Abstract
The primary cause of poor outcome following allogeneic hematopoietic cell transplantation (HCT) for chronic lymphocytic leukemia (CLL) is disease recurrence. Detection of increasing minimal residual disease (MRD) following HCT may permit early intervention to prevent clinical relapse; however, MRD quantification remains an uncommon diagnostic test because of logistical and financial barriers to widespread use. Here we describe a method for quantifying CLL MRD using widely available consensus primers for amplification of all Ig heavy chain (IGH) genes in a mixture of peripheral blood mononuclear cells, followed by high-throughput sequencing (HTS) for disease-specific IGH sequence quantification. To achieve accurate MRD quantification, we developed a systematic bioinformatic methodology to aggregate cancer clone sequence variants arising from systematic and random artifacts occurring during IGH-HTS. We then compared the sensitivity of IGH-HTS, flow cytometry, and allele-specific oligonucleotide PCR for MRD quantification in 28 samples collected from 6 CLL patients following allogeneic HCT. Using amplimer libraries generated with consensus primers from patient blood samples, we demonstrate the sensitivity of IGH-HTS with 454 pyrosequencing to be 10−5, with a high correlation between quantification by allele-specific oligonucleotide PCR and IGH-HTS (r = 0.85). From the same dataset used to quantify MRD, IGH-HTS also allowed us to profile IGH repertoire reconstitution after HCT—information not provided by the other MRD methods. IGH-HTS using consensus primers will broaden the availability of MRD quantification in CLL and other B cell malignancies, and this approach has potential for quantitative evaluation of immune diversification following transplant and nontransplant therapies.
Keywords: next-generation sequencing, consensus-primed polymerase chain reaction, immune reconstitution
Chronic lymphocytic leukemia (CLL) is the most common adult leukemia in the United States, with ∼15,500 new cases and 4,400 deaths per year (1). Despite improvements in treatment responses using multiagent therapy, CLL remains incurable with available immunochemotherapy regimens (2). Patients with relapsed CLL and those with high-risk features at presentation, such as 17p deletions or unmutated Ig heavy chain (IGH) regions, are generally referred for allogeneic hematopoietic cell transplantation (allo-HCT) (3, 4). Fifty percent of CLL patients undergoing allo-HCT experience long-term disease-free survival (DFS) and may be cured. Nevertheless, 50% of patients will experience disease recurrence (5, 6). Quantification of CLL MRD has prognostic value because achievement of MRD negativity 1 y after HCT is associated with long-term DFS (6–13). Furthermore, strategies for treating post-HCT relapse, including additional chemotherapy, donor lymphocyte infusions, and cell vaccines, may be more effective when CLL progression is detected with low tumor burden.
Validated methods for MRD assessment include allele-specific oligonucleotide PCR (ASO-PCR) and flow cytometry (FC). ASO-PCR affords high sensitivity but is laborious, time-intensive, and not widely available because of its dependence on the development and validation of patient-specific primers and probes for quantitative PCR. FC is widely available but is expensive and has lower sensitivity. High-throughput sequencing (HTS) of the IGH VDJ segment provides CLL clonotype quantification with “off-the-shelf” consensus primers that require no per-patient customization (14), thus combining the benefits of high sensitivity and universal applicability. Boyd et al. (15) evaluated this methodology in proof-of-concept experiments for detection of MRD in a variety of B cell malignancies. Here, we compare the performance characteristics of IGH-HTS, ASO-PCR, and FC for tracking disease burden in a group of CLL patients following HCT. We also demonstrate that the sequence data acquired through MRD quantification using this approach provide valuable information about the tempo of posttransplant immune reconstitution by profiling IGH repertoire diversification.
Results
The six patients evaluated in this study were treated with reduced intensity conditioning allo-HCT for high-risk CLL (Table S1). All patients had unmutated IGH loci. Two patients achieved complete disease remission before HCT, and all experienced complete clinical remission following HCT. Transplant characteristics and outcomes are summarized in Table S2. Twenty-eight cryopreserved peripheral blood mononuclear cells (PBMCs) acquired from these patients underwent MRD quantification using FC, ASO-PCR, and IGH-HTS.
Enumeration of CLL Clonotypes by IGH-HTS Requires Error Handling.
We limited our sequence analysis to the first 200 nt of sequence reads because of the uniformly high sequence quality scores up to this position using either BIOMED-2 framework 1 (FR1) or framework 2 (FR2) primer sets (Fig. S1). It is known that even in areas of high sequence quality, 454 pyrosequencing is prone to specific errors, particularly insertions or deletions (indels) near homopolymeric sequences (16). To maximize the sensitivity of CLL MRD detection using IGH-HTS, we developed a processing algorithm that accounted for common homopolymeric indels and computationally aggregated these variant representations of the CLL clonotype into the MRD quantification (Fig. S2).
