Abstract
Rational engineering to produce biologically active plant compounds has been greatly impeded by our poor understanding of the regulatory and metabolic pathways underlying the biosynthesis of these compounds. Here we capitalized on our previously described gene-to-metabolite network in order to engineer rosmarinic acid (RA) biosynthesis pathway for the production of beneficial RA and lithospermic acid B (LAB) in Salvia miltiorrhiza hairy root cultures. Results showed their production was greatly elevated by (1) overexpression of single gene, including cinnamic acid 4-hydroxylase (c4h), tyrosine aminotransferase (tat), and 4-hydroxyphenylpyruvate reductase (hppr), (2) overexpression of both tat and hppr, and (3) suppression of 4-hydroxyphenylpyruvate dioxygenase (hppd). Co-expression of tat/hppr produced the most abundant RA (906 mg/liter) and LAB (992 mg/liter), which were 4.3 and 3.2-fold more than in their wild-type (wt) counterparts respectively. And the value of RA concentration was also higher than that reported before, that produced by means of nutrient medium optimization or elicitor treatment. It is the first report of boosting RA and LAB biosynthesis through genetic manipulation, providing an effective approach for their large-scale commercial production by using hairy root culture systems as bioreactors.
Introduction
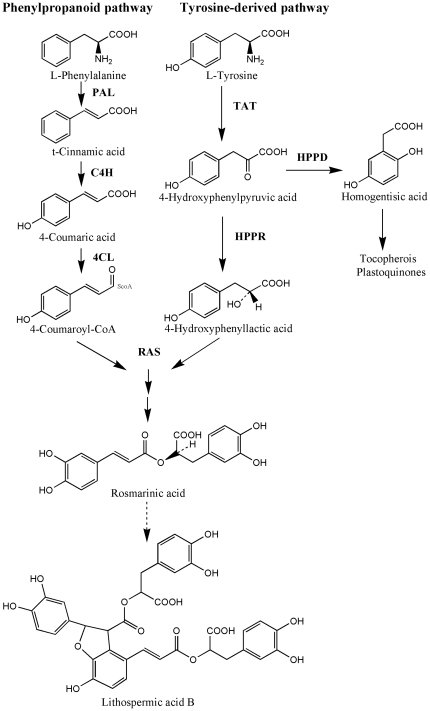
Rosmarinic acid (alpha-O-caffeoyl-3, 4-dihydroxyphenyllactic acid; RA) is a common hydroxycinnamoyl ester accumulated in the plant species of Boraginaceae and Lamiaceae. In recent years, RA has aroused scientists' interest because it exhibits various biological activities, including antibacterial, anti-oxidative, and antiviral effects [1], as well as has implications toward biofuel feedstock development [2]. These activities make it a potentially useful product for the pharmaceutical, cosmetic and bioenergy industries. The biosynthetic pathway leading to RA has been suggested to involve both the phenylpropanoid pathway and a tyrosine-derived pathway as depicted by Figure 1 [3]. RA formation provides an excellent model to investigate regulatory mechanisms of secondary metabolism because two parallel and presumably concertedly regulated pathways are involved in its biosynthesis [4].
Figure 1. The metabolic pathway leading to RA.
PAL; phenylalanine ammonia-lyase, C4H; cinnamic acid 4-hydroxylase, 4CL; hydroxycinnamate coenzyme A ligase, TAT; tyrosine aminotransferase, HPPR; 4-hydroxyphenylpyruvate reductase, RAS; RA synthase, HPPD; 4-hydroxyphenylpyruvate dioxygenase. The dotted line represents a putative biosynthesis process which has not yet been identified.
The RA biosynthetic pathway is under strict developmental and environmental control. The enzymology and regulation of RA biosynthesis have been actively investigated by using plant cell cultures. In Anchusa officinalis cell suspension cultures, tyrosine aminotransferase (TAT) activity was found positively correlated with the rate of RA biosynthesis during linear growth stage of the culture cycle [5]. In Lithospermum erythrorhizon cell suspension cultures, elicitation by methyl jasmonate (MeJA) led to a 10-fold stimulation of RA accumulation, and a strong transient increase in phenylalanine ammonia lyase (PAL) and 4-hydroxyphenylpyruvate reductase (HPPR) activity in the MeJA-treated cells was found to correlate well with the induced RA accumulation [4]. In Coleus blumei suspension cultures, a fungal elicitor enhanced RA accumulation ∼3-fold, and the specific activities of PAL and rosmarinic acid synthase (RAS) were coordinately induced [6]. Recently, Song and Wang [7] reported that RNAi-mediated suppression of PAL in S. miltiorrhiza caused a marked reduction in RA production. However, the specific role of enzyme(s) involved in activation of the RA pathway remains uncertain.
MeJA, a signal compound, is proposed to play an integral role as a second messenger in the elicitation process leading to the accumulation of secondary metabolites [8], thus it has been commonly used as an elicitor to explore the regulatory mechanisms underlying the biosynthesis of metabolites such as phenolic acid and alkaloids [4], [6], [9]. MeJA-induced plant cell culture provides a responsive model system to profile modulations in gene transcripts and related metabolites accumulations. In our previous study [10], a gene-to-metabolite network was constructed by searching for correlations between the transcripts of RA biosynthetic genes and the accumulations of both RA and its important derivative, lithospermic acid B (LAB) (Figure 1) [11], in MeJA-induced S. miltiorrhiza hairy root cultures. This gene-to-metabolite network indicated that pal, cinnamic acid 4-hydroxylase (c4h), tat and hppr transcripts were closely correlated with RA and LAB accumulation, implying a role for the corresponding enzymes as metabolic control points for RA and LAB production in S. miltiorrhiza. In addition, the successful cloning of tat, hppr, c4h and 4-hydroxyphenylpyruvate dioxygenase gene (hppd) from S. miltiorrhiza [12]–[14] in our previous study was another great motivation for the exploration of metabolic engineering as an effective approach to appraise the specific role of enzyme(s) in activation of the RA pathway by enhancing rate-limiting steps or by blocking competitive pathways, ultimately leading to an increase in the yield of target compounds (RA and LAB).
Here, we report the introduction of gene constructs containing cDNA clones of respective tat, hppr and c4h, binary tat and hppr, as well as antisense-hppd, driven by the CaMV 35S promoter (P35S) into S. miltiorrhiza hairy roots. The profile of gene transcripts and metabolites accumulations involved in the RA biosynthesis pathway, and phenolic acid production capacities of these engineered hairy root lines were investigated.
