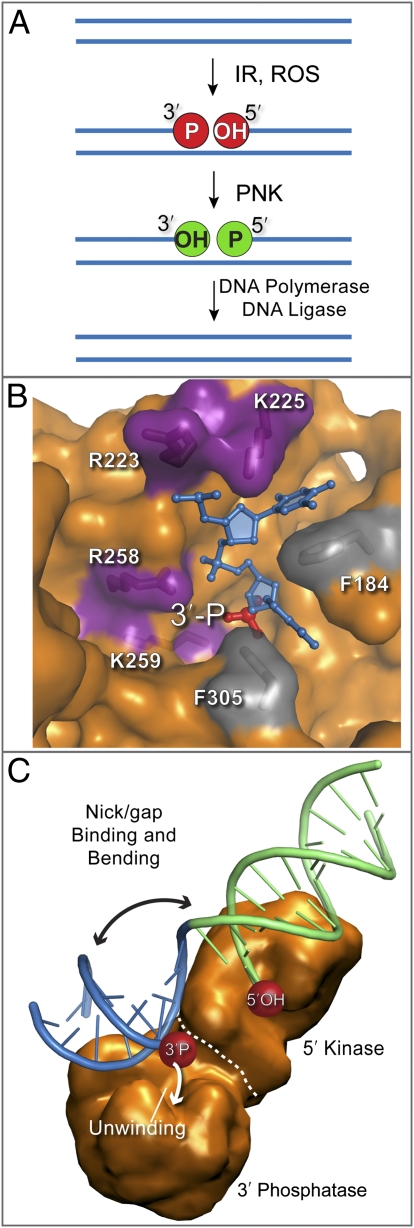
Scission of one or both strands of the DNA duplex occurs as ionizing radiation and oxidative DNA damage mount a constant assault our genomes. Like a broken limb, or split piece of wood, DNA strand breaks are typically not clean. That is, they lack the DNA 3′-hydroxyl and 5′-phosphate moieties needed for DNA repair synthesis by DNA polymerases and DNA end joining by DNA ligases (Fig. 1A). For instance, IR-induced DNA single-strand breaks (SSBs) or double-strand breaks (DSBs) harbor “dirty” chemically heterogeneous termini including 3′-phosphates (3′-PO4) and 5′-hydroxyl (5′-OH) DNA ends (Fig. 1A) (1). Variably adducted DNA termini also arise during the metabolism of DNA base damage by DNA glycosylases, from failed topoisomerase reactions (i.e., 3′ and 5′ topoisomerase protein adducts), and from aborted DNA ligation reactions (i.e., 5′-adenylate adducts) (2–4). To cope with structurally diverse DNA termini, eukaryotic cells have acquired an extensive array of enzymatic activities to tailor DNA ends for ligation. Now, in PNAS, key molecular insights into the mechanisms of DNA end recognition and processing are emerging (5).
Fig. 1.
Divide and conquer DNA end processing by PNK. (A) Cellular DNA is constantly assaulted by ionizing radiation (IR) and reactive oxygen species (ROS). This damage, along with the products of some DNA repair enzymes, may contain 5′ hydroxyls or 3′ phosphates (red circles). These are converted by PNK to 5′ phosphates and 3′ hydroxyls (green circles), which are required for DNA polymerases and DNA ligases to complete repair of the damaged DNA. (B) 3′-Phosphate recognition by PNK. The 3′ phosphate (red) of the DNA strand (blue) lies at the bottom of a pocket containing the active site of PNK (orange). Basic residues (magenta) interact with the negatively charged phosphates of the DNA and conserved phenylalanine residues (gray) provide platforms for DNA bases to stack upon. (C) PNK DNA bending shown schematically. A helical element at the kinase/phosphatase interface splays nicked DNA open, thereby directing 3′ and 5′ damaged ends into their respective active sites. DNA unwinding in the phosphatase domain facilitates 3′ end processing. Coupling of the kinase and phosphatase DNA binding surfaces may facilitate interdomain cooperation and functional crosstalk.
The bifunctional polynucleotide kinase/phosphatase (PNK or PNKP) contains both DNA 5′-kinase and 3′-phosphatase activities required for restoration of 3′-hydroxyls and 5′-phosphates needed to seal the broken DNA (Fig. 1A). PNK plays vital roles in mammalian SSB repair, DSB repair, and base excision repair (1, 4). The importance of PNK function in humans is underscored by the fact that PNK mutations are associated with the debilitating and heritable seizure syndrome MCSZ (i.e., microcephaly with seizures) (6). The basis for modular assembly of PNK domain architecture has been known for some time (7). Separate kinase and phosphatase catalytic activities are located in two halves of an interconnected bilobed catalytic domain that is flexibly linked to an aminoterminal phosphoprotein-binding forkhead-associated domain (8). However, how the enzyme engages strand breaks, interrogates damaged termini, and reverses damage has been unclear in the absence of enzyme–nucleic acid crystal structures. This raises the questions of how DNA end-damage first responders such as PNK identify their substrates, and how they participate in coordinated, multistep DNA repair pathways to prevent genome instability and cell death.
