Abstract
Essential for DNA biosynthesis and repair, ribonucleotide reductases (RNRs) convert ribonucleotides to deoxyribonucleotides via radical-based chemistry. Although long known that allosteric regulation of RNR activity is vital for cell health, the molecular basis of this regulation has been enigmatic, largely due to a lack of structural information about how the catalytic subunit (α2) and the radical-generation subunit (β2) interact. Here we present the first structure of a complex between α2 and β2 subunits for the prototypic RNR from Escherichia coli. Using four techniques (small-angle X-ray scattering, X-ray crystallography, electron microscopy, and analytical ultracentrifugation), we describe an unprecedented α4β4 ring-like structure in the presence of the negative activity effector dATP and provide structural support for an active α2β2 configuration. We demonstrate that, under physiological conditions, E. coli RNR exists as a mixture of transient α2β2 and α4β4 species whose distributions are modulated by allosteric effectors. We further show that this interconversion between α2β2 and α4β4 entails dramatic subunit rearrangements, providing a stunning molecular explanation for the allosteric regulation of RNR activity in E. coli.
Keywords: allostery, protein–protein interactions, conformational equilibria, nucleotide metabolism
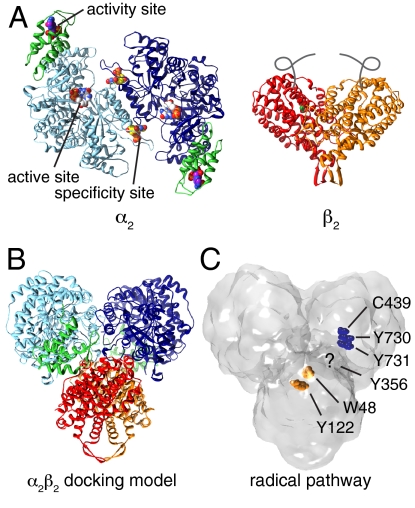
Important targets of anticancer and antiviral drugs, ribonucleotide reductases (RNRs) are classified by the metallocofactor used to generate a thiyl radical (1) that initiates reduction of ribonucleotides to deoxyribonucleotides (2, 3). Class Ia RNRs are found in all eukaryotes and many aerobic bacteria, with the Escherichia coli enzyme serving as the prototype. These RNRs reduce ribonucleoside 5′-diphosphates and are composed of two homodimeric subunits: α2 and β2 (Fig. 1A). The α2 subunit contains the active site, where ribonucleotide reduction occurs, and two types of allosteric effector binding sites (4, 5). One effector site tunes the specificity for all four ribonucleotide substrates in response to intracellular levels of deoxyribonucleoside 5′-triphosphates (dATP, dTTP, dGTP) and ATP (6, 7) such that balanced pools of deoxyribonucleotides are maintained (8). The second effector site controls the rate of reduction, binding ATP to turn the enzyme on or dATP to turn it off (4, 6). This activity site is housed in an N-terminal cone domain (5, 9) and provides a means for negative feedback regulation to safeguard against cytotoxic elevation of deoxyribonucleotide levels (2, 3, 10). The β2 subunit harbors the essential diferric-tyrosyl radical (Y122• in E. coli) cofactor (11) that initiates radical chemistry.
Fig. 1.
Previously determined structures and proposed models for E. coli class Ia RNR. (A) Homodimeric α2 with nucleotides bound (spheres) and the cone domains (residues 1–100) colored in green. Homodimeric β2 with diiron centers (green) and disordered C termini (gray lines). Protein Data Bank ID codes 4R1R, 3R1R, 1RIB (5, 15, 28). (B) Proposed α2β2 model in which the subunits are docked along their symmetry axis (15). (C) Docking model rendered as a surface. Radical pathway involving Y122 → W48 → Y356 in β2 (Y122 and W48 shown as orange spheres) and residues Y731 → Y730 → C439 in α2 (dark-blue spheres). Y356 in β2 lies in the disordered C termini (15).