During computational alignment of sequences with consensus germ-line IGH V and J segments, we rescued clonotypes with errors at homopolymeric sites of three or more nucleotides by recognizing alignment with the dominant CLL clonotype after shifting of the sequences upstream or downstream. Random single-nucleotide indels were next corrected by alignment to the dominant CLL clonotype. We also found it necessary to aggregate reads with up to one nucleotide substitution from the dominant CLL clonotype, because these are more likely to arise from PCR or sequencing artifacts than they are to be bona fide biologically derived clonotypes. This methodology strikes a balance between accepting the sequence of real subclonal populations as part of the CLL clone and providing room to count subclones that arose from PCR and/or sequencing error, but that originally derived from a true CLL clonotype. All corrections from raw reads to the final MRD count were processed in silico with minimal need for user supervision at the individual sequence level (Fig. S2).
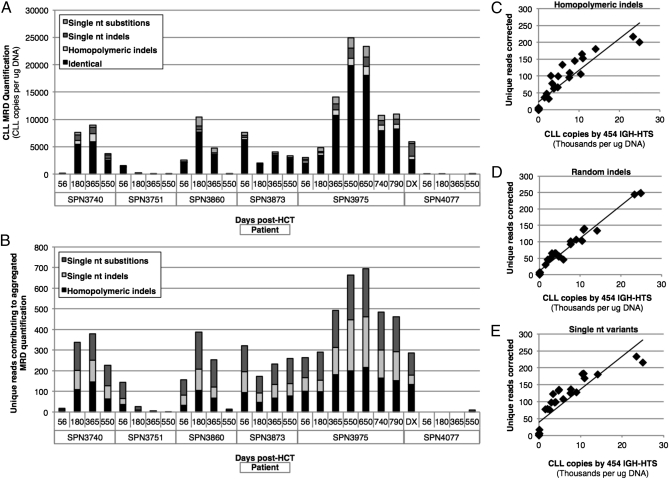
Overall, identical CLL clonotypes (i.e., those without sequencing errors) accounted for a median 74% of the final MRD quantifications; the contributions of each type of error correction to the final count are shown in Fig. 1A (and reported in Table S3). The median contribution of reads corrected for homopolymeric indels to the final MRD total was 6.1% (range 0–100%). Random single-nucleotide indels contributed a median of 7.5% (range 0–56%), and sequencing reads with single-nucleotide substitutions or gaps comprised a median of 8.3% (range 4.8–49%) of the final MRD quantification. In sum, the median contribution of corrected clonotypes to the final MRD count was 25.2% (range 0–100%) of the total. The number of unique reads requiring aggregation into the final MRD count ranged from 0 to 217 for homopolymeric indels, from 0 to 247 for single-nucleotide indels, and from 0 to 233 for single-nucleotide substitutions (Fig. 1B). There was a strong correlation between the abundance of disease and the number of unique reads requiring correction for homopolymeric indels (r = 0.92), random single-nucleotide indels (r = 0.98), and single-nucleotide substitutions (r = 0.88), consistent with expectation from nonbiased errors (Fig. 1 C–E).
Fig. 1.
454 pyrosequencing MRD quantification and artifact characteristics. (A) The final MRD count for each patient sample with the composition of identical reads, homopolymeric indels, random single-nucleotide indels, and single-nucleotide substitutions is shown. (B) The number of unique reads with each error type is indicated for each patient sample. (C–E) The correlation between IGH-HTS MRD quantification and the number of unique reads requiring correction for homopolymeric indels (C), random single-nucleotide indels (D), and single-nucleotide substitutions or gaps (E) are demonstrated.
HTS Reveals Oligoclonal IGH Sequences in CLL.
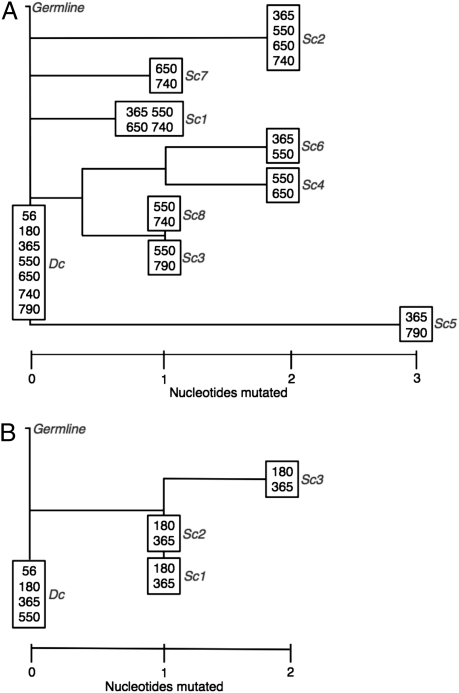
Campbell et al. (17) demonstrated IGH oligoclonality in somatically mutated CLL populations when evaluated by deep sequencing. In our somatically unmutated CLL dataset, after we aggregated clonotypes containing differences consistent with pyrosequencing errors, we also observed a generally small number of clonotypes closely resembling the dominant CLL clone that covaried with the dominant clone over multiple time points. In some patients, these apparent subclones of the dominant CLL clone were prevalent in larger concentrations than could be attributed to PCR or sequencing error. To consider a subclone with two or more nucleotide substitutions legitimate, we required clonotype frequency of two or more per sample and that the clonotype be seen in at least two time points (we call these “2 × 2 × 2 subclones”). Three patients had no identifiable CLL subclones. The other patients, all of whom had persistent molecular disease, had one or more legitimate subclones. We analyzed two patients with more than one subclone for phylogenetic relatedness to the dominant CLL clone (Fig. 2).
