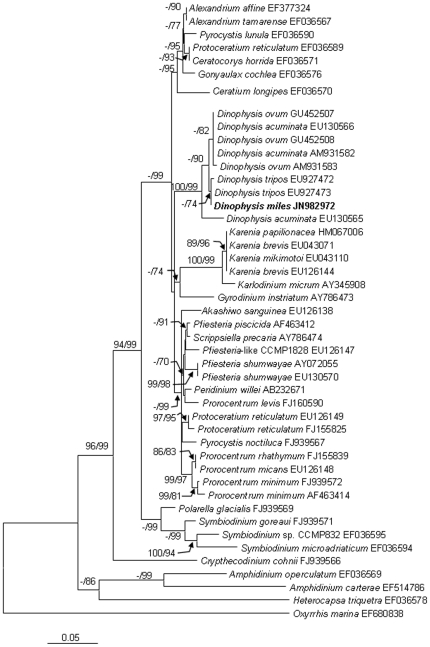
We obtained the nuclear-encoded ribosomal RNA sequence 2,824-bp (JN982970) from the sorted cells, composed of the partial sequence of SSU, ITS1, 5.8S, ITS2, and the partial sequence of LSU (D1–D2). Within the 2.824-kb sequence, the dinoflagellate SSU region spanned 1.59 kb (nucleotide positions 1–1593), the ITS1-5.8S-ITS2 region (abbreviated as ITS hereafter) 0.59 kb (positions 1557–2146), and the LSU region 0.68 kb (positions 2147–2824). The phylogenetic tree of SSU, ITS and LSU included 32, 40 and 36 sequences, respectively from Genbank, in addition to the sequences obtained in this study. The topologies of these trees inferred from the three datasets using Neighbor Joining (NJ) and Maximum Likelihood (ML) were similar and indicated clear separation of well-supported four genera, Phalacroma, Histioneis, Ornithocercus and Dinophysis (Figs. 2, 3, 4). In all three sets of trees, the genus of Dinophysis (such as D. acuminata and D. acuta) was distinct from other species. However, resolution of D. miles from other Dinophysis species varied among the three genes. In the LSU tree (Fig. 2), the South China Sea D. miles was identical to a sequence reported for D. miles from the Indian Ocean (FJ808688), but appeared to be identical also to D. tripos (FJ808692, AY040585) and D. odiosa (AY259241). Thus LSU was unable to resolve the three species. In the SSU tree (Fig. 3), D. miles could not be separated from D. caudata (EU780644) and D. norvegica (AF239261, AB073119, AJ506974). In contrast, ITS phylogeny placed D. miles as a distinct lineage, well separated from D. caudata (EU780642, EU780643, EU780644), D. tripos (AJ304806, EU927484, AY040585), and other Dinophysis species (Fig. 4). LSU and ITS results combined verified the morphological identification of the sorted cells as D. miles. Based on all the three sets of trees, D. miles appeared to be closely related to D. tripos and D. caudata.
Figure 2. Phylogenetic relationship of D. miles with other dinophysioid dinoflagellates inferred from LSU rDNA.
Sequence obtained in this study is bold-typed. Support of nodes is based on bootstrap values of ML/NJ with 1000 and 500 resamplings, respectively. Only values greater than 60 are shown. If only one of the two phylogenetic methods yielded significant support, the other is shown with “-”. Prorocentrum micans was used as the outgroup to root the tree. In this tree, D. miles cannot be separated from D. tripos and D. odiosa.
Figure 3. Phylogenetic relationship of D. miles with other dinoflagellates inferred from SSU rDNA.
Sequence obtained in this study is bold-typed. Support of nodes is based on bootstrap values of ML/NJ with 1000 and 500 resamplings, respectively. Only values greater than 60 are shown. If only one of the two phylogenetic methods yielded significant support, the other is shown with “-”. Cryptophyta sp. was used as the outgroup to root the tree. In this tree, D. miles cannot be separated from D. norvegica and D. caudata.
Figure 4. Phylogenetic relationship of D. miles with other dinophysioid dinoflagellates inferred from ITS1-5.8S-ITS2.
Sequence obtained in this study is bold-typed. Support of nodes is based on bootstrap values of ML/NJ with 1000 and 500 resamplings, respectively. Only values greater than 60 are shown. If only one of the two phylogenetic methods yielded significant support, the other is shown with “-”. Prorocentrum micans was used as the outgroup to root the tree. In this tree, D. miles appears as a distinct lineage, well separated from D. tripos, D. norvegica, D. caudata, and other Dinophysis species.