Active RNR has long been proposed to be a transient α2β2 complex (Fig. 1B) with enhanced subunit affinity upon binding of substrates and effectors (12–15). For each turnover, α2, β2, substrate and effector must interact, triggering long-range proton-coupled electron transfer (PCET) reducing Y122• in β2 and oxidizing C439 to a thiyl radical in the active site of α2, over an unprecedented distance of > 35 Å (13, 15, 16) (Fig. 1C). Once the thiyl radical is generated in the active site of α2, ribonucleotide reduction proceeds through a conserved mechanism (17). Allosteric regulation of this activity is key to cell survival and involves conformational changes as well as oligomeric state changes in both prokaryotic (12, 14, 18) and eukaryotic systems (19–23). For E. coli (12, 14, 18), mouse (20–22), yeast (19), and human (19, 23), the negative effector dATP has been linked to increases in oligomeric state with a recent gas-phase electrophoretic molecular mobility analysis (GEMMA) study estimating a molecular mass of 510 kDa for the dATP-inhibited E. coli RNR (most consistent with an α4β4 state) (18), whereas for human and yeast RNR, dATP has been linked to α-hexamerization (19, 22, 23).
To understand the role of oligomeric state in the activity regulation of this prototypic class Ia RNR from E. coli, we have combined data from four complementary structural techniques. Using small-angle X-ray scattering (SAXS) and analytical ultracentrifugation (AUC), we provide evidence that supports a compact α2β2 structure for the active complex that can be reversibly converted via a dynamic conformational rearrangement to an inactive α4β4 state in the presence of elevated dATP or protein concentrations. Additionally, using SAXS, single-particle EM, and X-ray crystallography, we demonstrate that the α4β4 complex induced by the negative effector, dATP, is an unexpected ring-like structure composed of alternating α2 and β2 subunits. Together, these results explain how activity can be modulated by oligomerization, which is induced by effector binding at an allosteric site that is approximately 42 Å from the active site.
Results
dATP Shifts Oligomeric Equilibrium.
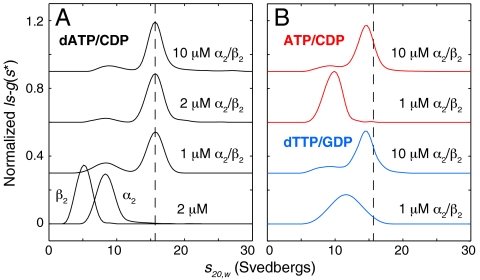
Sedimentation velocity AUC was employed to investigate the oligomeric distributions of E. coli RNR at multiple protein concentrations and in the presence of effectors at physiologically relevant concentrations (approximately 175 μM for dATP, 3 mM for ATP, and 0.1 mM for dTTP) (24–27). Using β2 that was pretreated with hydroxyurea to prevent turnover and thus oxidation of the active site during measurements, three different modes of RNR regulation were investigated: (i) under negative feedback regulation with dATP in both specificity and activity sites and CDP as the substrate; (ii) under enhanced CDP reduction with ATP in both specificity and activity sites and CDP as the substrate; and (iii) under GDP reduction with dTTP in the specificity site and no effector in the activity site.
In the presence of saturating dATP and CDP, α2 alone and β2 alone sediment at 5.2 and 8.4 S, respectively (Fig. 2A). These sedimentation coefficients are in good agreement with previously reported experimental values (12, 14) as well as theoretical values calculated from their crystal structures (5, 28) of 5.8 and 8.4 S. Similar results are obtained for the individual subunits with ATP/CDP and dTTP/GDP (SI Appendix, Fig. S1). When α2 and β2 are combined in the presence of saturating dATP/CDP, a distinct peak is observed at 15.6 S at a physiological RNR concentration of 1 μM (26, 27) (Fig. 2A). Increasing the protein concentration does not result in peak shifts, indicating that the 15.6-S peak can be attributed to a single slowly dissociating species (29). Gaussian fitting to the peak yields a molecular mass of 533 kDa by the Svedberg relation (30) (SI Appendix, Fig. S2), similar to the 510-kDa estimate for the dATP-inhibited E. coli enzyme reported by GEMMA (18), and consistent with an α4β4 complex. A typical globular protein of this molecular mass has a frictional ratio of 1.2, giving an expected sedimentation coefficient of 19 S (31). This value is much larger than the observed value for the dATP-inhibited complex, suggesting that this complex is highly nonglobular.
Fig. 2.