Fig. 2.
Phylogenetic analysis of CLL subclones. Neighbor-joining phylogenetic trees are depicted for patients SPN3975 (A) and SPN3860 (B). The number of nucleotide differences are represented by horizontal proximity to the dominant CLL clone (Dc). We required subclones (Sc) contributing to these phylogenetic trees to be seen at least twice in two or more time points. The specific time points in which each clonotype was observed is indicated at each node.
We identified eight subclones in a patient [Stanford patient number (SPN) 3975] with high-level persistent disease following allo-HCT. Alignments of subclone sequences in Fig. S3 demonstrate that few of the nucleotide variations occurred in the CDR3 region. Furthermore, when we evaluated where the ASO-PCR primers and probes for each patient annealed, we observed that none of the subclone mutations would interfere with the quantitative PCR, suggesting that these subclones were likely included in the ASO-PCR disease quantification. In our unmutated CLL dataset, the total prevalence of 2 × 2 × 2 subclones was uniformly <0.5% of each final CLL MRD count. We have elected not to aggregate these rare subclones into our CLL quantification, but future studies may need to consider subclone contributions, particularly if somatically mutated CLL is evaluated.
454 Pyrosequencing MRD Sensitivity Is 1:100,000.
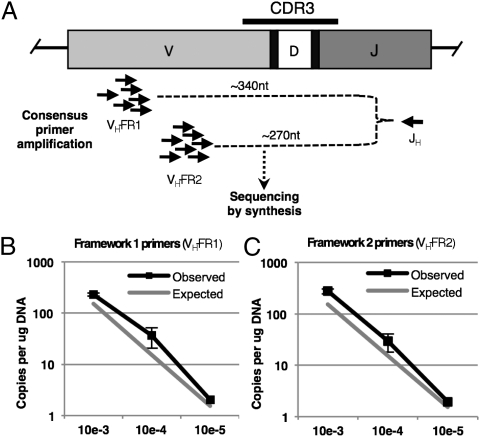
An advantage of 454 pyrosequencing over other sequence-by-synthesis methods is the ability to produce long sequence reads. BIOMED-2 FR1 and FR2 consensus primer sets (14) in conjunction with a consensus J primer give rise to amplimers of ∼340 and 270 nt in length, respectively. To first determine whether amplimer length affected sensitivity, we sequenced FR1-J and FR2-J amplification products from a normal donor with 150,000 reads each and found highly concordant IGH repertoire overlap and clonotype quantification (r = 0.92) (Fig. S4). We verified 1:100,000 (i.e., 10−5) clonotype-specific sensitivity using both primer sets by creating a serial dilution of FACS-sorted CLL into PBMC from the normal healthy donor of that same patient and amplifying the dilutions independently in two laboratories using FR1-J and FR2-J primer sets (Fig. 3A). We spiked purified CLL in log-series dilutions (10−3 to 10−5) into healthy donor PBMC with 14% B cell content. At the 10−5 dilution, we surveyed 450,000 PBMC genomes containing 63,000 B cell genomes with 150,000 dedicated IGH reads. This degree of oversequencing of the IGH genes in the mixture was estimated to yield a theoretical detection sensitivity of 10−5 with >99.9% certainty (Poisson probability). With FR1 consensus primers, we achieved 186,088 reads of productive IGH alleles and detected the CLL clone 6 out of 12 expected times, which is consistent with 2 × 10−5 sensitivity with a likelihood of failing to detect the CLL clone of 0.2%. With FR2 consensus primers, we achieved 98,877 reads and detected the CLL clone 5 out of 6 expected times, which is consistent with 10−5 sensitivity with a likelihood of failing to detect the CLL clone of 0.7%.
Fig. 3.
Sensitivity of CLL MRD detection by 454 pyrosequencing determined by limiting dilution. (A) Consensus primer groups in the FR1 or FR2 regions of VH were used in conjunction with a consensus JH primer to create IGH amplicon libraries by multiplexed PCR, which were then sequenced by synthesis using a 454 pyrosequencer. (B and C) CLL cells purified by FACS were diluted into PBMC from a healthy donor leukapheresis sample to a level of 1:1,000 (10−3), 1:10,000 (10−4), and 1:100,000 (10−5). Clonotypic IGH quantifications at each dilution are demonstrated for FR1 (B) and FR2 (C) primer sets with reference values for expected quantifications at ≥95% Poisson probability.
IGH-HTS MRD Sensitivity Compared with ASO-PCR and FC.
