Abstract
Effectors delivered into host cells by the Legionella pneumophila Dot/Icm type IV transporter are essential for the biogenesis of the specialized vacuole that permits its intracellular growth. The biochemical function of most of these effectors is unknown, making it difficult to assign their roles in the establishment of successful infection. We found that several yeast genes involved in membrane trafficking, including the small GTPase Ypt1, strongly suppress the cytotoxicity of Lpg0695(AnkX), a protein known to interfere severely with host vesicle trafficking when overexpressed. Mass spectrometry analysis of Rab1 purified from a yeast strain inducibly expressing AnkX revealed that this small GTPase is modified posttranslationally at Ser76 by a phosphorylcholine moiety. Using cytidine diphosphate-choline as the donor for phosphorylcholine, AnkX catalyzes the transfer of phosphorylcholine to Rab1 in a filamentation-induced by cAMP(Fic) domain-dependent manner. Further, we found that the activity of AnkX is regulated by the Dot/Icm substrate Lpg0696(Lem3), which functions as a dephosphorylcholinase to reverse AnkX-mediated modification on Rab1. Phosphorylcholination interfered with Rab1 activity by making it less accessible to the bacterial GTPase activation protein LepB; this interference can be alleviated fully by Lem3. Our results reveal reversible phosphorylcholination as a mechanism for balanced modulation of host cellular processes by a bacterial pathogen.
Keywords: vesicle trafficking, posttranslational modification, type IV secretion
Intravacuolar bacterial pathogens have evolved various strategies to engage selectively with specific cellular compartments to acquire membrane materials to accommodate the expansion of their phagosome. After phagocytosis, Legionella pneumophila, the etiological agent of the potentially fatal Legionnaires’ disease, initiates a unique trafficking route that bypasses the default phagosome maturation pathway. The Legionella-containing vacuole (LCV) sequentially engages in intimate interactions with several host organelles, including the endoplasmic reticulum (ER), mitochondria, and ribosomes (1). It now is well established that L. pneumophila actively converts its phagosomal membranes into membranes with characteristics of the ER. This model is supported by the observations that the LCV interacts intimately with the ER, and its membranes are enriched with several proteins such as binding immunoglobulin protein and calnexin specific for this organelle (2, 3). Furthermore, genetic and pharmaceutical interference with the formation of coat protein complex II vesicles blocks the maturation of LCVs (4, 5).
Successful conversion of the phagosome into a vacuole permissive of L. pneumophila replication is mediated by effectors translocated by its Dot/Icm type IV secretion system, which modulates various host cellular processes to coordinate the biogenesis of the LCV (6). More than 270 Dot/Icm substrates have been identified (7). However, with a few exceptions, the biochemical functions of most of these substrates are unknown, as are their contributions in the intracellular life cycle of L. pneumophila (6). Consistent with the notion that L. pneumophila actively intercepts membrane vesicles originating from the ER to remodel its phagosome (4), several bacterial proteins directly target key molecules critical for this phase of membrane transport. For example, the entire activity cycle of the small GTPase Rab1, which regulates many important events in membrane transport, is hijacked by bacterial virulence factors. The multifunctional protein SidM/DrrA extracts from and/or competes with the GDP dissociation inhibitor (GDI) and subsequently activates it by its guanine nucleotide exchange factor (GEF) activity (8–10). As infection proceeds to 2 h, Rab1 is inactivated by LepB, a L. pneumophila GTPase activation protein (GAP), leading to its removal from the bacterial phagosome by a yet-unidentified GDI (11).
Two lines of evidence suggest that other Dot/Icm substrates are involved in the modulation of host membrane transport. First, deletion mutants of genes known to interfere with Rab1 activity did not cause defects in intracellular bacterial growth. Second, a number of Dot/Icm substrates have been shown to inhibit membrane trafficking in yeast (12). However, the biochemical functions of these proteins are unknown.
In a screen to identify L. pneumophila proteins capable of killing eukaryotic cells, we isolated several L. pneumophila genes that are toxic to yeast (13). Among the identified toxic proteins, Lpg0695 (AnkX) is a Dot/Icm substrate that contains multiple ankyrin-repeat homology domains (14). Furthermore, ectopically expressed AnkX strongly disrupts the secretion pathways of mammalian cells in a process that requires presence of the filamentation-induced by cAMP (Fic) domain in its N-terminal domain (14). With a core sequence of HPFx(D/E)GN(G/K)R, the Fic domains are critical for the adenylyl transferase activity (AMPylation) that stably modifies substrates by transferring an AMP moiety from ATP to the target proteins (15). However, such enzymatic activity has not yet been demonstrated, and the full spectrum of its cellular targets is unknown. By using yeast genetics as a tool, we identified a number of yeast genes capable of efficiently suppressing the yeast toxicity of AnkX. By mass spectrometry analysis of Rab1 purified from a yeast strain inducibly expressing AnkX, we found that AnkX is a phosphorylcholine (PC) transferase that targets the small GTPase Rab1 and possibly other proteins involved in the docking of vesicles in membrane trafficking to the cis-Golgi compartment. Further, we found that the activity of AnkX is regulated by Lem3, a Dot/Icm substrate that possesses a biochemical activity opposite that of AnkX to remove the PC moiety from Rab1. Our results suggest the spatial regulation of Dot/Icm substrate activity by other substrates.