Sedimentation coefficient distributions of E. coli class Ia RNR: dATP and high protein concentrations shift the equilibrium toward a large complex. Physiologically relevant effector and substrate concentrations were chosen to saturate their respective sites, based on previously reported nucleotide-binding affinities (4, 24, 25, 34, 51). (A) In the presence of 175 μM dATP and 1 mM CDP, the individual subunits (each at 2 μM) sediment at 5.2 and 8.4 S. When mixed, a single slowly dissociating 15.6 S complex is observed with a molecular mass of 533 kDa. (B) In the presence of 3 mM ATP and 1 mM CDP (red curves) or 0.1 mM dTTP and 1 mM GDP (blue curves), broad, protein concentration-dependent peaks are observed, indicative of multiple species in rapid exchange. Raising the protein concentration leads to a peak shift toward 15.6 S (position indicated by dashed line). Curves are offset by a constant value for clarity.
In contrast, broad peaks with maxima near 10–11.5 S were observed in the presence of saturating dTTP/GDP or ATP/CDP at 1 μM protein concentration (Fig. 2B). Gaussian fitting of these peaks did not yield molecular mass estimates that were consistent with any one single species of RNR, including the expected α2β2 active complex. Furthermore, increasing the protein concentration by 10-fold shifted the dominant peak toward 15.6 S (dashed line in Fig. 2B), approaching the sedimentation coefficient of the dATP-inhibited complex. The observation of broad peaks that shift with increasing protein concentration is the hallmark of a mixture of rapidly exchanging species rather than a single species (29). Contrary to our previous understanding of E. coli class Ia RNR, the peak shifts observed here indicate that a large complex dominates at high protein concentration in the absence of dATP, with no effector in the activity site (Fig. 2B, dTTP/GDP) or even with a positive activity regulator occupying the activity site (Fig. 2B, ATP/CDP). Significantly, the peak shifts suggest that, under these conditions, this large complex is able to rapidly exchange with smaller species and is therefore in the equilibrium mixture even at physiological protein concentrations.
dATP-Inhibited Complex Contains Both α2 and β2.
To determine the subunit composition and stoichiometry of the dATP-inhibited complex, we used SAXS, a structural technique that provides protein size and shape information in solution. α2 was titrated into β2 under identical dATP/CDP conditions investigated by AUC, leading to a rapid and dramatic increase in radius of gyration (Rg) up to the equimolar point, consistent with the formation of a large complex with 1∶1 subunit stoichiometry (SI Appendix, Fig. S3). Given the 533-kDa molecular mass determined by AUC, a 1∶1 complex is most consistent with a 517 kDa α4β4 oligomerization state.
As a control, α2 was examined under identical conditions without β2. The Rg of α2 alone showed minimal concentration dependence with a value of 39.7 ± 0.3 Å when extrapolated to zero protein concentration to eliminate the effects of interparticle interactions (SI Appendix, Fig. S3B), in excellent agreement with the theoretical value of 39.3 Å calculated from the α2 crystal structure (5). The reverse titration and corresponding control with β2 alone yielded similar results (SI Appendix, Fig. S4), showing that both α2 and β2 must be present for the formation of higher order oligomers.
Structure of the dATP-Inhibited Complex Is an α4β4 Ring.
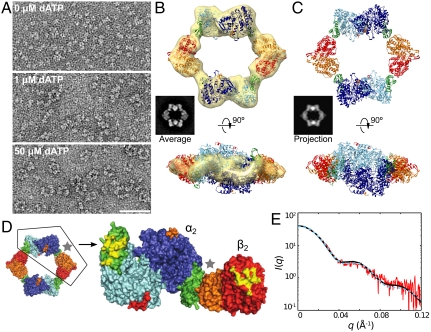
EM and X-ray crystallography were used to investigate the architecture of the dATP-induced α4β4 complex. EM images were acquired of 0.15 μM α2 and β2 in the presence of 1 mM CDP and 0, 1, or 50 μM dATP (Fig. 3A and SI Appendix, Fig. S5). In the absence of dATP, the subunits are largely dissociated, as expected from the measured micromolar subunit affinity (32). The addition of dATP drives the formation of distinct ring-like complexes approximately 200 Å in outer diameter. Images of these ring-shaped particles were aligned and classified (SI Appendix, Fig. S6) and the resulting class averages are consistent with α2 and β2 subunits arranged in an alternating pattern, forming an α4β4 complex (Fig. 3B, Average). Because the EM images were collected as tilted–untilted pairs, we were able to generate a 3D map of the α4β4 ring at approximately 23-Å resolution (Fig. 3B and SI Appendix, Fig. S7). Crystal structures of the individual dimeric subunits (5, 28) could be unambiguously fit to the 3D map of the α4β4 ring, confirming the alternating subunit arrangement suggested by the 2D class averages.
