Abstract
Abscisic acid (ABA) is an essential hormone that controls plant growth, development, and responses to abiotic stresses. Central for ABA signaling is the ABA-mediated autoactivation of three monomeric Snf1-related kinases (SnRK2.2, -2.3, and -2.6). In the absence of ABA, SnRK2s are kept in an inactive state by forming physical complexes with type 2C protein phosphatases (PP2Cs). Upon relief of this inhibition, SnRK2 kinases can autoactivate through unknown mechanisms. Here, we report the crystal structures of full-length Arabidopsis thaliana SnRK2.3 and SnRK2.6 at 1.9- and 2.3-Å resolution, respectively. The structures, in combination with biochemical studies, reveal a two-step mechanism of intramolecular kinase activation that resembles the intermolecular activation of cyclin-dependent kinases. First, release of inhibition by PP2C allows the SnRK2s to become partially active because of an intramolecular stabilization of the catalytic domain by a conserved helix in the kinase regulatory domain. This stabilization enables SnRK2s to gain full activity by activation loop autophosphorylation. Autophosphorylation is more efficient in SnRK2.6, which has higher stability than SnRK2.3 and has well-structured activation loop phosphate acceptor sites that are positioned next to the catalytic site. Together, these data provide a structural framework that links ABA-mediated release of PP2C inhibition to activation of SnRK2 kinases.
Abscisic acid (ABA) is a key hormone that plants use to regulate many important physiological processes, including seed germination and bud dormancy. ABA is also the central regulator to protect plants against abiotic stresses such as drought, cold, and salinity, which are the principal causes for crop losses worldwide (1). At the center of the ABA signaling network are the three Snf1-related kinases, SnRK2.2, -2.3, and -2.6 (2, 3), the kinase domains of which have high sequence homology to mammalian AMP-activated kinase (AMPK) and its yeast homolog Snf1 (SI Appendix, Fig. S1A). SnRK2.6/OST1 (open stomata 1) is critically important for the ABA-induced closure of stomata in response to drought. SnRK2.2 and SnRK2.3, which are functionally redundant and share almost identical sequences, are predominantly responsible to transduce the ABA response to the inhibition of seed germination and seedling growth. snrk2.2/2.3/2.6 triple mutants are severely compromised in all known ABA responses, implying their collectively essential role in ABA signaling (4). In addition to the conserved kinase domain, SnRK2 kinases contain a C-terminal regulatory region that encompasses two conserved motifs: (i) the SnRK2 box, which is required for kinase activity by an unknown mechanism; and (ii) the highly acidic ABA box, which is important to mediate SnRK2 interactions with type 2C protein phosphatases (PP2Cs) (SI Appendix, Fig. S1A) (5, 6).
The ABA signal is transmitted through a conserved core pathway that culminates in activation of SnRK2 kinases through inhibition of PP2Cs by ABA-bound receptors (SI Appendix, Fig. S1B) (7, 8). In the absence of ABA, the SnRK2 kinases are inactivated by clade A PP2Cs, including Homology to ABI1/ABI2 (HAB1) (3, 9–11), which physically interact with SnRK2 kinases and dephosphorylate a critical serine residue in the kinase activation loop (5, 6, 12). The presence of the ABA signal is perceived by the PYR/PYL/RCAR family of START domain receptors (10, 11). Upon ABA binding, these receptors undergo a conformational change that allows them to bind to the catalytic site of PP2Cs and inhibit their enzymatic activity (9, 13–16). In turn, ABA-induced inhibition of PP2Cs leads to autoactivation of SnRK2 kinases by activation loop autophosphorylation, which allows the SnRK2s to relay the ABA signal into several distinct pathways, leading to transcriptional activation, ion channel regulation, and generation of second messengers (SI Appendix, Fig. S1B) (7, 8). Recent structural studies have unraveled detailed molecular mechanisms of agonism and antagonism of ABA receptors, as well as receptor-mediated inhibition of PP2Cs (9, 13–19), but the mechanisms of SnRK2 autoactivation remain unclear.
Results
SnRK2 Kinases Have Basal, Phosphorylation-Independent Activity That Is Greatly Increased by Activation Loop Phosphorylation.
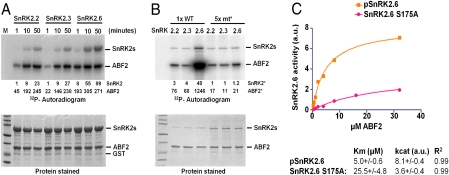
To characterize biochemical functions of SnRK2 kinases, we purified full-length SnRK2.2, -2.3, and -2.6 as recombinant proteins and determined their phosphorylation status by mass spectrometry. In agreement with a previous study on recombinant SnRK2.6 (5), we found clear evidence for phosphorylation of Ser7, Ser29, Ser175, and Thr176 (SI Appendix, Fig. S2A). Mutational studies have indicated that phosphorylation of Ser175 in the SnRK2.6 activation loop (5, 6, 12), but not of Ser7, Ser29, or Thr176 (5), is required for full activity of recombinant SnRK2.6. In SnRK2.2 and -2.3, the residues corresponding to Ser29 were phosphorylated as well, whereas phosphorylation of the residues corresponding to the critical Ser175 in SnRK2.2 (Ser177) and SnRK2.3 (Ser176) was below the limit of detection in the absence of ATP incubation (SI Appendix, Figs. S2A and S3) and low in the presence of ATP incubation (SI Appendix, Fig. S2B). Consistent with these data, the level of SnRK2.6 autophosphorylation upon incubation with ATP is higher by a factor of ∼5–10 than that of SnRK2.2 and 2.3 as determined by [32P]-γATP labeled proteins in SDS gels even though the activation loops are identical between SnRK2.3 and 2.6 (Fig. 1A, top band). We conclude that all three ABA-signaling SnRK2s can autophosphorylate the central serine residue in the activation loop but that the efficiency of activation loop autophosphorylation varies greatly between SnRK2.6 and SnRK2.2/2.3.
