Abstract
The p53-family member TAp73 is a transcription factor that plays a key role in many biological processes. Here, we show that p73 drives the expression of microRNA (miR)-34a, but not miR-34b and -c, by acting on specific binding sites on the miR-34a promoter. Expression of miR-34a is modulated in parallel with that of TAp73 during in vitro differentiation of neuroblastoma cells and cortical neurons. Retinoid-driven neuroblastoma differentiation is inhibited by knockdown of either p73 or miR-34a. Transcript expression of miR-34a is significantly reduced in vivo both in the cortex and hippocampus of p73−/− mice; miR-34a and TAp73 expression also increase during postnatal development of the brain and cerebellum when synaptogenesis occurs. Accordingly, overexpression or silencing of miR-34a inversely modulates expression of synaptic targets, including synaptotagmin-1 and syntaxin-1A. Notably, the axis TAp73/miR-34a/synaptotagmin-1 is conserved in brains from Alzheimer's patients. These data reinforce a role for TAp73 in neuronal development.
Keywords: cell death, neurodegeneration, Alzheimer disease
The p53-family member Trp73 is transcribed from two distinct promoters, resulting in isoforms containing or lacking the N-terminal TA domain, known as TAp73 (1, 2) and ΔNp73 (3), respectively; additionally, alternative 3′-splicing produces further variants (α, β, and so forth). In keeping with their sequence and structural similarities, TAp73 can mimic several p53 functions, including the transactivation of p21, Puma, and Bax, although p73 also has distinct transcriptional targets. Indeed, p73−/− mice show developmental defects of the CNS; for example, congenital hydrocephalus, hippocampal dysgenesis, and defects of pheromone detection rather than the enhanced tumor susceptibility of p53−/− mice (4). This development is not simply because of apoptosis, as the ectopic expression of p73 induces neurite outgrowth and expression of neuronal markers in neuroblastoma cell lines (5) and in primary oligodendrocytes (6).
Several microRNAs (miRs) are regulated by p53 (7), although p73-dependent miRs have been less well studied. In particular, the miR-34 family (miR-34a to -c) has been shown to be a direct target of p53 (8–10). Ectopic expression of miR-34 mimics several p53 effects, although in a cell type-specific manner. In mice, miR-34a is ubiquitous with the highest expression in brain, and overexpression of miR-34a in neuroblastoma cell lines modulates neuronal-specific genes (11); miR-34b and -c are mainly in the lung (12).
Because both p73 and miRs, including miR-34a, have been implicated in neuronal differentiation, we have investigated the possibility that p73 drives miR-34a expression using WT and p73-null mice. We demonstrate that miR-34a is transcriptionally regulated by TAp73 and that, in turn, miR-34a regulates the expression of a number of synaptic proteins, in particular synaptotagmin I and syntaxin 1A in cortical neurons. Moreover, neuronal architecture is disorganized in p73-null mice, and manipulation of miR-34a expression is associated with both morphological and electrophysiological changes. We highlight the importance of the TAp73/miR-34a axis in neuronal differentiation and synaptogenesis.
Results
TAp73 Drives the Expression of miR-34a.
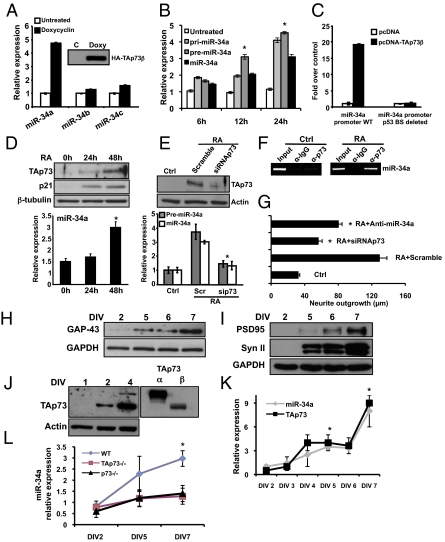
To investigate whether p73 modulates the expression of miR-34a, we initially used the SAOS-2-TAp73β inducible cell line. TAp73β induces miR-34a expression (P < 0.05), but not miR-34b or miR-34c (Fig. 1A) via a significant (P < 0.05) increase of its precursors (Fig. 1B). To investigate whether the expression of miR-34a was because of an induction of its precursors or because of an increase of primary transcript (pri-miR-34a) processing (13), we evaluated by real-time PCR the levels of pri-miR-34a and the intermediate precursor (pre-miR-34a). Results in Fig. 1B show that TAp73β induces a significant (P < 0.05) increase of both precursors as well as mature miR-34a, indicating that TAp73 acts at the transcriptional level. We next explored if miR-34a is directly regulated by TAp73. Therefore, we cloned the miR-34a promoter, which contains a p53-consensus binding site, upstream of a luciferase reporter gene. The construct was transfected together with TAp73 in SAOS-2 cells. TAp73 significantly increased the luciferase activity (Fig. 1C, miR-34a promoter WT), depending on the p53 binding site (Fig. 1C). In addition, TAp73β directly binds the miR-34 promoter (ChIP) (Fig. S1A). TAp73α also induces miR-34a expression as shown by real-time PCR (Fig. S1B), luciferase assay (Fig. S1C), and by ChIP (Fig. S1D).