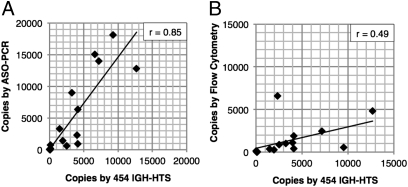
All patient samples in our series underwent MRD assessment by ASO-PCR and FC in addition to IGH-HTS. The concurrent measurements permitted us to directly compare the relative sensitivities of these different methodologies. The sensitivity of IGH-HTS is limited primarily by the number of sequence reads allocated to a given sample. We opted for 1:10,000 (10−4) sensitivity for most samples—the International Working Group on CLL consensus standard for CLL MRD assessment (2)—by allocating 15,000 reads to each sample. Samples with high disease burdens (i.e., >15,000 disease copies per microgram of genomic DNA) and clinically detectable CLL did not require quantification at the molecular level. Meaningful comparisons of sensitivity with the different methodologies are thus best made at lower disease burdens. When we restricted comparisons to samples with <15,000 disease copies by HTS (equivalent to <10% disease content in PBMC), we determined the correlation coefficient between HTS and ASO-PCR to be 0.85, whereas the correlation between HTS and FC was 0.49 (Fig. 4 A and B). For the purposes of this comparison, FC results were transformed into a genome equivalent scale as described in SI Materials and Methods.
Fig. 4.
Correlation of CLL MRD quantification by IGH-HTS, ASO-PCR, and FC. CLL MRD quantification for samples with 15,000 or fewer CLL copies per microgram of DNA using 454 IGH-HTS are graphed with paired analyses using ASO-PCR (A) and FC (B). The Pearson r correlation coefficient between the methods was 0.85 for 454 IGH-HTS vs. ASO-PCR and 0.49 for 454 IGH-HTS vs. FC. For this comparison, FC data were converted as described in SI Materials and Methods.
CLL Clonotypes in the Context of the IGH Repertoire.
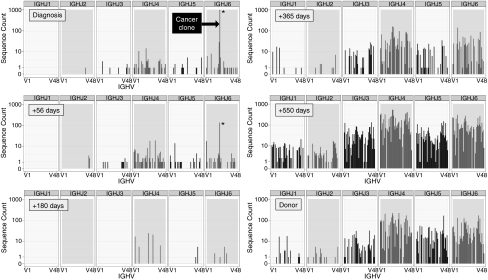
The datasets acquired with IGH-HTS represent samples of the donor IGH repertoire reconstituting after allo-HCT. Although our primary goal was to analyze IGH-HTS data to quantify MRD, we also note that this rich dataset can provide unique and timely insight into reconstitution kinetics and breadth in the IGH repertoire. Fig. 5 presents the V-J combinatorial diversity at diagnosis and at +56, +180, +365, and +550 d after allo-HCT in patient SPN4077. At diagnosis, the patient's IGH repertoire was dominated to near exclusivity by the CLL clonotype. The CLL clonotype was detectable at day +56 and then became undetectable. At days +56 and +180, the repertoire remained narrow, with only 21 and 12 unique CDR3 clonotypes with frequency of 2 or more detected, respectively. At days +365 and +550, the repertoire had reconstituted, with 232 and 3,122 unique legitimate CDR3 clonotypes, respectively, the latter of which was similar in number to that of the patient's healthy donor (1,464 unique clonotypes at the same sequencing depth). Patient SPN3751 exhibited a pattern similar to this patient, with 113, 26, 20, and 2,636 unique CDR3 clonotypes at days +56, +180, +365, and +550. The other four patients continued to have long-term domination of their IGH repertoire by the CLL clone (Fig. S5). In some cases, two coexisting dominant clonotypes were detected, representing a CLL clone with both IGH loci rearranged. The codominant allele was nonproductively rearranged in each case.
Fig. 5.
IGH-HTS reveals kinetics of IGH repertoire reconstitution following allo-HCT for CLL. All V-J recombinations detected in peripheral blood from patient SPN4077 at diagnosis and days +56, +180, +365, and +550 following allo-HCT are demonstrated. The x axis at the bottom of each section represents IGH V segments 1–49, which combined with IGH J segments 1–6 as defined along the x axis at the top of each section. The y axis represents the number of total reads for that recombination pair. The CLL clonotype is demarcated by an asterisk. The repertoire of this patient's donor is demonstrated for comparison.
Somatic Hypermutation of the IGH Repertoire Following Allo-HCT.
Given the long contiguous reads achieved with 454 pyrosequencing, it is possible to analyze each clonotype in comparison with the published consensus germ-line sequence for each V, D, and J to enumerate the degree of somatic hypermutation. Somatic hypermutation is a secondary mechanism of Ig diversification that occurs during affinity maturation after antigen encounter. Among our patients, four received post-HCT Rituximab at days +56, +63, +72, and +79 in a clinical trial assessing Rituximab (anti-CD20) for graft-versus-host disease (GVHD) prophylaxis. By eliminating the vast majority of mature B lymphocytes, it was expected that post-HCT Rituximab would delay repertoire reconstitution. Accordingly, we observed a significant deficiency of somatically mutated VDJ recombinants at day +365 following HCT in the patients who received Rituximab (7.9 ± 1.9%) compared with those not receiving peritransplant anti-B-cell therapy (17.8 ± 2.1%) (P = 0.03; unpaired two-tailed t test; Fig. S6).
