Abstract
Proteins are known to undergo a dynamical transition at around 200 K but the underlying mechanism, physical origin, and relationship to water are controversial. Here we report an observation of a protein dynamical transition as low as 110 K. This unexpected protein dynamical transition precisely correlated with the cryogenic phase transition of water from a high-density amorphous to a low-density amorphous state. The results suggest that the cryogenic protein dynamical transition might be directly related to the two liquid forms of water proposed at cryogenic temperatures.
Keywords: conformation fluctuations, water phase transition, high-pressure cryocooling, X-ray crystallography
It is known that a hydrated protein undergoes a dynamical transition at around 200 K (1). Proteins below the transition temperature are in a glassy state with little conformational flexibility and no appreciable biological function; above the transition temperature flexibility is restored and the protein becomes biologically active (2–4). This protein dynamical transition has been extensively studied by measuring the mean square atomic displacement, 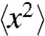 , of protein atoms as a function of temperature with various methods, including Mössbauer and terahertz time domain spectroscopies (5–7), X-ray crystallography (4, 8–11), neutron scattering (2, 12–19), and molecular dynamics simulations (20–25).
, of protein atoms as a function of temperature with various methods, including Mössbauer and terahertz time domain spectroscopies (5–7), X-ray crystallography (4, 8–11), neutron scattering (2, 12–19), and molecular dynamics simulations (20–25).
It is believed that the protein dynamical transition involves a strong coupling to the hydration water (10, 26, 27), as a protein dehydrated below a critical level (water/protein mass ratio of ∼0.2) does not show the dynamical transition (13, 14, 28). Several mechanisms have been proposed to explain the microscopic origin of protein dynamical transition, including α and β fluctuations in the bulk solvent and the hydration shell (29, 30), liquid-glass transition of hydration water (31), a frequency window scenario (32, 33), and a fragile to strong transition of the hydration water (15, 20).
In this work, we studied a protein dynamical transition inside protein crystals using a high-pressure cryocooling method (34) and temperature-dependent X-ray diffraction (8, 35, 36). Protein crystals contain large amounts of water in solvent channels between the proteins in the crystal lattice; water fractions of greater than 50% are very common. It has been reported that this intracrystalline water can be cryocooled under high pressure into a high-density amorphous (HDA) state that subsequently transforms upon heating to the low-density amorphous (LDA) (36) water typical of ambient-pressure flash-cooling [in bulk water the respective densities of HDA and LDA are 1.17 and 0.94 g cm-3 at 77 K and 0.1 MPa. (37, 38)]. In this study, the temperature of the HDA–LDA transition was adjusted by controlling solvent conditions inside the protein crystals. X-ray diffraction data from five thaumatin crystals were used for this study, including four high-pressure cryocooled crystals (Thau-0M-1, Thau-0M-2, Thau-0.45M, Thau-0.9M; 0.9M indicates water solvent that was 0.9 M salt) and one ambient-pressure cryocooled crystal (Thau-control). Details can be found in Materials and Methods and SI Appendix.
Results
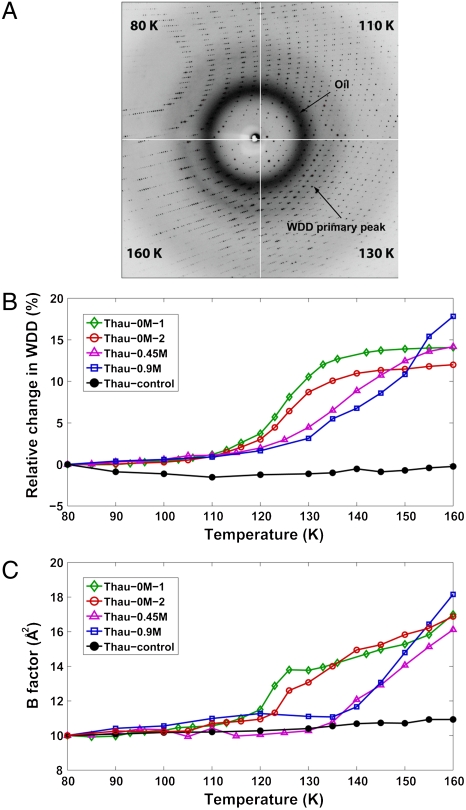
Fig. 1A shows the X-ray diffraction images from Thau-0M-1 at four different temperatures from 80 to 160 K. The diffraction image at each temperature provides two independent types of information: The crystal Bragg diffraction provides information on crystal unit-cell parameters, crystal mosaicity (a measure of crystal lattice disorder), protein structure, and the atomic temperature Debye–Waller factor (B-factor) of the protein. The superimposed diffuse diffraction provides the information on the state of the water in the roughly 2- to 4-nm wide solvent channels of the protein crystals.