Methods
Construction of vectors
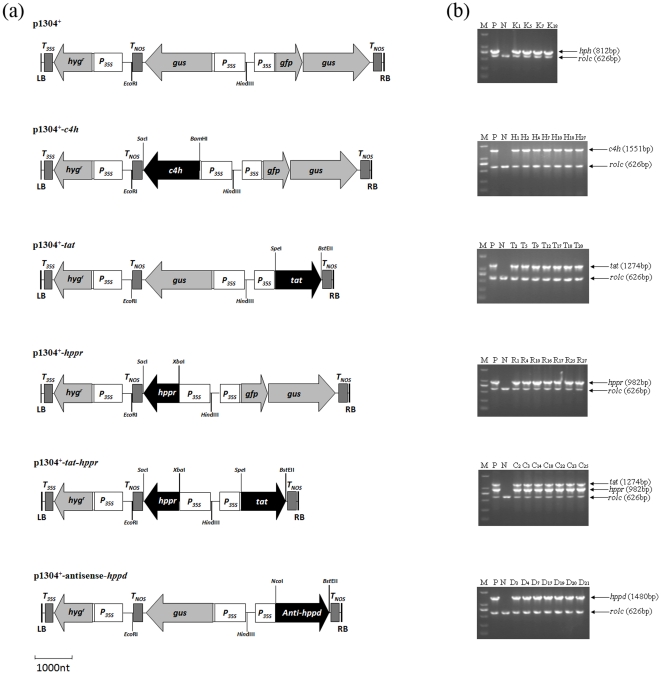
The vector pBI121 and pCAMBIA1304 were used to create an intermediate plasmid containing appropriate restriction sites for incorporating not only c4h, tat, hppr and hppd gene separately, but also for tat and hppr together. The pBI121 vector was digested by HindIII and EcoRI, and the 3,033-bp fragment containing the P35S, β-glucuronidase coding sequence (gus) and the nopaline synthase terminator (TNOS), was recovered. It was then ligated into a HindIII/EcoRI fragment, produced from the double digestion of pCAMBIA1304, to form the recombinant destination vector, designated as p1304+. It contained three separate expression cassettes driven by P35S, and the hygr cassette for conferring hygromycin resistance (Figure 2a).
Figure 2. The expression plasmids used in transformation and molecular analyses of transgenic hairy root lines.
(a) Schematic representation of transgene expression plasmids. P35S; CaMV 35S promoter, hygr; hygromycin-resistance gene, gus; β-glucuronidase coding sequence, gfp; green fluorescent protein coding sequence, T35S; CaMV 35S terminator, TNOS; nopaline synthase terminator, LB; T-DNA left border, RB; T-DNA right border. Restriction sites and target gene are marked. (b) Representative PCR analyses for the rolc gene and the specific genes in transgenic hairy root lines. M; DL-2000 Marker (100–2,000 bp), P; the corresponding engineered bacteria (positive control), N; the wild-type hairy root (negative control), K; vector transformed lines (ck), H; single c4h transgenic lines, T; single tat transgenic lines, R; single hppr transgenic lines, D; antisense-hppd transgenic lines, C; dual tat/hppr co-transgenic lines.
Full length open reading frames for each construct were cloned from cDNA by PCR, according to the published gene sequence information (see GenBank accession numbers in Table S1). Appropriate restriction sites were added to 5′ ends of the primers to amplify each gene coding sequence (see the resulting primer sequences in Table S1). After sequencing confirmation, the obtained DNA sequences of c4h, tat, hppr and hppd were digested using their corresponding restriction enzymes (Table S1) and then respectively subcloned into the p1304+ vector between P35S and TNOS, generating p1304+-c4h, p1304+-tat, p1304+-hppr and p1304+-antisense-hppd vectors (the bypass gene hppd coding sequence was inserted in an inverted orientation to create antisense-hppd structure). For tat-hppr binary expression vector, the XbaI/SacI hppr gene fragment was subcloned into the p1304+-tat vector giving plasmid p1304+-tat-hppr (Figure 2a). After PCR and sequencing confirmation, the above five plasmids, together with the construct without the target fragment (i.e. p1304+ empty plasmid), were separately introduced into the disarmed Agrobacterium tumefaciens C58C1 strain, which carried the pRiA4 of A. rhizogenes [15]. After confirmation by PCR, the positive clones were used to transform S. miltiorrhiza leaf disc explants.
Plant transformation and hairy root culture
Transformation of leaf explants from S. miltiorrhiza plants was carried out basically following the previously described method [10], with the simultaneous transformation with C58C1 strains as wild-type controls. Hairy roots developed at cut edges 2–3 weeks after co-cultivation were excised and cultured on solid, hormone-free, half-strength B5 medium supplemented with 30 g/l sucrose as the carbon source. All of the culture media contained 100 mg/l hygromycin and 500 mg/l cefotaxime. Root culture clones were maintained at 25°C in the dark and routinely subcultured every 25–30 days. The rapidly growing hygromycin-resistant lines with no bacterial contamination were used to establish hairy root lines. About 100 mg of fresh roots (3 cm in length) were inoculated into 150-ml conical flasks containing 40 ml of liquid and half-strength B5 medium, cultured on an orbital shaker (120 rpm) at 25°C in the dark and routinely refreshed the culture medium every 9 days. The fresh weight (fw) of root tissues from flasks of cultures were recorded individually at days 9, 18, 27, 36, and 45 after inoculation. At day 45, the roots were filtered and washed with 10 ml of sterile distilled water and lyophilized, for DNA extraction, RNA extraction and metabolites analysis.
PCR analysis
Genomic DNA was isolated from hairy root samples using the acetyl trimethyl ammonium bromide (CTAB) method [16]. The DNA was then used in PCR analysis for detecting the presence of the specific genes in transgenic lines. Primer sequences for amplifying these genes (these primers were particularly designed to cover the gene sequence and the vector sequence for detecting exogenous gene transformations) and the expected PCR product size are listed in Table S2. The selectable marker hygromycin resistance gene hph (Figure 2a) was used to check the p1304+ vector transformants, whereas Agrobacterium rolc gene was used to check the transformation of pRiA4 [17]. The PCR reactions were carried out in a total volume of 25 µl comprising 50 ng genomic DNA, 50 mM KCl, 10 mM Tris-HCl (pH 8.3), 1.5 mM MgCl2, 200 µM dNTPs, 1.25 units of Taq DNA polymerase and 25 pmol of each primer. For PCR analysis, DNA was denatured at 94°C for 3 min followed by 35 cycles of amplification (94°C for 10 s, 58°C for 30 s, 72°C for 1 min) with final extension at 72°C for 5 min.