Discussion
The utility of MRD assessment in CLL has been studied in several clinical contexts, but most reports have focused on MRD following hematopoietic cell transplantation because only allo-HCT achieves long-term remissions in high-risk CLL. PCR quantification of clonal IGH sequences was developed >20 y ago (18), and after demonstration that this technique has single cell sensitivity (19, 20), ASO-PCR was developed for MRD quantification (9, 21). Although it was predicted 2 decades ago that PCR quantification of MRD would become a routine practice (22), ASO-PCR remains restricted to research use, likely because of the financial and logistical barriers to routine development of these patient-specific assays. Van Dongen et al. (14) in the BIOMED-2 consortium developed groups of consensus primers for performing multiplexed PCR to recover all IGH gene sequences in a polyclonal mixture. These reagents can now be coupled with next-generation HTS to achieve universal MRD quantification without need for patient-specific reagents in any patient with CLL or other IGH clonal B cell malignancy.
One potential pitfall of all HTS platforms is their susceptibility to generating stereotypical and random artifacts introduced during the sequence-by-synthesis reaction (16, 23). 454 pyrosequencing is particularly prone to homopolymeric insertions and deletions. Although sequence reads with such errors can be discarded when IGH-HTS is used strictly for IGH repertoire analysis, a robust methodology that compensates for these errors is essential for accurate MRD quantification. We have developed a data processing pipeline that addresses systematic and random insertions, deletions, and substitutions introduced by the sequencing technology, thus permitting highly sensitive CLL MRD quantification with a high degree of correlation to ASO-PCR results.
ASO-PCR was first demonstrated to have predictive value by Provan et al. (7), who demonstrated that persistence of molecular disease was associated with increased probability of CLL relapse following autologous and allo-HCT. Molecular MRD assessment using ASO-PCR has since been evaluated by several clinical trials [reviewed by Dreger et al. (24)], the best of which is work by the German CLL Study Group (25, 26), which recently demonstrated that MRD negativity (10−4 or less) at 1 y following allo-HCT strongly correlated with decreased risk of relapse (HR 0.037; P < 0.00001). Four of 32 patients (13%) experienced conversion to MRD positivity subsequent to having undetectable MRD at 12 mo, however, which suggests that the predictive value of molecular MRD assessment could be improved with more sensitive techniques.
Although we demonstrate here that MRD sensitivity using 454 pyrosequencing IGH-HTS can approach 1:100,000 (10−5) and can be pushed to 1:1,000,000 (10−6) sensitivity by using other deep sequencing platforms with higher bandwidth, we do not propose that the major advantage of our technique is improved sensitivity. Rather, the wide applicability of this approach using consensus primers without the need for any patient customization is a significant development and will increase the availability of MRD quantification in patients with CLL and other B cell malignancies undergoing allo-HCT. Pretransplant CLL patients undergoing treatment with novel agents could also be assessed with IGH-HTS MRD quantification to determine not only the depth of remission on therapy, but also the impact that novel therapies could have on the remainder of the B cell compartment—information that cannot be determined from FC or ASO-PCR disease quantification.
Although the majority of patients undergoing allo-HCT for CLL have somatically unmutated IGH clonotypes associated with their disease, the broader applicability of IGH-HTS as an MRD assay in various clinical contexts will require methods for overcoming amplification failures owing to somatic mutations at the sites of consensus primer binding. Campbell et al. (17) successfully amplified and sequenced clonotypes from somatically mutated CLL—and in so doing, they identified greater subclonal heterogeneity than we identified in somatically unmutated CLL. We also identified somatic mutations in 15–20% of IGH clonotypes productively sequenced at 1 y or more following allo-HCT. Together, these data are encouraging for the application of this approach to somatically mutated CLL as well as other B lymphoid malignancies with somatically mutated IGH loci. If the technique should fail with one consensus primer set (e.g., FR1), it may be possible in many cases to salvage disease clonotype identification by using another established primer set (e.g., FR2).
In addition to highly sensitive and specific disease burden quantification, high-throughput IGH sequencing offers insight into the clonality of CLL and the IGH repertoire following allo-HCT. By using the same platform we used in this work, oligoclonality in two patients with somatically mutated CLL was demonstrated by Campbell et al. (17). All patients we evaluated here had unmutated CLL, and although we did find a small number of probable subclones with two or more nucleotide variants, these variants appear to be an extremely rare component of somatically unmutated CLL clades (groups of related clonotypes). Further application of HTS methodologies is needed to facilitate clarification of CLL clonality in larger patient series and will permit assessment for idiotype (Id) escape mutants following HCT or anti-Id vaccinations or immunotherapy.