Fig. 1.
TAp73 drives the expression of miR-34a and in vitro terminal differentiation of cortical neurons. (A) SAOS-2-TAp73β inducible cell lines were treated with Doxycyclin for 24 h to overexpress the human TAp73β protein (Inset, Western blot). Endogenous levels of miRs-34a, -34b, and -34c were assessed by real-time PCR. (B) SAOS-2-TAp73β inducible cell lines were treated with Doxycyclin for the indicate times and endogenous levels of pri-miR-34a, pre-miR-34a, and miR-34 were evaluated by real-time PCR. *P < 0.05. (C) The miR-34a promoter region between −1,472 and +551 bp, which contains a p53 consensus site was analyzed for TAp73β responsiveness. Insertion of this miR-34a promoter region in a luciferase reporter gene leads to increased luciferase activity in the presence of TAp73β in SAOS-2 cells. (D) TAp73 regulates miR-34 in RA-induced terminal differentiation of a neuroblastoma cell line. SHSY-5Y cells were treated with 10 μM RA and expression of TAp73, p21, and β-tubulin was assessed by Western blot. Expression of miR-34a was evaluated by real-time PCR (Lower, histogram). (E) p73siRNA prevents the up-regulation of precursor-miR-34a and miR-34a after RA treatment. *P < 0.05. (F) ChIP assay on SHSY-5Y cells untreated (ctrl) or treated with RA. Protein-chromatin complexes were immunoprecipitated with anti-TAp73 or IgG as control antibody. RT-PCR was performed with primers designed against the hsa-miR-34a promoter region containing the predicted and validated p53 binding site. (G) Neurite outgrowth in SHSY-5Y untransfected (ctrl), or transfected with scrambled, siRNAp73, or anti-miR-34a and then treated with RA for 48 h. *P < 0.05. (H and I) Western blot analysis of three different neuronal markers and GAPDH in cultured cortical neurons at DIV2, -5, -6, and -7. Syn II (Synapsin II). (J and K) Cortical neurons were collected at the indicated time points and expression of miR-34a and TAp73 was evaluated by Western blot and real-time PCR respectively. Data represent mean ± SD of three different experiments. *P = 0.016. (L) Relative expression of miR-34a compared between WT, TAp73−/−, and p73−/− mice during terminal neuronal differentiation. Data are normalized to the housekeeping gene Sno202 relative to DIV2. Data represent mean ± SD of three different experiments. *P = 0.045.
Retinoic acid (RA) is an inducer of terminal differentiation in neuroblastoma cells (14), and recently RA has been shown to induce miR-34a expression, as well as TAp73, in neuroblastoma cell lines (5, 15). We therefore hypothesized that the RA-induced increase in miR-34a expression may be secondary to the induction of TAp73. To this end, we treated SH-SY5Y neuroblastoma cells with RA and isolated RNA at different times during the differentiation. RA induces the expression of both TAp73 and miR-34a (Fig. 1D). To determine whether the increased expression of miR-34a required p73, we performed the same experiment in the presence of p73 siRNA. Fig. 1E shows that p73 knockdown abolishes the RA-induced increase in pre-miR-34a as well as that of mature miR-34a. Confirming the ability of p73 to directly regulate miR-34a expression after RA treatment, Fig. 1F shows that RA induces binding of TAp73 to the miR-34a promoter. To evaluate the functional significance of the TAp73/miR-34a axis in neuroblastoma differentiation, we assessed the degree of neurite outgrowth when either of these was inhibited. Knockdown of p73 by siRNA, and transfection of antagomiR-34a, both significantly (P < 0.0005) reduced RA-mediated neurite outgrowth (Fig. 1G and Fig. S1E).
miR-34a During Terminal Differentiation of Primary Cortical Neurons and in Vivo.