Results
Several Genes Involved in Membrane Trafficking Between the ER and the Golgi Apparatus Suppress Yeast Toxicity of AnkX.
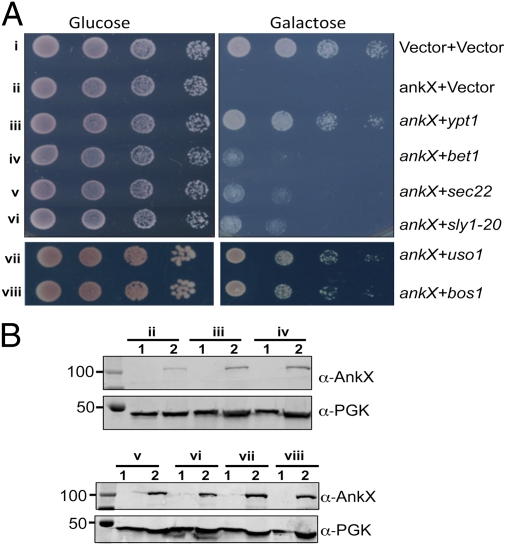
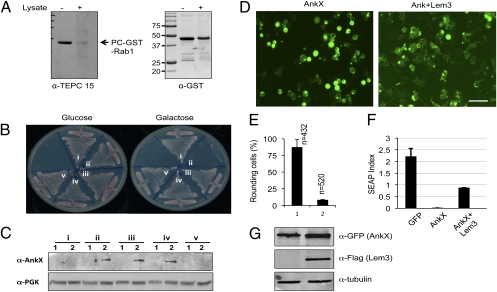
To exploit yeast genetics in the dissection of Dot/Icm effector activity, we isolated a number of L. pneumophila genes exhibiting toxicity to this organism (13). To identify the potential cellular target of AnkX, we introduced a yeast genomic library into a yeast strain inducibly expressing AnkX; a number of clones capable of suppressing AnkX cytotoxicity were isolated. Sequencing revealed that each of these clones contained at least one gene involved in membrane trafficking, including the small GTPase Ypt1 (Rab1 for mammalian cells); Bet1p, Sec22p, and Bos1p, three SNARES involved in the ER to Golgi membrane transport (16, 17); and the tethering factor Uso1p (16). Most of the genes are essential for yeast viability, and many of them can suppress defects caused by ypt1 mutation (18). When expressed independently, each of these genes exhibited suppressor phenotypes similar to the original clones without affecting the cellular levels of AnkX under inducing conditions (Fig. 1). To obtain a more complete list of suppressors, we constructed sly1-20p, a dominant mutant of the t-SNARE–associated protein Sly1, which suppresses ypt1 deficiency (19), and found that this allele was able to suppress the toxicity of AnkX (Fig. 1A, strain vi). These results suggest that one or more of these proteins is the potential cellular target of AnkX.
Fig. 1.
Suppression of AnkX yeast toxicity by yeast genes involved in membrane trafficking. (A) Yeast expressing chromosomally encoded (obtained by integration) AnkX from the galactose-inducible promoter was transformed with plasmids expressing the indicated genes from the alcohol dehydrogenase promoter. Yeast cells diluted with water were spotted onto glucose or galactose medium. The growth of yeast cells was documented 3 d after inoculation. Strains harboring only the vectors (Top row) or only ankX and vector (Second row) were used as controls. (B) Expression of ankX in yeast strains harboring the suppressor genes. Yeast strains grown in medium supplemented with raffinose (1) were induced with galactose (2) for 7 h, and total proteins resolved by SDS/PAGE were probed by immunoblotting with an AnkX-specific antibody. The 3-phosphoglycerate kinase (PGK) was probed as a loading control. Note that AnkX is expressed in all strains in a galactose-dependent manner.
AnkX is a PC Transferase That Modifies Rab1 at Ser76.
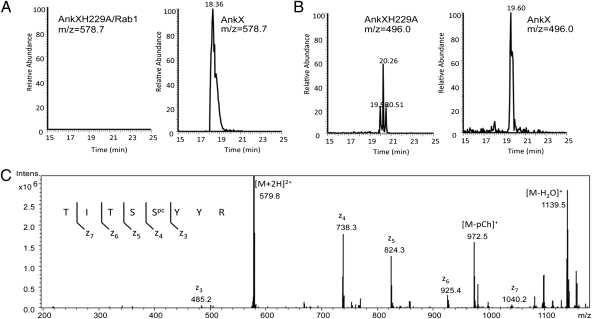
Although all characterized Fic domain proteins target small GTPases, and ypt1 displayed the strongest suppressing activity (Fig. 1) (15), we were unable to detect AnkX-mediated AMPylation of Rab1 or other suppressors. To determine the potential effect of AnkX on Rab1, we purified GST-Rab1 from yeast strains inducibly expressing AnkX or its Fic domain mutant AnkXH229A and subjected the protein to trypsin digestion and nanoflow liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. A mass shift of 165.1 (m/z = 578.7, z = 2) was observed in most (relative abundance >92%) of the tryptic fragment -T72ITSSYYR79- in GST-Rab1 coexpressed with wild-type AnkX (Fig. 2 A and B). Such increase was not observed in peptides from GST-Rab1 coexpressed with the AnkXH229A mutant (m/z = 496, z = 2) (Fig. 2 A and B). Further analysis revealed that this peptide contains a covalent modification of 165.1 D on Ser76, the residue next to Tyr77 (Fig. 2C), which was AMPylated by SidM/DrrA (20). This mass increase corresponds to the addition of a PC moiety, suggesting that AnkX functions as a PC transferase that potentially modifies Rab1 at Ser76.
