Abstract
Soil pH is a major determinant of microbial ecosystem processes and potentially a major driver of evolution, adaptation, and diversity of ammonia oxidizers, which control soil nitrification. Archaea are major components of soil microbial communities and contribute significantly to ammonia oxidation in some soils. To determine whether pH drives evolutionary adaptation and community structure of soil archaeal ammonia oxidizers, sequences of amoA, a key functional gene of ammonia oxidation, were examined in soils at global, regional, and local scales. Globally distributed database sequences clustered into 18 well-supported phylogenetic lineages that dominated specific soil pH ranges classified as acidic (pH <5), acido-neutral (5≤ pH <7), or alkalinophilic (pH ≥7). To determine whether patterns were reproduced at regional and local scales, amoA gene fragments were amplified from DNA extracted from 47 soils in the United Kingdom (pH 3.5–8.7), including a pH-gradient formed by seven soils at a single site (pH 4.5–7.5). High-throughput sequencing and analysis of amoA gene fragments identified an additional, previously undiscovered phylogenetic lineage and revealed similar pH-associated distribution patterns at global, regional, and local scales, which were most evident for the five most abundant clusters. Archaeal amoA abundance and diversity increased with soil pH, which was the only physicochemical characteristic measured that significantly influenced community structure. These results suggest evolution based on specific adaptations to soil pH and niche specialization, resulting in a global distribution of archaeal lineages that have important consequences for soil ecosystem function and nitrogen cycling.
Keywords: Thaumarchaeota, Nitrosotalea devanaterra, 454 pyrosequencing
Microorganisms are essential for maintaining soil ecosystem function and play a central role in the cycling of nutrients required for plant growth. Prediction and control of soil ecosystem functions require an understanding of the factors determining the diversity and community structure of particular functional groups and their responses to environmental change. In soil, these communities show considerable diversity (1, 2), universal mechanisms controlling diversity and abundance are difficult to determine, and ecological coherence of phylogenetic groups (3) and patterns of phylotype distribution determined by environmental traits are often difficult to discern. Investigation of ecological coherence in soil microbial communities has focused on bacteria (4), but phylogenomic analyses indicate greater coherence within cultivated archaea than within bacteria (5, 6), in which horizontal gene transfer significantly decreases links between phylogeny and function. Thus, archaea may be more appropriate for testing predictions of links between phylotype distribution and environmental traits, but have rarely been investigated in this context (7–11). Soil contains archaea belonging to both euryarchaea and the proposed thaumarchaea (12, 13) lineages (formerly described as crenarchaea), with the latter dominating archaeal communities in most soils. All currently known ammonia-oxidizing archaea are placed in this lineage. Although their physiology and soil ecosystem function are poorly characterized, increasing evidence suggests that these archaea play a major role in soil ammonia oxidation (14–18).
Soil pH appears to be an important driver of bacterial (19) and archaeal (20) ammonia oxidizer community structure, and pH selection may have a mechanistic basis. Cultivated autotrophic ammonia oxidizers demonstrate little or no growth in liquid batch culture below pH 6.5 (21–23). This is considered related to lower availability of the substrate, ammonia, due to increased ionization to ammonium as pH decreases. Most archaeal ammonia oxidizer cultures (24, 25) grow optimally at neutral pH, and one strain, Nitrosopumilus maritimus, has a high affinity for ammonia (26). Despite the lack of acidophilic growth in most laboratory cultures, nitrification occurs in many acid soils, and there is evidence of a slightly negative correlation between gross nitrification rate and pH (27). Ammonia oxidation in acid soils may be possible through growth in biofilms and aggregates (22, 23) and increased ureolytic activity (28, 29), which would enable growth below pH 5. However, adaptation to growth at low pH cannot be ruled out, as demonstrated recently by the cultivation of the first obligately acidophilic ammonia oxidizer, Nitrosotalea devanaterra (30).
The greater coherence indicated by archaeal genome studies, the importance of pH as a driver of soil bacterial community structure, and direct links between pH and cultivated ammonia oxidizer activity led us to test theories of ecological coherence by predicting soil pH selection of putative ammonia-oxidizing archaea. This was achieved by phylogenetic analysis of archaeal amoA genes, which encode ammonia monooxygenase, a key functional gene in ammonia oxidizers. Sequences from globally distributed soils were used to identify potential relationships between phylogeny and pH and to define groups adapted to acid (pH <5), acido-neutral (5≤ pH <7), and alkaline (pH ≥7) soils. This analysis was then used to predict the ecological coherence of phylotypes at a regional scale, in 47 well-characterized soils in the United Kingdom with a pH range of 3.5–8.7, and at the local scale, in a plot with a pH gradient of 4.5–7.5.