Fig. 1.
Protein dynamical transition inside a high-pressure cryocooled crystal. (A) X-ray diffraction images of a high-pressure cryocooled crystal (Thau-0M-1) at 80, 110, 130, and 160 K. The diffraction image at each temperature is a superposition of the protein crystallographic Bragg diffraction, diffuse scattering from oil coating the crystals, and WDD. The peak position of WDD shifts as temperature increases, indicating a transition from HDA to LDA states of water. Above 160 K, LDA water turns into cubic ice, leading to protein crystal disruption. (B) WDD primary peak position profiles of Thau-0M-1, Thau-0M-2, Thau-0.45M, Thau-0.9M, and Thau-control. Thau-0M-1 and Thau-0M-2 undergo a water transition mostly from 110 to 135 K. Thau-0.45M and Thau-0.9M show rather gradual transition up to higher temperatures. In contrast, Thau-control (black curve) shows little change, indicating no water transition is involved. The WDD primary peak positions can be found in tables in SI Appendix. (C) Average B-factor profiles (calculated from main chain atoms) of cryocooled thaumatin structures as temperature is raised. The profiles from high-pressure cryocooled crystals (Thau-0M-1, Thau-0M-2, Thau-0.45M, and Thau-0.9M) show changes from linearity indicative of a protein dynamical transition. For each solvent concentration, the temperature for the onset of this behavior roughly correlates with the shift of the WDD shown in Fig. 1B. By contrast, the B-factor profile of Thau-control shows no significant transition. The correlation between the WDD peak position profiles and the B-factor profiles suggests that the protein dynamical transition is related to the water phase transition inside the protein crystals. Note that the B-factor profiles are normalized to be 10 at 80 K for comparison (SI Appendix). The B-factor values can be found in tables in SI Appendix.
The crystal Bragg diffraction spots can be filtered out of the diffraction images to extract the water diffuse diffraction (WDD) profiles (see Materials and Methods); a shift in the WDD primary peak position is then an indicator of the water phase transition. Fig. 1B shows the relative changes in WDD peak positions for four high-pressure cryocooled crystals and one ambient-pressure cryocooled crystal as they were warmed from 80 to 160 K. The phase boundary, defined as the midpoint of the WDD shift moves to higher temperatures (from 125 to 145 K) as the added salt concentration increases in the high-pressure cryocooled crystals. No phase transition was observed with the ambient-pressure cryocooled crystal (Thau-control), which started out in the LDA state at low temperature.
The dynamical transition of the thaumatin molecules was studied by solving protein structures from the Bragg diffraction and monitoring the B-factors of the atomic structures from 80 to 160 K. A B-factor profile purely reflecting harmonic vibrational motions of atoms in a protein differs from one that includes the onset of the molecular fluctuations between different conformational states (8). At very low temperatures conformational freedom in the protein is frozen out and only local vibrational motions are allowed because conformational fluctuations involve activation over energy barriers. In this temperature regime, the B-factor increases almost linearly. As temperature increases, conformational fluctuations become enabled, depending on the height of the activation barrier relevant to the conformational states of a given residue in the protein. In this higher temperature regime, the B-factor increases more rapidly than in the lower temperature regime. A protein dynamical transition has been identified with a sudden change in the slope of the roughly linear increase of the average protein B-factor with temperature (1, 10), which is suggestive of the onset of the conformational fluctuations (see SI Appendix).
Fig. 1C shows the B-factor profiles of five cryocooled crystals while they were warmed from 80 to 160 K. The B-factor profiles of high-pressure cryocooled crystals (colored curves) clearly show transitions in the temperature range. The breaks from linearity in the B-factor profiles start at roughly 110–120 K for Thau-0M-1 and Thau-0M-2, 135 K for Thau-0.45M, and 140 K for Thau-0.9M. These correlate with the HDA–LDA transition, as seen by changes in the WDD peak positions shown in Fig. 1B. By contrast, the ambient-pressure cryocooled crystal (Thau-control; Fig. 1C, black curve) shows no such transition in the B-factor profile over the range from 80 to 160 K. This observation correlates with no change in the WDD (Fig. 1B, black curve), which starts and remains in the LDA state over the given temperature range. These observations suggest that protein structural fluctuations between different conformational states are enabled when the solvent undergoes an HDA–LDA transition and are suppressed if the solvent is in an LDA state throughout (SI Appendix).