Fig. 1.
Biochemical analysis of SnRK2 kinases. (A) Time course of SnRK2 auto- and transphosphorylation. Recombinant SnRK2.2, -2.3, and -2.6 were incubated with a fragment of the transcription factor ABF2 (GST-ABF2[73-120]) and with [32P]-γATP for the indicated length of time. Reactions were terminated by boiling in SDS sample buffer, separated by SDS/PAGE, and subjected to autoradiography. Numbers below autoradiogram: densitometry of autophosphorylation (top) and GST-ABF2(73-120) (bottom) phosphorylation bands. M, marker. (B) SnRK2 phosphorylation mutants retain basal kinase activity. To accommodate for the high activity of WT SnRK2.6, 0.4 μM WT SnRK2s and 2 μM, each, of SnRK2.2 S177A, 2.3 S176A, and 2.6 S175A mutated (mt) kinases were incubated with GST-ABF2(73-120) and [32P]-γATP for 30 min. *Reactions were separated by SDS/PAGE and subjected to autoradiography. Numbers below autoradiogram: densitometry of autophosphorylation (top) and GST-ABF2(73-120) (bottom) phosphorylation bands normalized for the amount of SnRK2. (C) Kinetic analyses of autophosphorylated SnRK2.6 (pSnRK2.6) and SnRK2.6 phosphorylation mutant (S175A). pSnRK2.6 was preincubated for 1 h with 1 mM ATP before dilution and incubation with ABF2 substrate.
Both SnRK2.2 and -2.3 can phosphorylate a fragment from the transcription factor ABA response element-binding factor (ABF)2 (2, 3), which is a natural substrate of SnRK2 kinases (20, 21), with an activity that is reduced by a factor of 5–10 relative to SnRK2.6 (compare the 10 min data of SnRK2.2 and 2.3 with the 1 min data of SnRK2.6 in Fig. 1A), suggesting that autophosphorylation is important for kinase activity. To corroborate this finding, we purified recombinant mutants of the three SnRK2 kinases in which the critical serine residue in the activation loop is replaced by alanine. As shown in Fig. 1B, the level of autophosphorylation of SnRK2.2/2.3 and SnRK2.6 is decreased by factors of 3–4 and ∼40, respectively, after normalization for the amount of proteins (Fig. 1B, bottom). The activity to phosphorylate ABF2 by the mutated kinases is also correspondingly decreased, particularly for SnRK2.6 S175A, which retains only ∼2% of WT kinase activity. We also analyzed the kinetic effects of activation loop phosphorylation by determining Km and kcat for both autophosphorylated SnRK2.6 (pSnRK2.6) and SnRK2.6 S175A. As shown in Fig. 1C, autophosphorylation increased the catalytic activity of SnRK2.6 by a factor of >10 (kcat/Km = 11.3), further supporting the critical role of this serine residue for autophosphorylation and kinase activation. In contrast to the nonphosphorylatable activation-loop serine mutants, SnRK2s are completely inactive in the presence of high concentrations of the PP2C HAB1 (SI Appendix, Fig. S2C and Ref. 22). Taken together, all three kinases have a similarly high level of phosphorylation-independent basal activity. In contrast, the level of autophosphorylation differs between the kinases and correlates with full kinase activity.
Activation Loop Autophosphorylation Is Likely to Occur Both Inter- and Intramolecularly.
Activation loop autophosphorylation is a widespread mechanism for kinase activation and can occur either intermolecularly (in trans) or intramolecularly (in cis) (23). To determine the mode of SnRK2 autophosphorylation, we generated six different catalytically inactive SnRK2.6 mutants. These mutants are unable to cis- (and trans-) autophosphorylate (SI Appendix, Fig. S4), yet their WT activation loops are expected to still be capable of being trans-phosphorylated. We expressed and purified WT and SnRK2.6 mutants with two different tags to allow us to distinguish the two proteins by size. When both proteins were mixed and incubated with [32P]-γATP, both WT and mutant SnRK2.6 proteins were phosphorylated (SI Appendix, Fig. S4B). This clearly demonstrates that SnRK2s can autophosphorylate in trans. Because the activation loops are identical between WT and mutant SnRK2s, we expected at least some mutant SnRK2s to become trans-phosphorylated with similar efficiency as WT SnRK2.6. However, the six mutant proteins with single amino acid replacements of key residues in the Mg2+-binding loop (D160R, F161A, and G162A), the ATP-binding loop (G33R), or the catalytic loop (D140A and K142A) were all to the same degree (by a factor of 1.6–2.2) less efficiently phosphorylated than the WT proteins, suggesting that autophosphorylation may occur both in trans and in cis (SI Appendix, Fig. S4B). As has been demonstrated for p21-activated protein kinase (PAK)2 kinase (24), autophosphorylation may be initiated in cis and then propagated in trans.