The alignment of cob consisted of the D. miles sequence obtained (JN982971) in the present study and 55 sequences from other dinoflagellates available in GenBank. The 913-bp cob sequence from D. miles var. indica differed by only 3 bp (0.33%) from that of D. acuminata (EU130568), the only Dinophysis cob sequence available in GenBank. The cox1 sequence obtained from D. miles var. indica (JN982972, 840-bp) contained the widely used DNA barcode region (∼650-bp) [40]. It was aligned with 46 homologous sequences from other dinoflagellates available in GenBank. The cox1 sequences from D. miles var. indica differed by only 3 or 4 bp (0.36% or 0.48%) from counterparts of D. ovum (AM931583, GU452507, GU452508), and also only 3 bp (0.36%) from a D. acuminata sequence (EU130566, mRNA sequence is EU130565), and 0 bp or only 1 bp (0.24%) from D. tripos sequences (EU927473, EU927472). Cob and cox1 molecular phylogenies showed that Dinophysis species formed strongly supported lineages (Fig. 5, 6).
Figure 5. Phylogenetic relationship of D. miles with other dinoflagellates inferred from cob.
Sequence obtained in this study is bold-typed. Support of nodes is based on bootstrap values of NJ/ML with 1000 and 500 resamplings, respectively. Only values greater than 60 are shown. If only one of the two phylogenetic methods yielded significant support, the other is shown with “-”. Oxyrrhis marina was used as the outgroup to root the tree. In this tree, D. miles is separated from D. acuminata, the only Dinophysis species whose cob sequence is available.
Figure 6. Phylogenetic relationship of D. miles with other dinoflagellates inferred from cox1.
Sequence obtained in this study is bold-typed. Support of nodes is based on bootstrap values of NJ/ML with 1000 and 500 resamplings, respectively. Only values greater than 60 are shown. If only one of the two phylogenetic methods yielded significant support, the other is shown with “-”. Oxyrrhis marina was used as the outgroup to root the tree. In this tree, D. miles is separated from D. ovum and D. acuminata.
Phylotypes of the plastid
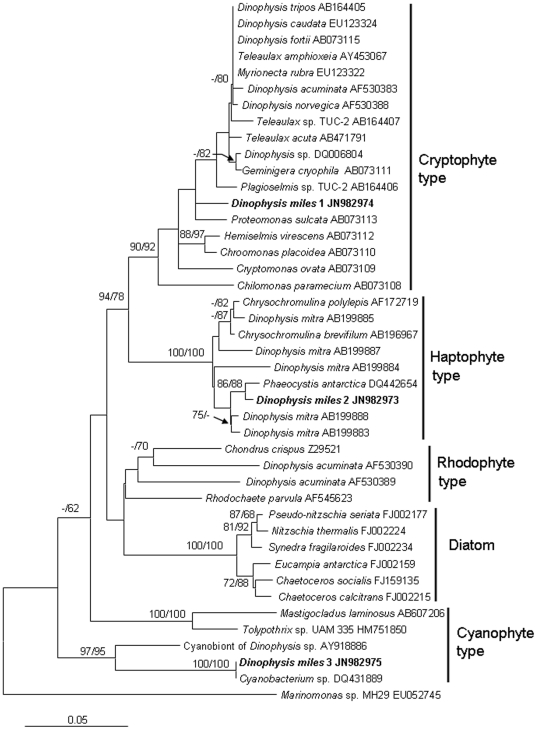
Sequencing results revealed three types of plastid SSU rDNA sequences (JN982973–JN982975) from colonies of D. miles var. indica. BLAST analyses of the 423-bp sequences indicated that they belonged to different lineages. One (JN982974) was 96% identical to the plastid SSU of the cryptophytes Teleaulax amphioxeia (AY453067) and Plagioselmis sp. TUC-2 (AB164407), one (JN982973) 98% identical to that of the haptophyte Phaeocystis antarctica (DQ442654) and the plastid SSU of D. mitra (AB199888), and the other (JN982975) 100% identical to that of an uncultured cyanobacterium (DQ431889) and 91% identical to that of the cyanobionts of Dinophysis sp. (AY918886). Phylogenetic analyses also showed that these D. miles var. indica sequences clustered with the plastid SSU of cryptophytes, haptophytes and cyanophytes, respectively (Fig. 7). Of these, the cryptophytes-type clade comprises cryptophytes and the majority of photosynthetic Dinophysis species; the haptophyte-type clade consists of haptophytes and several populations of D. mitra; the rhodophyte-type clade contains rhodophytes and D. acuminata; the cyanophyte-type clade is composed of cyanobacteria and Dinophysis sp.. While D. acuminata is represented in two (cryptophyte and rhodophyte) clades, only D. miles var. indica covers three clades.