Fig. 2.
Rab1 coexpressed with AnkX in yeast was modified on Ser76 by phosphorylcholination. (A) Extracted ion chromatograms for the -T72ITSSYYR79- peptide (m/z = 578.7) in GST-Rab1 coexpressed with the AnkXH229A mutant (1.6% relative to all forms of the peptide; Left) or with AnkX (92.5% relative abundance; Right). (B) Extracted ion chromatograms for the -T72ITSSYYR79- peptide (m/z = 496.0) of GST-Rab1 coexpressed with the AnkXH229A mutant (96.3% relative abundance) or AnkX (6.8% relative abundance). (C) Electron-transfer dissociation MS/MS spectrum of modified -TITSSYYR- peptide. The mass shift for fragment ions z4 through z7 and lack of shift for z3 shows that the phosphorylcholination occurred at Ser76. Labeled sequence-specific z-ions indicate the site of modification.
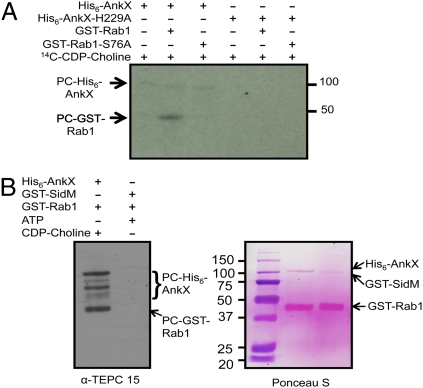
To examine directly whether AnkX functions as an enzyme to modify Rab1 with PC, we performed in vitro biochemical assays with cytidine diphosphorylcholine [methyl-14C] (14C-CDP-choline). Incubation of purified His6-AnkX with GST-Rab1 and 14C-CDP-choline led to the production of 14C-phosphoryl-choline-Rab1 (Fig. 3A, lane 2). Consistent with the observation that a mutation in the conserved histidine residue abolished its toxicity to eukaryotic cells (14), His6-AnkXH229A failed to catalyze the reaction, indicating the importance of the Fic domain in this enzymatic activity (Fig. 3A, lane 5). Further, His6-AnkX did not label GST-Rab1S76A, confirming that the PC modification is specific for Ser76. The fact that 14C-choline-AnkX was detected in all samples containing wild-type protein indicated that AnkX modifies itself (Fig. 3 A, lanes 1–3, and B, lane 1). Because detection of the 14C-choline incorporated into the target molecules requires at least 3-wk exposure of X-ray films to the gels, we established a Western blot-based detection with the PC-specific antibody TEPC 15 (21). This antibody recognizes both PC-Rab1 and PC-AnkX but not AMP-Rab1 (Fig. 3B and Fig. S1). Taken together, these results established that AnkX is a PC transferase that targets the Rab1 protein in a process that requires its Fic domain.
Fig. 3.
AnkX is a PC transferase that modifies Rab1. (A) PCylation of Rab1 by AnkX requires the Fic domain and Ser76 of the small GTPase. A series of reactions with the indicated components and 14C-CDP-choline was allowed to proceed for 30 min at 35 °C. After separation by SDS/PAGE, the incorporation of 14C-PC into the proteins was detected by autoradiography. The sizes of relevant protein markers (in kDa) are indicated. (B) The PC-specific antibody TEPC 15 can detect PCylated Rab1 and PCylated AnkX but not AMPylated Rab1. GST-Rab1 was incubated with His6-AnkX and CDP-choline or with GST-SidM and ATP. After SDS/PAGE, proteins on the same membrane were detected by Ponceau S staining (Right) or by Western blot with the TEPC 15 antibody (Left).
Phosphorylcholination Interferes with GTP Loading and LepB-Induced GTPase Activity of Rab1.