GUS assays
The genomic DNA of hairy root samples was used for detecting the presence of the gus gene (Figure 2a) by PCR reaction, with primers gus-F (5′-GCTGTGCCAGGCAGTTTTAAC-3′) and gus-R (5′-ATATCGTCCACCCAGGTGTTC-3′). The amplification condition was the same as those for the specific genes PCR described above. Histochemical localization of GUS activity was performed using 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc) as the chromogenic substrate. A reaction mixture consisting of 1 mM X-Gluc dissolved in 50 mM sodium phosphate buffer (pH 7.2) was used. Hairy root samples were incubated for 12 h at 37°C and pigments were removed by extraction with 100% ethanol prior to observation [18].
Real-time quantitative PCR analysis
Total RNAs from hairy root samples were extracted using TRIzol Reagent (GIBCO BRL, Gaithersburg, MD) according to the manufacturer's instruction [19]. The quality and concentration of RNA were examined by ethidium bromide (EB)-stained agarose gel electrophoresis and spectrophotometer analysis. Total RNA was reverse transcribed by using AMV (avian myeloblastosis virus) reserve transcriptase (Takara) to generate cDNA. All of the gene-specific primers were designed according to the conserved region of the corresponding sequences of S. miltiorrhiza. Part of the polyubiquitin gene was amplified with primers (5′-ACCCTCACGGGGAAGACCATC- 3′ and 5′-ACCACGGAGACGGAGGACAAG-3′) as a control. Real-time quantitative PCR (RT-qPCR) was performed according to the manufacturer's instructions (Takara) under the following conditions: 1 min pre-denaturation at 95°C, 1 cycle; 10 s denaturation at 95°C, 20 s annealing at a given temperature (set according to the character of the primers used as determined by Primer Premier 5.00 software); and 15 s for collecting fluorescence data at 72°C, 40 cycles. The products of RT-qPCR were subjected to 1.5% agarose-gel electrophoresis and showed an equal-sized band as the predicted product PCR. Quantification of the gene expression was done with the comparative CT method and each data point represents the average of three experiments.
Analysis of metabolites
Compound extraction and analysis followed the methods described by Yan et al. [20] with minor modifications. The dried hairy root sample (50 mg) was ground into powder, comminuted (100 mesh) and extracted twice with 30% ethanol (25 ml) under sonication for 10 min, and then centrifuged at 4500 rpm for 5 min. The supernatant was diluted with distilled water to 50 ml total volume, and the extract solution was then filtered through a 0.2-µm organic membrane before analysis. The concentration of the metabolites was determined by triple-quadrupole mass spectrometer (Agilent 6410, Agilent, Santa Clara, CA) equipped with a pump (Agilent 1200 G1311A, Agilent) and an autosampler (Agilent G1329A, Agilent). Chromatography separation was performed with a Thermo C18 column (150 mm×2.1 mm, i.d., 3.5 µm particle size, Agilent). A mobile phase consisting of acetonitrile: H2O (55∶45, v/v) was used, with the flow rate set at 0.3 ml min−1 and a 3.5 min run time. Multiple reaction monitoring (MRM) mode was used for the quantification, and the selected transitions of m/z were 359→161 for RA, 717→519 for LAB, 164→147 for L-phenylalanine, 147→102 for t-cinnamic acid, 163→119 for 4-coumaric acid, 180→119 for L-tyrosine, 179→107 for 4-hydroxyphenylpyruvic acid, and 167→123 for homogentisic acid. All standards were purchased from Sigma-Aldrich (St. Louis, MO) [21].
Statistical analysis
Statistical analysis was performed with SPSS 13.0 software. Analysis of variance (ANOVA) was followed by Tukey's pairwise comparison tests, at a level of p<0.05, to determine significant differences between means.
Results and Discussion
Gene transformation and confirmation of transgenic lines
Plant expression vectors, including p1304+-c4h, p1304+-tat, p1304+-hppr, p1304+-antisense-hppd and p1304+-tat-hppr, together with p1304+ plasmid as vector control (Figure 2a), were separately introduced into S. miltiorrhiza leaf explants by using A. tumefaciens C58C1 strain and generated hairy root lines were screened using hygromycin. Some transgenic roots turned brown and aged considerably faster than wild-type root cultures (hairy root lines generated through transformation with blank C58C1 strain, wt). These lines were discarded, and the remaining hairy root lines were subcultured for 45 days in hormone-free, half-strength B5 liquid medium, for PCR analysis (Table 1). All of the hairy roots contained the rolc gene, which was evidence of transformation by the pRiA4 [17]. To eliminate the interference from S. miltiorrhiza endogenous genes, the primers for the integration confirmation of the specific gene(s) into the transformed hairy root genome land on vector sequences (Table S2). PCR analyses confirmed the integration of the exogenous target gene(s) in the tested transgenic lines (Figure 2b). The GUS assays showed that ck, c4h, tat, hppr single-gene transformants, as well as antisense-hppd lines were positive for the presence of gus gene and GUS staining, whereas the tat-hppr co-transformed lines did not show any detectable PCR product and GUS staining (Figure S1). These results were in accordance with their respective introduced gene constructs (Figure 2a), further demonstrating the success of transgene. These confirmed transgenic lines that also simultaneously showed a good growth capability (Table 1) were selected for biomass growth statistical analysis, as well as further RT-qPCR analysis and metabolites determination.
Table 1. Gene constructs and derived root cultures.
| Numbers of established root lines | ||||
| T-DNA construct | Total | Antibiotic-resistant | PCR-positive | Established root cultures |
| p1304+-c4h | 30 | 18 | 14 | H1,H2,H4,H7,H10,H18,H27 |
| p1304+-tat | 25 | 16 | 13 | T2,T5,T9,T12,T17,T18,T20 |
| p1304+-hppr | 27 | 18 | 10 | R1,R4,R10,R16,R17,R25,R27 |
| p1304+-antisense-hppd | 22 | 10 | 7 | D3,D4,D7,D17,D19,D20,D21 |
| p1304+-tat-hppr | 27 | 20 | 18 | C2,C3,C14,C18,C22,C23,C25 |
| p1304+ vector control (CK) | 20 | 7 | 4 | K1,K5,K7,K18 |
T-DNA, portion of the Ti (tumor-inducing) plasmid that is transferred to plant cells. In addition, three lines transformed with C58C1 were also established, as wild-type control.