Results
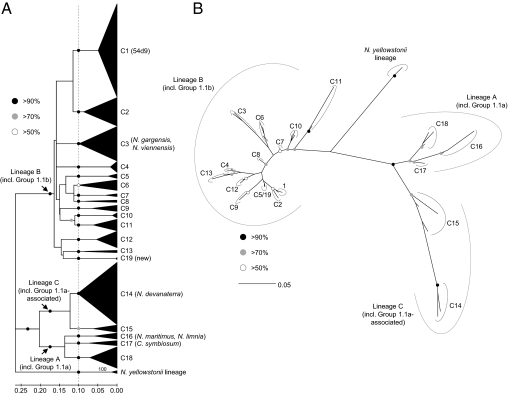
pH selection of globally distributed soil archaeal ammonia oxidizer communities was predicted by phylogenetic analysis of 606 archaeal amoA gene sequences retrieved from the National Center for Biotechnology Information (NCBI) database, comprising all those sequences for which pH data were available and with a minimum length of 586 bp. Other analyses showed no significant relationship between amoA gene distribution and country of origin or ecosystem function. Sequences from the pH dataset were derived from soils from 17 studies in nine countries (Tables S1 and S2) with a pH of 3.7–8.8, along with reference sequences. Phylogenetic analysis was performed using estimated variable sites only (74% of 586 positions). Four major clades were observed with robust phylogenetic support, including those with sequences from group 1.1a, group 1.1a-associated, group 1.1b, and Nitrosocaldus yellowstonii lineages. Preliminary observations indicated that pH-associated lineages could be identified within clusters of sequences with fewer than 0.2 change per nucleotide position, and 18 clusters of soil sequences with robust phylogenetic support were defined arbitrarily. Support for all lineages was >50%, 16 of 18 clusters had >90% support, and each cluster contained sequences with ≥83% pairwise similarity (Fig. 1). Phylogenetic analysis of inferred amino acid sequences found similar groupings but with less bootstrap support. Four different methods—distance, parsimony, maximum likelihood (ML), and Bayesian analysis—supported a topology with the same four major lineages. However, altering some parameters in ML analysis (constant vs. gamma-distributed site variation and modeled vs. empirical amino acid frequencies) of this restricted dataset indicated that the N. yellowstonii lineage can be placed within the group 1.1b lineage, with C11 the most basal lineage (Fig. 1B). The greater similarity at the amino acid vs. nucleotide level was also particularly high for some clusters; for example, the maximum similarities between any C5 and C19 sequence was 81% at the DNA level and 99% at the amino acid level, highlighting differences in codon use between these two groups.
Fig. 1.
Phylogenetic analysis of thaumarchaeal amoA gene sequences from an alignment of 606 sequences from soil from 17 studies and reference sequences, plus sequences of a newly described cluster (C19). (A) DNA distance analysis of all sequences (i.e., general time-reversible correction, gamma-distributed rates of site variation, ML-estimated variable sites only, minimum evolution optimality criterion, 1,000 bootstrap replicates). Circles describe bootstrap support from distance methods only. Cultivated organisms (24, 25, 30, 34–36) and those described by metagenomic analyses (32, 33) that are placed within specific clusters are highlighted. (B) Phylogenetic analysis of inferred amino acid sequences (194 positions) from a reduced representative selection of sequences from 19 clusters plus three sequences placed within the N. yellowstonii lineage (three sequences per cluster). Phylogenetic analyses were performed using four different methods: Bayesian (mixed model correction), ML [Le and Gascuel (LG), Jones, Taylor, and Thornton (JTT), Whelan and Goldman (WAG), and Dayhoff correction], maximum parsimony, and distance (JTT correction) using modeling of invariable sites and site rate heterogeneity where appropriate. Circles describing bootstrap support represent the most conservative value from bootstrap (1,000 replicates) or posterior probabilities (Bayesian) from all four methods.
The major soil archaeal ammonia oxidizer 16S rRNA gene-defined lineages, groups 1.1b, 1.1a, and 1.1a-associated (31), show congruence with archaeal amoA sequences, as determined from cultivated representatives or metagenomic fragments (24, 30–36) and environmental samples (20). The corresponding amoA lineages, referred to here as A (including group 1.1a), B (including group 1.1b), and C (including group 1.1a-associated), were represented by 62 (10.2%), 416 (68.6%), and 127 (21.2%) sequences, respectively. Individual clusters contained sequences from 1–10 sites and were represented by between 4 (0.7%) and 170 (28%) sequences. Thus, there was up to an ∼40-fold difference in the relative abundance of sequences within different clusters, and several were rare (<10 sequences) in the sites investigated.