In an exciting advance, Coquelle et al. (5) successfully crystallize an inactivated mouse PNK mutant protein with several 3′ phosphorylated DNAs of different sequence bound in the phosphatase active site (Fig. 1B). This structure shows how one side of the phosphatase catalytic center is lined with basic residues recognizing the DNA phosphate backbone and 3′-PO4. On the opposite side, terraced phenylalanine side chains provide a sequence independent binding platform upon which the bases of the DNA stack. Combined, these interactions hold the ssDNA substrate in a highly extended conformation, and precisely orient the 3′-PO4 into a deep active site pocket for dephosphorylation. The size and shape of this cleft may enable PNK to discriminate 3′-PO4 DNA from 3′-phoshorylated RNA or ribonucleotides, and most importantly, to exclude intact undamaged DNA phosphodiester linkages from the depths of the active site. From the structures, the authors hypothesize that PNK must separate the strands of the damaged dsDNA or nicked DNA duplex to promote 3′-PO4 processing. To test this, Coquelle et al. (5) use the DNA base analogue 2-aminopurine to monitor the terminal base pairing proximal to the 3′-PO4 end in duplex DNA. Their results show that, consistent with the structural observations, PNK induces strand separation of two or three terminal base pairs proximal to the 3′-PO4 end. Mutational analysis further reveals that the unwinding activity requires a conserved basic surface on the protein for engaging the complementary undamaged DNA strand. Thus, the energetic penalty of breaking Watson–Crick hydrogen bonds by PNK is likely offset by the protein–dsDNA binding energy. Together, these data point to a dynamic phosphatase–DNA interaction interface that enables DNA end recognition through localized unwinding of a 3′-PO4 terminus for active site access.
Whereas a stand-alone DNA 3′ phosphatase exists in budding yeast (i.e., Tpp1) (9), fused phosphatase/kinase catalytic domains are found in fission yeast (10) and mammalian PNK orthologues. This raises the question of why these activities are sometimes physically coupled. Hints to the reason for this come from biochemical studies showing that the DNA kinase activity is rate limiting for repair on doubly damaged 3′-phosphate/5′-hydroxyl SSBs (11). Further, mutations that ablate the phosphatase activity also block phosphoryl transfer by the 5′ kinase, implying that interdomain crosstalk between the two active sites is possible. Communication between these functionally intertwined activities makes biological sense. Phosphorylation of the 5′ terminus before removal of 3′ blocking adducts would produce intermediates that can be further metabolized by DNA ligases to generate added pathological 5′-adenylation damage, which then in turn must be reversed by the Aprataxin protein (2, 3) (Fig. 1A). Thus, intimate coupling of the kinase and phosphatase activities may have important implications for controlling the orderly progression of DNA end processing. Coquelle et al. (5) posit that productive engagement of a 3′-PO4
Coquelle et al. posit that productive engagement of a 3′-PO4 terminus may block access of a 5′-OH to the kinase active site.
terminus may block access of a 5′-OH to the kinase active site. So, this mechanism may ensure that 3′ dephosphorylation precedes 5′ phosphorylation, and suggests that the two DNA processing sites might use common DNA damage recognition elements.
How, then, are the kinase and phosphatase activities linked? Important insights into this issue come from a related study reporting an X-ray structure of PNK encountering a DNA nick-mimicking structure that assembled from multiple partially complementary short 5-mer ssDNA chains (12). Intriguingly, this work showed that, for DNA nick recognition, PNK drives a helical wedge into the DNA base stack at the interface between the kinase and phosphatase domains. Division of the DNA base stack segregates 3′ and 5′ termini into their respective active sites for direct damage reversal (Fig. 1C). The simultaneous interrogation of two DNA termini is reminiscent of the flap-endonuclease family nucleases, whose specificity and activities are similarly controlled by segregated 3′ and 5′ binding pockets (13, 14). For PNK, dissection of the mechanism of cooperation and regulation of 5′- and 3′-end processing activities will undoubtedly require additional characterization of PNK engaging intact, damaged nick duplex substrates.
