Abstract
Hepatitis C virus (HCV) infection is a global health burden with over 170 million people infected worldwide. In a significant portion of patients chronic hepatitis C infection leads to serious liver diseases, including fibrosis, cirrhosis, and hepatocellular carcinoma. The HCV NS3 protein is essential for viral polyprotein processing and RNA replication and hence viral replication. It is composed of an N-terminal serine protease domain and a C-terminal helicase/NTPase domain. For full activity, the protease requires the NS4A protein as a cofactor. HCV NS3/4A protease is a prime target for developing direct-acting antiviral agents. First-generation NS3/4A protease inhibitors have recently been introduced into clinical practice, markedly changing HCV treatment options. To date, crystal structures of HCV NS3/4A protease inhibitors have only been reported in complex with the protease domain alone. Here, we present a unique structure of an inhibitor bound to the full-length, bifunctional protease-helicase NS3/4A and show that parts of the P4 capping and P2 moieties of the inhibitor interact with both protease and helicase residues. The structure sheds light on inhibitor binding to the more physiologically relevant form of the enzyme and supports exploring inhibitor-helicase interactions in the design of the next generation of HCV NS3/4A protease inhibitors. In addition, small angle X-ray scattering confirmed the observed protease-helicase domain assembly in solution.
Keywords: structure-based drug design, X-ray structure, solution scattering, medicinal chemistry
Chronic hepatitis C virus (HCV) infection affects more than 3% of the world’s population and is a leading cause of chronic liver diseases (1). Therapeutic options have been suboptimal, especially for HCV genotype 1, the most prevalent genotype in developed countries. Recently, the addition of direct-acting antiviral agents (DAAs) to the previous standard of care (combination therapy with pegylated interferon and ribavirin) have demonstrated considerable improvement in sustained virological response rates in patients infected with HCV genotype 1 (2). With the first DAAs now having been introduced into clinical practice, it is to be expected that in the near future standard therapy will change to a triple therapy including an HCV NS3/4A protease inhibitor in combination with pegylated interferon and ribavirin.
The positive-strand RNA genome of HCV encodes a polyprotein precursor, which is proteolytically processed by host and viral proteases into 10 individual structural and nonstructural (NS) proteins. The viral NS3 protease in complex with the cofactor NS4A cleaves the polyprotein at four junctions releasing the NS proteins 4A, 4B, 5A, and 5B, and therefore is essential for viral replication (3, 4). A central, hydrophobic 14-mer peptide of the 54-residue NS4A, comprising residues 21–32, is necessary and sufficient for maximal activation in vitro (5). NS3/4A has also been shown to cleave cellular proteins leading to inhibition of interferon production, thereby impairing the innate immune response against viral infections (6). NS3 is unusual as it possesses in addition to the N-terminal protease domain (residues 1–180) a C-terminal, ATP-dependent superfamily 2 RNA helicase (residues 181–631). The isolated domains of NS3, i.e., the protease and helicase domains are functional on their own. It has been reported that in the full-length enzyme the NS3/4A protease enhances RNA binding and unwinding by the helicase and that also the helicase enhances protease activity (7, 8). In the past decade, HCV NS3/4A protease has emerged as an important drug target for treatment of HCV infection. Following intensive drug discovery efforts, several inhibitors are now in clinical development showing significant viral load reduction in patients (1) and two first-generation drugs have recently gained US Food and Drug Administration approval.
The crystal structures of the HCV NS3 protease domain in the absence and presence of an NS4A cofactor peptide were published in 1996 (9, 10). The protease has a chymotrypsin-like fold composed of two beta barrels with the catalytic triad located at the domain-interface. The NS4A peptide forms a beta strand which is part of the N-terminal beta barrel. This structural integration triggers the formation of a competent catalytic triad, explaining the role of the cofactor in protease activation. Currently approximately 50 HCV NS3/4A protease structures are available in the Protein Data Bank (PDB). In most cases the protease has been complexed with inhibitors, including two macrocyclic acylsulfonamides (11, 12). The NS3/4A full-length protease-helicase apo crystal structure was reported in 1999 (13) and recently several complexes with helicase ligands have been published (14). These structures show the C-terminal residues of the helicase occupying the nonprime side of the protease active site, corresponding to the product of cis-cleavage at the NS3-NS4A junction. Several additional helicase residues in the immediate vicinity of the protease active site participate in the domain interface leading to an overall compact assembly of the protease and helicase domains. The compact conformation is maintained in complexes of full-length HCV NS3/4A with helicase ligands (8-mer single-stranded RNA, nucleotides) permitting conformational changes within the helicase subdomains resulting from ATP binding and hydrolysis (14). Additional domain orientations are to be expected for the full repertoire of reactions catalyzed by the bifunctional enzyme in the membrane-associated replication complex. Ding et al. (15) have recently reported enzymatic data supporting a fully open form as proposed by Brass et al. (4).