Growth characteristics of transgenic lines
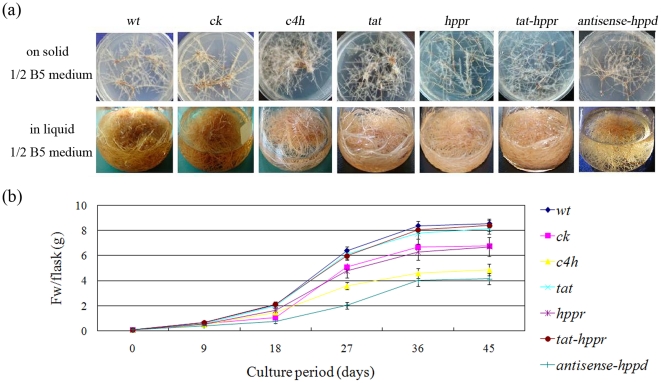
The morphology and biomass growth rate of transgenic lines were shown as Figure 3. Significant differences of growth characteristics were detected among independent transformed root lines. All of the wt, tat and tat/hppr lines grew fast and vigorously with thick branches whereas other lines (ck, c4h, hppr and antisense-hppd lines) grew slowly with slender branches (Figure 3a). Significant differences (p<0.05) were found between wt, tat, tat/hppr lines and other lines with respect to growth rate, but no significant difference (p>0.05) was detected among the tat, tat/hppr lines and hairy roots generated by blank transformation (wt) (Figure 3b). Generally, a fairly high content of secondary metabolites in the tissues is associated with poor growth, and the actual total productivity of secondary metabolites therefore remains low [22]. We found in this study, however, that the growth rate in the high phenolic acid-producing lines was not reduced as compared with those with low phenolic acid production. Line tat/hppr, for example, which produced the highest level of RA and LAB, grew very rapidly. All types of hairy root lines reached the highest growth rate at day 18 and achieved maximum fw at day 45 after inoculation (Figure 3b).
Figure 3. Analyses for the morphology and growth rate in transgenic hairy root lines.
(a) Phenotype of developed root lines on solid 1/2 B5 medium for 30 days, and their corresponding root culture in liquid 1/2 B5 medium for 45 days. (b) Time courses of growth of transgenic hairy root lines. Each set of lines corresponds to one transgenic line, and within each set the replicates of independent transgenic line are as depicted by Table 1. Error bars show the standard deviations of independent lines. wt; the untransformed hairy root lines (wild-type control), ck; empty vector transformed lines (vector control).
Gene expression in transgenic hairy roots
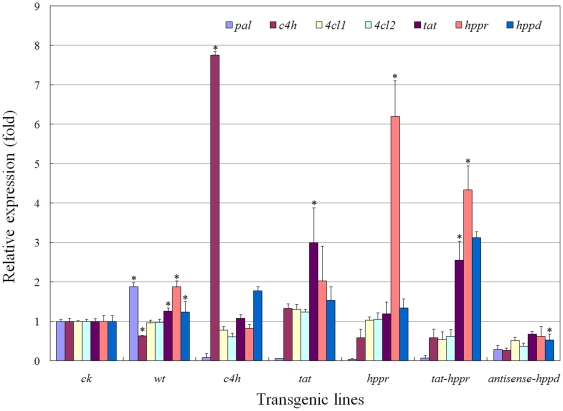
RT-qPCR analysis showed that the expression of the target gene(s) was successfully regulated through genetic manipulation. In comparison with levels of the empty vector transformed control (ck), c4h, tat and hppr transcript levels were coordinately enhanced (approx. 7.7, 3.0 and 6.2-fold, p<0.05) in their respective overexpression lines. The transcript levels of tat and hppr were up-regulated ∼2.5 and ∼4.3-fold (p<0.05) by simultaneous introduction, whereas hppd transcript was reduced ∼50% (p<0.05) by antisense approach (Figure 4). In addition, gene transcript fluctuations between ck and wt lines were generally observed, indicating the introduction of vector p1304+ made modulations in gene transcript profiles of S. miltiorrhiza hairy roots, a similar phenomenon was found in vector pKANNIBAL transformation of S. miltiorrhiza plantlets [7]. Meanwhile, the transcript profile analysis showed these specific transgenic manipulations also variably affected the transcript level of several other RA biosynthetic genes, implying the occurrence of regulatory crosstalk. For example, hppd suppression simultaneously down-regulated the overall gene transcripts when compared with ck, and pal transcript level was generally decreased by these bioengineering manipulations (Figure 4), which subsequently resulted in the high L-phenylalanine (the precursor of PAL) accumulation in the corresponding engineered lines (Figure 5). The transcript crosstalk phenomenon among RA synthesis-related genes has been widely observed in previous studies [7], [23]–[25], possibly occurring via a feedback loop involving coordinated regulation of a series of pathway enzymes [23].
Figure 4. Quantification of the transcripts of RA biosynthetic genes in transgenic hairy root lines.
Each set of bars corresponds to one transgenic line, and within each set the different color corresponds to a single gene. The replicates of independent transgenic line are as depicted by Table 1. Error bars show the standard deviations of independent lines. Asterisk indicates that difference is significant at p<0.05 compared with vector control. ck; empty vector transformed lines (vector control), wt; the untransformed hairy root lines (wild-type control).
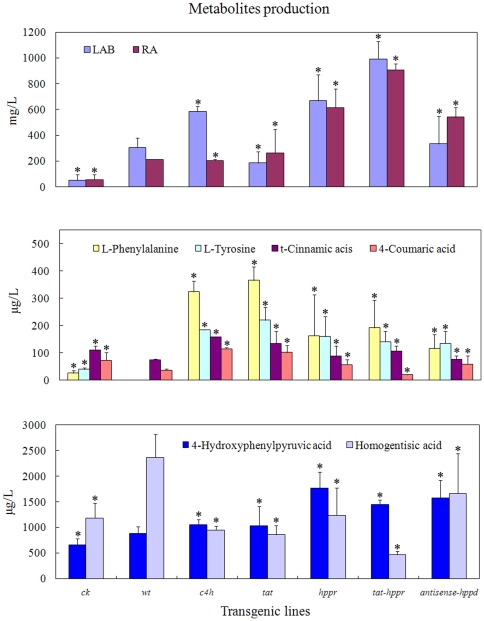
Figure 5. Metabolites production involved in phenolic acids biosynthetic pathway in transgenic hairy root lines.
Each set of bars corresponds to one transgenic line, and within each set the replicates of independent transgenic line are as depicted by Table 1. Error bars show the standard deviations of independent lines. Asterisk indicates that difference is significant at p<0.05 compared with vector control and untransformed control. ck; empty vector transformed lines (vector control), wt; the untransformed hairy root lines (wild-type control).