Figure 7. Phylogram of plastid SSU rDNA showing diverse types of plastids and symbionts in D. miles.
Sequence obtained in this study is bold-typed. Support of nodes is based on bootstrap values of NJ/ML with 1000 and 500 resamplings, respectively. Only values greater than 60 are shown. If only one of the two phylogenetic methods yielded significant support, the other is shown with “-”. Marinomonas sp. was used as the outgroup to root the tree.
Discussion
Analyzing natural populations of a dinoflagellate species alleviates the barrier of lack of cultures to study the species. The culture-independent approach also is the only way to gain understanding on physiological and molecular genetic characteristics in the natural populations. As the first study dedicated to D. miles, we have sequenced SSU and ITS in D. miles var. indica, and analyzed Dinophysis phylogenies based on nuclear SSU-ITS-LSU and mitochondrial cob and cox1 to compare their performance in distinguishing different species within this genus. The sequences obtained and the results of phylogenetic analyses will be useful for future phylogenetic and DNA barcoding studies for this and related species. Further, analysis of plastid SSU on the natural population of D. miles reveals multiple plastids (and cyanobionts) associated with this species, a finding that would be difficult to obtain using laboratory cultures. Therefore, taking advantage of culture-independent molecular techniques, research on natural populations of dinoflagellates has the potential of yielding more information. This potentially can be applied to other protists that are amenable to single cell (colony) isolation, which is becoming increasingly feasible with the aid of flow cytometry (e.g., [41]). However, working directly on natural populations of protists is challenging because it is often difficult to isolate the target species from the plankton assemblage and it is prone to contamination by co-existing organisms. In our study, D. miles is relatively large in cell size, and hence relatively easy to isolate. Careful washing and microscopic examination further minimized the chance of contamination by other phytoplankton.
Comparison of phylogenies based on the three regions in the nuclear rDNA sequences and mitochondrial cob and cox1
Morphological observations augmented by molecular analyses indicate that the Dinophysis population we detected was D. miles var. indica. Molecular phylogenies indicate that nuclear SSU, ITS, LSU rDNA and mitochondrial cob and cox1 all have sufficient resolving power to discriminate genera in Dinophysiales. Our results showed that among these gene regions, the ITS region offered the best resolution between D. miles and other Dinophysis species. The phylogenies of the nuclear rDNA regions showed varying interspecific distances in the genus of Dinophysis. LSU fails to differentiate the morphologically similar species D. miles, D. tripos, as well as the morphologically more distinct D. odiosa, and SSU could not distinguish D. miles from D. norvegica and D. caudata. Handy et al. (2009) indicated that the nuclear-encoded ITS1 and ITS2 have undergone higher evolutionary rate than LSU and SSU rDNA regions based on a comparison of percent identity among Histioneis sp., Ornithocercus magnificus, and Dinophysis spp. relative to Phalacroma rapa [4].
In the cob phylogenic tree, D. miles is closely related to, but different from, D. acuminata among other dinoflagellates. The sequence we obtained embraced a 334-bp region, which has been demonstrated to be a promising DNA barcoding marker for dinoflagellate species [40]. This gene sequence exhibit only three nucleotide difference between D. miles and D. acuminata, two of which are located within the 334-bp region. The separation of these two species is consistent with the result based on rRNA genes, but the overall resolving power of this gene for Dinophysis species remains to be determined in further studies with broader taxon sampling.
In the cox1 phylogenic tree, D. miles is well resolved from D. acuminata and D. ovum although their distances were short. D. miles and D. ovum only differed by 3 or 4 bp (0.36% or 0.48%). D. miles differed from a previously reported D. acuminata sequence (EU130566) by 3 bp (0.36%) yet from another (AM931582) by 9 bp (1.07%). These two reported D. acuminata cox1 sequences showed a difference of 93 bp (7.74%), which is unprecedented and highly unlikely for any dinoflagellates. Raho et al. (2008) based on their sequence of D. acuminata (AM931582) concluded that the cox1 region had higher resolving power than ITS [42]. Our results show that this is not the case, casting question on the accuracy of that reported sequence. Careful comparison of AM931582 with EU130566 and counterpart sequences from other Dinophysis species showed that the apparent variable sites in AM931582 were mostly in the 3′ end, suggesting possibility of sequencing errors toward the end of read length. Alternatively, host of the AM931582 might have been a totally unrelated organism misindentified as D. acuminata. Furthermore, previously reported cox1 sequence from D. tripos (EU927473) was identical to the D. miles sequence (JN982971) obtained in this study. Unlikely, this gene would separate the two species so well.