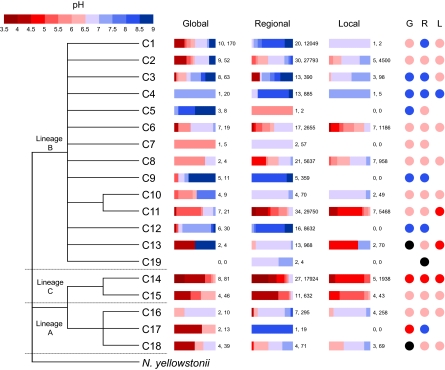
pH-associated distribution of clusters was assessed by determining the relative abundance of sequences within defined pH ranges (3.5–9 at intervals of 0.5) without normalizing for the number of sequences per study (Fig. 2). Clusters were classified as alkalinophilic (pH ≥7; C3-C5, C9, and C12), acido-neutral (5≤ pH <7; C1, C2, C6-C8, C10, C11, and C16), acidic (pH <5; C14, C15, and C17), or showing no pH preference (C13 and C18). Confidence in this classification decreases with sequence abundance and thus is weak for clusters C5, C7, C8, C10, C13, and C16, each of which was represented by ≤10 sequences. Clusters belonging to lineages A and C were restricted to acidic and acido-neutral soils, whereas lineage B contained clusters from both alkalinophilic and acido-neutral soils.
Fig. 2.
Dendrogram showing the distribution of amoA gene sequences derived from soil within one of 19 distinct clusters within three lineages, defined here as A, B, and C, plus sequences within the N. yellowstonii lineage (17 of 19 with >90% bootstrap support; details shown in Fig. 1). Clusters were determined from a meta-analysis of amoA gene sequences in the NCBI database. The relative pH distribution of 606 sequences deposited in the NCBI database from 17 studies (global), 108,192 sequences from 47 soils in the United Kingdom (regional), and 14,644 sequences from seven soil samples from one site with a pH range of 4.5–7.5 (local) are shown. Numbers at the right of each bar represent the number of sites and total number of sequences analyzed for that particular cluster. Columns of circles labeled G, R, and L (for global, regional, and local) indicate whether the pH distribution of sequences classifies the phylogenetic cluster as acidic (red), acido-neutral (pink), alkalinophilic (blue), or not pH-adapted (black). All sequences analyzed are 586 nt long.
To determine whether pH-associated groupings and distributions were similar at the regional scale, sequences were amplified from DNA extracted from 47 sites in the United Kingdom with a wide range of soil pH values (3.7–8.7), including multiple sites within each pH class (Table S3). These included seven samples from a pH gradient at one site (4.5–7.5, at intervals of 0.5), enabling analysis at the local scale. Sequencing was performed over the region of the amoA gene used for global analysis using high-throughput sequencing to increase coverage of amoA diversity, which generated 1.1 × 106 sequences. Sequences >400 nt in length were retained and screened using a strict series of filtering steps to ensure that only high-fidelity sequences were used. A pairwise assembly of forward and reverse sequences was then performed with a custom C program using exact Needleman–Wunsch alignments with a minimum overlap of 100 bp and 100% identity, resulting in a length of 629 nt (586 nt plus primer sequences). This resulted in 108,192 sequences from all sites being categorized into one of 18,858 unique assembled paired sequences, all of which translated perfectly to amino acid sequences. These represented 10% of all original sequences, 54.2% of the ≥400-nt sequences retained after the initial filtering steps, and 79–100% of the maximum number of pairs per site that could be formed by the bipartite matching assembler. Each sequence was compared with those in the global analysis and placed into one of the 18 previously identified clusters. Rarefaction analysis indicated high coverage of archaeal amoA diversity, with an estimated operational taxonomic unit richness of 19 at a 83% similarity level, the minimum level of similarity observed between two sequences in one cluster in the global analysis (Fig. S1). The library contained representative sequences from all clusters. Cluster C19, not represented in preexisting database sequences, was found in one grassland and one agricultural soil (pH 6.9–7.2) and was identified by BLASTn as a novel clade within lineage B.
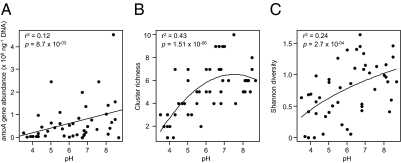
Quantitative PCR was used to determine amoA abundance in all 47 soils, which increased slightly but significantly with pH (Fig. 3A). Stronger relationships were observed between operational taxonomic unit richness and the Shannon diversity index with soil pH (Fig. 3 B and C). The most acidic soils contained only between one and three clusters, reflecting the lower number of phylotypes adapted to soils with pH <5. Richness was greatest at pH 6–8, the optimal pH for growth of cultivated ammonia oxidizers (21, 23–25), and decreased at pH >8. Soils of pH >8.7 were not examined.
Fig. 3.
Relationships between soil pH and amoA gene abundance (A), cluster richness (B), and diversity (C). Regression coefficients of the best-fitting model and associated P values are indicated.