Metabolites accumulation in transgenic hairy roots
The profiles of metabolites involved in the RA biosynthetic pathway in transgenic hairy root lines were determined by triple-quadrupole mass spectrometer. Results revealed a broad and dynamic reconfiguration of metabolic flux in engineered hairy root cultures that was similar to modulations in transcript profiles (Figure 5). All metabolic pathway strategies significantly enhanced target phenolic acids (RA and LAB) accumulations compared with ck (p<0.05), despite the lower transcript level of pal that was generally found in these engineered root lines (Figure 4). Previously pal was suggested to possess great importance to phenolic acids synthesis in S. miltiorrhiza [7], [10], [26], our result presented here implies that the pal gene plays an important role but not as a bottleneck in the RA biosynthesis pathway. The activation in the sequential biosynthetic steps could effectively compensate for the reduced flux resulted from the disruption of pal.
Interestingly the introduction of empty vector p1304+ into S. miltiorrhiza hairy roots greatly inhibited RA and LAB biosynthesis in comparison with wt. However, their accumulations increased following introduction of the target genes. Comparison of metabolites profile among these plant tissues demonstrated a strong correlation between 4-hydroxyphenylpyruvic acid accumulation and their capacity to produce phenolic acids. Elevated 4-hydroxyphenylpyruvic acid accumulation was widely detected in the highly productive transgene lines (compared with wt, p<0.05), while the lower ability of ck lines to accumulate phenolic acids may result from the lower 4-hydroxyphenylpyruvic acid accumulation (compared with wt, p<0.05) (Figure 5). For the unique role of 4-hydroxyphenylpyruvic acid not only as a product of TAT, but also as a co-substrate of HPPR and HPPD, this finding suggests that 4-hydroxyphenylpyruvic acid may represent regulatory bottlenecks in phenolic acid biosynthesis, or perhaps points of contact between separate metabolic channels, and that its accumulation probably has a positive influence on the flow of metabolites through the pathway. However, this remains to be investigated by further studies for stably producing and more efficient transformants.
C4H catalyses the second reaction of the phenylpropanoid pathway and has been one of the most studied cytochrome P450 enzymes in plants, exhibiting a key role in the synthesis of the phenylpropanoid polymer lignin [23], [27]–[29]. However, its effects on phenolic acids synthesis have not been as extensively studied. Here we observed overexpression of c4h activated two parallel pathways of RA biosynthesis, resulted in increased t-cinnamic acid and 4-coumaric acid accumulation in the phenylpropanoid pathway, increased L-tyrosine and 4-hydroxyphenylpyruvic acid in the tyrosine-derived pathway, and lastly the large production of end-product phenolic acids. The content of LAB in c4h transgenic lines was enhanced to 584 mg/liter, which was 11.1-fold of that in ck (52.7 mg/liter), and 1.9-fold of wt (307 mg/liter). Additionally, c4h transformation produced comparable RA concentration (201 mg/liter) with that in wt (211 mg/liter), which was ∼3.6-fold more than ck (56.1 mg/liter).
Overexpression of the tyrosine-derived pathway gene tat and hppr, either separately or simultaneously, activated the tyrosine-derived pathway, stimulating L-tyrosine and 4-hydroxyphenylpyruvic acid accumulation, and led to enhanced phenolic acids production compared with ck. However, the phenolic acids' production levels were highly variable. For example, the tat single-gene transgenic hairy root lines accumulated phenolic acids only at similar levels to those of wt, likely due to its relatively lower expression level (only 3 times of ck) (Figure 4), which was not sufficient to ensure maximal activity for promoting phenolic acids biosynthesis. In contrast, single hppr transgenic lines presented much higher RA (616 mg/liter) and LAB (669 mg/liter) production. As expected, the greatest abundance of target compounds was found in tat-hppr double gene-transformation lines, which produced RA and LAB with the content of 906 and 992 mg/liter, values that are more than 4.3 and 3.2 times that in wt lines, and over 16.1 and 18.8 times that in ck lines, respectively. Notably, although possessing a relatively low tat and hppr transcript level compared with the single gene transformed lines, the tat-hppr co-transformed lines produced the highest level of desirable metabolites. This result implies that the high phenolic acids level was associated with a coordinative effect of both tat and hppr transgenes. Meanwhile, tat-hppr transgenic lines was fascinatingly observed to possess the lowest accumulation level of homogentisic acid, a compound involved in the competitive branch of RA biosynthesis and produced by HPPD from 4-hydroxyphenylpyruvic acid precursor. This most likely suggests a metabolic flux shift from the homogentisic acid-branch towards the RA pathway. The strong pulling force from TAT and HPPR greatly boosted the overall RA pathway, and subsequently directed the flux from the homogentisic acid pool to restore the metabolic balance, leading to decreased homogentisic acid levels and increased phenolic acid accumulations. It was concluded that transgenic hairy roots harboring both tat and hppr possessed an increased flux in the phenolic acids biosynthetic pathway that enhanced RA and LAB yield, which was more efficient than that harboring only one of the two genes. A similar “push-pull” strategy has been reported in several previous studies, with a significant increase in provitamin A, limonene and artemisinin yields in plants such as Oryza sativa, Lavandula latifolia and Artemisia annua, respectively [30]–[32], as well as enhancement of scopolamine, tropane alkaloid and terpenoid indole alkaloid accumulation in hairy roots of Hyoscyamus niger, Anisodus acutangulus and Catharanthus roseus [33]–[35]. Currently, an increasing number of researchers are embracing this strategy as an approach to generate plants with more ambitious phenotypes instead of introducing single genes [36].
HPPD transforms 4-hydroxyphenylpyruvate to homogentisate, competing for the same substrate as HPPR. Homogentisate is then transformed to tocopherols, commonly known as vitamin E [37]. Because of the high nutrition value of vitamin E, previous investigation in HPPD usually focused on its contribution to the production of vitamin E [38]–[40]. Here, we firstly examined the role of HPPD as a side-branch enzyme of the RA biosynthesis pathway for phenolic acids accumulations. Suppression of hppd directly increased accumulation of its substrate (4-hydroxyphenylpyruvic acid), in comparison with ck, indicating this genetic manipulation successfully inhibited the substrate competition come from the bypass way. However, homogentisic acid (the catalysate of HPPD) was also enhanced by hppd suppression. This could be associated with the widespread suppression effect of antisense-hppd on the biosynthetic genes, such as shown in Figure 4. It may be envisaged that the lower gene expression-causing flux limitation upon the downstream of homogentisic acid pathway would make more homogentisic acid accumulation. The hppd-suppressed hairy roots produced higher levels of RA (542 mg/liter) and LAB (334 mg/liter) than both the wt and ck lines (p<0.05), which was in accordance with our anticipation that HPPD activity was of great importance to the synthesis of phenolic acids in S. miltiorrhiza [10]. However, the hppd engineered roots accumulated more RA than LAB, which contrasts with the native distribution of phenolic acids in S. miltiorrhiza, where the content of LAB was found to be much higher than that of RA [41]. This probably resulted from the directional engineering effect upon the specific RA pathway rather than LAB, demonstrating the power of genetic engineering to reprogram phenols metabolism and distribution in plant culture.