Although the B-factor analysis discussed above is a useful guideline for a protein dynamical transition, care should be taken in the interpretation of B-factor profiles. The X-ray crystallographic B-factor reflects the mean square atomic displacement 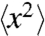 of atoms in a protein and consists of three dominant terms (8, 10) (SI Appendix). The first term accounts for lattice defects in crystalline packing (crystal lattice disorder) and can be monitored by crystal mosaicity. The other terms come from the internal structural motions of protein, which include both harmonic vibrations of individual atoms at a fixed position and molecular fluctuations between different conformational states. The lattice disorder in a protein crystal typically remains unchanged at cryogenic temperatures between 80 and 160 K for crystals that are cryocooled at ambient pressure. However, the lattice disorder in a high-pressure cryocooled protein crystal is changing at cryogenic temperatures through rearrangement in molecular packing (35) (SI Appendix). In those regions where the mosaicity is constant (or falling), the faster increase in the B-factor profiles can be attributed to new allowed fluctuations in the conformational states of the protein. However, a full quantitative analysis requires the separation of the lattice disorder term from the B-factor.
of atoms in a protein and consists of three dominant terms (8, 10) (SI Appendix). The first term accounts for lattice defects in crystalline packing (crystal lattice disorder) and can be monitored by crystal mosaicity. The other terms come from the internal structural motions of protein, which include both harmonic vibrations of individual atoms at a fixed position and molecular fluctuations between different conformational states. The lattice disorder in a protein crystal typically remains unchanged at cryogenic temperatures between 80 and 160 K for crystals that are cryocooled at ambient pressure. However, the lattice disorder in a high-pressure cryocooled protein crystal is changing at cryogenic temperatures through rearrangement in molecular packing (35) (SI Appendix). In those regions where the mosaicity is constant (or falling), the faster increase in the B-factor profiles can be attributed to new allowed fluctuations in the conformational states of the protein. However, a full quantitative analysis requires the separation of the lattice disorder term from the B-factor.
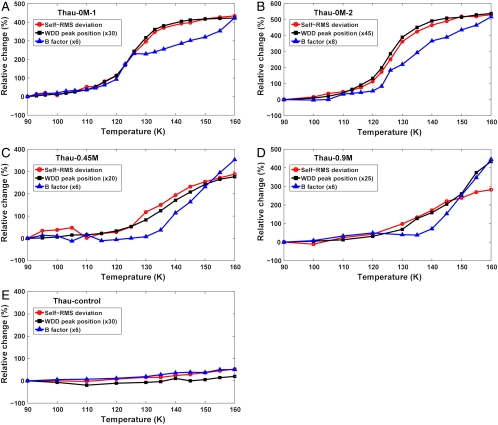
To more directly study changes in the protein conformational fluctuations at the HDA–LDA transition we investigated how the structure of the protein changed as temperature rose. The self root-mean-square (self-rms) deviation (see Materials and Methods) between the structure at the lowest temperature and the structures at higher temperatures from a single crystal was used as an additional metric for protein dynamical transitions. When previously prohibited structural fluctuations between different conformational states become allowed, one expects the averaged crystallographic structure to become slightly different from the structure before the protein dynamical transition. This structural change would then be reflected in an increase in the self-rms deviation profile.
The self-rms deviation profiles of five cryocooled crystals are plotted in Fig. 2 along with the WDD peak position profiles and B-factor profiles. It is obvious that the self-rms deviation profiles of all four high-pressure cryocooled crystals (Fig. 2 A–D) are highly correlated with the water phase transition indicated by the WDD peak position profiles. The B-factor profiles show some deviation from the WDD profiles. This deviation is due to the changes in the lattice disorder via molecular rearrangement during crystal warming (SI Appendix). The correlation between the self-rms deviation and the WDD peak position profiles suggests that the protein dynamical fluctuations were gradually turned on and the averaged protein structure was evolving in response to the water phase transition. By contrast, the self-rms deviation profile of the ambient-pressure cryocooled crystal (Fig. 2E) shows little change, along with no water phase transition over the temperature range. Therefore, we conclude that the observed protein dynamical transition inside a high-pressure cryocooled protein crystal is allowed by the water phase transition inside the crystal.
Fig. 2.