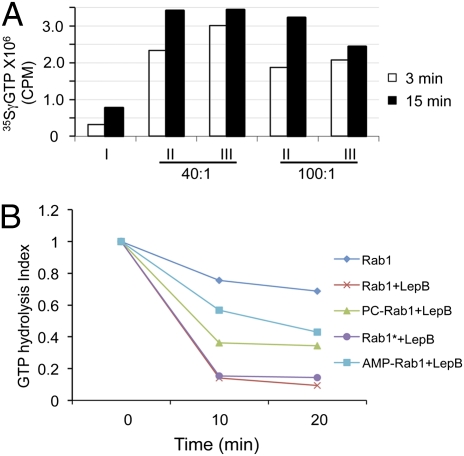
To determine the effects of phosphorylcholination (referred to as “PCylation” throughout the text) on the activity of Rab1, we first examined the loading of GTP to PCylated Rab1. Although at a low efficiency, incubation of 35SγGTP with GDP-Rab1 led to significant loading of GTP (Fig. 4A). Inclusion of SidM, the GEF for Rab1, in the reactions significantly increased GTP incorporation (Fig. 4A). When SidM and Rab1 were used at a 1:40 ratio, similar amounts of 35SγGTP associated with PCylated Rab1 were detected (Fig. 4A). However, PCylated Rab1 reproducibly received lower amounts of GTP when the molar ratio between these two proteins was increased to 100 (Fig. 4A). Thus, in these in vitro experiments, the ability of PCylated Rab1 to receive GTP was impaired, and this impairment could be compensated by higher amounts of SidM. Next, we examined the GTP hydrolysis activity of PCylated Rab1. The Legionella GAP protein LepB (11) induced efficient GTP hydrolysis by Rab1 or by Rab1 that had been incubated with AnkXH229A and CDP-choline (Fig. 4B). As expected, AMPylation of Rab1 by SidM/DrrA blocked its GTPase activity even in the presence of LepB (Fig. 4B) (11). Although less severe than AMPylation, PCylated Rab1 blocked the GTPase activity induced by LepB (Fig. 4B). On the other hand, PCylation did not interfere detectably with the binding of Rab1 to SidM/DrrA or LidA (Fig. S2). Thus, PCylation interferes with the GTPase activity of Rab1, and this interference, along with other potential alterations in the property of Rab1, such as the accessibility to endogenous GEF or GAP, may account for the inhibition of eukaryotic secretory pathway by AnkX.
Fig. 4.
PCylation of Rab1 affects its activity in GTP loading stimulated by SidM and hydrolysis induced by LepB. (A) GDP-loaded GST-Rab1 or GST-PC-Rab1 was incubated with 35SγGTP for the indicated time in reactions with different molar ratios of Rab1 and SidM. Radioactivity associated with the protein was determined by a scintillation counter. Reactions: I, Rab1 without SidM; II, Rab1 with SidM; III, PCylated Rab1 with SidM. (B) Differently modified GST-Rab1 was loaded with 32PγGTP, and GTPase activity was induced by adding LepB. Rab1* denotes the protein incubated with AnkXH229A and CDP-choline. The GTP hydrolysis index is the ratio of the radioactivity at the endpoint of the experiments and the radioactivity associated with Rab1 before LepB was added.
Lem3 Is a Dephosphorylcholinase That Modifies Rab1.
The strong effects of AnkX on the host secretory process prompted us to hypothesize that other bacterial factors may be involved in regulating its activity. We first tested whether the PC moiety on Rab1 can be removed by potential Legionella enzymes by incubating PCylated Rab1 with total bacterial lysate. Mixing PC-Rab1 with total Legionella cell lysate did not cause protein degradation but led to an almost complete loss of the PCylation signals (Fig. 5A), suggesting the existence of enzymes capable of reversing the effect of AnkX on Rab1. Therefore, we initiated a screening to identify L. pneumophila genes capable of suppressing the yeast toxicity of AnkX by introducing a L. pneumophila genomic library (22) into the yeast strain inducibly expressing AnkX. Sequencing of 86 clones reproducibly exhibiting the suppression phenotype revealed that, although the size of inserts in these clones varied greatly, all clones contained lpg0696, also called “lem3,” which encodes a 570-amino acid Dot/Icm substrate (23). In L. pneumophila genome, ankX and lem3 are separated by a 7-bp intergenic region and are transcribed in convergent orientations (24). Lem3 fully rescued the growth of the yeast strain harboring ankX on galactose medium (Fig. 5B, strains iii and iv). Such suppression is not caused by inhibition of AnkX expression in yeast, because AnkX in these yeast cells is readily detectable (Fig. 5C). In mammalian cells, Lem3 appeared to localize in the cytoplasm and did not cause any discernable change in cell morphology (Fig. S3). Lem3 was able to rescue the cell-rounding phenotype and the inhibition of secreted alkaline phosphatase (SEAP) secretion by AnkX (Fig. 5 D–G). Lem3 was unable to suppress the yeast toxicity of SidM (Fig. S4) (22), suggesting its specificity toward the activity of AnkX. Like ankX, lem3 is not essential for intracellular replication of L. pneumophila in bone marrow-derived macrophages or Dictyostelium discoideum, two commonly used host systems (Fig. S5). Together, these data suggest that Lem3 functions to antagonize the activity of AnkX.
Fig. 5.