The significance of the presumed interactions of helicase residues with protease inhibitors in the compact conformation as observed in the crystal structures has been addressed by several groups. Dahl et al. observed a 10-fold decrease in binding affinity of BILN2061 to the full-length NS3/4A harboring an H528S mutation (16). Thibeault et al., on the other hand, report that the full-length NS3/4A triple mutant M485A, V524A, Q526A did not lead to a significant change in KI for eight inhibitors tested (17). An example where the structural information of helicase residues was explicitly considered in the design of P2–P4 macrocyclic protease inhibitors was reported by Liverton et al. (18).
Several crystal structures of full-length NS3 proteins from another genus of the flaviviridae family, the flaviviruses, have been reported, those of Dengue virus and Murray Valley encephalitis virus (19–21). Flaviviral NS3 proteins employ NS2B rather than NS4A as protease cofactor (see ref. 22 for a review). Compared to the hepaciviral HCV NS3/4A, the flaviviral NS3/2B proteins show a different respective arrangement of protease and helicase domains with the protease active site fully solvent exposed. Two alternative conformations related by a ca. 160° rotation of the protease domain with respect to the helicase in the interdomain linker region have been reported (19–21). This interdomain flexibility may have mechanistic implications for flaviviral NS3 proteins. Small angle X-ray scattering (SAXS) data from full-length Dengue and Kunjin NS2B/NS3 proteins indicate an elongated shape and support the domain assembly observed in the flaviviral crystal structures (19, 23). Although there are significant differences between hapacivirus and flavivirus NS3 proteins, including the different cofactor, these results reinforced our interest to investigate if the domain orientation observed previously (13) is representative of the HCV NS3/4A domain organization in solution. To shed light on this question, we pursued SAXS experiments using full-length HCV NS3/4A.
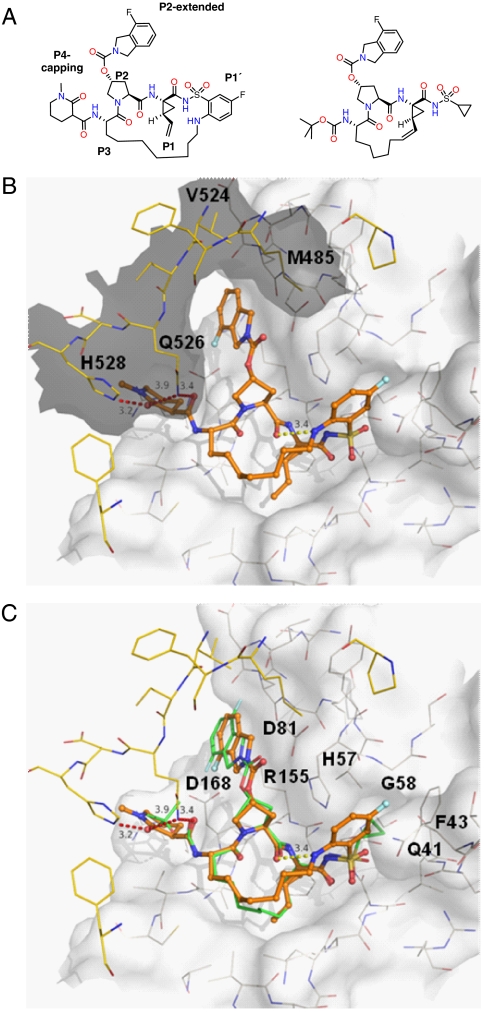
Based on feasibility and corroborated by the finding that protease inhibitors tested show comparable IC50 values toward the protease domain and the full-length protein, HCV protease structure-based drug design has focused almost exclusively on the interactions with the protease domain alone. An in-house X-ray structure of NS3 protease in complex with an acyclic acylsulfonamide inhibitor harboring a phenyl acetamide group in P1′ suggested a macrocyclization between the P1′ and P3 residue. Among different tethers and linker lengths the noncovalent, reversible acylsulfonamide inhibitor 1 revealed as the most potent analogue containing a 20-membered macrocycle (Fig. 1A, Left). To date, no structure of full-length HCV NS3/4A in complex with a protease inhibitor has been published. We have now determined such a structure in complex with inhibitor 1 at 2.7 Å resolution and observe the same domain orientation as in the apo structure (13) also when an inhibitor is bound.
Fig. 1.