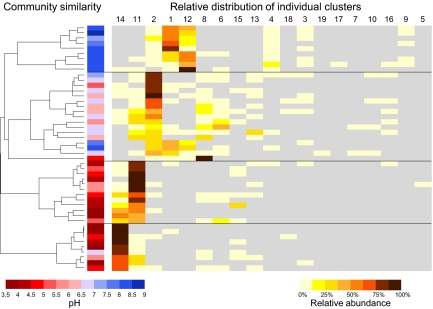
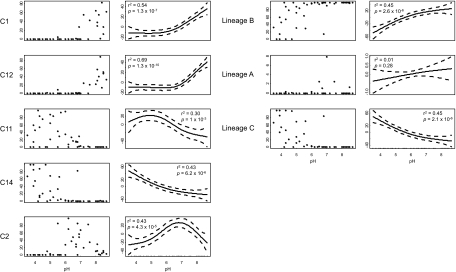
Sequence relative abundance again varied significantly between clusters, in a similar proportional range to database sequences (0.002–27.5%), and most sequences (82.5%) fell within lineage B. The proportion of sequences in lineage C clusters was similar to that in the initial NCBI collection database (17.1% vs. 21.1%), but fewer fell within lineage A (0.4% vs. 10.2%). After normalizing the number of sequences per soil, most clusters showed the predicted distributions with respect to soil pH (Fig. 2) and the same pH relationships observed with database sequences. Heat maps distinguishing pH-associated sequence types and clusters (Fig. 4) indicate four distinct community structures, with two acidophilic communities, one acido-neutrophilic and one alkalinophilic. These communities were dominated by five clusters: C14 and C11 (acidic soils), C2 (acido-neutral soils), and C1 and C12 (alkaline soils). Sequences from the remaining clusters generally comprised <38% sequences within a particular soil, and usually much less, except in one soil in which C8 was dominant (90.5% of sequences). Significant correlations (P < 0.01) between the relative abundance of the five dominant clusters and pH (Fig. 5A) are consistent with the data in the heat map. Lineages B and C were favored in acidic and alkaline soils, respectively (Fig. 5B), whereas 99.9% of the lineage C sequences were retrieved from acidic and acido-neutral soils, and 88.7% of the lineage B sequences from alkaline and acido-neutral soils. To analyze ammonia-oxidizing communities at a local scale, 7 of the 47 soil samples were taken from a single plot of soil, treated since 1961 to maintain seven discrete pH values. Thirteen of the original 18 amoA gene clusters were found at this site, including four of the five dominant regional clusters. This local scale analysis showed a very similar distribution to that seen in the regional analysis (Fig. 2).
Fig. 4.
Dendrogram showing the relatedness of archaeal ammonia oxidizer community structures in a regional analysis of 47 soils (pH 3.5–8.7) based on the relative abundance of distinct lineages of archaeal amoA genes analyzed using 454 sequencing. The relative abundance of each lineage (clusters 1–19) in each soil is displayed in a heat map representation. The pH of soil from each site is indicated by red-blue panels, in the range 3.5–9 at intervals of 0.5. The relative abundance of sequences within an individual cluster in soil is indicated by yellow-brown shading at 12.5% intervals.
Fig. 5.
Relative abundances of selected amoA gene-defined lineages as a function of pH in a regional analysis of 47 UK soils. (Left) Influence of pH on the relative abundance of each of the five dominant archaeal amoA gene clusters. (Right) Relationships between pH and the distribution of the three major lineages A, B, and C. In each row, the first panel represents the percentage relative abundance of sequences belonging to a particular cluster, and the second panel represents the best-fitting model of this distribution. Regression coefficients and associated P values are indicated.
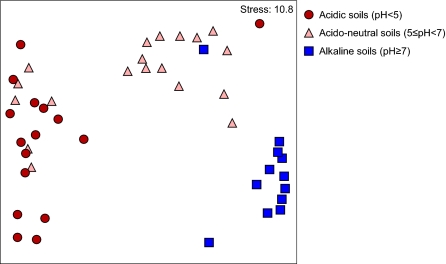
Although soils were chosen to provide a range of pH values, the influences of carbon, nitrogen, C:N ratio, moisture content, organic matter, and vegetation were investigated by canonical correspondence analysis (CCA). A combination of these factors explains archaeal ammonia oxidizer community structure (P = 0.005). Of the characteristics evaluated, pH was the major driver of the archaeal ammonia oxidizer community structure (CCA; P = 0.01) (Table S4). Nonmetric multidimensional scaling analysis also demonstrated the importance of pH for community structure, with the first axis dominated by soil pH effects (Fig. 6). None of the other environmental factors significantly influenced amoA-defined archaeal community structure (CCA; P > 0.13) (Table S4).
Fig. 6.