The rich datasets acquired through IGH-HTS also provide the possibility of novel quantitative assessment of posttransplant immune reconstitution, which remains a critical outcome with difficult-to-quantify measures of success. We demonstrate the utility of analyzing IGH-HTS data to depict the kinetics of repertoire reconstitution. We found that our allo-HCT recipients generally had narrow (i.e., small number of unique clonotypes) and shallow (i.e., small number of total IGH reads per microgram of PBMC DNA) IGH repertoire reconstitution until 1–1.5 y following transplant. Our finding is consistent with total CD19+ B cell reconstitution observed in allograft patients receiving posttransplant Rituximab for chronic GVHD (27, 28). We also found that our patients who received posttransplant Rituximab for GVHD prophylaxis had a significantly diminished degree of somatic mutation across the IGH repertoire at 1 y following HCT compared with those who received no additional anti-B-cell therapy. Similarly, we suggest that this quantitative assessment of immune repertoire diversity may be a useful outcome to follow in allo-HCT clinical trials to determine whether repertoire reconstitution correlates with important clinical outcomes such as progression-free survival, overall survival, and the incidence of GVHD and infections.
The use of IGH-HTS to quantify MRD in IGH clonal malignancies promises to have a significant role in patient management, due simply to the fact that this technique will dramatically broaden the scope of patients eligible for routine MRD monitoring. Because relapse following allo-HCT for CLL remains common, there is a developing consensus in favor of acting upon progressive molecular disease in CLL (26, 29>–32). Although this treatment approach has been a goal of the CLL management community for a decade or more, it has been difficult to realize because of the barriers to practical application that personalized assays require. HTS using consensus primers overcomes these barriers and permits widespread use of molecular MRD assessment for the management of CLL and other diseases with clonally rearranged immunoreceptor genes. The added benefit that IGH-HTS provides by simultaneously permitting clonality assessment, Ig repertoire profiling, and potential quantification and tracking of adoptively transferred B cells are areas for significant discovery regarding the biology of B cell malignancies and allo-HCT therapy.
Materials and Methods
Patients.
To compare IGH-HTS, ASO-PCR, and FC at a range of disease burdens, we selected samples from six patients with CLL who underwent nonmyeloablative HCT at Stanford University. All patients underwent conditioning with total lymphoid irradiation and anti-thymocyte globulin (TLI/ATG) as described (33, 34). All patient and donor samples were obtained with explicit authorization and monitoring by the Stanford University School of Medicine Institutional Review Board.
CLL Cell Isolation, FC, and DNA Isolation.
Peripheral blood samples were acquired from patients at indicated time points following HCT, cryopreserved, and analyzed for the CLL immunophenotype by using standard procedures (SI Materials and Methods). DNA was harvested after washing with buffered saline (pH 7.4), cellular disruption in lysis buffer (50 mM Tris⋅HCl, 50 mM EDTA, and 1% SDS; pH 7.4) containing proteinase K, followed by phenol extraction, ethanol precipitation, and resuspension in Tris⋅EDTA buffer (pH 7.4).
ASO-PCR and High-Throughput VDJ Sequencing.
Quantitative real-time PCR was performed by using patient ASO primers as described (SI Materials and Methods; ref. 35). For IGH-HTS, BIOMED-2 FR1 or FR2 consensus primers were used in conjunction with a consensus J segment primer to amplify the IGH locus (Fig. 3A) (14). The 28 patient samples sequenced for this study arose from amplification with FR1-J primer sets after we demonstrated highly concordant repertoire coverage using FR1-J and FR2-J primer sets (Fig, S4). PCR was performed with AmpliTaq Gold DNA polymerase with 250 ng of genomic DNA template per reaction and 35 cycles of amplification. A sufficient number of replicate reactions were performed to cover the degree of sensitivity indicated for each experiment. Consensus J segment primers containing sextamer, septamer, or decamer oligonucleotide barcodes for disambiguation of multipatient data were used to prime the sequence-by-synthesis reaction on the 454 Life Sciences GS20 platform by using Titanium chemistry (454 Life Sciences, Roche) (36).
Data Analysis.
Sequence reads were mapped to germ-line V and J reference sequences downloaded from the IMGT Web site (www.imgt.org) (37). For MRD quantification, reads with identical V and J segment use and with <20-nt differences from the dominant CLL clone, previously determined for every CLL patient from a traditional Sanger sequencing read performed for CLL prognostication, were retained for further analysis using an algorithm for correcting artifactual sequencing errors (SI Materials and Methods).
Supplementary Material
Acknowledgments
We thank the patients who participated in this study and the bone marrow transplant nurses, patient coordinators, and staff at Stanford University Medical Center who made this work possible. This work was supported by Leukemia and Lymphoma Translational Research Award R618-09, National Cancer Institute Grant P01 CA049605, and the Stanford University Cancer Institute. H.G. was supported by Stanford Genome Technology Center National Institutes of Health Grant P01 HG000205. I.B. was supported by an Instituto de Salud Carlos III Bolsas de Ampliación de Estudios grant (Ministerio de Ciencia e Innovación, Spain). A.C.L. is an American Society of Hematology Research Training Award Fellow.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118357109/-/DCSupplemental.
References
- 1.National Cancer Institute SEER Cancer Statistics Review. 2011. Available at http://seer.cancer.gov/csr. Accessed May 13, 2011.