Because ITS as a non-coding region has higher variability than the coding regions SSU and LSU, it is expected to have greater resolving power for all eukaryotes. The usefulness of ITS in resolving dinoflagellate species has been demonstrated [43]. Consistent with these findings, our results also showed that the ITS region separated D. miles from D. tripos, D. acuminata, and other Dinophysis species with strong bootstrap support (Fig. 4), indicating its greater resolving power for D. miles and related species. In contrast, as shown above, the SSU, LSU, and the two mitochondrial genes, overall show lower, albeit varying, levels of resolving power between Dinophysis species. Therefore, ITS1-5.8S-ITS2 region seems to be the most effective region to distinguish D. miles from other Dinophysis species among these five gene loci. In addition, based on all the current phylogenies inferred from the five gene loci, D. miles is closely related to D. tripos and D. caudata and more distant from D. acuminata.
“Plastid” consortium in D. miles
In this study, we retrieved three different types of plastid SSU rDNA sequences from D. miles var. indica. Based on the phylogenetic analyses of the plastid genes, two plastid sequences are of crytophyte and haptophyte origin, the third sequence is closely related to cyanobacterial SSU. These different plastid SSU sequences are unlikely to be a result of contamination. First, microscopic examination of our net tow samples showed predominance of diatoms (Chaetoceros, Rhizosolenia and other genera); any cryptophytes, haptophytes, or cyanobacteria cells present in the study ocean area would have been mostly lost through the 55-µm mesh during the net tow. Second, our picked cell colonies were extensively rinsed in filtered seawater before DNA extraction. Furthermore, cryptophyte and haptophyte plastids have both been demonstrated to be plastids in Dinophysis spp. and cyanobacteria have been reported to associate with some dinophysioids. Our microscopic observation on some of the cells we isolated revealed the intracellular plastid stained intensely with iodide, indicative of starch storage, and phycoerythrin-like fluorescence, indicating presence of cryptophyte type of plastid or cyanobacteria inside D. miles var. indica cells. Therefore, the D. miles var. indica population in the South China Sea likely possesses a consortium of plastids and cyanobionts previously documented separately in different dinophysioids species.
One of the plastid SSU sequences retrieved in our study is most closely related to that in Proteomonas sulcata. One the one hand, this agrees with the previous results that most of the Dinophysis species contain plastids originated from cryptophytes [16], [18], [22] (Table 2); on the other hand, this distinguishes D. miles from most of Dinophysis spp. which have plastids originating from a different cryptophyte [15], [16], [19]. The second plastid SSU sequence found from D. miles var. indica is of haptophyte origin, similar to D. mitra from Okkirai Bay, Japan [23] (Table 2). Intriguingly, the D. mitra population harbors plastids of different haptophyte lineages, including those closely related to Phaeocystis and Chrysochromulina, respectively, suggesting that these are kleptoplastids retained from prey algae, in contrast to the more controversial status of cryptophyte-derived plastids in other Dinophysis species. The haptophyte-type plastid of D. miles var. indica is most closely related to plastids of Phaeocystis antarctica (Fig. 7). Interestingly, Gast et al. (2007) showed that a haptophyte alga closely related to Phaeocystis antarctica was grazed by a dinoflagellate in the Ross Sea, Antarctica, and its plastid was retained in the dinoflagellate cell for temporary photosynthesis [44]. This suggests that grazing and retention of haptophyte plastids by dinoflagellates occur in both polar and tropical waters, and are likely a widespread phenomenon in dinoflagellates.
Table 2. Types of plastids found in Dinophysis spp.