To better understand, in a more physiological context, whether miR-34a might play a role in terminal neuronal differentiation, we tested the expression of miR-34a during terminal differentiation of mouse primary cortical neurons. After 7 d of in vitro culture (DIV7), embryonic day (E) 17.5 cortical neurons were fully differentiated morphologically (Fig. S1F), and expressed GAP-43 (Fig. 1H) and synaptic proteins (Fig. 1I). We next examined expression of miR-34a and TAp73 during cortical neuron differentiation. Fig. 1J shows an increase in the expression of both α- and β-isoforms of TAp73 protein, together with an increase in miR-34a expression by DIV4 (Fig. 1K). No significant changes were observed in ΔNp73 and p53 expression (Fig. S1G).
To address whether the up-regulation of miR-34a observed during terminal neuronal differentiation required p73, we compared the expression of miR-34a during in vitro differentiation of primary cortical neurons from WT, p73−/−, and TAp73−/− mice. As shown in Fig. 1L, the increase of miR-34a is significantly less pronounced in cells from both total and TAp73−/− mice compared with WT littermates.
To validate the in vitro relationship between p73 and miR-34a expression, we investigated miR-34a expression in vivo. real time PCR analysis shows a significant reduction (∼50% in the cortex and 60% in the hippocampus) of miR-34a expression in p73−/− mice compared with control mice (Fig. 2A). To further verify the reduction of miR-34a level in the p73−/− mice, we performed laser-capture microdissection analysis of three different regions of the cortex, and two regions of the hippocampus (CA1+CA2 and dentate gyrus), known to show developmental defects in p73−/− mice (Fig. S2A). Fig. 2B shows a significant reduction of mir-34a in the p73−/− mice in all of the regions analyzed. Because miR-34a can also be induced by p53, we analyzed p53 expression in the same samples. Fig. S2B shows no significant differences in p53 mRNA levels in cortex and hippocampus between WT, heterozygous, or homozygous p73 knockout mice, in contrast to the partial or total reduction in p73 levels in p73+/− and p73−/− mice, respectively (Fig. S2C). Moreover, DAPI staining of two layers of the cortex (Fig. S2D) and H&E staining of the hippocampus (Fig. S2E) shows no significant differences in cell number between p73+/+ and p73−/− mice and in hippocampal morphology at postnatal day (P) 2, when these experiments were performed.
Fig. 2.
Expression of miR-34a and TAp73 in vivo. (A) Expression of miR-34a is reduced in p73−/− mice. Cortex and hippocampus were isolated from p73+/+, p73+/−, and p73−/− mice (n = 5–9; age, P2), and levels of miR-34a were evaluated by real-time PCR. **P < 0.01. (B) Different regions of the cortex and hippocampus were isolated by laser-capture microdissection from p73+/+ and p73−/− mice and levels of miR-34a were evaluated by real-time PCR. Data represent mean ± SD of three different experiments. *P < 0.05. (C) The cerebellum was isolated from WT mice (n = 4 per each point) and expression of miR-34a and TAp73 was evaluated during cerebellar development by real-time PCR at birth (P0) and subsequent postnatal days. (D) Relative expression of miR-34a in the cerebellum from p73+/+ and p73−/− mice during development. Data are normalized to the housekeeping gene Sno202 relative to P1. Data represent mean ± SD of three different experiments. *P = 0.045.
To further explore whether similar temporal changes in expression of TAp73 and miR-34a also occurred during cerebellar development, we isolated RNA from whole cerebellum at the immediate postnatal stage (P0) and subsequently. Fig. 2C shows that miR-34a expression increased in parallel with that of TAp73 up to P4, after which other factors seem to contribute. An increase in miR-34a levels was also detected during postnatal development of the whole brain (minus the olfactory bulb), although this was more prolonged than in the cerebellum (Fig. S3A). In addition, p73−/− mice show a delay in the increase of miR-34a, in particular at P3 (Fig. 2D), further suggesting that TAp73 is one factor driving miR-34a expression during brain development in vivo.
mir-34a Modulates Expression of Synaptic Genes.
To investigate the potential mechanisms whereby miR-34a affects terminal neuronal differentiation, we performed overexpression experiments. Bioinformatics analysis (Fig. S4A) suggested that several synaptic genes contain miR-34a consensus sites in their 3′ untranslated region (3′UTR), and we therefore focused on the potential regulation of synaptic genes by miR-34a. Thus, when DIV5 cortical neurons were transfected with pre-miR-34a, we found reduced mRNA levels of synaptotagmin I, and synapsin I and II 24 h posttransfection (Fig. 3A) as predicted targets (Fig. S4B).