Above all we conclude as follows: (1) By using genetic transformation, the present study demonstrates previous gene-to-metabolite network described role of those enzymes as critical metabolic control points for the production of target phenolic acids in S. miltiorrhiza hairy root cultures. Furthermore, phenolic acid production capacities of these engineered root lines paralleled their corresponding gene-to-metabolite correlation coefficients drawn by previous study. For instance, the correlation coefficient corresponding to hppr was much more than tat. That is, hppr transgene lines exhibited higher level of phenolic acids than their tat counterparts. (2) Compared with those MeJA-treated lines (494 mg/liter RA and 1585 mg/liter LAB production) [10], hppr single-gene, tat-hppr double-gene transformants, as well as antisense-hppd lines reported here were all observed to accumulate more RA but less LAB. On the one hand, these results fully display the power of genetic engineering for direct regulation of the specific RA pathway versus elicitor induction. However, on the other hand the results also prompt us to speculate combining transgenic technology with elicitor treatments as another feasible strategy to further improve phenolic acids yield in S. miltiorrhiza hairy roots in the near future [42], [43]. (3) It appears that the pulling force from the phenylpropanoid pathway involved genes, such as c4h, play a more important role in stimulating LAB accumulation, whereas genes involved in the tyrosine-derived pathway, e.g. tat, hppr, hppd, make a greater contribution to RA biosynthesis regulation. At the extreme, hppd engineered hairy roots contained much more RA than LAB, which completely contrasted with their natural distribution in S. miltiorrhiza. (4) Gene activation of the RA pathway was found to not only boost RA biosynthesis, but also promote accumulation of its derivative LAB. This result further strengthens our previously reported hypothesis that RA is a precursor leading to LAB synthesis [11].
With our ever increasing knowledge of complex plant metabolic networks involved in the production of biologically active plant compounds [44], a deeper understanding of the regulatory system governing secondary metabolism attracts particular interest and could eventually allow for the possibility of successful metabolic engineering of target compound biosynthesis [9]. In the current study, by using attractive targets enabled by previously described gene-to-metabolite network, we successfully engineered the RA biosynthesis pathway for the production of beneficial RA and its derivative LAB in S. miltiorrhiza hairy root cultures, shedding light on how to effectively increase the end products of secondary metabolic pathways by more accurate and appropriate genetic engineering strategies. Moreover, it is the first report of boosting the RA biosynthesis pathway through genetic manipulation. Co-expression of tat/hppr dramatically activates the RA pathway, produces high concentration of RA (906 mg/liter), a value more than that produced by means of nutrient medium optimization or elicitor treatment reported previously, in plant cell cultures such as cell suspensions of L. erythrorhizon [4], C. blumei [6], S. miltiorrhiza [26], and Lavandula vera MM [45], [46], as well as hairy roots of S. miltiorrhiza [10], [11], [20], Hyssopus officinalis [47], Salvia officinalis [48], and Coleus forskohlii [49]. The current study provides an effective approach for commercially large-scale production of RA by using the S. miltiorrhiza hairy root systems as bioreactors.
Supporting Information
PCR primers of the coding sequences of c4h, tat, hppr and hppd genes.
(DOC)
PCR primers used for detecting the specific genes in transgenic lines.
(DOC)
The transgenic hairy roots were verified by GUS assays. (a) PCR analysis of hairy root DNA using the primers to amplify a 425-bp fragment of the gus gene. M; DL-2000 Marker (100–2,000 bp), P; the pCAMBIA1304 plasmid (positive control), N; the wild-type hairy root (negative control). (b) Corresponding GUS histochemical staining of transgenic hairy roots.
(TIF)
Acknowledgments
We thank Modern Research Center for Traditional Chinese Medicine, Second Military Medical University, Shanghai, for excellent technical assistance.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was funded by grants from National Natural Science Foundation of China (31100221, 31160059, 30900786) and Program of Shanghai Subject Chief Scientist (09XD1405900), Shanghai Science and Technology Committee. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Petersen M, Abdullah Y, Benner J, Eberle D, Gehlen K, et al. Evolution of rosmarinic acid biosynthesis. Phytochem. 2009;70:1663–1679. doi: 10.1016/j.phytochem.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 2.Vega-Sánchez ME, Ronald PC. Genetic and biotechnological approaches for biofuel crop improvement. Curr Opin Biotech. 2010;21:218–224. doi: 10.1016/j.copbio.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 3.Petersen M, Simmonds MS. Rosmarinic acid. Phytochem. 2003;62:121–125. doi: 10.1016/s0031-9422(02)00513-7. [DOI] [PubMed] [Google Scholar]
- 4.Mizukami H, Tabira Y, Ellis BE. Methyl jasmonate-induced rosmarinic acid biosynthesis in Lithospermum erythrorhizon cell suspension cultures. Plant Cell Rep. 1993;12:706–709. doi: 10.1007/BF00233424. [DOI] [PubMed] [Google Scholar]
- 5.Mizukami H, Ellis BE. Rosmarinic acid formation and differential expression of tyrosine aminotransferase isoforms in Anchusa officinalis cell suspension cultures. Plant Cell Rep. 1991;10:321–324. doi: 10.1007/BF00193150. [DOI] [PubMed] [Google Scholar]