Structure and active-site view of inhibitor 1 bound to full-length HCV NS3/4A. For comparison, ITMN191 for which the NS3/4A protease domain complex is reported is shown (12). (A) Structure of inhibitor 1 (Left) and of ITMN-191 (Right) (PDB ID code 3M5L) (12). The IC50 inhibition data of inhibitor 1 are [μM]: gt1a NS3FL/4A: 0.010; gt1a NS3PD/4A: 0.005; gt1b NS3FL/4A: 0.032; gt1b NS3PD/4A: 0.0073; gt1b NS3FL/4A E628A_T631L: 0.028 (gt, genotype; FL, full-length; PD, protease domain). (B) Active-site view with inhibitor 1 bound. The inhibitor is shown with orange carbon atoms, protease residues with gray and helicase residues with yellow carbon atoms in a 7-Å shell around the inhibitor. The protease and helicase surfaces are shown in white and gray, respectively. (C) Active-site view of complex with inhibitor 1 (color scheme as in Fig. 1B) showing ITMN-191 superimposed [green carbon atoms (PDB ID code 3M5L)] (12). The surface is only shown for protease domain residues. (B) and (C) were prepared with PyMOL (36).
Results and Discussion
For crystal soaking to be successful, the inhibitor needs to displace the C-terminal helicase residues occupying the nonprime side of the protease active site. To destabilize these interactions, we have introduced two C-terminal point mutations, E628A and T631L. Apo crystals of the double-mutant could be soaked with inhibitor 1. The crystals are isomorphous to the previously published crystal structures of full-length HCV NS3/4A containing a dimer in the asymmetric unit. The inhibitor binds to the protease active site employing a substrate-like binding mode as seen for related peptidomimetic inhibitors in complex with the HCV NS3/4A protease domain and interacts in the S4 to S1′ subsites (Fig. 1 B and C).
Interactions with Helicase Residues.
The P4-capping and P2 moieties of inhibitor 1 are exposed toward the helicase interface and interact both with protease and helicase residues. The inhibitor buries 170 Å2 of accessible surface area (ASA) of helicase residues involving in particular residues Met485 and the segment Val524 to His528. Upon inhibitor binding there are two significant changes in the crystal involving helicase residues in the active site. Residues beyond Ala625 are disordered, i.e., the helicase C terminus is not represented by electron density. The nonprime portions of the inhibitor take on the position occupied by helicase residues Glu628, Val629, Val630, and Thr631 in the apo structure (13). In addition, residue Gln526, which in the apo structure is in H-bonding distance to Glu628, adopts a different rotamer conformation to accomodate the P4-capping and P2-extended portion of the inhibitor. In its new orientation it is positioned with its Nϵ at a distance of 3.3 Å to the inhibitor P4 carbonyl and at a distance of 3.2 Å to the piperidine oxygens, forming weak helicase-inhibitor interactions. In addition, the piperidine oxygen is involved in an H-bond to His528-Nϵ. The P2-proline and the isoindole ring are in van der Waals contact with helicase residues Met485 and Gln526. The IC50 values determined for inhibitor 1, a 1∶1 mixture of diastereoisomers, with the genotype 1a and 1b protease domain and full-length enzymes indicate a slightly higher potency with the former (Fig. 1A, legend). It is assumed that the enantiomer with the S-configuration, modeled in the X-ray structure, would inhibit the full-length NS3/4A protease more potently. Whereas for inhibitor 1 we observe rather weak helicase interactions, we expect, based on our structure, that inhibitors with additional helicase interactions should offer a route to gain potency.
Interactions with Protease Residues and Comparison to HCV NS3/4A Protease in Complex with ITMN-191.
A clearly more intimate interaction of the inhibitor is observed with protease domain residues, also reflected in an ASA burial of 447 Å2. The aliphatic portion of the piperidine ring is in van der Waals distance to the protease domain Ala156 and Val158 side chains. The P3 backbone amide nitrogen and carbonyl oxygen form hydrogen bonds to the Ala157 backbone, which is part of the canonical antiparallel beta strand involved in substrate recognition. The aliphatic P3 side chain runs along the S3-patch formed by Ala157, Cys159, and Val132 superimposing up to Cγ with ITMN-191 (Fig. 1A, Right), a macrocyclic acylsulfonamide inhibitor, the structure of which has been described bound to the isolated NS3/4A protease domain harboring an active-site Ser139Ala mutation (12) (Fig. 1C). Following the path of the Lys136 side chain, the P3 moiety continues to form the linker to the inhibitor P1′ residue. In the apo structure and in complexes with inhibitors lacking P2-extensions, Arg155 is part of an electrostatic network with alternating charges extending from Arg123, Asp168, Arg155 to the catalytic residues Glu81, and His57. Upon binding of inhibitor 1, Arg155 switches conformation to allow positioning of the inhibitor P2-extended isoindoline ring, breaking side-chain hydrogen bonds formed with the catalytic Glu81 and with helicase residues Gln526 and Glu628. In its new orientation, the Arg155 side chain is still hydrogen bonded with Asp168 and newly bonded to the carbonyl oxygen of Gln80 and acts as a binding platform for the inhibitor isoindoline moiety. Corresponding Arg155 rotamers have been reported for NS3/4A protease in complex with TMC435 (11), ITMN-191 (12), and BILN-261 (24). It is interesting to note that the P2-extended isoindoline portion in ITMN-191, identical to inhibitor 1, binds with a slight distal-shift of about 0.75 Å in the absence of the helicase. In the ITMN-191 complex, solved at 1.25 Å resolution, a second orientation for the isoindole ring is modeled, rotated by 180° with the fluorine pointing up (Fig. 1C). Although the protease active site subsites are in general relatively flat and featureless, the cyclopropylvinyl side chain of the P1 residue snugly fits into a surface depression bounded by residues Phe154, Ala157, and Leu135. The P1 amide nitrogen forms the third conserved H-bond with the backbone carbonyl of Arg155 and the P1 carbonyl oxygen is placed in the oxyanion hole formed by the amide nitrogens of Gly137 and Ser139. The inhibitor sulfonyl oxygens form H-bonds with the backbone amide nitrogen of Gly135 and with the Oγ of Ser139, which changes rotamer upon inhibitor binding, loosing the interaction with His57. The corresponding inactive conformation of Ser139 is also observed in the TMC435 complex.