Fig. 3.
Structure of the dATP α4β4 complex by EM and X-ray crystallography. (A) EM images of RNR in the presence of 1 mM CDP and increasing dATP concentrations show the formation of α4β4 rings. (B) A class average with 1039 particles (Average) is representative of the ring structures observed at 50 μM dATP and is composed of alternating α2 and β2 subunits as indicated by its close resemblance to the 2D projection of the α4β4 crystal structure (Projection in C). These insets are 314-Å wide. Crystal structures of individual α2 and β2 subunits (5, 39), colored as in Fig. 1, fit to a 3D EM map of the α4β4 ring structure. (C) Crystal structure of dATP-bound RNR at 5.65-Å resolution with the asymmetric unit containing an α4β4 ring that agrees well with the EM model (5–9 Å Cα rmsd). (D) Surface rendering of the crystal structure with half of the ring removed, revealing the areas on α and β that are buried (yellow) upon formation of the α4β4 ring. (E) Experimental solution scattering (red) of 2 μM RNR in the presence of saturating dATP/CDP superimposed with the theoretical scattering curves calculated from the EM model (black solid) and crystal structure (cyan dashed).
To obtain a more detailed picture of the α4β4 complex, a 5.65-Å resolution crystal structure was obtained by cocrystallizing α2 and β2 with 10 mM dATP (Fig. 3C and SI Appendix, Table S1). The structure, which was solved by molecular replacement, displays electron density for dATP bound at both activity and specificity sites (SI Appendix, Fig. S8 D and E) and reveals the same α4β4 ring-shaped species in the asymmetric unit of the crystal as that observed by EM (Fig. 3 B and C). The individual α2 and β2 subunits of the α4β4 structure align well with structures of uncomplexed α2 and β2 subunits, showing only small variations in the α2 cone domain, and sharing overall average root-mean-square derivations of 1.11 and 1.01 Å for α2 and β2 subunits, respectively (SI Appendix, Fig. S8 B and C). In the α4β4 structure, α2 and β2 contact each other at two points. In addition to the previously identified (15) binding pocket on α2 for the C terminus of β2, we find a second interaction between the activity site, housed in the N-terminal cone domain of α2 (Fig. 3 C and D, green), and each lobe of β2 (Fig. 3 C and D, red/orange). This direct contact between the dATP-bound α2 activity site and the β2 subunits provides a molecular explanation for how the presence of activity effectors can be sensed, leading to the regulation of activity. From these structural data, we can also rationalize why a dATP-induced α4β4 complex would be inactive. We find that residues on the radical propagation pathway (Fig. 1C) are not aligned appropriately for electron transfer, instead facing a large central opening approximately 100 Å in diameter (Fig. 3C) such that W48 of β2 and Y731 of α2 are solvent exposed, and Y356 on the flexible C-terminal tail of β2 remains disordered. Also, the distance between W48 and Y731 is too long (ca. 55–58 Å) for PCET via only a single residue in the middle (Y356). In contrast, for active E. coli RNR, expected W48-Y731 distances are approximately 23–25 Å based on the proposed docking model (15) (Fig. 1B), which has been supported experimentally using distance restraints from pulsed electron–electron double resonance (PELDOR) spectroscopy (13).
The unanticipated α4β4 structure observed by EM and crystallography is supported by SAXS and AUC. Importantly, the use of these four methods allows for the interrogation of this RNR structure at a wide range of protein and dATP concentrations. Protein concentrations of 0.15 μM (EM) to 25 μM (X-ray), and dATP concentrations of 50 μM (EM) to 10 mM (X-ray) were examined. In addition, physiological concentrations of protein (1–2 μM) and dATP (175 μM) were investigated by AUC and SAXS in solution. Over this wide range of conditions, we find a consistent view of the α4β4 structure. The theoretical sedimentation coefficient calculated from the α4β4 structure is 15.6 S, in excellent agreement with that observed experimentally by AUC (Fig. 2A). The extrapolated Rg of 71.1 ± 1.1 Å for α4β4 determined by SAXS (SI Appendix, Fig. S9) is a larger value than expected for a globular protein of this molecular mass, but consistent with an open ring structure. Moreover, theoretical scattering curves calculated from the α4β4 X-ray and EM structures agree well with the experimental scattering obtained by SAXS (Fig. 3E), demonstrating that the α4β4 ring is not only stable in solution, but also the dominant form of the complex under negative feedback regulation by dATP near physiological protein and effector concentrations.
dATP Promotes Subunit Rearrangement.