| Source | Study sites | Cryptophyte origin | Rhodophyte origin | Haptophyte origin | References |
| D. norvegica | Baltic Sea; Okkirai Bay and Funka Bay, Japan; Clam Cove, Maine; Masfjord and North Sea | Y | Carpenter et al., 1995; Takahashi et al., 2002, 2005; Hackett et al., 2003; Minnhagen and Janson, 2005 | ||
| D. tripos | Okkirai Bay and Funka Bay, Japan | Y | Takahashi et al., 2005; Nishitani et al., 2010 | ||
| D. caudata | Near Namhae, Korea; Yatsushiro Sea, Japan | Y | Park et al., 2008; Nishitani et al., 2010 | ||
| D. infundibulus | Funka Bay, Japan | Y | Nishitani et al.,2010 | ||
| D. fortii | Okkirai Bay, Hiroshima Bay, Yatsushiro Sea and Notoro saline lake, Japan | Y | Takahashi et al., 2002, 2005; Nishitani et al., 2010 | ||
| D. acuta | Ninigret Pond, USA | Y | Hackett et al., 2003 | ||
| D. acuminata | Kesennuma Bay, Yatsushiro Sea, Funka Bay and Okkirai Bay, Japan; Greenwich Cove and Watch Hill Cove, Rhode Island; Near Frederikssund, Denmark; Masfjord and Baltic Sea | Y | Takishita et al., 2002; Hackett et al., 2003; Nishitani et al., 2010; Garcia-Cuetos et al., 2010; Minnhagen and Janson, 2005 | ||
| D. acuminata | Greenwich Cove, Rhode Island | Y | Y | Hackett et al., 2003 | |
| D. mitra | Okkirai Bay, Japan | Y | Koike et al., 2005 | ||
| D. miles | South China Sea | Y | Y | This study |
The third plastid-like SSU sequence from D. miles var. indica belongs to the lineage of cyanobacteria. While cyanobacteria have been shown to be endosymbionts of some dinophysioid species [12]–[14], most cyanobacterial associations are believed to behave as extracellular symbionts (cyanobionts). Cyanobionts occur in three genera of Dinophysiaceae, Citharistes, Histioneis, and Ornithocercus and our finding extends that to the genus of Dinophysis [4], [8]–[10]. It was thought that the lists that develop from extended cingulum and sulcus provide a habitat for the cyanobionts in some dinophysioids [4], [45]–[47]. Histioneis and Ornithocercus possess prominent lists on the epicone or cingulum for the ectophytic cyanobionts to reside in [13]. It was postulated that in Phalacroma and Dinophysis both the cingular and sulcal lists are not so elaborate and as a result no cyanobionts occur on them [4], [46], [47]. It is unclear if the cyanobionts detected in D. miles are endosymbiotic or ectosymbiotic. Our microscopic observations showed that D. miles cells had a well-developed anterior cingular list, sulcal list and rib systems (Fig. 1 C, D), suggesting that it is suited for cyanobionts to inhabit. Handy et al. (2009) showed, based on SSU phylogeny, that Histioneis and Ornithocercus cluster together and both have cyanobionts; in contrast, Dinophysis and Phalacroma were separated from those two genera and did not have cyanobionts [4]. However, in our nuclear SSU, ITS, and LSU phylogenetic trees, Dinophysis, Histioneis, and Ornithocercus consistently clustered together, and the clade was distinct from Phalacroma. Citharistes was not included in our analyses due to the unavailability of SSU and ITS sequences and its phylogenetic relationship with those lineages could not be confirmed. Nevertheless, our nuclear rDNA phylogenetic analysis results consistently show that Dinophysis as well as Histoneis and Orthithocercus can host cyanobionts. It is noteworthy that our detected cyanobacterial sequence is 91% identical to recently reported cyanobionts of Dinophysis sp. cells [48].
Three plastid-types suggest a possibility that D. miles has cryptic species that acquire different types of plastids. They can also be indication that Dinophysis nutritional physiology is more complicated than currently understood. The cryptophyte-type plastid seems to be the most common among Dinophysis spp., although whether it is a permanent or temporary (kleptoplastid) plastid is still being debated [17]–[19], [21]. The only exception is in D. mitra, if verified by further research. The different type of cryptophytes found in D. miles var. indica suggest that the cryptophyte plastid is probably not a permanent and universal plastid for the genus of Dinophysis. The failure to detect plastid-maintaining gene transcripts in D. acuminata [21] further supports the case for kleptoplastidy. The more variable and spotty presence of haptophyte (D. mitra, D. miles), rhodophyte (D. acuminata, Table 2), and cyanobacteria (Dinophysis sp., D. miles) most likely indicate the availability and the selection (if any) in the environment by the different Dinophysis species. This remains a question that can be answered only by systematic investigation on Dinophysis species and their sympatric phytoplankton assemblages in the natural environments. Further studies are also needed to determine whether all these photosynthetic entities are present in every single D. miles cell in the population, and whether they all are functional for photosynthesis and benefit the growth of the D. miles var. indica population.