The P1′ fluorophenyl ring is sandwiched in the cleft formed by His57, pointing with the edge of its imidazole toward the ring-center, and Gln41, engaged in a π-π-stacking interaction with the other face of the phenyl ring. Compared to the structure of ITMN-191 bound to the isolated NS3/4A protease (12), Gln41 adapts its rotamer conformation to engage in this interaction (Fig. 1C). The phenyl moiety does not project as deeply into the S1′ pocket as observed for the cyclopropyl group of ITMN-191 keeping a distance of ca. 5 Å from Phe43 at the base of the pocket. The fluorine is at 3.2 Å from the Gly58 Cα atom and its presence does not affect potency. The phenyl aminoalkyl substituent is part of the macrocyclic tether to the P3 moiety and forms with its nitrogen an internal H-bond with the P2 carbonyl oxygen.
Small Angle X-ray Scattering.
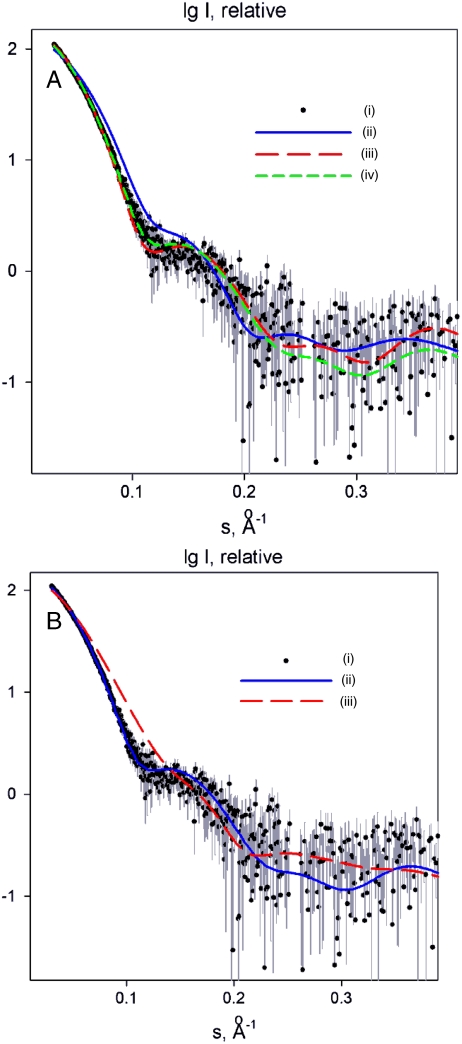
To answer the question whether the domain arrangement observed in the crystal structure is preserved in solution, SAXS analysis was performed. The solution SAXS data from the recombinant wild-type full-length NS3/4A were compared with available crystallographic models. The scattering calculated from the coordinates of the full-length HCV NS3/4A dimer constituting the asymmetric unit [PDB ID code 1CU1 (13)] fits the experimental data very well with discrepancy χ = 2.0 (Fig. 2A). It is interesting to note that although the fit by the monomer (PDB ID code 1CU1) is much worse than that of the dimer (discrepancy χ = 7.8), allowing for a monomer-dimer equilibrium gives an even better agreement with the experiment than for the dimer alone. The best mixture of ca. 25% monomeric and 75% dimeric forms yields an excellent fit with χ = 1.3, indicating that the solute is largely dimeric but partially dissociates into monomers. The SAXS data unambiguously prove the domain orientation as in the crystal structure. The experimental data cannot be reconciled with the scattering patterns computed from a full-length flaviviral NS2B/3 structure with an extended domain orientation, exemplified by the Dengue virus structure 2VBC (17) for which no satisfactory fit could be obtained (discrepancy χ = 7.5; Fig. 2B). These results further support the relevance of the HCV-NS3/4A full-length crystal structures and of the different domain assembly of flaviviral and hepaciviral NS3 full-length proteins in solution.