Crystal Structures of SnRK2.3 and SnRK2.6 Reveal Canonical Kinase Folds.
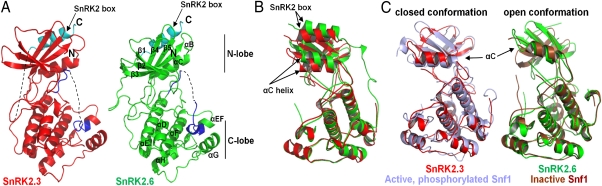
To determine the mechanism of autophosphorylation and high basal activity of SnRK2 kinases, we solved the crystal structures of SnRK2.3 by molecular replacement with the kinase domain of human AMPK α2 (25) (2H6D) as an initial model and the structure of SnRK2.6 using the SnRK2.3 structure as model. In addition, we analyzed their dynamic properties by hydrogen/deuterium exchange (HDX), followed by mass spectrometry, which has become a powerful technique to determine stability and flexibility of proteins and protein–protein interactions (26). WT SnRK2.6 did not crystallize under any condition tested, whereas SnRK2.3 yielded crystals under a single condition that diffracted poorly even after extensive optimization. Introduction of localized surface entropy-reduction mutations (27, 28) allowed us to crystallize and solve the structure of SnRK2.6 D59A/E60A at a resolution of 2.3 Å and of SnRK2.3 D57A/K58A at 1.9 Å (statistics of structure refinement shown in SI Appendix, Table S2). The above crystallization mutations lie at the crystal packing interface and do not affect SnRK2 kinase activity or its binding and inhibition by HAB1 (SI Appendix, Fig. S5). In the crystal structures, the nonphosphorylated SnRK2.3 and -2.6 adopted canonical bilobal kinase folds very similar to those of AMPK and the yeast AMPK homolog Snf1 (25, 29, 30). A standout feature is the well-ordered SnRK2 box, which forms a single α-helix and is packed parallel against the αC-helix in the N-terminal lobe (Fig. 2A). Not visible in the SnRK2.3 structure were the N-terminal 10 aa, the activation loop, a 4-aa segment between the αF- and αG-helices, the linkers to both sides of the SnRK2 box, and the entire ABA box. SnRK2.6 appears to be more stable and contains more structural elements, including parts of the activation loop and the linker between the kinase domain and the SnRK2 box (Fig. 2A and SI Appendix, Fig. S6). In perfect agreement with the HDX data, the structured and unstructured regions in the crystal structures match well with the conformational flexibility map of SnRK2.6 kinase generated by HDX analysis (SI Appendix, Fig. S7).
Fig. 2.
SnRK2.3 and 2.6 structures. Structure overview of SnRK2.3 and 2.6 monomer A (for monomer B see SI Appendix, Fig. S6). (A) The SnRK2 boxes are highlighted in cyan, and the activation loop segment is highlighted in blue. Parts that are not resolved in the structures are the C-terminal 44 residues harboring the ABA box and the segments indicated by dotted lines. (B) Overlay of the SnRK2.3 (red) and 2.6 (green) structures, indicating that SnRK2.3 and SnRK2.6 adopt closed and open conformations, respectively. (C) Overlays of SnRK2.3 (red) with the active Snf1 kinase domain (light blue) (PDB code 3DAE) and of SnRK2.6 (green) with the kinase domain of Snf1 in the inactive, open conformation (brown) (PDB code 2FH9). The SnRK2 boxes were omitted from the overlay for clarity.
Catalytic Centers of SnRK2.3 and -2.6 Show Characteristics of Active and Inactive Kinases.
At the junction of the larger C-terminal lobe and the smaller N-terminal lobe resides the catalytic cleft, which contains the binding sites for substrate and ATP. The two lobes are connected by a flexible hinge that allows kinase domains to adopt two alternative ensembles of conformations: open conformations that are indicative for inactive kinases and closed conformations that are adopted by active kinases (29, 31, 32). Many kinases, including AMPK, require activation loop phosphorylation for the conformational switch from inactive to active conformation. The kinase domain of SnRK2.6 adopted an open conformation that resembles that of unphosphorylated, inactive Snf1 (30). In contrast, SnRK2.3 adopted a closed conformation very similar to that found in phosphorylated, active Snf1 (29) (Fig. 2 B and C) and showed other hallmarks of active or partially active kinases (see below). Crystal structures of kinase domains typically provide snapshots within the ensemble of the different conformations that may be adopted in physiological settings (32). Because both SnRK2.3 and -2.6 share a similar level of basal activity, it is reasonable to assume that both kinases can adopt partially active conformations in their nonphosphorylated state.