Acknowledgments
We thank Dr. Huan Zhang and Ms. Yunyun Zhuang from the Department of Marine Sciences, University of Connecticut, for their technical assistance in our work, and the two anonymous reviewers for their valuable comments that led to significant improvement of the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This research is supported by the National Science Foundation of China-Overseas Collaboration Fund (41129001, 40828006) (http://www.nsfc.gov.cn/), Chinese Academy of Sciences/SAFEA International Partnership Program for Creative Research Teams grants KZCX2-YW-T001 (http://www.cas.cn/), the National Science Foundation of China (41006067, 41076096) (http://www.nsfc.gov.cn/) and the US National Science Foundation “Assembly of a Tree of Life” grant EF -0629624 (http://www.nsf.gov/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of manuscript.
References
- 1.Marasigan AN, Sato S, Fukuyo Y, Kodama M. Accumulation of a high level of diarrhetic shellfish toxins in the green mussel Perna viridis during a bloom of Dinophysis caudata and Dinophysis miles in Saipan Bay, Panay Island, the Philippines. Fisheries Science. 2001;67:994–996. [Google Scholar]
- 2.Nishitani G, Nagai S, Takano Y, Baba K, Kiyokawa S, et al. High-Level Congruence of Myrionecta rubra Prey and Dinophysis Species Plastid Identities as Revealed by Genetic Analyses of Isolates from Japanese Coastal Waters. Appl Environ Microbiol. 2010;76:2791–2798. doi: 10.1128/AEM.02566-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang H, Bhattacharya D, Maranda L, Lin S. Mitochondrial cob and cox1 and their mRNA editing in Dinophysis acuminata from Narragansett Bay, with special reference to the phylogenetic position of Dinophysis. Appl Envion Microbiol. 2008;74:1546–1554. doi: 10.1128/AEM.02103-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Handy SM, Bachvaroff TR, Timme RE, Coats DW, Kim S, et al. Phylogeny of four dinophysiacean genera (Dinophyceae, Dinophysiales) based on rDNA sequences from single cells and environmental samples. J Phycol. 2009;45:1163–1173. doi: 10.1111/j.1529-8817.2009.00738.x. [DOI] [PubMed] [Google Scholar]
- 5.Jensen MH, Daugbjerg N. Molecular phylogeny of selected species of the order Dinophysiales (Dinophyceae): Testing the hypothesis of a Dinophysioid radiation. J Phycol. 2009;45:1136–1152. doi: 10.1111/j.1529-8817.2009.00741.x. [DOI] [PubMed] [Google Scholar]
- 6.Lessard EJ, Swift E. Dinoflagellates from the North Atlantic classified as phototrophic or heterotrophic by epifluorescence microscopy. J Plankton Res. 1986;8:1209–1215. [Google Scholar]
- 7.Carpenter EJ, Janson S, Boje R, Pollehne F, Chang J. The dinoflagellate Dinophysis norvegica: biological and ecological observations in the Baltic Sea. Europ J Phycol. 1995;30:1–9. [Google Scholar]
- 8.Taylor FJR. Topography of cell-division in structurally complex dinoflagellate genus Ornithocercus. J Phycol. 1973;9:1–10. [Google Scholar]
- 9.Steidinger KA. Tomas CR, editor. Dinoflagellates. Identifying Marine Phytoplankton. 1997. pp. 387–584. Academic Press, Oxford, UK.
- 10.Tarangkoon W, Hansen G, Hansen PJ. Dinoflagellate cyanobacteria consortia in the tropical Indian ocean and the north west Australian Sea. J Phycol. 2007;43:38. [Google Scholar]
- 11.Kataoka T, Suzuki K, Hayakawa M, Higashi S, Tsuda A. Temporal changes in community composition of heterotorophic bacteria during an in situ iron enrichment experiment in the western subarctic Pacific (SEEDS-II). Deep Sea Res Part II. 2009;56:2779–2787. [Google Scholar]
- 12.Lucas IAN. Symbionts of the tropical Dinophysiales (Dinophyceae). Ophelia. 1991;33:213–224. [Google Scholar]
- 13.Fensome RA, Taylor FJR, Norris G, Sargeant WAS, Wharton DI, et al. A Classification of Living and Fossil Dinoflagellates. 1993. 355 Micropaleontology Special Publication No. 7. Sheridan Press, Hanover.