Fig. 2.
SAXS analysis of the conformation of full-length NS3/4A in solution. (A, i) Experimental scattering; (ii) calculated scattering from PDB ID code 1CU1 (12) monomer, discrepancy of the fit χ = 7.8; (iii) calculated scattering from PDB ID code 1CU1 dimer, χ = 2.0; (iv) calculated scattering from the best mixture of 25% monomers and 75% dimers, χ = 1.3. The logarithm of the scattering intensity is plotted as a function of momentum transfer s = 4π sin(θ)/λ, where θ is the scattering angle and λ at 0.15 nm is the X-ray wavelength. (B, i) Experimental scattering; (ii) calculated scattering from the best mixture of PDB ID code 1CU1 monomers and dimers (13), discrepancy of the fit χ = 1.3; (iii) calculated scattering from the crystal structure of full-length NS3/2B from Dengue virus [PDB ID code 2VBC (19)], χ = 7.5.
Conclusion
In summary, we have shown that in addition to interactions with protease residues, inhibitor 1 is forming weak interactions also with helicase residues. We are not aware of any systematic efforts in the field to include helicase interactions in the design of NS3/4A protease inhibitors. The structure reported here suggests that the latter approach could be viable in the pursuit of next generation HCV-protease inhibitors.
Methods
Inhibitor 1.
Details regarding the synthesis of macrocyclic inhibitor 1 are described in patent WO2008101665(A1) (25). The inhibitor was obtained as a 1∶1 mixture of diastereoisomers. It is assumed that the S-configuration at the 1-methyl-2-oxo-piperidine-3-(S)-carboxylic acid amide should be more potent, however, this assumption could not be proven because separation of the two isomers was not feasible due to rapid epimerization at the piperidinyl 3-position.
Cloning and Expression of Single-Chain Recombinant HCV NS3/4A Constructs.
The single-chain constructs encoding HCV NS3/4A full-length (gt1a/HCV-1 strain) and HCV NS3/4A protease domain (gt1a/HCV-1 strain and gt1b/BK strain) in which the activation sequences of the NS4A peptides (residues 21–32) followed by a Gly-Ser linker are covalently tethered to the N terminus of full-length NS3 (residues 3–631) were cloned as described. The corresponding single-chain NS3/4A protease construct had previously been shown to have equivalent enzymatic activity as the authentic NS3 protease NS4A complex (5). The single-chain construct encoding full-length HCV NS3/4A_E628A_T631L (gt1b/BK strain) used to determine the complex structure was obtained from the gt1a/HCV-1 strain full-length construct by site-directed mutagenesis to match the sequence as published by Yao et al. (13) with two mutations at the C terminus, E628A and T631L (Fig. S1). All cDNAs were cloned in pET28a for expression in Escherichia coli.
E. coli strain BL21(DE3) harboring the expression plasmid was cultivated at 37 °C in LB medium containing chloramphenicol and kanamycin and induced at OD600 = 0.8 with 0.4 mM IPTG after cooling to 16 °C. After 18 h at 16 °C cells were harvested by centrifugation.
Purification.
Both wild-type and E628A-T631L mutant proteins were purified using nickel chelate and size-exclusion chromatography. The His-tag was not removed. The major peak from a Superdex 200PG Hiload 26/60 column in 25 mM Tris (pH 7.5), 1 M NaCl, 10% glycerol, 1 mM TCEP, 0.1% β-octyl glucoside was pooled and concentrated to 8.8 mg/mL for crystallization trials. All reagents were from GE Healthcare.
Determination of IC50 Values.
For the determination of IC50 values, the assay was performed at 22 °C in 384-well plates using a TECAN Ultra plate reader. Total assay volume was 30 μL. Test compounds were dissolved in 90% (vol/vol) DMSO/water and diluted in water [containing 0.05% (wt/vol) CHAPS] to three times the desired assay concentration. For the assay, 10 μL water/CHAPS (± test compound) were added per well, followed by 10 μL HCV single-chain NS4A/NS3 protease solution diluted with assay buffer [50 mM Tris·HCl (pH 7.4), 150 mM NaCl, 0.05% CHAPS, 1 mM dithioerythritol]; final assay concentration was 2 nM (according to the enzyme concentrations determined by the Bradford method). After 1 h of incubation at room temperature the assay was started by addition of 10 μL substrate solution. The final concentration was 2 μM, fluorescence lifetime substrate Ac-D.E.W.E.E-Abu-ψ-[COO]↓A.S.C(PT14)-NH2, Biosyntan. The effect of compound on the enzymatic activity was obtained from the linear progress curves and determined from two readings, the first one taken directly after the addition of substrate (t = 0 min) and the second one after 60 min. The apparent inhibition constant, IC50, was calculated from the plot of percentage of inhibition vs. inhibitor concentration using nonlinear regression analysis software (XLfit, Version 3.0.5; ID Business Solution Ltd.).