- 6.Szabo E, Thelen A, Petersen M. Fungal elicitor preparations and methyl jasmonate enhance rosmarinic acid accumulation in suspension cultures of Coleus blumei. . Plant Cell Rep. 1999;18:485–489. [Google Scholar]
- 7.Song J, Wang Z. RNAi-mediated suppression of the phenylalanine ammonia-lyase gene in Salvia miltiorrhiza causes abnormal phenotypes and a reduction in rosmarinic acid biosynthesis. J Plant Res. 2011;124:183–192. doi: 10.1007/s10265-010-0350-5. [DOI] [PubMed] [Google Scholar]
- 8.Gundlach H, Müller MJ, Kutchan TM, Zenk MH. Jasmonic acid is a signal transducer in elicitor-induced plant cell cultures. Proc Natl Acad Sci U S A. 1992;89:2389–2393. doi: 10.1073/pnas.89.6.2389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rischer H, Oresic M, Seppanen-Laakso T, Katajamaa M, Lammertyn F, et al. Gene-tometabolite networks for terpenoid indole alkaloid biosynthesis in Catharanthus roseus cells. Proc Natl Acad Sci U S A. 2006;103:5614–5619. doi: 10.1073/pnas.0601027103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xiao Y, Gao S, Di P, Chen J, Chen W, et al. Methyl jasmonate dramatically enhances the accumulation of phenolic acids in Salvia miltiorrhiza hairy root cultures. Physiol Plantarum. 2009;137:1–9. doi: 10.1111/j.1399-3054.2009.01257.x. [DOI] [PubMed] [Google Scholar]
- 11.Xiao Y, Gao S, Di P, Chen J, Chen W. Lithospermic acid B is more responsive to silver ion (Ag+) than rosmarinic acid in Salvia miltiorrhiza hairy root cultures. Biosci Rep. 2010;30:33–40. doi: 10.1042/BSR20080124. [DOI] [PubMed] [Google Scholar]
- 12.Huang B, Duan Y, Yi B, Sun L, Lu B, et al. Characterization and expression profiling of Cinnamate 4-hydroxylase gene from Salvia miltiorrhiza in rosmarinic acid biosynthesis pathway. Russ J Plant Physl. 2008;55:390–399. doi: 10.1007/s11033-007-9130-2. [DOI] [PubMed] [Google Scholar]
- 13.Huang B, Yi B, Duan Y, Sun L, Yu X, et al. Characterization and expression profiling of tyrosine aminotransferase gene from Salvia miltiorrhiza (Dan-shen) in rosmarinic acid biosynthesis pathway. Mol Biol Rep. 2008;35:601–612. doi: 10.1007/s11033-007-9130-2. [DOI] [PubMed] [Google Scholar]
- 14.Xiao Y, Di P, Chen J, Chen W, Zhang L. Characterization and expression profiling of 4-hydroxyphenylpyruvate dioxygenase gene (Smhppd) from Salvia miltiorrhiza hairy root cultures. Mol Biol Rep. 2009;36:2019–2029. doi: 10.1007/s11033-008-9413-2. [DOI] [PubMed] [Google Scholar]
- 15.Mozo T, Hooykaas PJ. Electroporation of megaplasmids into Agrobacterium. Plant Mol Biol. 1991;16:917–918. doi: 10.1007/BF00015085. [DOI] [PubMed] [Google Scholar]
- 16.Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12:13–15. [Google Scholar]
- 17.Chilton MD, Tepfer DA, Petit A, David C, Casse-Delbart F, et al. Agrobacterium rhizogenes inserts T-DNA into the genome of the host plant root cells. Nat. 1982;295:432–434. [Google Scholar]
- 18.Jefferson R. Assaying chimeric genes in plants: the GUS gene fusion system. Plant Mol Biol Rep. 1987;5:387–405. [Google Scholar]
- 19.Jaakola L, Pirttila AM, Halonen M, Hohtola A. Isolation of high quality RNA from bilberry (Vaccinium myrtillus L.) fruit. Mol Biotechnol. 2001;19:201–203. doi: 10.1385/MB:19:2:201. [DOI] [PubMed] [Google Scholar]
- 20.Yan Q, Shi M, Ng J, Wu J. Elicitor-induced rosmarinic acid accumulation and secondary metabolism enzyme activities in Salvia miltiorrhiza hairy roots. Plant Sci. 2006;170:853–858. [Google Scholar]
- 21.Lu W, Gao S, Xiao Y, Zhang L, Li J, et al. A liquid chromatographic–tandem mass spectrometric method for the quantitation of eight components involved in lithospermic acid B biosynthesis pathway in Salvia miltiorrhiza hairy root cultures. J Med Plants Res. 2011;5:1664–1672. [Google Scholar]
- 22.Jouhikainen K, Lindgren L, Jokelainen T, Hiltunen R, Teeri TH, et al. Enhancement of scopolamine production in Hyscyamus muticus L. hairy root cultures by genetic engineering. Planta. 1999;208:545–551. [Google Scholar]
- 23.Blount JW, Korth KL, Masoud SA, Rasmussen S, Lamb C, et al. Altering expression of cinnamic acid 4-hydroxylase in transgenic plants provides evidence for a feedback loop at the entry point into the phenylpropanoid pathway. Plant Physiol. 2000;122:107–116. doi: 10.1104/pp.122.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Achnine L, Blancaflor EB, Rasmussen S, Dixon RA. Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell. 2004;16:3098–3109. doi: 10.1105/tpc.104.024406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rohde A, Morreel K, Ralph J, Goeminne G, Hostyn V, et al. Molecular phenotyping of the pal1 and pal2 mutants of Arabidopsis thaliana reveals far-reaching consequences on phenylpropanoid, amino acid, and carbohydrate metabolism. Plant Cell. 2004;16:2749–2771. doi: 10.1105/tpc.104.023705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chen H, Chen F. Effect of yeast elicitor on the secondary metabolism of Ti-transformed Salvia miltiorrhiza cell suspension cultures. Plant Cell Rep. 2000;19:710–717. doi: 10.1007/s002999900166. [DOI] [PubMed] [Google Scholar]
- 27.Sewalt V, Ni W, Blount JW, Jung HG, Masoud SA, et al. Reduced lignin content and altered lignin composition in transgenic tobacco down-regulated in expression of phenylalanine ammonia-lyase or cinnamate 4-hydroxylase. Plant Physiol. 1997;115:41–50. doi: 10.1104/pp.115.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blee K, Choi JW, O'Connell AP, Jupe SC, Schuch W, et al. Antisense and sense expression of cDNA coding for CYP73A15, a class II cinnamate 4-hydroxylase, leads to a delayed and reduced production of lignin in tobacco. Phytochem. 2001;57:1159–1166. doi: 10.1016/s0031-9422(01)00150-9. [DOI] [PubMed] [Google Scholar]