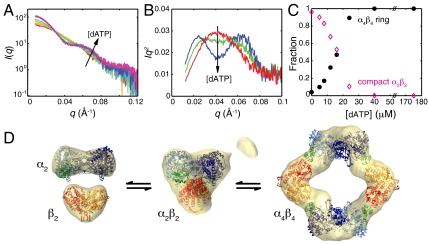
With the structures of free α2 and β2 determined previously (5, 28) and the structure of the dATP-inhibited α4β4 complex now determined by both EM and crystallography, SAXS was used to study the interconversion between different oligomeric states in solution. SAXS is ideal for probing structural interconversions as the relative fractions of individual states can be deconvoluted from mixtures (33). Titration of dATP into RNR in the presence of CDP leads to a dramatic change in the shape of the scattering curves and their corresponding Kratky curves, indicative of a transition from a predominantly compact state to a nonglobular state (Fig. 4 A and B). Above 24 μM dATP, or 4 molar equivalents of dATP per α2, the scattering curves are nearly superimposable and the Rg approaches that of the α4β4 ring (SI Appendix, Fig. S10). This result is consistent with full dATP occupancy in the effector sites of α4β4, as expected from the reported dATP binding affinities for the specificity and activity sites of 0.5 and 5 μM, respectively (4, 34).
Fig. 4.
SAXS investigation of the transition between lower and higher order structures of E. coli class Ia RNR. (A) Scattering curves measured as 0–175 μM dATP was titrated into a 6 μM solution of α2 and β2 in the presence of 1 mM CDP (red to violet) display isointensity points, suggesting a two-state transition. (B) Kratky representations of the scattering curves (Iq2 vs. q) at 0 μM (red), 12 μM (green), and 40 μM (blue) dATP show a transition from a compact globular state, as indicated by the monomodal peak, to a large nonglobular state, indicated by a bimodal curve (36). (C) Fitting linear combinations of the α2β2 docking model (15) and the α4β4 ring to the titration data provided relative fractions of the two states. (D) Ab initio SAXS reconstructions of free subunits aligned with deposited crystal structures (5, 39), a compact α2β2 state aligned with the proposed docking model (15), and the α4β4 ring aligned with the crystal structure (SI Appendix, Tables S2 and S3, and Fig. S10). The small additional density observed in the molecular envelope of α2β2 can be explained by the presence of approximately 3% α4β4 (Fig. 4C).
Isointensity points in the scattering curves are observed, indicating that this structural change can be explained by a two-state process (35) (Fig. 4A). Consistent with this interpretation, singular value decomposition shows that the scattering curves can be fit to linear combinations of two independent states with low residuals (SI Appendix, Fig. S11). The two-state transition is accompanied by an increase in the zero-angle scattering intensity I(0), which is a function of protein concentration and the electron density contrast between the hydrated protein and buffer (36). Because the protein concentration was fixed and the micromolar dATP concentration does not significantly affect the buffer density, the large increase in I(0) can only be explained by an increase in protein mass. Using experimental I(0) values from the individual subunits as calibrants, the average molecular mass in the dATP-driven transition was calculated to increase from 223 kDa in the absence of dATP to 512 kDa at saturating dATP (SI Appendix, Fig. S12). These values are in close agreement with the 259 kDa α2β2 expected in the absence of dATP and the 517 kDa α4β4 expected under saturating dATP. The excellent molecular mass estimation of α4β4 by this method suggests that the subunits have similar excluded volumes, and hence similar hydration layers, as the free subunits (37). In contrast, the underestimation in molecular mass of α2β2 can be at least in part explained by diminished hydration around the individual subunits due to the compact arrangement as determined by Kratky analysis (Fig. 4B).
An ab initio shape reconstruction was performed on the scattering curve measured in the absence of dATP, yielding a globular, three-lobed molecular envelope (Fig. 4D, Center). Consistent with the molecular mass estimation and compact shape, this molecular envelope aligns well with the proposed α2β2 “docking” model for active RNR (15). The fractions of this compact α2β2 and of α4β4 in the dATP titration series were determined (Fig. 4C) from a two-state fit to the scattering curves with low residuals (SI Appendix, Fig. S13B). By comparison, poor fits were obtained with open configurations of α2β2, including an arrangement based on the previously reported asymmetric structure of a class Ib RNR from Salmonella typhimurium (38) (SI Appendix, Fig. S13C). Together, these results indicate that the addition of dATP drives a two-state transition from a compact state, in which the subunits are closely associated, to that in which the subunits form an open ring (Fig. 4 C and D, and SI Appendix, Fig. S10).