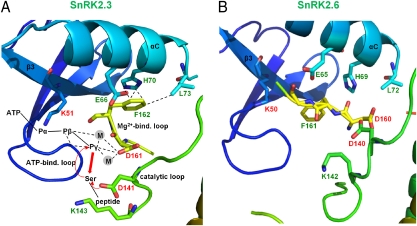
Although cleft opening and closing can also be affected by crystal packing, other important features of active kinase structures include the presence of a helix αC-orchestrated network of interactions that stabilize the binding of ATP and Mg2+ in the catalytic cleft, as well as the conformation of a phosphate-acceptor-binding aspartate in the catalytic loop (31, 33, 34). In the closed conformation of SnRK2.3, the catalytic loop and the Mg2+-binding loop adopted the highly conserved conformations found in active kinases (Fig. 3A). The ATP-binding K51 and the K51-stabilizing residue E66 have their side chains rotated away from each other, yet sterically unconstrained rotations around two C–C single bonds are sufficient to correctly align these residues upon ATP/Mg2+ binding (Fig. 3A), suggesting a partially active state, which presents a structural rationale for the basal activity of SnRK2s. In contrast, in the open conformational snapshot of SnRK2.6, E65 is positioned too far away from the ATP-binding K50 for a productive interaction and the Mg2+-binding loop is misfolded with the Mg2+-binding D160 facing away from the ATP-binding site (Fig. 3B and SI Appendix, Fig. S6B). Taken together, our crystal structures illustrate that unphosphorylated SnRK2s can adopt conformational characteristics of both inactive and active kinases in agreement with the basal activities that we observed in our functional assays (Fig. 1). Full activation of SnRK2.3 and -2.6, similar to other Ser/Thr kinases (34), would require the closed conformation of the αC-helix and phosphorylation of the activation loop to permit full alignment of catalytic residues in the active conformation.
Fig. 3.
Close view of the intramolecular interaction network in the SnRK2.3 and SnRK2.6 (monomer A) active sites. (A) SnRK2.3 adopts a partially active conformation. To illustrate active kinase features in the absence of any ATP-bound SnRK2 structure, we schematically indicated the highly conserved position of ATP with its α-, β-, and γ-phosphates (Pα, Pβ, Pγ), of the Mg2+-ions (gray spheres), and the phosphate-acceptor serine based on Kornev et al. (31). The Mg2+-binding DFG motif is stabilized by a hydrophobic interaction between αC and the DFG phenylalanine. ATP is positioned by three structural elements: (i) the ATP-binding loop (G-loop); (ii) the Mg2+-ions complexed by the DFG aspartate; and (iii) by the α- and β-phosphate-binding K51, which, in turn, requires orientation by forming a salt bridge with αC E66. Note that the catalytic loop and the Mg2+-binding loop adopt conserved, active positions, whereas αC E66 and K51 are at a suitable distance and on the correct side to form a salt bridge but rotated away from each other. (B) SnRK2.6 adopts an inactive conformation in which the K50-orienting E65, the Mg2+-binding D160, and the catalytic D140 face away from the active center.
SnRK2 Box Stabilizes the Regulatory αC-Helix.
How do SnRK2s achieve formation of partially active conformations in the absence of activation loop phosphorylation? Cleft closing and the alignment of catalytic residues is often determined by the position of the αC-helix, which, in many kinases, is modulated by the binding of regulatory proteins to the kinase domain (e.g., 34, 35, 36–41). The SnRK2 box is positioned roughly parallel to the αC-helix and makes extensive contacts with this helix to stabilize its conformation (SI Appendix, Fig. S8). This suggests that the SnRK2 box–αC interaction is important for kinase activity, which we tested by introducing mutations into key interaction residues. Single-point mutations that change interacting residues in the SnRK2 box (SnRK2.3 I313A, I309R, Q304A; SnRK2.6 I312A, I308R) or the αC-helix (SnRK2.3 R71A, I68R, I67R; SnRK2.6 I66R) nearly abolished kinase activity, supporting an essential role of the SnRK2 box in αC positioning (SI Appendix, Fig. S8, panels on the right). SnRK2.6 is noticeably less sensitive to several SnRK2 box mutations than SnRK2.3, consistent with its higher level of activation loop autophosphorylation, which is known to help stabilize the active kinase conformation (34).
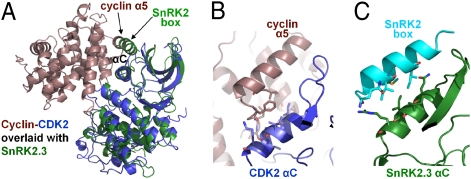
Outside of the AMPK family, the SnRK2 kinase domain shows greatest homology with cyclin-dependent kinase (Cdk)2. Apo Cdk2 is completely inactive and becomes partially active upon cyclin binding, which rearranges the Cdk2 αC-helix to move toward the catalytic cleft (33, 35). This conformational change correctly aligns key residues for ATP- and Mg2+-binding and enables activation loop phosphorylation to complete reorganization of the catalytic cleft to gain full activity (33, 35). As shown in Fig. 4, the intramolecular SnRK2 box–αC interaction structurally mimics the intermolecular stabilization of the Cdk2 αC helix by the cyclin α5-helix. As in the case of Cdk2/cyclin A (33, 35), the SnRK2 box–αC interaction is required for basal activity, because mutations designed to disrupt this interaction inactivate the kinase (Fig. 4). Therefore, the essential function of the SnRK2 box (5) appears to be the stabilization of αC, which is required for correct folding of the catalytic center and kinase activation, in a way that functionally resembles cyclins.