Identification of a L. pneumophila gene capable of suppressing AnkX toxicity to eukaryotic cells. (A) Removal of PCylation signals from Rab1 by total-cell lysate of L. pneumophila. Soluble bacterial cell lysate was incubated with PCylated GST-Rab1, and proteins resolved by SDS/PAGE were detected for the levels of PCylation with the TEPC 15 antibody (Left) or with a GST-specific antibody (for GST-Rab1). The sizes of relevant protein markers (in kDa) are indicated. (B) The yeast strain W303(pGal::AnkX was transformed with empty vector (ii), one of the original identified suppressor clones (iii), or plasmid harboring the lem3 gene (iv). Yeast cells were streaked onto selective medium containing glucose (Left) or galactose (Right). Strains carrying the vector (i) or the lem3 plasmid (v) were included as additional controls. Plates were incubated at 30 °C for 3 d before image acquisition. (C) Expression of ankX in yeast. SDS/PAGE-resolved lysates of indicated yeast cells uninduced (1) or induced (2) with galactose were detected with an AnkX specific antibody. PGK was probed as a loading control (Lower). (D and E) Lem3 rescued the cell-rounding phenotype caused by AnkX in mammalian cells. (D) 293T cells were transfected with pEGFP::AnkX/pFlag (Left) or with pEGFP::AnkX/pFlag::Lem3 (1:1 molar ratio) (Right) for 24 h before image acquisition. (Scale bar, 50 μm.) (E) The same samples were used to quantitate the percentage of cells exhibiting the rounding phenotype. (F and G). Lem3 rescued the AnkX-mediated inhibition of SEAP secretion by mammalian cells. The indicated combinations of plasmids were used to cotransfect 293T cells with a plasmid that directs SEAP biosynthesis. Twenty-four hours after transfection, extracellular and intracellular SEAP levels were measured. (F) The SEAP index is the ratio between extracellular and intracellular SEAP activity. (G) The same samples were used to evaluate the expression of GFP-AnkX and Flag-Lem3. For quantitative assays, each experiment was performed in triplicate, and data from the samples were used to calculate SD. Similar results were obtained in at least three independent experiments.
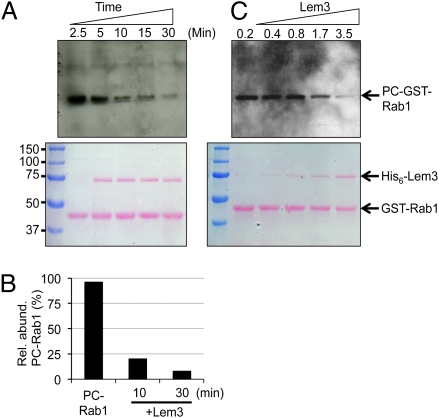
The activity associated with total bacterial cell lysate in removing the PCylation signal from Rab1 suggested that Lem3 biochemically reverses the posttranslational modification on the target of AnkX. Because His6-AnkX present in the samples may interfere with Lem3 activity, we first removed the PC transferase from the reactions by antibody neutralization and extensive washing (Fig. S6). When cleaned PCylated GST-Rab1 was incubated with His6-Lem3 at a molar ratio of 5:1, about 80% of the PCylated substrate was dePCylated in 10 min (Fig. 6A, lane 3). Further incubation for 30 min removed the PC moiety from Rab1 almost completely (Fig. 6B, lane 5). Similar kinetics of Lem3-mediated PC moiety loss from PC-GST-Rab1 were observed when the samples were analyzed by mass spectrometry, in which the relative abundance of PCylated peptide -T72ITSSYYR79- dropped to about 20% after 10-min incubation with Lem3 and to less than 8% when incubation was extended to 30 min (Fig. 6B). In experiments to determine dose-dependent activity of Lem3, incubation of PC-GST-Rab1 with His6-Lem3 at a similar molar ratio for 10 min led to an ∼75% reduction of PCylated GST-Rab1 (Fig. 6C, lane 5). Importantly, the mass of the dePCylated peptide was identical to that of untreated Rab1 (Table S1), indicating that Lem3 catalyzed a simple hydrolytic reaction to remove the PC moiety from Rab1. Consistent with this observation, dePCylated GST-Rab1 can be remodified by AnkX and CDP-choline (Fig. S7). Further, Lem3 did not detectably affect LepB-induced GTP hydrolysis by Rab1, but treatment of PCylated Rab1 with Lem3 restored the accessibility of LepB to the small GTPase, leading to full GTP hydrolysis activity (Fig. S8). Taken together, these results establish that Lem3 is a dephosphorylcholinase that functions to regulate the activity of AnkX by reversing its posttranslational modification on Rab1.
Fig. 6.
Lem3 is a dephosphorycholinase that removes the PC moiety from PCylated Rab1. (A and B) GST-Rab1 modified by AnkX and CDP-choline was incubated with His6-Lem3 at a 5:1 molar ratio for the indicated time. (A) After SDS/PAGE, the levels of PCylated GST-Rab1 were analyzed by immunoblotting with the TEPC 15 antibody (Upper). Protein levels were detected by Ponceau S staining (Lower). (B) After SDS/PAGE, the levels of PCylated GST-Rab1 were analyzed by mass spectrometry of the GST-Rab1 protein bands. (C) Dose-dependent activity of Lem3. Ten micrograms of PCylated GST-Rab1 were incubated with the indicated amounts of His6-Lem3 for 10 min. Proteins resolved by SDS/PAGE were detected first by Ponceau S staining (Lower), and the levels of PCylation were detected by Western blot (Upper). The sizes of relevant protein markers (in kDa) are indicated.