Nonmetric multidimensional scaling plot of archaeal ammonia oxidizer community structures in a regional analysis of 47 soils based on the relative abundance of sequences within clusters 1–19 in each soil. The first principal axis is dominated by soil pH effect, and each soil is placed within the acidic, acido-neutral, or alkaline category.
Discussion
Distribution of Major Archaeal amoA Lineages in Soil.
Previous phylogenetic analyses of environmental and cultivated archaeal amoA sequences demonstrated substantial congruence among phylogenies based on 16S rRNA and amoA genes (20, 34), indicating nonlateral inheritance. Lineage B sequences (which include those of group 1.1b) had the greatest overall relative abundance in both global and regional collections (68.6% and 82.5%, respectively), whereas lineage A sequences (which include group 1.1a sequences) had abundances of only 10.2% and 0.4%, respectively. This finding agrees broadly with previous 16S rRNA gene-based archaeal surveys (10, 11). However, one potential difference is the large relative abundance of lineage C sequences, which dominated in acidic soils. This group is rarely a significant component of 16S rRNA-defined ammonia-oxidizing archaeal communities, and the data may indicate a bias toward nonacidic soils in previous studies or some level of differential primer bias between commonly used 16S rRNA and amoA gene primer sets. N. devanaterra, an obligately acidophilic ammonia oxidizer recently cultivated from acidic soil, is also placed within lineage C (30). The discovery of this organism and demonstration of its growth and activity in acid soil provide strong evidence that lineage C ammonia oxidizers play a major role in ammonia oxidation in a large proportion of the world's acidic soils.
Analysis of Archaeal amoA Gene Sequences Using High-Throughput Sequencing.
Despite a general lack of contextual data for database sequences from soil, sufficient numbers of globally distributed sequences were available for characterization of 18 arbitrarily defined clusters. These were divided into four groups based on dominance of sequences from acid (pH <5), acido-neutral (5≤ pH <7), and alkaline (pH ≥7.0) soils and those lacking significant association with pH. At the phylogenetic resolution investigated, high-throughput sequencing identified only one additional group at the regional scale, despite the analysis of approximately one order of magnitude more sequences than at the global scale. This additional B lineage represented only a very minor component of the amoA sequences in the two soils in which it was found (<1%), possibly explaining the previous lack of discovery. Thus, high-throughput sequencing slightly increased coverage, but its major benefit was in providing greater confidence in relative abundance values.
To use amoA gene sequences of maximum possible length and facilitate the use of high-fidelity sequences, forward and reverse sequences were assembled into contiguous fragments of 629 bp, representing the assumed length of the original PCR amplicons. Stringent filtering steps were used to remove those individual sequences in which the confidence in fidelity was low as errors are observed with 454 titanium reads (37).
Ecological Coherence.
The results provide strong evidence for ecological coherence with respect to soil pH. The majority of clusters could be defined as acidophilic, acido-neutrophilic, or alkalinophilic, and these definitions applied for a large number of soils at local, regional, and global scales. Two clusters (lineage B, C11 and lineage C, C14) dominated acid soils (pH <5), with their combined relative abundance comprising 93.9% of all sequences, compared with only 47.5% in acido-neutral soils and 0.45% in alkaline soils. These two clusters are relatively divergent from each other, and their dominance strongly suggests that they are specifically adapted to growth at low pH. The adaptive mechanisms used by archaeal ammonia oxidizers at low pH, where ammonia concentration is negligible, are unknown, due in part to the absence of cultivated representatives. However, ecological coherence across local, regional, and global scales suggests that adaptive mechanisms identified locally will be broadly applicable. Similarly, organisms placed within two lineage B clusters (C1 and C12) are adapted to growth at high pH and compose 71.1% of the sequences in alkaline soils. Adaptation to high pH may require mechanisms to overcome the inhibitory effects of high ammonia concentration seen in autotrophic bacterial ammonia oxidizers. Given the relatively low frequency of alkaline soils, they have been largely ignored, and there have been few studies of ammonia oxidizer growth at pH >7.5. However, activity at such high pH, and the associated greater ammonia availability, rejects the prevailing paradigm, based on N. maritimus physiology (26), that all archaeal ammonia oxidizers prefer growth at low ammonia concentrations. Most clusters dominated in soils of pH 5.5–7, in which ammonia-oxidizing archaea have been shown to be active and to potentially contribute more to nitrification (17).
Following isolation of the first ammonia-oxidizing archaeon N. maritimus (24), several members of this functional group have been cultivated, including the thermophilic archaeon N. yellowstonii (34), group 1.1b archaea Nitrososphaera gargensis (35) and Nitrososphaera viennensis (25), group 1.1a archaeon Nitrosoarchaeum limnia (36), and group 1.1a-associated archaeon N. devanaterra (30). Although all are representatives of clusters found in soil, only the obligate acidophilic soil archaeon N. devanaterra belongs to one of the five dominant lineages, with C14 sequences also dominating in acidic soils only.