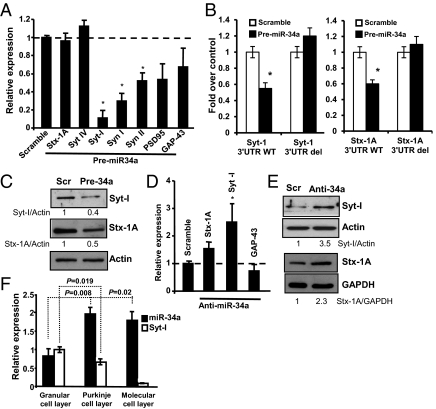
Fig. 3.
miR-34a modulates the expression of synaptic markers. (A) WT cortical neurons (DIV3) were transfected with pre-miR-34a or a scrambled control (Scramble) and after 24 h, mRNA levels of several neuronal markers were determined by real-time PCR. Stx (syntaxin), Syn (synapsin), Syt (synaptotagmin). Data represent mean ± SD of three different experiments. *P = 0.01. (B) Insertion of Syt-I 3′ UTR or Stx1A 3′ UTR (WT or deleted) upstream of a luciferase reporter gene shows diminished luciferase activity in the presence precursor miR-34a. The assay was performed in HEK 293E cells. Data represent mean ± SD of three different experiments. *P = 0.03. (C) Cortical neurons were transfected with precursor miR-34a (Pre-34a) or scrambled control (Scramble) for 72 h and the cell lysates were analyzed for Syt-I and Stx-1A expression. (D) WT cortical neurons (DIV3) were transfected with anti–miR-34a or scrambled control (Scramble) and, after 24 h, mRNA levels of the indicated synaptic proteins were determined by real-time PCR. Data represent mean SD of three different experiments. *P = 0.03. (E) Western blot analysis of proteins from cortical neurons transfected at DIV2 with anti–miR-34a or scrambled control. Actin or GAPDH were used as loading controls. Numbers below the Western blot indicate the relative levels of synaptic protein normalized to actin or GAPDH related to the scrambled control. (F) The indicated cell layers of the cerebellum were isolated by laser capture microdissection (Fig. S7) and levels of miR-34a and Syt-I were evaluated by real-time PCR. Data are normalized to the housekeeping gene Sno202 or GAPDH relative to granular cell layer expression. Data represent mean ± SD of three different experiments.
Computational analysis of mouse 3′ UTR of these proteins show the presence of three binding sites for miR34a in Syt-I, in positions 187 (poorly conserved), 1543, and 1563 (highly conserved), respectively (Fig. S4B), and one conserved binding site in the 3′ UTR of Stx-1A (position 516) (Fig. S4C). We therefore concentrated on two synaptic proteins: synaptotagmin I (Syt-I) and syntaxin 1A (Stx-1A). Fig. 3B shows that miR-34a reduces luciferase activity of both promoters by roughly 50%. Similar results were obtained with the other miR-34 family members or a combination of these (Fig. S4 B and C). Deletion of the miR-34a target sites abolished the 3′ UTR responsiveness to miR-34a overexpression (Fig. 3B). The effect of miR-34a overexpression on Syt-I and Stx-1A transcripts was confirmed at the protein level because (Fig. 3C) cortical neurons transfected with pre-miR-34a showed a significant reduction in expression of the two proteins after 72 h. No significant differences were observed when membranes were reprobed with GAP-43 antibody (Fig. S5A), indicating the selective effect of miR-34a on these synaptic proteins.
In addition, the absence of p73 impairs the expression of the two miR-34a validated targets. Thus, during terminal differentiation in vitro we found that cortical neurons from p73−/− mice show an altered kinetics, particularly of Syt-I during differentiation (Fig. S5B, lanes 3 and 4). No significant differences were found in GAP-43 expression between WT and p73−/− mice (Fig. S5C). We also investigated whether TAp73 could regulate Bcl-2 expression, a well-established miR-34a target (12). Results in Fig. S5D show that the absence of TAp73 does not significantly affect the level of Bcl-2 protein during in vitro cortical neurons differentiation (Fig. S5D), suggesting that Bcl-2 is regulated by distinct pathways.
Role of Endogenous miR-34 in Neurons.
On this basis, we asked whether the endogenous level of miR-34a is able to regulate per se the expression of these two targets. First, to suppress miR-34a function, we transfected cortical neurons either with a specific miR-34a inhibitor or with a scrambled control, and measured the mRNA levels of Syt-I and Stx-1A by real-time PCR after 24 h. When miR-34a function was suppressed by Locked Nucleic Acid (LNA)-miR-34a, we found significant up-regulation of Syt-I, and to a lesser extent Stx-1A, but not GAP-43 mRNA (Fig. 3D).