Self-rms deviation profiles of cryocooled thaumatin structures as an alternative indicator for the protein dynamical transition. The self-rms deviation of each crystal was calculated between the structure at 80 K and the structures at higher temperatures. Plotted is the relative change from the self-rms value at 90 K (see tables in SI Appendix). The WDD primary peak position and B-factor profiles (scaled for comparison with self-rms deviation profiles) are superimposed to clarify the correlation between the water transition and the protein dynamical transition. (A–D) Self-rms deviation profiles of high-pressure cryocooled crystals (Thau-0M-1, Thau-0M-2, Thau-0.45M, and Thau-0.9M). The self-rms deviation profiles of high-pressure cryocooled crystals show high correlation with the WDD peak position profiles. The B-factor profiles show significant correlation but are less precise, which is due to changes in the crystalline lattice disorder (see main text and SI Appendix). (E) Self-rms deviation profile of ambient-pressure cryocooled crystal (Thau-control). The profile of Thau-control shows no features of a dynamical transition.
Discussion
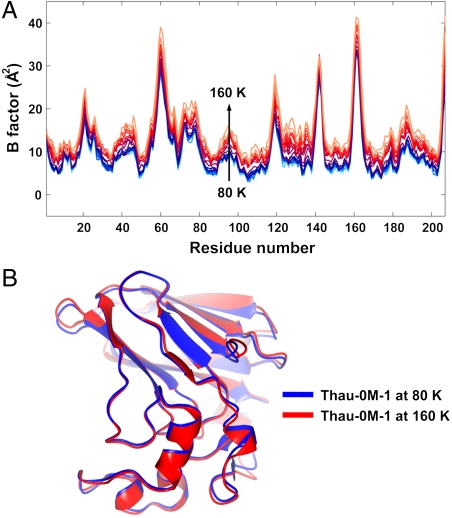
We have presented evidence that thaumatin protein undergoes a dynamical transition as low as 110 K, which is significantly lower than the typically accepted temperature range for the transition (180 ∼ 240 K). It has been reported that some proteins exhibit a rapid increase in the mean square atomic displacement below 150 K, but it was attributed to the onset of local structural motions (such as rotational motions of methyl group) rather than the global protein molecular motions (14, 16, 28, 39). Fig. 3A shows the averaged B-factor values along the main chain of a high-pressure cryocooled thaumatin, Thau-0M-1. As the temperature is raised the B-factor values rise for the entire protein molecule, suggesting that the observed dynamical transition in this study is not due to the increased local motions of a few specific side chains, but is rather due to the global motions over the whole protein molecule. The structural displacements of Thau-0M-1 between 80 and 160 K are illustrated in Fig. 3B. The displacements are small, but well outside of measurement error.
Fig. 3.
Global conformational fluctuations during protein dynamical transition. (A) The average B-factor values of the main chain atoms along the 207 thaumatin residues. As temperature is raised from 80 to 160 K, the B-factor profiles rise over the whole length of the chain. (B) Superposition of the structures of Thau-0M-1 at 80 K (blue) and 160 K (red). The rms atomic displacement in main chain atoms between two structures is 0.235 Å (magnified three times in the figure for visual clarity), which is well above the noise level (∼0.05 Å). This result demonstrates that the structural displacements are spread out over the whole protein molecule, indicating that the protein dynamical transition is due to global protein dynamical motions rather than a local motion such as a rotation of methyl group. A site-specific conformational relaxation in the disulfide bond during the dynamical transition can be found in the earlier work (35). Note that the structure at 160 K cannot be generated by the simple rigid-body motions of the structure at 80 K. This observation supports that the rms deviation is not the consequence of the crystal lattice disorder.
During the protein conformational fluctuations, the water surrounding the protein must accommodate changes in the shape of the protein and movement of at least those protein residues in contact with the water. Indeed, solvent mobility, or the translational dynamics of water, was shown to be the dominant factor in determining protein fluctuations in molecular dynamics simulations (21–23). Some proposed mechanisms for the protein dynamical transition at around 200 K (15, 20, 29–31) relate a protein dynamical transition with dynamical motions in hydration water. The fact that we observe thaumatin protein to undergo a cryogenic dynamical transition as low as 110 K suggests that water around the protein gains increased mobility at the HDA–LDA transition, even though it occurs at a deeply cryogenic temperature. This interpretation is consistent with the previous suggestions that protein conformational fluctuations are mainly restricted by the solvent viscosity rather than the potential energy barriers of the protein even at cryogenic temperatures (40–42).