Discussion
Allosteric regulation of activity in class Ia RNR provides a mechanism to prevent the accumulation of cytotoxic levels of deoxyribonucleotides. dATP is unique among allosteric effectors in its ability to down-regulate RNR activity (4, 6). Here, using SAXS, EM, and X-ray crystallography, we present a structural model to explain dATP inhibition of activity for the prototypic class Ia RNR. We find that the dATP-inhibited complex has an α4β4 stoichiometry as predicted by GEMMA (18), but with an unprecedented arrangement of alternating subunits in an open ring structure. Importantly, we have confirmed this surprising structure using four different techniques and have shown that this state is the predominant species stabilized by dATP under physiological solution conditions. Using the same concentration of protein in the absence of dATP, the SAXS data agree well with the predicted α2β2 docking model of the active RNR (15), providing a structural depiction of the elusive α2β2 oligomerization state.
In addition to high dATP concentrations driving the formation of the α4β4 state of E. coli RNR, AUC results show that increasing protein concentrations also shift the conformational equilibrium toward the higher molecular mass α4β4 (Fig. 2B). At lower protein concentrations and in the absence of dATP, α4β4 is still present but to a smaller degree, in rapid equilibrium with the active α2β2 state. Taken together, the SAXS and AUC results suggest that the active RNR complex is an intermediate between two inactive states, the dissociated α2 and β2 and the α4β4 ring (Fig. 4D), and that the equilibrium between these states is sensitive to both protein and effector concentrations.
Although it has long been known that the α2 cone domain binds the activity effectors ATP and dATP (5, 19), this α4β4 structure provides a molecular explanation for how the cone domain regulates the activity of E. coli RNR. We find that each α2 cone domain contacts a lobe of β2, burying a surface area (556 Å2 at each of four interfaces; Fig. 3D) that is large enough to stabilize the ring structure as the dominant species under physiologically relevant protein (1–2 μM) and dATP (175 μM) concentrations in the absence of ATP, although not so large as to inhibit interconversion back to the active α2β2 state in the presence of ATP and other effectors. In the α4β4 state, the radical transfer pathway from β2 to α2 is disrupted and solvent exposed, unable to propagate radicals. In contrast, in a compact α2β2 state, these residues are expected to be closer together, buried, and shielded from bulk water (15). Thus the cone domain of α2 “communicates” the nucleotide levels of the cell to β2, and the equilibrium of conformers can be shifted toward the α4β4 state, disrupting the radical pathway and inhibiting activity, or toward α2β2, restoring the radical pathway and enzyme activity, as nucleotide levels dictate.
Interestingly, a recently published 6.6-Å structure of yeast RNR also shows the α2 cone domain to be present at a protein–protein interface. In this case, however, the interface connects two α-subunits in a dATP-induced α6 hexamer (19). Although it is still early in terms of understanding structure/function for eukaryotic RNRs, this recent structural work along with the results presented here provide compelling support for the involvement of the cone domain in the formation of high-order RNR oligomers and pave the way for future studies.
Twenty-one years after the structure of β2 was published (39), a structure of an αβ-complex for this prototypic class Ia RNR is now available. Through the power of multiple structural techniques and combined efforts of multiple laboratories, the structures of all dominant forms of this enzyme can now be described. From crystallography and SAXS, views of the dissociated subunits are available (this work and refs. 5 and 28), from SAXS and PELDOR, low-resolution structural data support an active α2β2 model (this work and ref. 13), and from crystallography, EM, and SAXS, the structure of an allosterically inhibited α4β4 ring has been determined (this work). These structures have broad applications, providing a molecular framework for considering the relationship between in vivo E. coli nucleotide concentrations and RNR activity, as well as offering a basis for the rational design of a class of inhibitors that could act by stabilizing inactive oligomeric states of the enzyme. This open ring structure of alternating subunits is the latest surprise as this prototypic RNR enzyme, in all of its states, comes into focus.