Fig. 4.
The intramolecular interaction between the SnRK2 box and SnRK2.3 αC-helices mimics the intermolecular CDK2 αC stabilization by cyclin α5. (A) Overlay of SnRK2.3 (green) with CDK2 (blue)/cyclin A (brown). Details of the CDK2 αC–α5 (B) and SnRK2.3 αC–SnRK2 box (C) interactions. Both B and C show the same perspective from whole molecule structure alignments.
Although the SnRK2 box lacks sequence homology with cyclin α5, it shows clear homology with the α2-helix of the three-helical autoinhibitory domain (AID) of AMPK and the closely related ubiquitin-associated (UBA) domain of AMPK-related kinases (36) (SI Appendix, Fig. S1). Despite their sequence similarity, these domains differ in both their binding modes and regulatory functions. As shown in SI Appendix, Fig. S9, the AMPK AID packs against the backside of the catalytic cleft to keep the cleft in a wide open, inactive conformation (29). In contrast, the MARK UBA domain binds to the N-terminal lobe and was speculated to stabilize both inactive and active conformations (36), which would resemble the αC stabilization by the SnRK2 box seen in the SnRK2.3 and -2.6 structures.
SnRK2.6 Activation Loop Phosphate-Acceptor Residues Are in Close Proximity to the Catalytic Center.
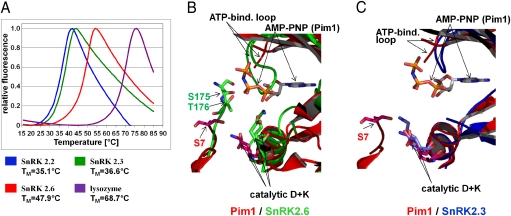
In the SnRK2.6 structure, part of the activation loop, including the autophosphorylated residues S175 and T176, were visible (SI Appendix, Fig. S6A). In contrast, they were not visible in the SnRK2.3 structure. Thermoshift assays also revealed that SnRK2.6 is a much more stable protein than SnRK2.2 and 2.3 (Fig. 5A). Together, these results suggest that the activation loop of SnRK2.6 is more stable than that of SnRK2.3 and -2.2, a possible explanation for the much higher level of autophosphorylation in SnRK2.6 (Fig. 1). To gain further insight into the mechanism of autophosphorylation, we overlaid the SnRK2.6 catalytic cleft with the structure of Pim-1, which, like the SnRK2s, is a member of the calmodulin-dependent protein kinase-related (CAMK) kinase family. The Pim-1 structure is in active conformation with a bound substrate peptide and the ATP analog AMP-PNP (42). As shown in Fig. 5B, SnRK2.6 S175 and T176 assume positions very close to that of the phosphorylated serine (S7) in the Pim-1 substrate peptide. Whereas the position of AMP-PNP clashes with the position of the flexible SnRK2.6 ATP-binding loop in the open conformation (Fig. 5B), there are no clashes in the closed conformation seen in the SnRK2.3 structure (Fig. 5C). Equilibrium between these conformations is, therefore, likely to rearrange this loop to allow ATP-binding and activation loop autophosphorylation of SnRK2.6. The correct positions of SnRK2.6 S175 and T176 toward the catalytic center thus provide a basis for the efficient autophosphorylation of SnRK2.6, which would then lead to its full activation, as we observed in solution (Fig. 1).
Fig. 5.
SnRK2.6 is more stable than SnRK2.3 and has its phosphate-acceptor residues positioned at the catalytic site. (A) Melting curves of SnRK2.2, 2.3 and 2.6 with calculated TM values. As reference, the melting curve of lysozyme is shown as well. (B) Overlay of the catalytic centers of SnRK2.6 (green) and Pim-1 (red), bound to a substrate peptide and the nonhydrolyzable ATP analog AMP-PNP (PDB code 2BZK). Shown as stick models are the phosphate-acceptor residues S175 and T176 (SnRK2.6 monomer B) and S7 (Pim-1 substrate peptide), AMP-PNP, and the catalytic aspartate and arginine residues from both kinases. Note that the Pim-1 ATP-binding loop is not completely resolved. (C) Overlay of SnRK2.3 (blue) and Pim-1, using the same orientation and crop as in (B).
Discussion
Autoactivation of the SnRK2 kinases, triggered by receptor-mediated inhibition of PP2C, is at the center of the ABA signaling network. The structures and biochemical studies presented here help to establish a mechanism for autoactivation of SnRK2 kinases and to complete a structural framework for understanding how ABA signaling is transmitted from hormone perception by the PYR/PYL receptors to inhibition of PP2Cs and to autoactivation of the downstream effectors: SnRK2 kinases.