We next performed Western blot analysis to investigate whether the knock-down of miR-34a also affected the steady-state protein levels. After 72 and 96 h, we observed a significant increase of Syt-I and Stx-1A, respectively (Fig. 3E). To investigate whether miR-34a could regulate these synaptic proteins in vivo, we performed laser-capture microdissection of the cerebellum. Because Stx-1A mRNA is only slightly affected by miR-34a modulation, we focused on Syt-I. In particular, the granular, molecular and Purkinje cell layers were microdissected (Fig. S5E) and expression of miR-34a and Syt-I was evaluated by real-time PCR. Results in Fig. 3F show that miR-34a is more highly expressed in the molecular and Purkinje layers than in the granular cell layer. In parallel, Syt-I expression is significantly lower in the molecular and Purkinje layers, suggesting that miR-34a negatively regulates Syt-I expression in the cerebellum in vivo.
TAp73 and miR-34a in Alzheimer's Disease.
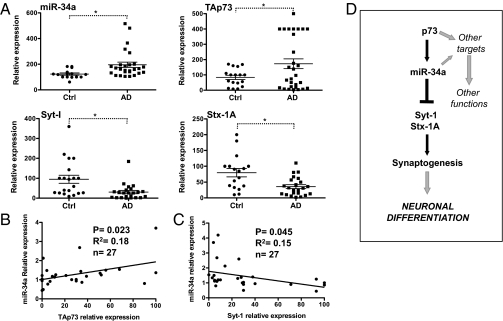
To investigate whether the experimental changes induced by manipulation of the p73/miR-34a axis were also conserved in neuronal pathology, we investigated the levels of TAp73, miR-34a, and two of their targets, Stx-1a and Syt-1, in Alzheimer's disease (AD) brains. We extracted total RNA from 20 control and 29 AD brains (hippocampal region) and used real-time PCR to assess the level of expression of these four transcripts. As shown in Fig. 4A, we found a significant increase of both miR-34a (control 123.3 ± 8.8 vs. AD 195.7 ± 20.7) and TAp73 (control 84.09 ± 13.6 vs. AD 172.8 ± 31.9) in the AD hippocampus. In addition, the levels of TAp73 and miR-34a in individual AD brains showed a significant positive correlation (Fig. 4B). In contrast, the levels of the synaptic proteins were reduced (Syt-I control 95.2 ± 20.28 vs. AD 30.6 ± 8.2 and Stx-1A control 79.9 ± 13.2 vs. AD 35.8 ± 6.0), and the expression of Syt-1 was significantly negatively correlated with that of miR-34a (Fig. 4C and Fig. S6).
Fig. 4.
Expression of miR-34a, TAp73, and synaptic proteins in AD brain. (A) Total RNA was extracted from postmortem control and AD hippocampus. Expression of the indicated genes was evaluated by real-time PCR. Data represent mean ± SEM. *P < 0.05. (B) Pearson's test on individual AD samples show significant positive correlation between TAp73 and miR-34a expression. (C) A significant negative correlation between miR-34a and Syt-1. (D) Schematic model for the role of TAp73/miR-34a axis in developmental neuronal differentiation.
miR-34a and Apoptosis in Cortical Neurons.
Because miR-34a induced by p53 can regulate apoptosis (9, 10), we investigated whether miR-34a could also play a role in the apoptosis of neurons treated with etoposide or staurosporine. Both treatments induced apoptosis in cortical neurons (Fig. S7A, Left), but only etoposide treatment induced a significant increase of miR-34a (Fig. S7A, Right). Moreover, etoposide up-regulated the expression of p53 but not of TAp73, suggesting that p53, and not TAp73, is the regulator of miR-34a following this apoptotic stimulus (Fig. S7B). Importantly, the absence of p73 did not impair the apoptotic response to etoposide (Fig. S7C) nor did it prevent the increase in expression of miR-34a (Fig. S7D). To investigate the functional significance of miR-34a up-regulation during DNA damage, DIV4 cortical neurons were transfected with pre-miR-34a. Ectopic expression of miR-34a by itself did not induce apoptosis as evaluated by flow cytometry analysis and Western blot with antibody against cleaved caspase-3 (Fig. S7E). However, etoposide treatment did result in increased binding of TAp73 to the miR-34a promoter by ChIP (Fig. S7F, Upper), as well as producing increased levels of pri-miR, pre-miR, and mature miR-34a (Fig. S7F, Lower).