Materials and Methods
E. coli α2 and β2 were isolated as previously described (40, 41). All of the β2 used in these structural studies was treated with hydroxyurea (HU) to reduce the essential tyrosyl radical to prevent substrate turnover. All experiments were performed in the standard assay buffer (50 mM Hepes, pH 7.6, 15 mM MgCl2 1 mM EDTA) with 5 mM DTT and the nucleotide concentrations adjusted to the indicated levels.
Crystallization of α2 and β2 with dATP was performed by hanging drop vapor diffusion at pH 7.5. Data were collected at the Advanced Light Source beamline 8.2.2, and the structure was solved by molecular replacement using the coordinates 2R1R (5) and 1MXR (42) to 5.65-Å resolution and R factors of 25.7 (work) and 30.3 (free) (SI Appendix, Table S1).
SAXS images were collected at the Cornell High Energy Synchrotron Source G1 station and processed following previously described protocols (43). Data analyses were performed using the ATSAS package (44) and MATLAB (MathWorks). The momentum transfer variable, q, is defined as q = 4π/λ sin θ, where 2θ is the scattering angle, and λ is the X-ray wavelength.
Sedimentation velocity AUC was performed using a Beckman XL-I analytical ultracentrifuge equipped with interference optics. Sedimentation coefficient distributions g(s∗) and ls-g∗(s) were generated in DCDT+ (30) and Sedfit (45), respectively. The s values were corrected to standard values (s20,w) using Sednterp (46). Theoretical s20,w values were calculated with HYDROPRO (47).
EM specimens were prepared by staining with uranyl acetate. Eighty-nine pairs of CCD images of untilted and -55° tilted specimens were collected on a Tecnai F20 electron microscope (FEI). Particles were selected in untilted images using e2boxer.py (48) and were matched with particles from tilted images using TiltPicker (49). A final dataset of 13,895 untilted particles were iteratively aligned and classified using SPIDER as described previously (50) and the corresponding set of tilted images used to generate a 3D reconstruction for each class.
Supplementary Material
Acknowledgments.
We thank Deborah Pheasant [Massachusetts Institute of Technology (MIT) Biophysical Instrumentation Facility] and Dr. Walter Stafford (Boston Biomedical Research Institute) for helpful discussions on AUC. For assistance with SAXS data collection, we thank Mackenzie Firer-Sherwood (Boston University), Yan Kung (MIT), and Cornell High Energy Synchrotron Source (CHESS) scientists, Drs. Arthur Woll and Richard Gillilan. We thank Prof. Sol Gruner (Cornell) for access to SAXS equipment and wet lab space. CHESS is supported by the National Science Foundation (NSF) and National Institutes of Health/National Institute of General Medical Sciences (NIH/NIGMS) via NSF award DMR-0936384, and the Macromolecular Diffraction Facility at CHESS resource is supported by NIH/National Center for Research Resources (NCRR) award RR-01646. Electron microscopy was performed at the National Resource for Automated Molecular Microscopy, which is supported by the NIH though the NCRR P41 program (RR017573). Crystallographic data collection was conducted at Advanced Light Source, a Department of Energy (DOE) national user facility (Contract DE-AC02-05CH11231), at beamline 8.2.2 operated by the Berkeley Center for Structural Biology, which is supported in part by the DOE and NIH/NIGMS. This work was supported by NIH grants F32GM904862 (to N.A.), F32DK080622 (to E.J.B.), T32GM08334 (to C.M.Z.), GM67167 (to F.J.A.), and GM29595 (to J.S.), and the NSF Graduate Research Fellowship under Grant 0645960 (to M.A.F.). C.L.D. is a Howard Hughes Medical Institute Investigator.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 3UUS).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1112715108/-/DCSupplemental.