Discussion
Posttranslational modification conferred by Fic domain proteins, particularly via AMPylation, has emerged as a recently recognized mechanism of signal transduction (15, 25). Our findings and the recent discoveries by Mukerjee and colleagues (26) have expanded our knowledge about the biochemical activity of the Fic domain, and our understanding of the biochemical capacity of proteins harboring this catalytic motif likely will continue to expand. For example, some enzymes may use CTP, GTP, or UTP as the substrate in the reaction (27). Our identification of Lem3 as a dephosphorylcholinase showed that, like other commonly used posttranslational modifications such as phosphorylation, ubiquitination, and the more recently recognized AMPylation, PCylation is a reversible process regulated by specific enzymes.
AnkX has been shown to be necessary for efficient inhibition of the acquisition of lysosomal-associated membrane protein 1 by the LCV but not to be essential for intracellular growth of L. pneumophila under standard laboratory conditions (14). Similarly, Lem3 is dispensable for intracellular replication in two tested hosts. As suggested by a recent study, L. pneumophila appears to acquire clusters of Dot/Icm substrates for the expansion of its host range (28). These proteins may be required for maximal virulence under certain conditions or during the interactions of L. pneumophila with some hosts. The synteny of ankX and lem3 provided another example of the regulation of the activity of the Dot/Icm substrate by closely linked genes. The ubiquitin ligase LubX targets the product of its neighboring gene sidH for proteasome degradation (29), presumably to prevent the latter from causing yet-unidentified detrimental effects in the development of LCVs. Similarly, SidD down-regulates the effects of SidM/DrrA by an enzymatic activity that is the opposite of AMPylase to allow efficient removal of Rab1 from the bacterial phagosome (22, 30). The close linkage of these gene pairs in the L. pneumophila genomes emphasizes the importance of properly regulating the activity of the proteins that directly target host processes. Many genes coding for Dot/Icm substrates form clusters, with some organized into operon-like structures (7). Investigation of the potential functional relationships among these closely linked genes could facilitate the study of their biochemical activities and their roles in L. pneumophila infection.
Although the functional relationship between AnkX and Lem3 is reminiscent of the relationshp between SidM/DrrA and SidD, the role of Lem3 during L. pneumophila infection is not as clear as that of SidD. Furthermore, SidD appears to regulate the effects of SidM/DrrA temporally, but such regulation is not apparent for Lem3 on AnkX. Unlike many Dot/Icm substrates whose expression is induced when the bacterium reaches postexponential phase, ankX and lem3 appear to express constitutively throughout the entire bacterial growth cycle in broth (Fig. S9). Consistently, their protein levels in the cytosol of infected macrophages were constant during the first 8 h of infection (Fig. S10). Deletion of lem3 did not cause detectable changes in infected cells, suggesting that the effect of AnkX during infection is subtle and probably highly specific for certain cellular compartments. Most of the yeast genes capable of suppressing AnkX toxicity are involved in the docking of ER-derived trafficking vesicles at the cis-Golgi compartment (16). Thus, it is tempting to speculate that Lem3 spatially regulates the activity of AnkX. It is possible that one of the functions of AnkX is to inhibit membrane-fusion events at the cis-Golgi compartment. Such cellular compartment-specific regulation can be achieved by specific targeting of the protein to its sites of action. The multiple ankyrin repeat in AnkX may be involved in localizing the protein to specific cellular compartments by interacting with proper resident proteins. Clearly, to substantiate this speculation, it is of paramount importance to determine the precise cellular localization of AnkX during L. pneumophila infection. Such inhibition could be beneficial to the pathogen because it might make more vesicles available for fusion with its phagosome, thus facilitating the membrane-remodeling process. On the other hand, Lem3 may localize at cellular sites such as the LCV, which are different from those at which AnkX localizes, to ensure that mislocalized AnkX does not “accidentally” prevent membrane fusion at the bacterial phagosome. Regardless of the mechanism, the activities of AnkX and Lem3 emphasize the importance of a balanced manipulation of host function during L. pneumophila infection and may explain the necessity for this pathogen to code for such a large effector repertoire.
Phosphorylcholination has been described in all domains of organisms, mostly on antigens of both prokaryotic and eukaryotic pathogens that play important roles in the modulation of the host's immune system (31, 32). Because the modification leads to alternations in the activity of its target proteins, PCylation can be considered a form of signaling mechanism. This modification appears to play an important role in development and reproduction of Caenorhabditis elegans (33). Despite its wide use and the potentially important roles in many essential biological processes, few PC transferases that directly modify the backbone of target proteins have been identified, nor have enzymes that specifically reverse the modification (32). Further study of proteins with biochemical activities similar to those of AnkX and Lem3 likely will unravel novel cellular processes regulated by phosphorylcholination.
Materials and Methods
Bacterial, Yeast Strains and Plasmid Construction.