Interestingly, the glass transition temperature of bulk water has been estimated to be 136 K or higher (43). It was reported that confined water shows a glass transition at even higher temperatures than bulk water (44). The viscosity of water diverges as one approaches the water glass transition temperature; therefore, translational diffusive motions of water molecules are restricted. Considering that a protein dynamical transition is triggered by the increased mobility of water around protein, how can we understand the protein dynamical transition at 110 K, well below the known glass transition temperature of water? It is important to note that the cryogenic protein dynamical transition in our study is observed along with the phase transition from HDA to LDA states of water. It has been proposed that the water phase transition from HDA to LDA may involve a high mobility, liquid form of water: The HDA ice first undergoes a glass-liquid transition, then continuously converts to LDA ice (35).
One theoretical explanation for the relationship between the cryogenic protein dynamical transition and the water phase transition from HDA to LDA states can be found from the liquid-liquid (LL) critical point theory (45) for water. The LL critical theory was originally proposed to explain the anomalous thermodynamic properties of the supercooled water (46–48). The theory predicts a first-order phase transition between HDA and LDA ice (35, 46). The theory also requires the liquid state counterparts of HDA and LDA ice (46–48). Based on the LL critical theory, the observed protein dynamical transition at cryogenic temperatures might be related to the two proposed liquid forms of water (high-density liquid and low-density liquid) existing well below the homogeneous nucleation temperature of water (35, 46–49). It remains to be seen how the cryogenic protein dynamical transition is related to the conventional protein dynamical transition at around 200 K, which is thought to be a precondition for a biologically active state of proteins.
Materials and Methods
Protein Crystallization and Crystal Handling.
Lyophilized thaumatin powder from Thaumatococcus daniell (catalog no. T7638, Sigma) was used for crystallization without further purification. Crystals were grown at 20 °C by the hanging-drop method with 25 mg/mL thaumatin solution in 50 mM Hepes buffer at pH 7 and crystallization solution containing 0.9 M sodium potassium tartrate (NaK tartrate) as a precipitant. The crystal space group was determined to be P41212 (a = b = ∼ 58 Å, c = ∼ 150 Å), having a solvent content of 60% by volume. To adjust solvent concentrations in protein crystals, the fully grown thaumatin crystals were equilibrated to 0.9 M, 0.45 M sodium potassium tartrate solutions (labeled as Thau-0.9M and Thau-0.45M, respectively), and to deionized water (labeled as Thau-0M-1 and Thau-0M-2), before high-pressure cryocooling at 200 MPa (34). To reduce osmotic shock, crystals were gradually transferred to the target concentration in 0.1 M steps. We found that thaumatin crystals are stable in deionized water over several hours.
Crystal Cryocooling.
High-pressure cryocooling of crystal samples was carried out as described by Kim et al. (34). In brief, samples were loaded into the high-pressure cryocooling apparatus, which was then pressurized with helium gas to 200 MPa at ambient temperature. Once at high pressure, the samples were cryocooled to liquid-nitrogen temperature (77 K). Helium pressure was then released. Thereafter crystal samples were handled/stored at ambient pressure and at liquid-nitrogen temperature before X-ray diffraction measurements. For crystals cryocooled at ambient pressure, 20% glycerol (vol/vol) was added to the crystal as a cryoprotectant to reduce crystal damage upon cryocooling. Then the crystal was cryocooled by directly plunging into liquid nitrogen at ambient pressure. An X-ray diffraction study confirmed that the added glycerol does not affect the protein dynamical transition at cryogenic temperatures (SI Appendix). Before cryocooling of crystals, liquid surrounding the crystals was carefully removed during crystal-coating with a mineral oil (34). Therefore the water diffuse diffraction is almost entirely from the fluid inside the crystal.
X-Ray Diffraction Data Collection.
The X-ray diffraction data were collected at the macromolecular crystallography stations F1 (λ = 0.9179 Å, Area Detector Systems Corporation (ADSC) Quantum 270 CCD detector, beam size of 100 μm), and F2 (λ = 0.9795 Å, ADSC Quantum 210 CCD detector, beam size of 150 μm) at the Cornell High Energy Synchrotron Source (CHESS). During data collection, the sample temperature ranging from 80 to 160 K was controlled by a Cryostream 700 series cryocooler (Oxford Cryosystems). The sample temperature was raised at the rate of 6 K/ min. After reaching a desired temperature, samples were left at the temperature for 3–5 min for equilibration. The X-ray diffraction data of the protein crystals were collected with temperature steps from 3 to 10 K (see tables in SI Appendix). At each temperature, a complete dataset was collected covering 50 to 90° of crystal rotation (50° for Thau-0M-2, 60° for Thau-0.45M/Thau-0.9M/Thau-control, and 90° for Thau-0M-1). The X-ray exposure time was 1 s with oscillation angle of 1°. The data collection parameters were the same for all the complete datasets from a single crystal except for the crystal temperature adjustment.