- 29.Vanholme R, Morreel K, Ralph J, Boerjan W. Lignin engineering. Curr Opin Plant Biol. 2008;11:278–285. doi: 10.1016/j.pbi.2008.03.005. [DOI] [PubMed] [Google Scholar]
- 30.Ye X, Al-Babili S, Klöti A, Zhang J, Lucca P, et al. Engineering the provitamin A (beta-carotene) biosynthetic pathway into (carotenoid-free) rice endosperm. Sci. 2000;287:303–305. doi: 10.1126/science.287.5451.303. [DOI] [PubMed] [Google Scholar]
- 31.Muñoz-Bertomeu J, Ros R, Arrillaga I, Segura J. Expression of spearmint limonene synthase in transgenic spike lavender results in an altered monoterpene composition in developing leaves. Metab Eng. 2008;10:166–177. doi: 10.1016/j.ymben.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 32.Aquil S, Husaini AM, Abdin MZ, Rather GM. Overexpression of the HMG-CoA reductase gene leads to enhanced artemisinin biosynthesis in transgenic Artemisia annua plants. Planta Med. 2009;75:1453–1458. doi: 10.1055/s-0029-1185775. [DOI] [PubMed] [Google Scholar]
- 33.Zhang L, Ding R, Chai Y, Bonfill M, Moyano E, et al. Engineering tropane biosynthetic pathway in Hyoscyamus niger hairy root cultures. Proc Natl Acad Sci U S A. 2004;101:6786–6791. doi: 10.1073/pnas.0401391101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kai GY, Li L, Jiang YX, Yan XM, Zhang Y, et al. Molecular cloning and characterization of two tropinone reductases in Anisodus acutangulus and enhancement of tropane alkaloid production in AaTRI-transformed hairy roots. Biotechnol Appl Biochem. 2009;54:177–186. doi: 10.1042/BA20090171. [DOI] [PubMed] [Google Scholar]
- 35.Peebles CA, Sander GW, Hughes EH, Peacock R, Shanks JV, et al. The expression of 1-deoxy-D-xylulose synthase and geraniol-10-hydroxylase or anthranilate synthase increases terpenoid indole alkaloid accumulation in Catharanthus roseus hairy roots. Metab Eng. 2011;13:234–240. doi: 10.1016/j.ymben.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 36.Naqvi S, Farre G, Sanahuja G, Capell T, Zhu C, et al. When more is better: multigene engineering in plants. Trends Plant Sci. 2009;15:48–56. doi: 10.1016/j.tplants.2009.09.010. [DOI] [PubMed] [Google Scholar]
- 37.Kim KH, Petersen M. cDNA-cloning and functional expression of hydroxyphenylpyruvate dioxygenase from cell suspension cultures of Coleus blumei. Plant Sci. 2002;163:1001–1009. [Google Scholar]
- 38.Tsegaye Y, Shintani DK, DellaPenna D. Overexpression of the enzyme p-hydroxyphenolpyruvate dioxygenase in Arabidopsis and its relation to tocopherol biosynthesis. Plant Physiol Biochem. 2002;40:913–920. [Google Scholar]
- 39.Falk J, Andersen G, Kernebeck B, Krupinska K. Constitutive overexpression of barely 4-hydroxyphenylpyruvate dioxygenase in tobacco results in elevation of the vitamin E content in seeds but not in leaves. FEBS Lett. 2003;540:35–40. doi: 10.1016/s0014-5793(03)00166-2. [DOI] [PubMed] [Google Scholar]
- 40.Crowell EF, McGrath JM, Douches DS. Accumulation of vitamin E in potato (Solanum tuberosum) tubers. Transgenic Res. 2008;17:205–217. doi: 10.1007/s11248-007-9091-1. [DOI] [PubMed] [Google Scholar]
- 41.Omoto T, Murakami Y, Shimomura K, Yoshihira K, Mori K, et al. Caffeic acid derivatives in Lamiaceae and Boraginaceae plants. Jpn J Food Chem. 1997;4:11–16. [Google Scholar]
- 42.Peebles CA, Hughes EH, Shanks JV, San KY. Transcriptional response of the terpenoid indole alkaloid pathway to the overexpression of ORCA3 along with jasmonic acid elicitation of Catharanth usroseus hairy roots over time. Metab Eng. 2009;11:76–86. doi: 10.1016/j.ymben.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 43.Zhang HC, Liu JM, Lu HY, Gao SL. Enhanced flavonoid production in hairy root cultures of Glycyrrhiza uralensis Fisch by combining the over-expression of chalcone isomerase gene with the elicitation treatment. Plant Cell Rep. 2009;28:1205–1213. doi: 10.1007/s00299-009-0721-3. [DOI] [PubMed] [Google Scholar]
- 44.Allen DK, Libourel IG, Shachar-Hill Y. Metabolic flux analysis in plants: coping with complexity. Plant Cell Environ. 2009;32:1241–1257. doi: 10.1111/j.1365-3040.2009.01992.x. [DOI] [PubMed] [Google Scholar]
- 45.Pavlov AI, Georgiev MI, Panchev IN, Ilieva MP. Optimization of rosmarinic acid production by Lavandula vera MM plant cell suspension in a laboratory bioreactor. Biotechnol Prog. 2005;21:394–396. doi: 10.1021/bp049678z. [DOI] [PubMed] [Google Scholar]
- 46.Georgiev MI, Kuzeva SL, Pavlov AI, Kovacheva EG, Ilieva MP. Elicitation of rosmarinic acid by Lavandula vera MM cell suspension culture with abiotic elicitors. World J Microbiol Biotechnol. 2007;23:301–304. [Google Scholar]
- 47.Murakami Y, Omoto T, Asai I, Shimomura K, Yoshihira K, et al. Rosmarinic acid and related phenolics in transformed root cultures of Hyssopus officinalis. Plant Cell Tiss Org. 1998;53:75–78. [Google Scholar]
- 48.Grzegorczyk I, Królicka A, Wysokińska H. Establishment of Salvia officinalis L. hairy root cultures for production of rosmarinic acid. Z Naturforsch [C] 2006;61:351–356. doi: 10.1515/znc-2006-5-609. [DOI] [PubMed] [Google Scholar]
- 49.Li W, Koike K, Asada Y, Yoshikawa T, Nikaido T. Rosmarinic acid production by Coleus forskohlii hairy root cultures. Plant Cell Tiss Org. 2005;80:151–155. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PCR primers of the coding sequences of c4h, tat, hppr and hppd genes.
(DOC)
PCR primers used for detecting the specific genes in transgenic lines.
(DOC)
The transgenic hairy roots were verified by GUS assays. (a) PCR analysis of hairy root DNA using the primers to amplify a 425-bp fragment of the gus gene. M; DL-2000 Marker (100–2,000 bp), P; the pCAMBIA1304 plasmid (positive control), N; the wild-type hairy root (negative control). (b) Corresponding GUS histochemical staining of transgenic hairy roots.
(TIF)