Crystallization.
Initial crystallization experiments were performed using an Oryx liquid handling system, 0.3 μL protein was mixed with 0.3 μL reservoir from commercially available screens. Only one condition, condition 12, produced crystals of the PEGs Suite screen (Qiagen) which contained 25% PEG monomethyl ether (MME) 2000, 0.1 M MES (pH 6.5).
These crystals could be reproduced and were used as seeds to obtain diffraction quality wild-type apo crystals employing a microseed matrix seeding technique. [D’Arcy et al. (26) and Fig. 1 therein]. Crystals were isomorphous to the published apo structure (PDB ID code 1CU1) (13). Crystal-to-crystal variability of diffraction quality was observed, even when the crystals were from the same drop. Cocrystallization with ketoamide or acylsulfonamide-type inhibitors was not successful and initial soaking attempts with inhibitors from different chemotypes failed, generally leading to insufficient diffraction of the soaked crystals. Apo crystals of the E628A, T631L double-mutant isomorphous to the wild-type crystals were obtained by vapor diffusion in hanging drops at room temperature. Two microliters of the protein at a concentration of 8.8 mg/mL in 25 mM Tris (pH 7.5), 10% glycerol, 1 M NaCl, 1 mM TCEP, 0.1% β-octylglucoside were mixed on siliconized glass slides with 2 μL of reservoir solution. The drops were sealed over 500 μL reservoir containing 20% to 25% PEG MME 2000 and 200 mM sodium thiocyanate. The electron density showed the C terminus of the helicase occupying the protease nonprime side as in the wild type structure.
Data Collection, Structure Elucidation, and Refinement.
A crystal was soaked overnight in 25% PEG MME 2000, 200 mM sodium thiocyanate, and 1 mM of inhibitor 1, a concentratrion at which most of the inhibitor was soluble. For data collection the crystal was transferred to 30% PEG MME 2000 and directly flash cooled in the nitrogen gas stream at 100 K.
X-ray diffraction data were collected at the PXII beamline of the Swiss Light Source at a wavelength of 0.99988 Å at 100 K using a Mar225 CCD detector and processed with XDS as implemented in the program package APRV (27). The crystal diffracted to 2.7 Å resolution and belonged to the orthorhombic space group P212121 with two monomers in the asymmetric unit. The structure was determined by using as a starting model the coordinates of the isomorphous PDB ID code 1CU1 (13). Model rebuilding was performed in coot (28) and refmac (29) was employed for structure refinement using noncrystallographic symmetry restraints. We have modeled for inhibitor 1 the S-enantiomer based on the electron density fit and chemical rationale. A final round of refinement was performed using BUSTER (30). The final model has good geometry (Table 1) with 96.6% of residues in the preferred and 3.2% in the allowed regions of the Ramachandran plot. Residues 183 and 184, forming part of the linker between protease and helicase domain, are not represented by electron density for monomer B, indicating flexibility. Fig. S2 shows the active site electron density including a 7-Å sphere around the inhibitor. AREAIMOL (31) was used to determine the accessible surface area. Possibly because of improved crystal properties in the course of the work, it subsequently proved possible to soak a related inhibitor also into crystals of the wild-type full-length enzyme confirming the result described for the E628A-T631L mutant enzyme and showing that the C-terminal residues 626–631 have become disordered as well (Fig. S3).
Table 1.
Data collection and refinement statistics
| Inhibitor 1 complex | |
| Data collection | |
| Space group | P212121 |
| Cell dimensions | |
| a, b, c (Å) | 91.9, 110.5, 137.2 |
| α, β, γ (°) | 90, 90, 90 |
| Resolution (Å) | 76.4-2.73 (2.81-2.73) |
| Rsym | 0.088 (0.464) |
| I/σI | 17.2 (4.6) |
| Completeness (%) | 99.8 (100.0) |
| Redundancy | 4.5 (4.6) |
| Refinement | |
| Resolution (Å) | 76.4 − 2.73 |
| No. reflections | 37797 |
| Rwork/Rfree | 0.178/0.231 |
| No. atoms | |
| Protein | 9522 |
| Ligand/ion | 124 |
| Water | 202 |
| B-factors | |
| Protein | 55.5 |
| Ligand/ion | 50.1 |
| Water | 41.1 |
| R.m.s. deviations | |
| Bond lengths (Å) | 0.010 |
| Bond angles (°) | 1.19 |
Values in parentheses are for highest-resolution shell.
Small Angle X-ray Scattering.