Data Analysis on Water Diffuse Diffraction.
The underlying diffuse diffraction from the diffraction image was isolated from the Bragg spots by applying a custom polar coordinate median filter to the intensity values of the image (36). The sample to detector distance was calibrated based on the known Bragg peaks of hexagonal ice. To determine the position of the WDD peak, the median-filtered diffuse scattering curves were fit to a series of three Voigt functions plus linear background; one Voigt function at the position of the oil scattering peak, one at the main WDD peak, and a third function at the secondary WDD peak.
Structure Refinement.
The complete thaumatin datasets were processed and refined with HKL2000 (50) and CCP4 program suite (51). To minimize errors in structural refinement, the structure at 80 K was used as a starting model for the structural refinement at higher temperatures. The refined structures were proofread with COOT (52) and structural errors were carefully corrected. The B-factor values of main chain atoms in the final refined structures were calculated with CCP4 program suite (51). The self-rms deviation between structures at 80 K and higher temperatures were calculated using CCP4 program suite by aligning main chain atoms from residues 1–204 of thaumatin molecules. The final three residues (205–207) were excluded in the self-rms deviation calculation because those residues were highly disordered in all temperatures. Details in data processing and structure refinement can be found in tables in SI Appendix.
Supplementary Material
Acknowledgments.
We thank Marian Szebenyi and Dave Schuller for useful comments, Ji-Won Park for assistance in manuscript preparation, and the Cornell High Energy Synchrotron Source (CHESS) staff for support in data collection. This work is based upon research conducted at CHESS, which is supported by the National Science Foundation (NSF) and the National Institutes of Health (NIH)/National Institute of General Medical Sciences under NSF award DMR-0936384, using the Macromolecular Diffraction at CHESS (MacCHESS) facility, which is supported by award RR-01646 from NIH, through its National Center for Research Resources.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110840108/-/DCSupplemental.
References
- 1.Ringe D, Petsko GA. The ‘glass transition’ in protein dynamics: What it is, why it occurs, and how to exploit it. Biophys Chem. 2003;105:667–680. doi: 10.1016/s0301-4622(03)00096-6. [DOI] [PubMed] [Google Scholar]
- 2.Ferrand M, Dianoux AJ, Petry W, Zaccai G. Thermal motions and function of bacteriorhodopsin in purple membranes: Effects of temperature and hydration studied by neutron scattering. Proc Natl Acad Sci USA. 1993;90:9668–9672. doi: 10.1073/pnas.90.20.9668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ostermann A, Waschipky R, Parak FG, Nienhaus GU. Ligand binding and conformational motions in myoglobin. Nature. 2000;404:205–208. doi: 10.1038/35004622. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen BF, Stock AM, Ringe D, Petsko GA. Crystalline ribonuclease A loses function below the dynamic transition at 220 K. Nature. 1992;357:423–424. doi: 10.1038/357423a0. [DOI] [PubMed] [Google Scholar]
- 5.Knapp EW, Fischer SF, Parak F. Protein dynamics from mossbauer spectra. The temperature dependence. J Phys Chem. 1982;86:5042–5047. [Google Scholar]
- 6.He Y, Ku PI, Knab JR, Chen JY, Markelz AG. Protein dynamical transition does not require protein structure. Phys Rev Lett. 2008;101:178103. doi: 10.1103/PhysRevLett.101.178103. [DOI] [PubMed] [Google Scholar]
- 7.Chong SH, et al. Dynamical transition of myoglobin in a crystal: Comparative studies of X-ray crystallography and Mössbauer spectroscopy. Eur Biophys J. 2001;30:319–329. doi: 10.1007/s002490100152. [DOI] [PubMed] [Google Scholar]
- 8.Frauenfelder H, Petsko GA, Tsernoglou D. Temperature-dependent X-ray diffraction as a probe of protein structural dynamics. Nature. 1979;280:558–563. doi: 10.1038/280558a0. [DOI] [PubMed] [Google Scholar]
- 9.Tilton RF, Dewan JC, Petsko GA. Effects of temperature on protein structure and dynamics: X-ray crystallographic studies of the protein ribonuclease-A at 9 different temperatures from 98 to 320 K. Biochemistry. 1992;31:2469–2481. doi: 10.1021/bi00124a006. [DOI] [PubMed] [Google Scholar]