It is attractive to view the pH selection of different archaeal amoA phylotypes in terms of the influence of pH on ammonia concentration and consequent adaptations to low (growth-limiting) and high (inhibitory) concentrations of ammonia. There is strong evidence of ammonia oxidation by archaea in soils, particularly in acid soils, where adaptation to low ammonia concentrations is essential. However, it is dangerous to assume that all organisms containing amoA genes are actively oxidizing ammonia in these environments. There is evidence for mixotrophy (25) in soil archaea and even limited evidence for increased amoA abundance when nitrification is inhibited (38). In addition, other studies have shown pH to be a major driver of bacterial community structure (4, 39–43), and ecotype coherence within ammonia-oxidizing archaea may merely reflect general mechanisms for adaptation to pH. Regardless of the physiological mechanisms involved, this study provides strong evidence that individual archaeal lineages found in soil are adapted to specific pH ranges. This finding is of significance with respect to both the existence of uncharacterized mechanisms for their growth and activity at low and high ammonia concentrations and the consequences for nitrification rates in these soils and responses to management strategies, such as liming, that will influence their respective activities.
Materials and Methods
Sequence Database of Archaeal amoA Gene Sequences.
Sequences were retrieved from the NCBI GenBank database. Sequences recovered from soils of unknown pH or that did not translate perfectly, were of ambiguous phylogenetic placement, or were too short were removed. Details are provided in SI Materials and Methods.
Phylogenetic Analysis and Affiliation of Database Sequences to Clusters.
Phylogenetic clusters were defined after distance and ML analyses on aligned DNA sequences. Analyses were also performed on inferred amino acid sequences using distance, ML, parsimony, and Bayesian approaches. Details of all phylogenetic analyses are provided in SI Materials and Methods.
Soil Sampling and Analysis.
Soil cores were selected from the UK Countryside Survey (www.countrysidesurvey.org.uk/) and from across an individual pH gradient (20). Details of sampling and physicochemical analysis are described in SI Materials and Methods.
DNA Extraction, Amplification, and Analysis.
Total nucleic acids were extracted from soil and archaeal amoA gene abundance was determined by quantitative PCR (SI Materials and Methods). Pyrosequencing of amoA gene sequences in 47 soil samples was performed using multiplexed, bar-coded amplicons using fusion primers. After amplification, purification, and pooling, PCR products were sequenced on a second-generation pyrosequencer (454 GS FLX titanium; Roche). A series of strict quality filtering steps were performed before assembling forward and reverse sequences and affiliation to phylogenetic clusters (SI Materials and Methods).
Statistical Analysis.
Statistical analyses were performed using a series of R packages, as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Thomas Rattei (Department of Computation and Systems Biology, University of Vienna) for discussions on sequence analysis. C.G.-R. is funded by a research grant (NE/F021909/1) and G.W.N. by an Advanced Fellowship (NE/D010195/1) from the Natural Environment Research Council. C.Q. is supported by an Engineering and Physical Sciences Research Council Career Acceleration Fellowship (EP/H003851/1). We acknowledge assistance in molecular analysis by Mrs. Zena Smith, to whom this paper is dedicated in recognition of 25 years of nitrification research at the University of Aberdeen until her death in November, 2011.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1109000108/-/DCSupplemental.