- 2.Hallek M. State-of-the-art treatment of chronic lymphocytic leukemia. Hematology (Am Soc Hematol Educ Program) 2009;2009:440–449. doi: 10.1182/asheducation-2009.1.440. [DOI] [PubMed] [Google Scholar]
- 3.Hallek M, et al. International Workshop on Chronic Lymphocytic Leukemia Guidelines for the diagnosis and treatment of chronic lymphocytic leukemia: A report from the International Workshop on Chronic Lymphocytic Leukemia updating the National Cancer Institute-Working Group 1996 guidelines. Blood. 2008;111:5446–5456. doi: 10.1182/blood-2007-06-093906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gribben JG. How I treat CLL up front. Blood. 2010;115:187–197. doi: 10.1182/blood-2009-08-207126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Khouri IF, et al. Nonablative allogeneic stem cell transplantation for chronic lymphocytic leukemia: Impact of rituximab on immunomodulation and survival. Exp Hematol. 2004;32:28–35. doi: 10.1016/j.exphem.2003.09.021. [DOI] [PubMed] [Google Scholar]
- 6.Sorror ML, et al. Hematopoietic cell transplantation after nonmyeloablative conditioning for advanced chronic lymphocytic leukemia. J Clin Oncol. 2005;23:3819–3829. doi: 10.1200/JCO.2005.04.569. [DOI] [PubMed] [Google Scholar]
- 7.Provan D, et al. Eradication of polymerase chain reaction-detectable chronic lymphocytic leukemia cells is associated with improved outcome after bone marrow transplantation. Blood. 1996;88:2228–2235. [PubMed] [Google Scholar]
- 8.Esteve J, et al. Stem cell transplantation for chronic lymphocytic leukemia: Different outcome after autologous and allogeneic transplantation and correlation with minimal residual disease status. Leukemia. 2001;15:445–451. doi: 10.1038/sj.leu.2402036. [DOI] [PubMed] [Google Scholar]
- 9.Böttcher S, et al. Comparative analysis of minimal residual disease detection using four-color flow cytometry, consensus IgH-PCR, and quantitative IgH PCR in CLL after allogeneic and autologous stem cell transplantation. Leukemia. 2004;18:1637–1645. doi: 10.1038/sj.leu.2403478. [DOI] [PubMed] [Google Scholar]
- 10.Rawstron AC, de Tute R, Jack AS, Hillmen P. Flow cytometric protein expression profiling as a systematic approach for developing disease-specific assays: Identification of a chronic lymphocytic leukaemia-specific assay for use in rituximab-containing regimens. Leukemia. 2006;20:2102–2110. doi: 10.1038/sj.leu.2404416. [DOI] [PubMed] [Google Scholar]
- 11.Ritgen M, et al. Graft-versus-leukemia activity may overcome therapeutic resistance of chronic lymphocytic leukemia with unmutated immunoglobulin variable heavy-chain gene status: Implications of minimal residual disease measurement with quantitative PCR. Blood. 2004;104:2600–2602. doi: 10.1182/blood-2003-12-4321. [DOI] [PubMed] [Google Scholar]
- 12.Moreton P, et al. Eradication of minimal residual disease in B-cell chronic lymphocytic leukemia after alemtuzumab therapy is associated with prolonged survival. J Clin Oncol. 2005;23:2971–2979. doi: 10.1200/JCO.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 13.Farina L, et al. Qualitative and quantitative polymerase chain reaction monitoring of minimal residual disease in relapsed chronic lymphocytic leukemia: Early assessment can predict long-term outcome after reduced intensity allogeneic transplantation. Haematologica. 2009;94:654–662. doi: 10.3324/haematol.2008.000273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Dongen JJM, et al. Design and standardization of PCR primers and protocols for detection of clonal immunoglobulin and T-cell receptor gene Recombinations in suspect lymphoproliferations: report of the BIOMED-2 Concerted Action BMH4-CT98-3936. Leukemia. 2003;17:2257–2317. doi: 10.1038/sj.leu.2403202. [DOI] [PubMed] [Google Scholar]
- 15.Boyd SD, et al. Measurement and clinical monitoring of human lymphocyte clonality by massively parallel V-D-J pyrosequencing. Sci Transl Med. 2009;1(12):12ra23. doi: 10.1126/scitranslmed.3000540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C, Mitsuya Y, Gharizadeh B, Ronaghi M, Shafer RW. Characterization of mutation spectra with ultra-deep pyrosequencing: Application to HIV-1 drug resistance. Genome Res. 2007;17:1195–1201. doi: 10.1101/gr.6468307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Campbell PJ, et al. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc Natl Acad Sci USA. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brisco MJ, Tan LW, Orsborn AM, Morley AA. Development of a highly sensitive assay, based on the polymerase chain reaction, for rare B-lymphocyte clones in a polyclonal population. Br J Haematol. 1990;75:163–167. doi: 10.1111/j.1365-2141.1990.tb02643.x. [DOI] [PubMed] [Google Scholar]