Discussion
p73 is implicated in neuronal stemness (16–19), differentiation, and survival. Indeed, p73 deletion results in neuronal and immunological pathologies, including hydrocephalus and hippocampal dysgenesis, with abnormalities in the CA1-CA3 pyramidal cell layers and in the dentate gyrus (4). The hippocampal defects are largely present in the selective TAp73−/− mice (20), suggesting that TAp73 affects neuronal differentiation. Overexpression of TAp73 also increases spontaneous differentiation of oligodendrocytes from precursor cells (6), and its role in differentiation of the nervous system may therefore not be confined to neurons (5). In contrast, ΔNp73 has been shown to act together with ΔNp63 in regulating the survival of cortical precursor cells (21), and both p73+/− and selective ΔNp73−/− mice are more susceptible to neurodegeneration (22, 23).
Here, we demonstrate that TAp73 acts, at least in part, through miR-34a to regulate neuronal differentiation. TAp73 isoforms selectively transcribe miR-34a, and RA-induced in vitro neuroblastoma differentiation and terminal differentiation of cortical neurons are accompanied by increased expression of both TAp73 and miR-34a. Finally, RA-induced differentiation of neuroblastoma cells is inhibited both by knockdown of p73 or by antagomiR-34a. In vivo, miR-34a expression is reduced in the brains of p73−/− mice, and the induction of miR-34a expression is delayed in the cerebellum of the p73-null mice. Overall, our data suggest that TAp73 is a relevant regulator of miR-34a. Because an increase of miR-34a is also observed in a p73-null context, we cannot exclude the possibility that other members of the p53 family can participate in this regulation (Fig. S3B). In addition, we demonstrated a possible TAp73/p53 independent mechanism in miR-34a regulation. Indeed, here we show that the expression of mir-34a is also up-regulated by neuronal activity (Fig. S3C), as has already been dem-onstrated for other miRs (24). We have also identified two synaptic proteins as TAp73/miR-34a targets, and show that miR-34a overexpression produces changes in dendritic spine morphology together with electrophysiological abnormalities, as shown in Agostini et al. (25). Finally, we demonstrate that levels of TAp73 and miR-34a are increased in brains of patients with AD, with a corresponding reduction in the levels of the same synaptic targets.
Both TAp73 and p53 (26) induce the expression of miR-34a by acting on the p53-binding sites present in the miR-34a promoter, although in completely distinct contexts. This finding indicates that miRs can be involved in different pathways, depending on the stimuli and the cellular context. Indeed, in our experimental model, the expression of TAp73 and miR-34a, both in vivo and in vitro, is particularly evident at times when synaptogenesis is taking place. Using gain-of-function experiments as well as luciferase assays, we identified Syt-I and Stx-1A as direct targets of miR-34a. In accordance with previous work (27), we did not observe any effect of miR-34a overexpression on apoptosis of cortical neurons, suggesting that the effect of miR-34a on Syt-I as well as on Stx-1A in neurons is not linked to the apoptotic response.
Syt-I is a member of a protein family, which shows different patterns of spatiotemporal distribution during development (28, 29) and which is expressed on synaptic vesicles where it regulates fast synchronous Ca2+-mediated-exocytosis of neurotransmitters. Stx-1A is also a regulator of ion channels and neurotransmitter transporters, although it clearly has other functions because Stx-1A null mice die in utero (30). Our data show that the complexity of the dendritic tree of cortical neurons, as well as the morphology of hippocampal dendritic spines, is affected by miR-34a, possibly resulting, at least in part, from altered expression of Syt-1 and Stx-1A (25).
Preliminary evidence already implicates p73 and miR-34a in the pathogenesis of AD: an altered subcellular distribution of p73 has been described in the hippocampus of AD brains with increased nuclear localization and accumulation of neurofibrillary tangles, although the specific isoform has not been defined (31). p73 polymorphisms have also been suggested to influence the phenotypic manifestations of AD (32), and TAp73α has been shown to phosphorylate Tau (33), possibly by a JNK-dependent mechanism. Indeed, p73+/− mice are more susceptible to AD and have increased phospho-Tau within neurofibrillary tangles (22). Expression of miR-34a has also been reported to be increased in the cortex in mouse models of AD (34). Our data would appear to confirm these data and establish a relationship between p73, miR-34a, and their targets in AD pathology. Although, these data do not establish a causal relationship between p73 and disease, its aberrant expression, localization, and effects upon synaptic proteins, described here, may be a relevant consequence of the disease process. Intriguingly, recent data has suggested the involvement of the entire p53 family in AD.
Together, these data therefore provide a mechanism, involving miR-34a and its synaptic protein targets, for the role of p73 in neuronal differentiation and degeneration, exemplified by the phenotype of total and TA-selective p73 knockout mice.
Methods
Mice.
The p73−/− and TAp73−/− mice were generated as previously described (4, 20). Mice were bred and subjected to listed procedures under the project license released from the United Kingdom Home Office.
Cell Culture and Transfection.