References
- 1.Curtis TP, Sloan WT, Scannell JW. Estimating prokaryotic diversity and its limits. Proc Natl Acad Sci USA. 2002;99:10494–10499. doi: 10.1073/pnas.142680199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gans J, Wolinsky M, Dunbar J. Computational improvements reveal great bacterial diversity and high toxicity in soil. Science. 2005;309:1387–1390. doi: 10.1126/science.1112665. [DOI] [PubMed] [Google Scholar]
- 3.Philippot L, et al. The ecological coherence of high bacterial taxonomic ranks. Nat Rev Microbiol. 2010;8:523–529. doi: 10.1038/nrmicro2367. [DOI] [PubMed] [Google Scholar]
- 4.Griffiths RI, et al. The bacterial biogeography of British soils. Environ Microbiol. 2011;13:1642–1654. doi: 10.1111/j.1462-2920.2011.02480.x. [DOI] [PubMed] [Google Scholar]
- 5.Valentine DL. Adaptations to energy stress dictate the ecology and evolution of the Archaea. Nat Rev Microbiol. 2007;5:316–323. doi: 10.1038/nrmicro1619. [DOI] [PubMed] [Google Scholar]
- 6.Kelly S, Wickstead B, Gull K. Archaeal phylogenomics provides evidence in support of a methanogenic origin of the Archaea and a thaumarchaeal origin for the eukaryotes. Proc R Soc Lond B Biol Sci. 2011;278:1009–1018. doi: 10.1098/rspb.2010.1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whitaker RJ, Grogan DW, Taylor JW. Geographic barriers isolate endemic populations of hyperthermophilic archaea. Science. 2003;301:976–978. doi: 10.1126/science.1086909. [DOI] [PubMed] [Google Scholar]
- 8.Oline DK, Schmidt SK, Grant MC. Biogeography and landscape-scale diversity of the dominant Crenarchaeota of soil. Microb Ecol. 2006;52:480–490. doi: 10.1007/s00248-006-9101-5. [DOI] [PubMed] [Google Scholar]
- 9.Angel R, Soares MIM, Ungar ED, Gillor O. Biogeography of soil archaea and bacteria along a steep precipitation gradient. ISME J. 2010;4:553–563. doi: 10.1038/ismej.2009.136. [DOI] [PubMed] [Google Scholar]
- 10.Auguet J-C, Barberan A, Casamayor EO. Global ecological patterns in uncultured Archaea. ISME J. 2010;4:182–190. doi: 10.1038/ismej.2009.109. [DOI] [PubMed] [Google Scholar]
- 11.Bates ST, et al. Examining the global distribution of dominant archaeal populations in soil. ISME J. 2011;5:908–917. doi: 10.1038/ismej.2010.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brochier-Armanet C, Boussau B, Gribaldo S, Forterre P. Mesophilic Crenarchaeota: Proposal for a third archaeal phylum, the Thaumarchaeota. Nat Rev Microbiol. 2008;6:245–252. doi: 10.1038/nrmicro1852. [DOI] [PubMed] [Google Scholar]
- 13.Spang A, et al. Distinct gene set in two different lineages of ammonia-oxidizing archaea supports the phylum Thaumarchaeota. Trends Microbiol. 2010;18:331–340. doi: 10.1016/j.tim.2010.06.003. [DOI] [PubMed] [Google Scholar]
- 14.Offre P, Prosser JI, Nicol GW. Growth of ammonia-oxidizing archaea in soil microcosms is inhibited by acetylene. FEMS Microbiol Ecol. 2009;70:99–108. doi: 10.1111/j.1574-6941.2009.00725.x. [DOI] [PubMed] [Google Scholar]
- 15.Prosser JI, Nicol GW. Relative contributions of archaea and bacteria to aerobic ammonia oxidation in the environment. Environ Microbiol. 2008;10:2931–2941. doi: 10.1111/j.1462-2920.2008.01775.x. [DOI] [PubMed] [Google Scholar]
- 16.Schauss K, et al. Dynamics and functional relevance of ammonia-oxidizing archaea in two agricultural soils. Environ Microbiol. 2009;11:446–456. doi: 10.1111/j.1462-2920.2008.01783.x. [DOI] [PubMed] [Google Scholar]
- 17.Gubry-Rangin C, Nicol GW, Prosser JI. Archaea rather than bacteria control nitrification in two agricultural acidic soils. FEMS Microbiol Ecol. 2010;74:566–574. doi: 10.1111/j.1574-6941.2010.00971.x. [DOI] [PubMed] [Google Scholar]
- 18.Zhang L-M, et al. Autotrophic ammonia oxidation by soil thaumarchaea. Proc Natl Acad Sci USA. 2010;107:17240–17245. doi: 10.1073/pnas.1004947107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stephen JR, et al. Analysis of β-subgroup proteobacterial ammonia oxidizer populations in soil by denaturing gradient gel electrophoresis analysis and hierarchical phylogenetic probing. Appl Environ Microbiol. 1998;64:2958–2965. doi: 10.1128/aem.64.8.2958-2965.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nicol GW, Leininger S, Schleper C, Prosser JI. The influence of soil pH on the diversity, abundance and transcriptional activity of ammonia-oxidizing archaea and bacteria. Environ Microbiol. 2008;10:2966–2978. doi: 10.1111/j.1462-2920.2008.01701.x. [DOI] [PubMed] [Google Scholar]
- 21.Jiang QQ, Bakken LR. Comparison of Nitrosospira strains isolated from terrestrial environments. FEMS Microbiol Ecol. 1999;30:171–186. doi: 10.1111/j.1574-6941.1999.tb00646.x. [DOI] [PubMed] [Google Scholar]