X-ray scattering data were collected following standard procedures, on the X33 beamline (32) of the European Molecular Biology Laboratory, Hamburg at the Deutsches Elektronen Synchrotron using a Pilatus 500 K detector (Dectris). The scattering patterns from wild-type full-length NS3 protease-helicase at protein concentrations of 7 and 3.5 mg/mL in 50 mM Tris (pH 7.5), 1 M NaCl, 10% glycerol, 3 mM DTT were measured using a sample detector distance of 2.4 m and a wavelength of λ = 1.5 Å, covering the range of momentum transfer: 0.1 < s < 4.5 nm-1 [s = 4π sin(θ)/λ, where θ is the scattering angle and λ at 0.15 nm is the X-ray wavelength]. The noise at higher angles (s > 0.2 Å-1) can be explained by rather harsh sample conditions for SAXS studies (1 M NaCl, 10% glycerol), required to avoid aggregation but leading to high absorption and reduced contrast. The higher-angle data are however not critical for assessing the quaternary and tertiary structure. The data for s < 0.2 Å-1 are sufficiently accurate to unambiguously prove the domain orientation as in the crystal structure. The data were normalized to the intensity of the transmitted beam, and the scattering of the buffer was subtracted as background. These difference curves were scaled for concentration, and practically no concentration effect was observed. All data-processing steps were performed with the programs PRIMUS (33) and GNOM (34).
The scattering from the crystallographic models was computed by CRYSOL (35). The optimal volume fractions in the equilibrium mixtures were estimated by OLIGOMER (32).
Supplementary Material
Acknowledgments.
The authors thank Paulus Erbel, Robert Pearlstein, Michael Schaefer, Lilian Steiner, Ulrich Hommel, Christian Wiesmann, and Sylvain Cottens at Novartis Institutes for BioMedical Research for discussion and support, as well as the staff of beamline PXII at the Swiss Light Source for help with data collection.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 4a92 and r4a92sf).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1110534108/-/DCSupplemental.
References
- 1.Melnikova I. Hepatitis C—pipeline update. Nat Rev Drug Discovery. 2011;10:93–94. doi: 10.1038/nrd3361. [DOI] [PubMed] [Google Scholar]
- 2.Hofmann WP, Zeuzem S. A new standard of care for the treatment of chronic HCV infection. Nat Rev Gastroenterol Hepatol. 2011;8:257–264. doi: 10.1038/nrgastro.2011.49. [DOI] [PubMed] [Google Scholar]
- 3.Bartenschlager R, Lohmann V, Wilkinson T, Koch JO. Complex formation between the NS3 serine-type proteinase of the hepatitis C virus and NS4A and its importance for polyprotein maturation. J Virol. 1995;69:7519–7528. doi: 10.1128/jvi.69.12.7519-7528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brass V, et al. Structural determinants for membrane association and dynamic organization of the hepatitis C virus NS3-4A complex. Proc Natl Acad Sci USA. 2008;105:14545–14550. doi: 10.1073/pnas.0807298105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Taremi SS, et al. Construction, expression, and characterization of a novel fully activated recombinant single-chain hepatitis C virus protease. Protein Sci. 1998;7:2143–2149. doi: 10.1002/pro.5560071011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gale M, Jr, Foy EM. Evasion of intracellular host defence by hepatitis C virus. Nature. 2005;436:939–945. doi: 10.1038/nature04078. [DOI] [PubMed] [Google Scholar]
- 7.Beran RK, Serebrov V, Pyle AM. The serine protease domain of hepatitis C viral NS3 activates RNA helicase activity by promoting the binding of RNA substrate. J Biol Chem. 2007;282:34913–34920. doi: 10.1074/jbc.M707165200. [DOI] [PubMed] [Google Scholar]
- 8.Beran RK, Pyle AM. Hepatitis C viral NS3-4A protease activity is enhanced by the NS3 helicase. J Biol Chem. 2008;283:29929–29937. doi: 10.1074/jbc.M804065200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim JL, et al. Crystal structure of the hepatitis C virus NS3 protease domain complexed with a synthetic NS4A cofactor peptide. Cell. 1996;87:343–355. doi: 10.1016/s0092-8674(00)81351-3. [DOI] [PubMed] [Google Scholar]
- 10.Love RA, et al. The crystal structure of hepatitis C virus NS3 proteinase reveals a trypsin-like fold and a structural zinc binding site. Cell. 1996;87:331–342. doi: 10.1016/s0092-8674(00)81350-1. [DOI] [PubMed] [Google Scholar]
- 11.Cummings MD, et al. Induced-fit binding of the macrocyclic noncovalent inhibitor TMC435 to its HCV NS3/NS4A protease target. Angew Chem Int Ed Engl. 2010;49:1652–1655. doi: 10.1002/anie.200906696. [DOI] [PubMed] [Google Scholar]