All L. pneumophila strains used in this study were derivatives of the Philadelphia 1 strain Lp02 (34). Bacteria were grown and maintained with standard procedures (35). In-frame deletion mutants of ankX and lem3 were constructed as described (36). In each case, the primers were designed so that the target gene was replaced by a 32-amino acid polypeptide consisting of the first and the last 15 residues and two amino acids encoded by recognition element of the restriction enzyme BamHI (36). For complementation experiments, the gene was expressed on the RSF1010-derived plasmid pJB908 (37). When necessary, antibiotics were used as described (35). For infection experiments, L. pneumophila strains were grown in AYE broth to the postexponential phase as judged by optical density of the cultures (OD600 = 3.3–3.8) as well as by an increase of bacterial motility. For expression in mammalian cells, genes were cloned into pEGFPC1 (Clontech) or p4XFlag (Sigma). All constructs were verified by sequencing analysis. The sequences of primers used in this study are available upon request. Intracellular bacterial growth was performed with established methods (35). Other materials and methods used are listed in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Tony Hazbun (Purdue University, West Lafayette, IN) for yeast strains, plasmids, and helpful discussion. We also thank Dr. Craig Roy (Yale University Medical School, New Haven, CT) for plasmid expressing SEAP and Dr. Phil Lowry (University of Reading, Berkshire, United Kingdom) for suggesting the TEPC 15 antibody. This work was supported by National Institutes of Health-National Institute of Allergy and Infectious Diseases Grants R01AI069344, K02AI085403, and R21AI092043 (to Z.-Q.L.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1114023109/-/DCSupplemental.
References
- 1.Tilney LG, Harb OS, Connelly PS, Robinson CG, Roy CR. How the parasitic bacterium Legionella pneumophila modifies its phagosome and transforms it into rough ER: Implications for conversion of plasma membrane to the ER membrane. J Cell Sci. 2001;114:4637–4650. doi: 10.1242/jcs.114.24.4637. [DOI] [PubMed] [Google Scholar]
- 2.Derré I, Isberg RR. Legionella pneumophila replication vacuole formation involves rapid recruitment of proteins of the early secretory system. Infect Immun. 2004;72:3048–3053. doi: 10.1128/IAI.72.5.3048-3053.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kagan JC, Stein MP, Pypaert M, Roy CR. Legionella subvert the functions of Rab1 and Sec22b to create a replicative organelle. J Exp Med. 2004;199:1201–1211. doi: 10.1084/jem.20031706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kagan JC, Roy CR. Legionella phagosomes intercept vesicular traffic from endoplasmic reticulum exit sites. Nat Cell Biol. 2002;4:945–954. doi: 10.1038/ncb883. [DOI] [PubMed] [Google Scholar]
- 5.Dorer MS, Kirton D, Bader JS, Isberg RR. RNA interference analysis of Legionella in Drosophila cells: Exploitation of early secretory apparatus dynamics. PLoS Pathog. 2006;2:e34. doi: 10.1371/journal.ppat.0020034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubber A, Roy CR. Modulation of host cell function by Legionella pneumophila type IV effectors. Annu Rev Cell Dev Biol. 2010;26:261–283. doi: 10.1146/annurev-cellbio-100109-104034. [DOI] [PubMed] [Google Scholar]
- 7.Zhu W, et al. Comprehensive identification of protein substrates of the Dot/Icm type IV transporter of Legionella pneumophila. PLoS ONE. 2011;6:e17638. doi: 10.1371/journal.pone.0017638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Machner MP, Isberg RR. Targeting of host Rab GTPase function by the intravacuolar pathogen Legionella pneumophila. Dev Cell. 2006;11:47–56. doi: 10.1016/j.devcel.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 9.Machner MP, Isberg RR. A bifunctional bacterial protein links GDI displacement to Rab1 activation. Science. 2007;318:974–977. doi: 10.1126/science.1149121. [DOI] [PubMed] [Google Scholar]
- 10.Schoebel S, Oesterlin LK, Blankenfeldt W, Goody RS, Itzen A. RabGDI displacement by DrrA from Legionella is a consequence of its guanine nucleotide exchange activity. Mol Cell. 2009;36:1060–1072. doi: 10.1016/j.molcel.2009.11.014. [DOI] [PubMed] [Google Scholar]
- 11.Ingmundson A, Delprato A, Lambright DG, Roy CR. Legionella pneumophila proteins that regulate Rab1 membrane cycling. Nature. 2007;450:365–369. doi: 10.1038/nature06336. [DOI] [PubMed] [Google Scholar]
- 12.Heidtman M, Chen EJ, Moy MY, Isberg RR. Large-scale identification of Legionella pneumophila Dot/Icm substrates that modulate host cell vesicle trafficking pathways. Cell Microbiol. 2009;11:230–248. doi: 10.1111/j.1462-5822.2008.01249.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen X, et al. Targeting eEF1A by a Legionella pneumophila effector leads to inhibition of protein synthesis and induction of host stress response. Cell Microbiol. 2009;11:911–926. doi: 10.1111/j.1462-5822.2009.01301.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pan X, Lührmann A, Satoh A, Laskowski-Arce MA, Roy CR. Ankyrin repeat proteins comprise a diverse family of bacterial type IV effectors. Science. 2008;320:1651–1654. doi: 10.1126/science.1158160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Woolery AR, Luong P, Broberg CA, Orth K. AMPylation: Something old is new again. Front Microbiol. 2010;1:113. doi: 10.3389/fmicb.2010.00113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hay JC, Chao DS, Kuo CS, Scheller RH. Protein interactions regulating vesicle transport between the endoplasmic reticulum and Golgi apparatus in mammalian cells. Cell. 1997;89:149–158. doi: 10.1016/s0092-8674(00)80191-9. [DOI] [PubMed] [Google Scholar]