With these new insights in hand, many important questions remain. For instance, how are PNK activities regulated? The observation of DNA-induced fit conformational changes (5, 12), interdomain kinase/phosphatase flexibility (7), and PNK posttranslational modifications such as phosphorylation (15, 16) suggests multiple avenues for modulating PNK activity. PNK is further integrated into multienzyme SSB and DSB repair pathways complexes by binding scaffolding proteins such as Xrcc1 (17) and Xrcc4 (18). A major challenge for future studies will be to understand how PNK activities are further coordinated with additional enzymatic effectors to promote the orderly progression of multistep DNA repair pathways.
Acknowledgments
Research on the DNA damage response in the R.S.W. laboratory is supported by National Institutes of Health Intramural Research Program 1Z01-ES102765-01 (to R.S.W.).
Footnotes
The authors declare no conflict of interest.
See companion article on page 21022.
References
- 1.Weinfeld M, Mani RS, Abdou I, Aceytuno RD, Glover JN. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36:262–271. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443:713–716. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 3.Tumbale P, et al. Structure of an aprataxin-DNA complex with insights into AOA1 neurodegenerative disease. Nat Struct Mol Biol. 2011;18:1189–1195. doi: 10.1038/nsmb.2146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caldecott KW. Single-strand break repair and genetic disease. Nat Rev Genet. 2008;9:619–631. doi: 10.1038/nrg2380. [DOI] [PubMed] [Google Scholar]
- 5.Coquelle N, Havali-Shahriari Z, Bernstein N, Green R, Glover JNM. Structural basis for the phosphatase activity of PNKP on single- and double-stranded DNA substrates. Proc Natl Acad Sci USA. 2011;108:21022–21027. doi: 10.1073/pnas.1112036108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shen J, et al. Mutations in PNKP cause microcephaly, seizures and defects in DNA repair. Nat Genet. 2010;42:245–249. doi: 10.1038/ng.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bernstein NK, et al. The molecular architecture of the mammalian DNA repair enzyme, polynucleotide kinase. Mol Cell. 2005;17:657–670. doi: 10.1016/j.molcel.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein NK, et al. Mechanism of DNA substrate recognition by the mammalian DNA repair enzyme, Polynucleotide Kinase. Nucleic Acids Res. 2009;37:6161–6173. doi: 10.1093/nar/gkp597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Deshpande RA, Wilson TE. Identification of DNA 3′-phosphatase active site residues and their differential role in DNA binding, Mg2+ coordination, and catalysis. Biochemistry. 2004;43:8579–8589. doi: 10.1021/bi049434n. [DOI] [PubMed] [Google Scholar]
- 10.Meijer M, Karimi-Busheri F, Huang TY, Weinfeld M, Young D. Pnk1, a DNA kinase/phosphatase required for normal response to DNA damage by gamma-radiation or camptothecin in Schizosaccharomyces pombe. J Biol Chem. 2002;277:4050–4055. doi: 10.1074/jbc.M109383200. [DOI] [PubMed] [Google Scholar]
- 11.Dobson CJ, Allinson SL. The phosphatase activity of mammalian polynucleotide kinase takes precedence over its kinase activity in repair of single strand breaks. Nucleic Acids Res. 2006;34:2230–2237. doi: 10.1093/nar/gkl275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garces F, Pearl LH, Oliver AW. The structural basis for substrate recognition by Mammalian polynucleotide kinase 3′ phosphatase. Mol Cell. 2011;44:385–396. doi: 10.1016/j.molcel.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tsutakawa SE, et al. Human flap endonuclease structures, DNA double-base flipping, and a unified understanding of the FEN1 superfamily. Cell. 2011;145:198–211. doi: 10.1016/j.cell.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Orans J, et al. Structures of human exonuclease 1 DNA complexes suggest a unified mechanism for nuclease family. Cell. 2011;145:212–223. doi: 10.1016/j.cell.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Segal-Raz H, et al. ATM-mediated phosphorylation of polynucleotide kinase/phosphatase is required for effective DNA double-strand break repair. EMBO Rep. 2011;12:713–719. doi: 10.1038/embor.2011.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zolner AE, et al. Phosphorylation of polynucleotide kinase/ phosphatase by DNA-dependent protein kinase and ataxia-telangiectasia mutated regulates its association with sites of DNA damage. Nucleic Acids Res. 2011;39:9224–9237. doi: 10.1093/nar/gkr647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitehouse CJ, et al. XRCC1 stimulates human polynucleotide kinase activity at damaged DNA termini and accelerates DNA single-strand break repair. Cell. 2001;104:107–117. doi: 10.1016/s0092-8674(01)00195-7. [DOI] [PubMed] [Google Scholar]
- 18.Koch CA, et al. Xrcc4 physically links DNA end processing by polynucleotide kinase to DNA ligation by DNA ligase IV. EMBO J. 2004;23:3874–3885. doi: 10.1038/sj.emboj.7600375. [DOI] [PMC free article] [PubMed] [Google Scholar]