- 10.Ringe D, Petsko GA. Study of protein dynamics by X-ray diffraction. Methods Enzymol. 1986;131:389–433. doi: 10.1016/0076-6879(86)31050-4. [DOI] [PubMed] [Google Scholar]
- 11.Hartmann H, et al. Conformational substates in a protein: Structure and dynamics of metmyoglobin at 80 K. Proc Natl Acad Sci USA. 1982;79:4967–4971. doi: 10.1073/pnas.79.16.4967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doster W, Cusack S, Petry W. Dynamical transition of myoglobin revealed by inelastic neutron scattering. Nature. 1989;337:754–756. doi: 10.1038/337754a0. [DOI] [PubMed] [Google Scholar]
- 13.Nakagawa H, Joti Y, Kitao A, Kataoka M. Hydration affects both harmonic and anharmonic nature of protein dynamics. Biophys J. 2008;95:2916–2923. doi: 10.1529/biophysj.107.128546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roh JH, et al. Onsets of anharmonicity in protein dynamics. Phys Rev Lett. 2005;95:038101. doi: 10.1103/PhysRevLett.95.038101. [DOI] [PubMed] [Google Scholar]
- 15.Chen SH, et al. Observation of fragile-to-strong dynamic crossover in protein hydration water. Proc Natl Acad Sci USA. 2006;103:9012–9016. doi: 10.1073/pnas.0602474103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schiro G, Caronna C, Natali F, Cupane A. Molecular origin and hydration dependence of protein anharmonicity: An elastic neutron scattering study. Phys Chem Chem Phys. 2010;12:10215–10220. doi: 10.1039/c003482g. [DOI] [PubMed] [Google Scholar]
- 17.Doster W, et al. Dynamical transition of protein hydration water. Phys Rev Lett. 2010;104:098101. doi: 10.1103/PhysRevLett.104.098101. [DOI] [PubMed] [Google Scholar]
- 18.Wood K, et al. Coupling of protein and hydration-water dynamics in biological membranes. Proc Natl Acad Sci USA. 2007;104:18049–18054. doi: 10.1073/pnas.0706566104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chu XQ, et al. Proteins remain soft at lower temperatures under pressure. J Phys Chem B. 2009;113:5001–5006. doi: 10.1021/jp900557w. [DOI] [PubMed] [Google Scholar]
- 20.Kumar P, et al. Glass transition in biomolecules and the liquid-liquid critical point of water. Phys Rev Lett. 2006;97:177802. doi: 10.1103/PhysRevLett.97.177802. [DOI] [PubMed] [Google Scholar]
- 21.Tarek M, Tobias DJ. Role of protein-water hydrogen bond dynamics in the protein dynamical transition. Phys Rev Lett. 2002;88:138101. doi: 10.1103/PhysRevLett.88.138101. [DOI] [PubMed] [Google Scholar]
- 22.Tournier AL, Xu JC, Smith JC. Translational hydration water dynamics drives the protein glass transition. Biophys J. 2003;85:1871–1875. doi: 10.1016/S0006-3495(03)74614-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vitkup D, Ringe D, Petsko GA, Karplus M. Solvent mobility and the protein ‘glass’ transition. Nat Struct Biol. 2000;7:34–38. doi: 10.1038/71231. [DOI] [PubMed] [Google Scholar]
- 24.Guo JG, Budarz T, Ward JM, Prohofsky EW. Dynamical transition in proteins and non-Gaussian behavior of low-frequency modes in self-consistent normal mode analysis. Phys Rev E. 2010;82:041917. doi: 10.1103/PhysRevE.82.041917. [DOI] [PubMed] [Google Scholar]
- 25.Kohn JE, Afonine PV, Ruscio JZ, Adams PD, Head-Gordon T. Evidence of functional protein dynamics from X-ray crystallographic ensembles. PLoS Comput Biol. 2010;6:e1000911. doi: 10.1371/journal.pcbi.1000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fenimore PW, Frauenfelder H, McMahon BH, Parak FG. Slaving: Solvent fluctuations dominate protein dynamics and functions. Proc Natl Acad Sci USA. 2002;99:16047–16051. doi: 10.1073/pnas.212637899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teeter MM, Yamano A, Stec B, Mohanty U. On the nature of a glassy state of matter in a hydrated protein: Relation to protein function. Proc Natl Acad Sci USA. 2001;98:11242–11247. doi: 10.1073/pnas.201404398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roh JH, et al. Influence of hydration on the dynamics of lysozyme. Biophys J. 2006;91:2573–2588. doi: 10.1529/biophysj.106.082214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Frauenfelder H, et