- 22.De Boer W, Gunnewiek PJAK, Veenhuis M, Bock E, Laanbroek HJ. Nitrification at low pH by aggregated chemolithotrophic bacteria. Appl Environ Microbiol. 1991;57:3600–3604. doi: 10.1128/aem.57.12.3600-3604.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Allison SM, Prosser JI. Ammonia oxidation at low pH by attached populations of nitrifying bacteria. Soil Biol Biochem. 1993;25:935–941. [Google Scholar]
- 24.Könneke M, et al. Isolation of an autotrophic ammonia-oxidizing marine archaeon. Nature. 2005;437:543–546. doi: 10.1038/nature03911. [DOI] [PubMed] [Google Scholar]
- 25.Tourna M, et al. Nitrososphaera viennensis, an ammonia-oxidizing archaeon from soil. Proc Natl Acad Sci USA. 2011;108:8420–8425. doi: 10.1073/pnas.1013488108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature. 2009;461:976–979. doi: 10.1038/nature08465. [DOI] [PubMed] [Google Scholar]
- 27.Booth MS, Stark JM, Rastetter E. Controls on nitrogen cycling in terrestrial ecosystems: A synthetic analysis of literature data. Ecol Monogr. 2005;75:139–157. [Google Scholar]
- 28.Burton SAQ, Prosser JI. Autotrophic ammonia oxidation at low pH through urea hydrolysis. Appl Environ Microbiol. 2001;67:2952–2957. doi: 10.1128/AEM.67.7.2952-2957.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Boer W, Duyts H, Laanbroek HJ. Urea stimulated autotrophic nitrification in suspensions of fertilized, acid heath soil. Soil Biol Biochem. 1989;21:349–354. [Google Scholar]
- 30.Lehtovirta-Morley LE, Stoecker K, Vilcinskas A, Prosser JI, Nicol GW. Cultivation of an obligate acidophilic ammonia oxidizer from a nitrifying acid soil. Proc Natl Acad Sci USA. 2011;108:15892–15897. doi: 10.1073/pnas.1107196108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeLong EF. Everything in moderation: Archaea as “non-extremophiles.”. Curr Opin Genet Dev. 1998;8:649–654. doi: 10.1016/s0959-437x(98)80032-4. [DOI] [PubMed] [Google Scholar]
- 32.Treusch AH, et al. Novel genes for nitrite reductase and Amo-related proteins indicate a role of uncultivated mesophilic crenarchaeota in nitrogen cycling. Environ Microbiol. 2005;7:1985–1995. doi: 10.1111/j.1462-2920.2005.00906.x. [DOI] [PubMed] [Google Scholar]
- 33.Hallam SJ, et al. Pathways of carbon assimilation and ammonia oxidation suggested by environmental genomic analyses of marine Crenarchaeota. PLoS Biol. 2006;4:e95. doi: 10.1371/journal.pbio.0040095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de la Torre JR, Walker CB, Ingalls AE, Könneke M, Stahl DA. Cultivation of a thermophilic ammonia-oxidizing archaeon synthesizing crenarchaeol. Environ Microbiol. 2008;10:810–818. doi: 10.1111/j.1462-2920.2007.01506.x. [DOI] [PubMed] [Google Scholar]
- 35.Hatzenpichler R, et al. A moderately thermophilic ammonia-oxidizing crenarchaeote from a hot spring. Proc Natl Acad Sci USA. 2008;105:2134–2139. doi: 10.1073/pnas.0708857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blainey PC, Mosier AC, Potanina A, Francis CA, Quake SR. Genome of a low-salinity ammonia-oxidizing archaeon determined by single-cell and metagenomic analysis. PLoS ONE. 2011;6:e16626. doi: 10.1371/journal.pone.0016626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gilles A, et al. Accuracy and quality assessment of 454 GS-FLX Titanium pyrosequencing. BMC Genomics. 2011;12:245. doi: 10.1186/1471-2164-12-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jia Z, Conrad R. Bacteria rather than Archaea dominate microbial ammonia oxidation in an agricultural soil. Environ Microbiol. 2009;11:1658–1671. doi: 10.1111/j.1462-2920.2009.01891.x. [DOI] [PubMed] [Google Scholar]
- 39.Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. Proc Natl Acad Sci USA. 2006;103:626–631. doi: 10.1073/pnas.0507535103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hartman WH, Richardson CJ, Vilgalys R, Bruland GL. Environmental and anthropogenic controls over bacterial communities in wetland soils. Proc Natl Acad Sci USA. 2008;105:17842–17847. doi: 10.1073/pnas.0808254105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.da C Jesus E, Marsh TL, Tiedje JM, de S Moreira FM. Changes in land use alter the structure of bacterial communities in Western Amazon soils. ISME J. 2009;3:1004–1011. doi: 10.1038/ismej.2009.47. [DOI] [PubMed] [Google Scholar]
- 42.Lauber CL, Hamady M, Knight R, Fierer N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl Environ Microbiol. 2009;75:5111–5120. doi: 10.1128/AEM.00335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baker KL, et al. Environmental and spatial characterisation of bacterial community composition in soil to inform sampling strategies. Soil Biol Biochem. 2009;41:2292–2298. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.