After completion of this work, Yunta et al. reported the crystal structures of two catalytically inactive mutants (D160A and D160A/S175A) of SnRK2.6 (43). Despite the mutations and different space groups, these structures were, as expected, in similar open, inactive conformations as the SnRK2.6 structure reported here (SI Appendix, Fig. S6B; relative to molecule A in the asymmetric unit, the root mean square deviation for molecule B is 0.817 Å, for the two molecule of the D160A mutant 0.87 and 0.76 Å, and for the two molecules of the D160A/S175A mutant 0.86 and 0.79 Å). In contrast, both SnRK2.3 and -2.6 used in our crystallization studies are fully active kinases, and our structures illustrate that the kinases can adopt either close or open conformations.
The extensive structural and biochemical data presented here illustrate a two-step activation mechanism of SnRK2 kinases. The first step is the ABA-mediated conversion of SnRK2 kinase from a PP2C-inhibited state to a partially active state, which is then further converted into a fully active state by activation loop phosphorylation. In both SnRK2.3 and -2.6 structures, the conserved SnRK2 box adopts a helical structure that forms extensive interactions with αC, which we showed is important for kinase activity. This intramolecular stabilization of αC is both structurally and functionally analogous to the intermolecular αC interaction seen in cyclin-dependent kinases, which are related to the AMPK family of kinases. However, another variation of αC displacement occurs in the tyrosine kinase Src, where an intramolecular interaction stabilizes an inactive αC conformation and disruption of that interaction allows αC to adopt a position very similar to that of SnRK2.3 and cyclin-bound Cdk2 (41). Full activation of the kinases requires phosphorylation of the activation loop, which SnRK2.6 can efficiently achieve on its own. We suggest that this is attributable to the higher stability of SnRK2.6 and the well-structured S175 and T176 residues from the activation loop, which are in close proximity to the active site. Autophosphorylation of recombinant SnRK2.2 and -2.3, which are less stable, is ∼5–10 times less efficient than for SnRK2.6, suggesting that these kinases may also be phosphorylated by as yet unknown upstream kinases, as has been suggested for several members of the SnRK2 family (6, 44). The mechanism of SnRK2 kinase activation by the SnRK2 box and phosphorylation of the activation loop resemble that of AMP-activated kinase and cyclin-dependent kinase, thus further unifying the conserved mechanism of kinase activation despite their diverse cellular functions.
Materials and Methods
Protein Preparation.
WT and mutant SnRK2s were expressed in Escherichia coli BL21 (DE3) as H6Sumo fusion proteins and purified following the same general method as described previously for the purification of PYL1 (13). For more details, see SI Appendix, SI Materials and Methods.
Crystallization.
SnRK2.3 D57A/K58A crystals were grown at room temperature in hanging drops containing 1 μL of the purified protein at a concentration of 15.9 mg mL−1 and 1 μL of well solution containing 0.2 M ammonium sulfate, 0.1 M sodium cacodylate trihydrate (pH 6.5), 25% PEG 8000, and 0.01M Hexamine cobalt (III) chloride. Crystals of ∼50–90 μm in length appeared within 2–3 d. Crystals were serially transferred to well buffer with increasing glycerol concentration [10% (vol/vol) final] before flash-freezing in liquid nitrogen.
SnRK2.6 D59A/E60A crystals were grown at room temperature in sitting drops containing 0.2 μL of the purified protein at a concentration of 6.75 mg mL−1 and 0.2 μL of well solution containing 5% Tacsimate (Hampton Research) (pH 7.0) and 27% PEG 3350. Crystals appeared within 1–2 d and grew to a dimension of ∼50 μm in length over a period of 10 d. Crystals were soaked in well solution with a final concentration of 40% PEG 3350 before flash-freezing in liquid nitrogen.
Data Collection and Structure Determination.
All diffraction datasets were collected at 100 K using an X-ray beam at 0.97872 Å wavelength with Rayomics300 or Rayomics225 (Rayonix, Inc) CCD detectors at the ID-D and ID-F beam lines of sector 21 [Life Sciences Collaborative Access Team (LS-CAT)] at the Advanced Photon Source at Argonne National Laboratory. The observed reflections were reduced, merged, and scaled with DENZO and SCALEPACK in the HKL2000 package (45). The structure of SnRK2.3 was solved by molecular replacement using the kinase domain of human AMPK α2 (PDB code 2H6D, with loop regions deleted) as a search model. The SnRK2.6 structure was solved by molecular replacement using SnRK2.3 as a search model. Molecular replacement was performed by using the Collaborative Computational Project 4 (CCP4) program Phaser (46). Programs O and Coot were used to manually fit the protein models (47, 48). Model refinement was performed with CNS and the CCP4 program Refmac5 (49, 50).