- 12.Romano KP, Ali A, Royer WE, Schiffer CA. Drug resistance against HCV NS3/4A inhibitors is defined by the balance of substrate recognition versus inhibitor binding. Proc Natl Acad Sci USA. 2010;107:20986–20991. doi: 10.1073/pnas.1006370107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yao N, Reichert P, Taremi SS, Prosise WW, Weber PC. Molecular views of viral polyprotein processing revealed by the crystal structure of the hepatitis C virus bifunctional protease-helicase. Structure. 1999;7:1353–1363. doi: 10.1016/s0969-2126(00)80025-8. [DOI] [PubMed] [Google Scholar]
- 14.Appleby TC, et al. Visualizing ATP-dependent RNA translocation by the NS3 helicase from HCV. J Mol Biol. 2011;405:1139–1153. doi: 10.1016/j.jmb.2010.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ding SC, Kohlway AS, Pyle AM. Unmasking the active helicase conformation of nonstructural protein 3 from hepatitis C virus. J Virol. 2011;85:4343–4353. doi: 10.1128/JVI.02130-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dahl G, Sandström A, Akerblom E, Danielson UH. Effects on protease inhibition by modifying of helicase residues in hepatitis C virus nonstructural protein 3. FEBS J. 2007;274:5979–5986. doi: 10.1111/j.1742-4658.2007.06120.x. [DOI] [PubMed] [Google Scholar]
- 17.Thibeault D, et al. Use of the fused NS4A peptide-NS3 protease domain to study the importance of the helicase domain for protease inhibitor binding to hepatitis C virus NS3-NS4A. Biochemistry. 2009;48:744–753. doi: 10.1021/bi801931e. [DOI] [PubMed] [Google Scholar]
- 18.Liverton NJ, et al. Molecular modeling based approach to potent P2-P4 macrocyclic inhibitors of hepatitis C NS3/4A protease. J Am Chem Soc. 2008;130:4607–4609. doi: 10.1021/ja711120r. [DOI] [PubMed] [Google Scholar]
- 19.Luo D, et al. Crystal structure of the NS3 protease-helicase from dengue virus. J Virol. 2008;82:173–183. doi: 10.1128/JVI.01788-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo D, et al. Flexibility between the protease and helicase domains of the dengue virus NS3 protein conferred by the linker region and its functional implications. J Biol Chem. 2010;285:18817–18827. doi: 10.1074/jbc.M109.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Assenberg R, et al. Crystal structure of a novel conformational state of the flavivirus NS3 protein: Implications for polyprotein processing and viral replication. J Virol. 2009;83:12895–12906. doi: 10.1128/JVI.00942-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bollati M, et al. Structure and functionality in flavivirus NS-proteins: Perspectives for drug design. Antiviral Res. 2010;87:125–148. doi: 10.1016/j.antiviral.2009.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mastrangelo E, et al. Crystal structure and activity of Kunjin virus NS3 helicase; protease and helicase domain assembly in the full-length NS3 protein. J Mol Biol. 2007;372:444–455. doi: 10.1016/j.jmb.2007.06.055. [DOI] [PubMed] [Google Scholar]
- 24.Tsantrizos YS, et al. Macrocyclic inhibitors of the NS3 protease as potential therapeutic agents of hepatitis C virus infection. Angew Chem Int Ed. 2003;42:1356–1360. doi: 10.1002/anie.200390347. [DOI] [PubMed] [Google Scholar]
- 25.Shawn B, et al. Macrocyclic compounds as HCV NS3 protease inhibitors. WO2008101665. US Patent. 2008
- 26.D’Arcy A, Villard F, Marsh M. An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr. 2007;D63:550–554. doi: 10.1107/S0907444907007652. [DOI] [PubMed] [Google Scholar]
- 27.Kroemer M, Dreyer MK, Wendt KU. APRV—a program for automated data processing, refinement and visualization. Acta Crystallogr. 2004;D60:1679–1682. doi: 10.1107/S0907444904015215. [DOI] [PubMed] [Google Scholar]
- 28.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 29.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 30.Bricogne G, et al. Cambridge, UK: Global Phasing Ltd.; 2010. BUSTER. Version 2.9. [Google Scholar]
- 31.Lee B, Richards FM. The interpretation of protein structures: Estimation of static accessibility. J Mol Biol. 1971;55:379–400. doi: 10.1016/0022-2836(71)90324-x. [DOI] [PubMed] [Google Scholar]
- 32.Roessle MW, et al. Upgrade of the small angle X-ray scattering beamline X33 at the European Molecular Biology Laboratory, Hamburg. J Appl Crystallogr. 2007;40:190–194. [Google Scholar]
- 33.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS—a Windows-PC based system for small-angle scattering data analysis. J Appl Crystallogr. 2003;36:1277–1282. [Google Scholar]
- 34.Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J Appl Crystallogr. 1992;25:495–503. [Google Scholar]
- 35.Svergun DI, Barberato C, Koch MHJ. CRYSOL—a program to evaluate X-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Crystallogr. 1995;28:768–773. [Google Scholar]
- 36.DeLano WL. San Carlos, CA: DeLano Scientific; 2002. The PyMOL Molecular Graphics System . [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.