al. A unified model of protein dynamics. Proc Natl Acad Sci USA. 2009;106:5129–5134. doi: 10.1073/pnas.0900336106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fenimore PW, Frauenfelder H, McMahon BH, Young RD. Bulk-solvent and hydration-shell fluctuations, similar to alpha- and beta-fluctuations in glasses, control protein motions and functions. Proc Natl Acad Sci USA. 2004;101:14408–14413. doi: 10.1073/pnas.0405573101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Doster W. The protein-solvent glass transition. Biochim Biophys Acta. 2010;1804:3–14. doi: 10.1016/j.bbapap.2009.06.019. [DOI] [PubMed] [Google Scholar]
- 32.Becker T, Hayward JA, Finney JL, Daniel RM, Smith JC. Neutron frequency windows and the protein dynamical transition. Biophys J. 2004;87:1436–1444. doi: 10.1529/biophysj.104.042226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Khodadadi S, et al. The origin of the dynamic transition in proteins. J Chem Phys. 2008;128:195106. doi: 10.1063/1.2927871. [DOI] [PubMed] [Google Scholar]
- 34.Kim CU, Kapfer R, Gruner SM. High pressure cooling of protein crystals without cryoprotectants. Acta Crystallogr Sect D Biol Crystallogr. 2005;61:881–890. doi: 10.1107/S090744490500836X. [DOI] [PubMed] [Google Scholar]
- 35.Kim CU, Barstow B, Tate MW, Gruner SM. Evidence for liquid water during the high-density to low-density amorphous ice transition. Proc Natl Acad Sci USA. 2009;106:4596–4600. doi: 10.1073/pnas.0812481106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim CU, Chen YF, Tate MW, Gruner SM. Pressure induced high-density amorphous ice in protein crystals. J Appl Crystallogr. 2008;41:1–7. [Google Scholar]
- 37.Mishima O, Calvert LD, Whalley E. ‘Melting ice’ I at 77 K and 10 kbar: A new method of making amorphous solids. Nature. 1984;310:393–395. [Google Scholar]
- 38.Ghormley JA, Hochanad CJ. Amorphous ice—density and reflectivity. Science. 1971;171:62–64. doi: 10.1126/science.171.3966.62. [DOI] [PubMed] [Google Scholar]
- 39.Lee AL, Wand AJ. Microscopic origins of entropy, heat capacity and the glass transition in proteins. Nature. 2001;411:501–504. doi: 10.1038/35078119. [DOI] [PubMed] [Google Scholar]
- 40.Ansari A, Jones CM, Henry ER, Hofrichter J, Eaton WA. The role of solvent viscosity in the dynamics of protein conformational changes. Science. 1992;256:1796–1798. doi: 10.1126/science.1615323. [DOI] [PubMed] [Google Scholar]
- 41.Ansari A, Jones CM, Henry ER, Hofrichter J, Eaton WA. Conformational relaxation and ligand binding in myoglobin. Biochemistry. 1994;33:5128–5145. doi: 10.1021/bi00183a017. [DOI] [PubMed] [Google Scholar]
- 42.Hagen SJ, Hofrichter J, Eaton WA. Geminate rebinding and conformational dynamics of myoglobin embedded in a glass at room temperature. J Phys Chem. 1996;100:12008–12021. [Google Scholar]
- 43.Velikov V, Borick S, Angell CA. The glass transition of water, based on hyperquenching experiments. Science. 2001;294:2335–2338. doi: 10.1126/science.1061757. [DOI] [PubMed] [Google Scholar]
- 44.Cerveny S, Schwartz GA, Bergman R, Swenson J. Glass transition and relaxation processes in supercooled water. Phys Rev Lett. 2004;93:245702. doi: 10.1103/PhysRevLett.93.245702. [DOI] [PubMed] [Google Scholar]
- 45.Poole PH, Sciortino F, Essmann U, Stanley HE. Phase behavior of metastable water. Nature. 1992;360:324–328. [Google Scholar]
- 46.Mishima O, Stanley HE. The relationship between liquid, supercooled and glassy water. Nature. 1998;396:329–335. [Google Scholar]
- 47.Debenedetti PG. Supercooled and glassy water. J Phys Condens Matter. 2003;15:R1669–R1726. [Google Scholar]
- 48.Debenedetti PG, Stanley HE. Supercooled and glassy water. Phys Today. 2003;56:40–46. [Google Scholar]
- 49.Stanley HE, et al. The puzzling unsolved mysteries of liquid water: Some recent progress. Physica A. 2007;386:729–743. [Google Scholar]
- 50.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 51.Collaborative Computational Project No. 4. The ccp4 suite: Programs for protein crystallography. Acta Crystallogr Sect D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 52.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr Sect D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.