Cells were transfected by Lipofectamine 2000 or Lipofectamine LTX and PLUS reagent according to the manufacturer's protocols (Invitrogen). Details are provided in SI Methods.
Luc Assay.
Details are provided in SI Methods.
Chromatin Immunoprecipitation.
Details are provided in SI Methods.
RNA Extraction and Real-Time PCR.
Total RNA from cells or tissue was isolated using TRIzol (Invitrogen) according to the manufacturer's instructions. Details are provided in SI Methods.
Laser-Capture Microdissection.
Details are provided in SI Methods.
Bioinformatics.
Details are provided in SI Methods.
Western Blot.
Proteins were extracted with RIPA buffer containing mixture inhibitors (Roche) and separated on SDS-PAGE. Details are provided in SI Methods.
Alzheimer's Disease Samples.
RNA was extracted from snap-frozen brain tissue from the London Neurodegenerative Disease brain bank. Details are provided in SI Methods.
Statistical Analysis.
All results are expressed as means ± SD. P < 0.05 was considered significant.
Supplementary Material
Acknowledgments
We thank Moshe Oren (The Weizmann Institute, Rehovot, Israel) for the miR-34a promoter, and the London Neurodegenerative Disease brain bank and Brains for Dementia Research. This work was supported by the Medical Research Council, United Kingdom; Odysseus and VIB, Belgium; grants from, “Alleanza Contro il Cancro” (ACC12), Ministero dell'Istruzione, dell'Università e della Ricerca/Progetti di Ricerca d'Interesse Nazionale/Fondo per gli Investimenti della Ricerca di Base (RBIP06LCA9_0023, RBIP06LCA9_0C), Associazione Italiana per la Ricerca sul Cancro (2008-2010_33-08), (#5471) (2011-IG11955), AIRC 5xmille (#9979) and Italian Human ProteomeNet RBRN07BMCT (to G.M.); and in part by Ministry of Health (Ricerca Oncologica 26/07) and Istituto Dermopatico dell'Immacolata-Istituto di Ricovero e Cura a Carattere Scientifico RF06 (Conv. 73), RF07 (Conv. 57), and RF08 (Conv. 15) (to G.M. and E.C.). Work was supported by Ministry of Education and Science of the Russian Federation (11.G34.31.0069).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112061109/-/DCSupplemental.
References
- 1.Müller M, et al. TAp73/Delta Np73 influences apoptotic response, chemosensitivity and prognosis in hepatocellular carcinoma. Cell Death Differ. 2005;12:1564–1577. doi: 10.1038/sj.cdd.4401774. [DOI] [PubMed] [Google Scholar]
- 2.Wang J, et al. TAp73 is a downstream target of p53 in controlling the cellular defense against stress. J Biol Chem. 2007;282:29152–29162. doi: 10.1074/jbc.M703408200. [DOI] [PubMed] [Google Scholar]
- 3.Grob T-J, et al. Human delta Np73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 2001;8:1213–1223. doi: 10.1038/sj.cdd.4400962. [DOI] [PubMed] [Google Scholar]
- 4.Yang A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 5.De Laurenzi V, et al. Induction of neuronal differentiation by p73 in a neuroblastoma cell line. J Biol Chem. 2000;275:15226–15231. doi: 10.1074/jbc.275.20.15226. [DOI] [PubMed] [Google Scholar]
- 6.Billon N, et al. Roles for p53 and p73 during oligodendrocyte development. Development. 2004;131:1211–1220. doi: 10.1242/dev.01035. [DOI] [PubMed] [Google Scholar]
- 7.Shin S, et al. MicroRNAs that respond to histone deacetylase inhibitor SAHA and p53 in HCT116 human colon carcinoma cells. Int J Oncol. 2009;35:1343–1352. doi: 10.3892/ijo_00000452. [DOI] [PubMed] [Google Scholar]
- 8.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang T-C, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 11.Wei J-S, et al. The MYCN oncogene is a direct target of miR-34a. Oncogene. 2008;27:5204–5213. doi: 10.1038/onc.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bommer G-T, et al. p53-mediated activation of miRNA34 candidate tumor-suppressor genes. Curr Biol. 2007;17:1298–1307. doi: 10.1016/j.cub.2007.06.068. [DOI] [PubMed] [Google Scholar]
- 13.Thiele C-J, Reynolds C-P, Israel M-A. Decreased expression of N-myc precedes retinoic acid-induced morphological differentiation of human neuroblastoma. Nature. 1985;313:404–406. doi: 10.1038/313404a0. [DOI] [PubMed] [Google Scholar]