- 17.Stone S, et al. Bet1p activates the v-SNARE Bos1p. Mol Biol Cell. 1997;8:1175–1181. doi: 10.1091/mbc.8.7.1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sapperstein SK, Lupashin VV, Schmitt HD, Waters MG. Assembly of the ER to Golgi SNARE complex requires Uso1p. J Cell Biol. 1996;132:755–767. doi: 10.1083/jcb.132.5.755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dascher C, Ossig R, Gallwitz D, Schmitt HD. Identification and structure of four yeast genes (SLY) that are able to suppress the functional loss of YPT1, a member of the RAS superfamily. Mol Cell Biol. 1991;11:872–885. doi: 10.1128/mcb.11.2.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Müller MP, et al. The Legionella effector protein DrrA AMPylates the membrane traffic regulator Rab1b. Science. 2010;329:946–949. doi: 10.1126/science.1192276. [DOI] [PubMed] [Google Scholar]
- 21.Lovell TM, et al. Identification of a novel mammalian post-translational modification, phosphocholine, on placental secretory polypeptides. J Mol Endocrinol. 2007;39:189–198. doi: 10.1677/JME-07-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tan Y, Luo ZQ. Legionella pneumophila SidD is a deAMPylase that modifies Rab1. Nature. 2011;475:506–509. doi: 10.1038/nature10307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burstein D, et al. Genome-scale identification of Legionella pneumophila effectors using a machine learning approach. PLoS Pathog. 2009;5:e1000508. doi: 10.1371/journal.ppat.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chien M, et al. The genomic sequence of the accidental pathogen Legionella pneumophila. Science. 2004;305:1966–1968. doi: 10.1126/science.1099776. [DOI] [PubMed] [Google Scholar]
- 25.Itzen A, Blankenfeldt W, Goody RS. Adenylylation: Renaissance of a forgotten post-translational modification. Trends Biochem Sci. 2011;36:221–228. doi: 10.1016/j.tibs.2010.12.004. [DOI] [PubMed] [Google Scholar]
- 26.Mukherjee S, et al. Modulation of Rab GTPase function by a protein phosphocholine transferase. Nature. 2011;477:103–106. doi: 10.1038/nature10335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mattoo S, et al. Comparative analysis of Histophilus somni IbpA with other FIC enzymes reveals differences in substrate and nucleotide specificities. J Biol Chem. 2011;286:32834–32842. doi: 10.1074/jbc.M111.227603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.O'Connor TJ, Adepoju Y, Boyd D, Isberg RR. Minimization of the Legionella pneumophila genome reveals chromosomal regions involved in host range expansion. Proc Natl Acad Sci USA. 2011;108:14733–14740. doi: 10.1073/pnas.1111678108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kubori T, Shinzawa N, Kanuka H, Nagai H. Legionella metaeffector exploits host proteasome to temporally regulate cognate effector. PLoS Pathog. 2010;6:e1001216. doi: 10.1371/journal.ppat.1001216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Neunuebel MR, et al. De-AMPylation of the small GTPase Rab1 by the pathogen Legionella pneumophila. Science. 2011;333:453–456. doi: 10.1126/science.1207193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Grabitzki J, Lochnit G. Immunomodulation by phosphocholine—biosynthesis, structures and immunological implications of parasitic PC-epitopes. Mol Immunol. 2009;47:149–163. doi: 10.1016/j.molimm.2009.09.035. [DOI] [PubMed] [Google Scholar]
- 32.Lochnit G, Bongaarts R, Geyer R. Searching new targets for anthelminthic strategies: Interference with glycosphingolipid biosynthesis and phosphorylcholine metabolism affects development of Caenorhabditis elegans. Int J Parasitol. 2005;35:911–923. doi: 10.1016/j.ijpara.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 33.Berger KH, Isberg RR. Two distinct defects in intracellular growth complemented by a single genetic locus in Legionella pneumophila. Mol Microbiol. 1993;7:7–19. doi: 10.1111/j.1365-2958.1993.tb01092.x. [DOI] [PubMed] [Google Scholar]
- 34.Conover GM, Derré I, Vogel JP, Isberg RR. The Legionella pneumophila LidA protein: A translocated substrate of the Dot/Icm system associated with maintenance of bacterial integrity. Mol Microbiol. 2003;48:305–321. doi: 10.1046/j.1365-2958.2003.03400.x. [DOI] [PubMed] [Google Scholar]
- 35.Luo ZQ, Isberg RR. Multiple substrates of the Legionella pneumophila Dot/Icm system identified by interbacterial protein transfer. Proc Natl Acad Sci USA. 2004;101:841–846. doi: 10.1073/pnas.0304916101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu L, et al. Inhibition of host vacuolar H+-ATPase activity by a Legionella pneumophila effector. PLoS Pathog. 2010;6:e1000822. doi: 10.1371/journal.ppat.1000822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sexton JA, et al. The Legionella pneumophila PilT homologue DotB exhibits ATPase activity that is critical for intracellular growth. J Bacteriol. 2004;186:1658–1666. doi: 10.1128/JB.186.6.1658-1666.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.