- 19.Sioutos N, et al. Polymerase chain reaction versus Southern blot hybridization. Detection of immunoglobulin heavy-chain gene rearrangements. Diagn Mol Pathol. 1995;4:8–13. doi: 10.1097/00019606-199503000-00004. [DOI] [PubMed] [Google Scholar]
- 20.Magnac C, et al. Detection of minimal residual disease in B chronic lymphocytic leukemia (CLL) Hematol Cell Ther. 1999;41:13–18. doi: 10.1007/s00282-999-0013-y. [DOI] [PubMed] [Google Scholar]
- 21.Böttcher S, et al. Standardized MRD flow and ASO IGH RQ-PCR for MRD quantification in CLL patients after rituximab-containing immunochemotherapy: A comparative analysis. Leukemia. 2009;23:2007–2017. doi: 10.1038/leu.2009.140. [DOI] [PubMed] [Google Scholar]
- 22.Pollard P, Owen G, Worwood M. PCR-based immunogenotyping at the Ig heavy chain CDR3 locus: Improvements in resolution. Br J Haematol. 1993;84:169–171. doi: 10.1111/j.1365-2141.1993.tb03043.x. [DOI] [PubMed] [Google Scholar]
- 23.Nguyen P, et al. Identification of errors introduced during high throughput sequencing of the T cell receptor repertoire. BMC Genomics. 2011;12:106. doi: 10.1186/1471-2164-12-106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dreger P, Ritgen M, Böttcher S, Schmitz N, Kneba M. The prognostic impact of minimal residual disease assessment after stem cell transplantation for chronic lymphocytic leukemia: Is achievement of molecular remission worthwhile? Leukemia. 2005;19:1135–1138. doi: 10.1038/sj.leu.2403800. [DOI] [PubMed] [Google Scholar]
- 25.Ritgen M, et al. German CLL Study Group Quantitative MRD monitoring identifies distinct GVL response patterns after allogeneic stem cell transplantation for chronic lymphocytic leukemia: Results from the GCLLSG CLL3X trial. Leukemia. 2008;22:1377–1386. doi: 10.1038/leu.2008.96. [DOI] [PubMed] [Google Scholar]
- 26.Böttcher S, Ritgen M, Dreger P. Allogeneic stem cell transplantation for chronic lymphocytic leukemia: Lessons to be learned from minimal residual disease studies. Blood Rev. 2011;25:91–96. doi: 10.1016/j.blre.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Sarantopoulos S, et al. Recovery of B-cell homeostasis after rituximab in chronic graft-versus-host disease. Blood. 2011;117:2275–2283. doi: 10.1182/blood-2010-10-307819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arai S, et al. Prophylactic Rituximab after reduced intensity conditioning transplantation results in low chronic GVHD. Blood. 2008;112:466. [Google Scholar]
- 29.Itälä M, et al. Stem cell transplantation in poor-risk chronic lymphocytic leukemia: assessment of post-transplant minimal residual disease using four- and six-color flow cytometry and allele-specific RQ-PCR. Eur J Haematol. 2008;81:100–106. doi: 10.1111/j.1600-0609.2008.01082.x. [DOI] [PubMed] [Google Scholar]
- 30.Nabhan C, Coutré S, Hillmen P. Minimal residual disease in chronic lymphocytic leukaemia: Is it ready for primetime? Br J Haematol. 2007;136:379–392. doi: 10.1111/j.1365-2141.2006.06428.x. [DOI] [PubMed] [Google Scholar]
- 31.Moreno C, Ritgen M, Rawstron A. Is MRD eradication a desirable goal in CLL? Best Pract Res Clin Haematol. 2010;23:97–107. doi: 10.1016/j.beha.2010.01.005. [DOI] [PubMed] [Google Scholar]
- 32.Varghese AM, Rawstron AC, Hillmen P. Eradicating minimal residual disease in chronic lymphocytic leukemia: Should this be the goal of treatment? Curr Hematol Malig Rep. 2010;5:35–44. doi: 10.1007/s11899-009-0041-2. [DOI] [PubMed] [Google Scholar]
- 33.Lowsky R, et al. Protective conditioning for acute graft-versus-host disease. N Engl J Med. 2005;353:1321–1331. doi: 10.1056/NEJMoa050642. [DOI] [PubMed] [Google Scholar]
- 34.Kohrt HE, et al. TLI and ATG conditioning with low risk of graft-versus-host disease retains antitumor reactions after allogeneic hematopoietic cell transplantation from related and unrelated donors. Blood. 2009;114:1099–1109. doi: 10.1182/blood-2009-03-211441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ladetto M, et al. Real-time polymerase chain reaction of immunoglobulin rearrangements for quantitative evaluation of minimal residual disease in multiple myeloma. Biol Blood Marrow Transplant. 2000;6:241–253. doi: 10.1016/s1083-8791(00)70006-1. [DOI] [PubMed] [Google Scholar]
- 36.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Giudicelli V, Chaume D, Lefranc MP. IMGT/GENE-DB: A comprehensive database for human and mouse immunoglobulin and T cell receptor genes. Nucleic Acids Res. 2005;33(Database issue):D256–D261. doi: 10.1093/nar/gki010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.