- 14.Escalera L, Reguera B, Takishita K, Yoshimatsu S, Koike K. Cyanobacterial endosymbionts in the benthic dinoflagellate Sinophysis canaliculata (Dinophysiales, Dinophyceae). Protist. 2011;162:304–314. doi: 10.1016/j.protis.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 15.Janson S. Molecular evidence that plastids in the toxin-producing dinoflagellate genus Dinophysis originate from the free-living cryptophyte Teleaulax amphioxeia. Environ Microbiol. 2004;6:1102–1106. doi: 10.1111/j.1462-2920.2004.00646.x. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi Y, Takishita K, Koike K, Maruyama T, Nakayama T, et al. Development of molecular probes for Dinophysis (Dinophyceae) plastid: a tool to predict blooming and explore plastid origin. Mar Biotechnol. 2005;7:95–103. doi: 10.1007/s10126-004-0482-5. [DOI] [PubMed] [Google Scholar]
- 17.Garcia-Cuetos L, Moestrup Ø, Hansen P, Daugbjerg N. The toxic dinoflagellate Dinophysis acuminata harbors permanent chloroplasts of cryptomonad origin, not kleptochloroplasts. Harmful Algae. 2009;8:196–211. [Google Scholar]
- 18.Takishita K, Kolke K, Maruyama T, Ogata T. Molecular evidence for plastid robbery (kleptoplastidy) in Dinophysis, a dinoflagellate causing diarrhetic shellfish poisoning. Protist. 2002;153:293–302. doi: 10.1078/1434-4610-00106. [DOI] [PubMed] [Google Scholar]
- 19.Minnhagen S, Janson S. Genetic analyses of Dinophysis spp. Support 463 kleptoplastidy. FEMS Microbiol Ecol. 2006;57:47–54. doi: 10.1111/j.1574-6941.2006.00096.x. [DOI] [PubMed] [Google Scholar]
- 20.Park MG, Park JS, Kim M, Yih W. Plastid dynamics during survival of Dinophysis caudata without its ciliate prey. J Phycol. 2008;44:1154–1163. doi: 10.1111/j.1529-8817.2008.00579.x. [DOI] [PubMed] [Google Scholar]
- 21.Wisecaver JH, Hackett JD. Transcriptome analysis reveals nuclear-encoded proteins for the maintenance of temporary plastids in the dinoflagellate Dinophysis acuminata. BMC Genomics. 2010;11:366. doi: 10.1186/1471-2164-11-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hackett JD, Maranda L, Yoon HS, Bhattacharya D. Phylogenetic evidence for the cryptophyte origin of the plastid of Dinophysis (Dinophysiales, Dinophyceae). J Phycol. 2003;39:440–448. [Google Scholar]
- 23.Koike K, Sekiguchi H, Kobiyama A, Takishita K, Kawachi M, et al. A novel type of kleptoplastidy in Dinophysis (Dinophceae): presence of haptophyte-type plastid in Dinophysis mitra. Protist. 2005;156:225–237. doi: 10.1016/j.protis.2005.04.002. [DOI] [PubMed] [Google Scholar]
- 24.Park MG, Kim S, Kim HS, Myung G, Kang YG, et al. First successful culture of the marine dinoflagellate Dinophysis acuminata. Aquat Microb Ecol. 2006;45:101–106. [Google Scholar]
- 25.Nagai S, Nishitani G, Tomaru Y, Sakiyama S, Kamiyama T. Predation on the ciliate Myrionecta rubra by the toxic dinoflagellate Dinophysis fortii and observation of sequestration of ciliate chloroplasts. J Phycol. 2008;44:909–922. doi: 10.1111/j.1529-8817.2008.00544.x. [DOI] [PubMed] [Google Scholar]
- 26.Nishitani G, Nagai S, Takano Y, Sakiyama S, Baba K, et al. Growth characteristics and phylogenetic analysis of the marine dinoflagellate Dinophysis infundibulus (Dinophyceae). Aquat Microb Ecol. 2008;52:209–221. [Google Scholar]
- 27.Hackett JD, Tong M, Kulis DM, Fux E, Hess P, et al. DSP toxin production de novo in cultures of Dinophysis acuminata (Dinophyceae) from North America. Harmful Algae. 2009;8:873–879. [Google Scholar]
- 28.Lin S, Zhang H, Zhuang Y, Bao T, Gill J. Spliced leader-based metatranscriptomic analyses lead to recognition of hidden genomic features in dinoflagellates. Proc Natl Acad Sci USA. 2010;107:20033–20038. doi: 10.1073/pnas.1007246107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Taylor FJR, Hoppenrath M, Saldarriaga JF. Dinoflagellate diversity and distribution. Biodivers Conserv. 2008;17:407–418. [Google Scholar]
- 30.Lin Y, Zhou J. Dinoflagellates in the South China Sea I. Science Press, Beijing, China. 1993. pp. 30–31. (In Chinese with English subtitle)