Supplementary Material
Acknowledgments
We thank the staff of LS-CAT for assistance in data collection at the beam lines of sector 21, which is funded, in part, by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor. Use of the Advanced Photon Source was supported by the Office of Science of the US Department of Energy. This work was supported by The Jay and Betty Van Andel Foundation and Amway (China) Limited (H.E.X.), the National Institutes of Health (H.E.X., P.R.G., and J.-K.Z.), and the Singapore Biomedical Research Council (E.-L.Y.). L.-M.N. and F.-F.S. were supported by an overseas PhD scholarship from the National University of Singapore Graduate School for Integrative Sciences and Engineering (NGS).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates reported in this paper have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3UC3 and 3UC4).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1118651109/-/DCSupplemental.
References
- 1.Tuteja N. Abscisic Acid and abiotic stress signaling. Plant Signal Behav. 2007;2:135–138. doi: 10.4161/psb.2.3.4156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fujii H, et al. In vitro reconstitution of an abscisic acid signalling pathway. Nature. 2009;462:660–664. doi: 10.1038/nature08599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Umezawa T, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci USA. 2009;106:17588–17593. doi: 10.1073/pnas.0907095106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci USA. 2009;106:8380–8385. doi: 10.1073/pnas.0903144106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belin C, et al. Identification of features regulating OST1 kinase activity and OST1 function in guard cells. Plant Physiol. 2006;141:1316–1327. doi: 10.1104/pp.106.079327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boudsocq M, Droillard MJ, Barbier-Brygoo H, Laurière C. Different phosphorylation mechanisms are involved in the activation of sucrose non-fermenting 1 related protein kinases 2 by osmotic stresses and abscisic acid. Plant Mol Biol. 2007;63:491–503. doi: 10.1007/s11103-006-9103-1. [DOI] [PubMed] [Google Scholar]
- 7.Cutler SR, Rodriguez PL, Finkelstein RR, Abrams SR. Abscisic acid: Emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679. doi: 10.1146/annurev-arplant-042809-112122. [DOI] [PubMed] [Google Scholar]
- 8.Hubbard KE, Nishimura N, Hitomi K, Getzoff ED, Schroeder JI. Early abscisic acid signal transduction mechanisms: Newly discovered components and newly emerging questions. Genes Dev. 2010;24:1695–1708. doi: 10.1101/gad.1953910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yin P, et al. Structural insights into the mechanism of abscisic acid signaling by PYL proteins. Nat Struct Mol Biol. 2009;16:1230–1236. doi: 10.1038/nsmb.1730. [DOI] [PubMed] [Google Scholar]
- 10.Ma Y, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324:1064–1068. doi: 10.1126/science.1172408. [DOI] [PubMed] [Google Scholar]
- 11.Park SY, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324:1068–1071. doi: 10.1126/science.1173041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vlad F, et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21:3170–3184. doi: 10.1105/tpc.109.069179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melcher K, et al. A gate-latch-lock mechanism for hormone signalling by abscisic acid receptors. Nature. 2009;462:602–608. doi: 10.1038/nature08613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Miyazono K, et al. Structural basis of abscisic acid signalling. Nature. 2009;462:609–614. doi: 10.1038/nature08583. [DOI] [PubMed] [Google Scholar]
- 15.Nishimura N, et al. Structural mechanism of abscisic acid binding and signaling by dimeric PYR1. Science. 2009;326:1373–1379. doi: 10.1126/science.1181829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santiago J, et al. The abscisic acid receptor PYR1 in complex with abscisic acid. Nature. 2009;462:665–668. doi: 10.1038/nature08591. [DOI] [PubMed] [Google Scholar]
- 17.Hao Q, et al. Functional mechanism of the abscisic acid agonist pyrabactin. J Biol Chem. 2010;285:28946–28952. doi: 10.1074/jbc.M110.149005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Melcher K, et al. Identification and mechanism of ABA receptor antagonism. Nat Struct Mol Biol. 2010;17:1102–1108. doi: 10.1038/nsmb.1887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peterson FC, et al. Structural basis for selective activation of ABA receptors. Nat Struct Mol Biol. 2010;17:1109–1113. doi: 10.1038/nsmb.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Furihata T, et al. Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA. 2006;103:1988–1993. doi: 10.1073/pnas.0505667103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnson RR, Wagner RL, Verhey SD, Walker-Simmons MK. The abscisic acid-responsive kinase PKABA1 interacts with a seed-specific abscisic acid response element-binding factor, TaABF, and phosphorylates TaABF peptide sequences. Plant Physiol. 2002;130:837–846. doi: 10.1104/pp.001354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soon F.-F., et al. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science. 2011 doi: 10.1126/science.1215106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lochhead PA. Protein kinase activation loop autophosphorylation in cis: Overcoming a Catch-22 situation. Sci Signal. 2009;2:pe4. doi: 10.1126/scisignal.254pe4. [DOI] [PubMed] [Google Scholar]