- 14.Welch C, Chen Y, Stallings R-L. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 15.Mody M, et al. Genome-wide gene expression profiles of the developing mouse hippocampus. Proc Natl Acad Sci USA. 2001;98:8862–8867. doi: 10.1073/pnas.141244998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Agostini M, et al. p73 regulates maintenance of neural stem cell. Biochem Biophys Res Commun. 2010;403:13–17. doi: 10.1016/j.bbrc.2010.10.087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Talos F, et al. p73 is an essential regulator of neural stem cell maintenance in embryonal and adult CNS neurogenesis. Cell Death Differ. 2010;17:1816–1829. doi: 10.1038/cdd.2010.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fujitani M, et al. TAp73 acts via the bHLH Hey2 to promote long-term maintenance of neural precursors. Curr Biol. 2010;20:2058–2065. doi: 10.1016/j.cub.2010.10.029. [DOI] [PubMed] [Google Scholar]
- 19.González–Cano L, et al. p73 deficiency results in impaired self renewal and premature neuronal differentiation of mouse neural progenitors independently of p53. Cell Death Dis. 2010;1 doi: 10.1038/cddis.2010.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tomasini R, et al. TAp73 knockout shows genomic instability with infertility and tumor suppressor functions. Genes Dev. 2008;22:2677–2691. doi: 10.1101/gad.1695308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dugani C-B, Paquin A, Fujitani M, Kaplan D-R, Miller F-D. p63 antagonizes p53 to promote the survival of embryonic neural precursor cells. J Neurosci. 2009;29:6710–6721. doi: 10.1523/JNEUROSCI.5878-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wetzel M-K, et al. p73 regulates neurodegeneration and phospho-tau accumulation during aging and Alzheimer's disease. Neuron. 2008;59:708–721. doi: 10.1016/j.neuron.2008.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Wilhelm M-T, et al. Isoform-specific p73 knockout mice reveal a novel role for delta Np73 in the DNA damage response pathway. Genes Dev. 2010;24:549–560. doi: 10.1101/gad.1873910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fiore R, et al. Mef2-mediated transcription of the miR379-410 cluster regulates activity-dependent dendritogenesis by fine-tuning Pumilio2 protein levels. EMBO J. 2009;28:697–710. doi: 10.1038/emboj.2009.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Agostini M, et al. microRNA-34a regulates neurite outgrowth, spinal morphology and function. Proc Natl Acad Sci USA. 2011;108:21092–21097. doi: 10.1073/pnas.1112063108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hermeking H. The miR-34 family in cancer and apoptosis. Cell Death Differ. 2010;17:193–199. doi: 10.1038/cdd.2009.56. [DOI] [PubMed] [Google Scholar]
- 27.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berton F, Iborra C, Boudier J-A, Seagar M-J, Marquèze B. Developmental regulation of synaptotagmin I, II, III, and IV mRNAs in the rat CNS. J Neurosci. 1997;17:1206–1216. doi: 10.1523/JNEUROSCI.17-04-01206.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fox M-A, Sanes J-R. Synaptotagmin I and II are present in distinct subsets of central synapses. J Comp Neurol. 2007;503:280–296. doi: 10.1002/cne.21381. [DOI] [PubMed] [Google Scholar]
- 30.McRory JE, et al. Syntaxin 1A is required for normal in utero development. Biochem Biophys Res Commun. 2008;375:372–377. doi: 10.1016/j.bbrc.2008.08.031. [DOI] [PubMed] [Google Scholar]
- 31.Wilson C, Henry S, Smith M-A, Bowser R. The p53 homologue p73 accumulates in the nucleus and localizes to neurites and neurofibrillary tangles in Alzheimer disease brain. Neuropathol Appl Neurobiol. 2004;30:19–29. doi: 10.1046/j.0305-1846.2003.00496.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scacchi R, Gambina G, Moretto G, Corbo R-M. Association study between P53 and P73 gene polymorphisms and the sporadic late-onset form of Alzheimer's disease. J Neural Transm. 2009;116:1179–1184. doi: 10.1007/s00702-009-0276-z. [DOI] [PubMed] [Google Scholar]
- 33.Hooper C, Killick R, Tavassoli M, Melino G, Lovestone S. TAp73alpha induces tau phosphorylation in HEK293a cells via a transcription-dependent mechanism. Neurosci Lett. 2006;401:30–34. doi: 10.1016/j.neulet.2006.02.082. [DOI] [PubMed] [Google Scholar]
- 34.Wang X, et al. miR-34a, a microRNA up-regulated in a double transgenic mouse model of Alzheimer's disease, inhibits bcl2 translation. Brain Res Bull. 2009;80:268–273. doi: 10.1016/j.brainresbull.2009.08.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.