- 31.Zhang H, Lin S. Detection and quantification of Pfiesteria piscicida by using the mitochondrial cytochrome b gene. Appl Environ Microbiol. 2002;68:989–994. doi: 10.1128/AEM.68.2.989-994.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wood EJF. Dinoflagellates in the Australian region. Division of Fisheries and Oceanography. Common wealth Scientific and Industrial Research Organization. Australia. 1963:193–203. [Google Scholar]
- 33.Morey-Gaines G. Gymnodinium catenatum Graham (Dinophyceae): morphology and affinities with armoured forms. Phycologia. 1982;21:154–163. [Google Scholar]
- 34.Marie D, Partensky F, Jacquet S, Vaulot D. Enumeration and cell cycle analysis of natural populations of marine picoplankton by flow cytometry using the nucleic acid stain SYBR Green I. Appl Environ Microbiol. 1997;63:186–193. doi: 10.1128/aem.63.1.186-193.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang H, Lin S. Development of a cob-18S rDNA Real-Time PCR assay for quantifying Pfiesteria shumwayae in the natural environment. Appl Environ Microbiol. 2005;71:7053–7063. doi: 10.1128/AEM.71.11.7053-7063.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Handy SM, Demir E, Hutchins DA, Portune KJ, Whereat EB, et al. Using quantitative real-time PCR to study competition and community dynamics among Delaware Inland Bays harmful algae in field and laboratory studies. Harmful Algae. 2008;7:599–613. [Google Scholar]
- 37.Yamaguchi A, Horiguchi T. Molecular phylogenetic study of the heterotrophic dinoflagellate genus Protoperidinium (Dinophyceae) inferred from small subunit rRNA gene sequences. Phycol Res. 2005;53:30–42. [Google Scholar]
- 38.Mühling M, Woolven-Allen J, Murrell JC, Joint I. Improved group-specific PCR primers for denaturing gradient gel electrophoresis analysis of the genetic diversity of complex microbial communities. ISME J. 2008;2:379–392. doi: 10.1038/ismej.2007.97. [DOI] [PubMed] [Google Scholar]
- 39.Dereeper A, Guignon V, Blanc G, Audic S, Buffet S, et al. Phylogeny.fr:robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008;2008 Jul 1; 36(Web Server issue):W465–9. doi: 10.1093/nar/gkn180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin S, Zhang H, Hou Y, Zhuang Y, Miranda L. High-level diversity of dinoflagellates in the natural environment, revealed by assessment of mitochondrial cox1 and cob for dinoflagellate DNA barcoding. Appl Environ Microbiol. 2009;75:1279–1290. doi: 10.1128/AEM.01578-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Woyke T, Xie G, Copeland A, González JM, Han C, et al. Assembling the marine metagenome, one cell at a time. PLoS ONE. 2009;4:e5299. doi: 10.1371/journal.pone.0005299. doi: 10.1371/journal.pone.0005299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Raho N, Pizarro G, Escalera L, Reguera B, Marin I. Morphology, toxin composition, and molecular analysis of Dinophysis ovum Schutt, a dinoflagellate of the “Dinophysis acuminata complex”. Harmful Algae. 2008;7:839–848. [Google Scholar]
- 43.Litaker RW, Vandersea MW, Kibler SR, Steven R, Reece KS, et al. Recognizing dinoflagellate species using ITS rDNA sequences. J Phycol. 2007;43:344–355. [Google Scholar]
- 44.Gast RJ, Moran DM, Dennett MR, Caron DA. Kleptoplasty in an Antarctic dinoflagellate: caught in evolutionary transition?. Environ Microbiol. 2007;9:39–45. doi: 10.1111/j.1462-2920.2006.01109.x. [DOI] [PubMed] [Google Scholar]
- 45.Taylor FJR. On dinoflagellate evolution. BioSystems. 1980;13:65–108. doi: 10.1016/0303-2647(80)90006-4. [DOI] [PubMed] [Google Scholar]
- 46.Larsen J, Moestrup Ø. Potentially toxic phytoplankton 2. Genus Dinophysis (Dinophyceae). ICES Identification Leaflets for Plankton. 1992;180:1–12. [Google Scholar]
- 47.Gòmez F. Synonymy and biogeography of the dinoflagellate genus Histioneis (Dinophysiales: Dinophyceae). Rev Biol Trop. 2007;55:59–477. doi: 10.15517/rbt.v55i2.6025. [DOI] [PubMed] [Google Scholar]
- 48.Foster RA, Collier JL, Carpenter EJ. Reverse transcription PCR amplification of cyanobacterial symbiont 16S rRNA sequences from single non-photosynthetic eukaryotic marine planktonic host cells. J Phycol. 2006;42:243–250. [Google Scholar]