- 24.Wang J, Wu JW, Wang ZX. Mechanistic studies of the autoactivation of PAK2: A two-step model of cis initiation followed by trans amplification. J Biol Chem. 2011;286:2689–2695. doi: 10.1074/jbc.M110.156505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Littler DR, et al. A conserved mechanism of autoinhibition for the AMPK kinase domain: ATP-binding site and catalytic loop refolding as a means of regulation. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:143–151. doi: 10.1107/S1744309109052543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chalmers MJ, Busby SA, Pascal BD, Southern MR, Griffin PR. A two-stage differential hydrogen deuterium exchange method for the rapid characterization of protein/ligand interactions. J Biomol Tech. 2007;18:194–204. [PMC free article] [PubMed] [Google Scholar]
- 27.Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 28.Derewenda ZS, Vekilov PG. Entropy and surface engineering in protein crystallization. Acta Crystallogr D Biol Crystallogr. 2006;62:116–124. doi: 10.1107/S0907444905035237. [DOI] [PubMed] [Google Scholar]
- 29.Chen L, et al. Structural insight into the autoinhibition mechanism of AMP-activated protein kinase. Nature. 2009;459:1146–1149. doi: 10.1038/nature08075. [DOI] [PubMed] [Google Scholar]
- 30.Nayak V, et al. Structure and dimerization of the kinase domain from yeast Snf1, a member of the Snf1/AMPK protein family. Structure. 2006;14:477–485. doi: 10.1016/j.str.2005.12.008. [DOI] [PubMed] [Google Scholar]
- 31.Kornev AP, Haste NM, Taylor SS, Eyck LF. Surface comparison of active and inactive protein kinases identifies a conserved activation mechanism. Proc Natl Acad Sci USA. 2006;103:17783–17788. doi: 10.1073/pnas.0607656103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rabiller M, et al. Proteus in the world of proteins: Conformational changes in protein kinases. Arch Pharm (Weinheim) 2010;343:193–206. doi: 10.1002/ardp.201000028. [DOI] [PubMed] [Google Scholar]
- 33.Pavletich NP. Mechanisms of cyclin-dependent kinase regulation: Structures of Cdks, their cyclin activators, and Cip and INK4 inhibitors. J Mol Biol. 1999;287:821–828. doi: 10.1006/jmbi.1999.2640. [DOI] [PubMed] [Google Scholar]
- 34.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15:661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 35.Jeffrey PD, et al. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 36.Marx A, Nugoor C, Panneerselvam S, Mandelkow E. Structure and function of polarity-inducing kinase family MARK/Par-1 within the branch of AMPK/Snf1-related kinases. FASEB J. 2010;24:1637–1648. doi: 10.1096/fj.09-148064. [DOI] [PubMed] [Google Scholar]
- 37.Kannan N, Haste N, Taylor SS, Neuwald AF. The hallmark of AGC kinase functional divergence is its C-terminal tail, a cis-acting regulatory module. Proc Natl Acad Sci USA. 2007;104:1272–1277. doi: 10.1073/pnas.0610251104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang X, Gureasko J, Shen K, Cole PA, Kuriyan J. An allosteric mechanism for activation of the kinase domain of epidermal growth factor receptor. Cell. 2006;125:1137–1149. doi: 10.1016/j.cell.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 39.Lamers MB, Antson AA, Hubbard RE, Scott RK, Williams DH. Structure of the protein tyrosine kinase domain of C-terminal Src kinase (CSK) in complex with staurosporine. J Mol Biol. 1999;285:713–725. doi: 10.1006/jmbi.1998.2369. [DOI] [PubMed] [Google Scholar]
- 40.Bayliss R, Sardon T, Vernos I, Conti E. Structural basis of Aurora-A activation by TPX2 at the mitotic spindle. Mol Cell. 2003;12:851–862. doi: 10.1016/s1097-2765(03)00392-7. [DOI] [PubMed] [Google Scholar]
- 41.Huse M, Kuriyan J. The conformational plasticity of protein kinases. Cell. 2002;109:275–282. doi: 10.1016/s0092-8674(02)00741-9. [DOI] [PubMed] [Google Scholar]
- 42.Bullock AN, Debreczeni J, Amos AL, Knapp S, Turk BE. Structure and substrate specificity of the Pim-1 kinase. J Biol Chem. 2005;280:41675–41682. doi: 10.1074/jbc.M510711200. [DOI] [PubMed] [Google Scholar]
- 43.Yunta C, Martínez-Ripoll M, Zhu JK, Albert A. The structure of Arabidopsis thaliana OST1 provides insights into the kinase regulation mechanism in response to osmotic stress. J Mol Biol. 2011;414:135–144. doi: 10.1016/j.jmb.2011.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burza AM, et al. Nicotiana tabacum osmotic stress-activated kinase is regulated by phosphorylation on Ser-154 and Ser-158 in the kinase activation loop. J Biol Chem. 2006;281:34299–34311. doi: 10.1074/jbc.M601977200. [DOI] [PubMed] [Google Scholar]
- 45.Otwinowski Z, Borek D, Majewski W, Minor W. Multiparametric scaling of diffraction intensities. Acta Crystallogr A. 2003;59:228–234. doi: 10.1107/s0108767303005488. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 48.Kleywegt GJ, Jones TA. Efficient rebuilding of protein structures. Acta Crystallogr D Biol Crystallogr. 1996;52:829–832. doi: 10.1107/S0907444996001783. [DOI] [PubMed] [Google Scholar]
- 49.Brünger AT, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 50.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr D Biol Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.