References
- 1.Licht S, Gerfen GJ, Stubbe J. Thiyl radicals in ribonucleotide reductases. Science. 1996;271:477–481. doi: 10.1126/science.271.5248.477. [DOI] [PubMed] [Google Scholar]
- 2.Jordan A, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 1998;67:71–98. doi: 10.1146/annurev.biochem.67.1.71. [DOI] [PubMed] [Google Scholar]
- 3.Nordlund P, Reichard P. Ribonucleotide reductases. Annu Rev Biochem. 2006;75:681–706. doi: 10.1146/annurev.biochem.75.103004.142443. [DOI] [PubMed] [Google Scholar]
- 4.Brown NC, Reichard P. Role of effector binding in allosteric control of ribonucleoside diphosphate reductase. J Mol Biol. 1969;46:39–55. doi: 10.1016/0022-2836(69)90056-4. [DOI] [PubMed] [Google Scholar]
- 5.Eriksson M, et al. Binding of allosteric effectors to ribonucleotide reductase protein R1: Reduction of active-site cysteines promotes substrate binding. Structure. 1997;5:1077–1092. doi: 10.1016/s0969-2126(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 6.Larsson A, Reichard P. Enzymatic synthesis of deoxyribonucleotides. IX. Allosteric effects in the reduction of pyrimidine ribonucleotides by the ribonucleoside diphosphate reductase system of Escherichia coli. J Biol Chem. 1966;241:2533–2539. [PubMed] [Google Scholar]
- 7.Larsson A, Reichard P. Enzymatic synthesis of deoxyribonucleotides. X. Reduction of purine ribonucleotides; allosteric behavior and substrate specificity of the enzyme system from Escherichia coli B. J Biol Chem. 1966;241:2540–2549. [PubMed] [Google Scholar]
- 8.Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- 9.Aravind L, Wolf YI, Koonin EV. The ATP-cone: An evolutionarily mobile, ATP-binding regulatory domain. J Mol Microbiol Biotechnol. 2000;2:191–194. [PubMed] [Google Scholar]
- 10.Wheeler LJ, Rajagopal I, Mathews CK. Stimulation of mutagenesis by proportional deoxyribonucleoside triphosphate accumulation in Escherichia coli. DNA Repair (Amst) 2005;4:1450–1456. doi: 10.1016/j.dnarep.2005.09.003. [DOI] [PubMed] [Google Scholar]
- 11.Sjöberg B-M, Reichard P. Nature of the free radical in ribonucleotide reductase from Escherichia coli. J Biol Chem. 1977;252:536–541. [PubMed] [Google Scholar]
- 12.Brown NC, Reichard P. Ribonucleoside diphosphate reductase. Formation of active and inactive complexes of proteins B1 and B2. J Mol Biol. 1969;46:25–38. doi: 10.1016/0022-2836(69)90055-2. [DOI] [PubMed] [Google Scholar]
- 13.Seyedsayamdost MR, Chan CT, Mugnaini V, Stubbe J, Bennati M. PELDOR spectroscopy with DOPA-beta2 and NH2Y-alpha2s: Distance measurements between residues involved in the radical propagation pathway of E. coli ribonucleotide reductase. J Am Chem Soc. 2007;129:15748–15749. doi: 10.1021/ja076459b. [DOI] [PubMed] [Google Scholar]
- 14.Thelander L. Physicochemical characterization of ribonucleoside diphosphate reductase from Escherichia coli. J Biol Chem. 1973;248:4591–4601. [PubMed] [Google Scholar]
- 15.Uhlin U, Eklund H. Structure of ribonucleotide reductase protein R1. Nature. 1994;370:533–539. doi: 10.1038/370533a0. [DOI] [PubMed] [Google Scholar]
- 16.Reece SY, Hodgkiss JM, Stubbe J, Nocera DG. Proton-coupled electron transfer: The mechanistic underpinning for radical transport and catalysis in biology. Philos Trans R Soc Lond B Biol Sci. 2006;361:1351–1364. doi: 10.1098/rstb.2006.1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stubbe J. Ribonucleotide reductases in the twenty-first century. Proc Natl Acad Sci USA. 1998;95:2723–2724. doi: 10.1073/pnas.95.6.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rofougaran R, Crona M, Vodnala M, Sjöberg B-M, Hofer A. Oligomerization status directs overall activity regulation of the Escherichia coli class Ia ribonucleotide reductase. J Biol Chem. 2008;283:35310–35318. doi: 10.1074/jbc.M806738200. [DOI] [PubMed] [Google Scholar]
- 19.Fairman JW, et al. Structural basis for allosteric regulation of human ribonucleotide reductase by nucleotide-induced oligomerization. Nat Struct Mol Biol. 2011;18:316–322. doi: 10.1038/nsmb.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashlan OB, Cooperman BS. Comprehensive model for allosteric regulation of mammalian ribonucleotide reductase: Refinements and consequences. Biochemistry. 2003;42:1696–1706. doi: 10.1021/bi020634d. [DOI] [PubMed] [Google Scholar]
- 21.Kashlan OB, Scott CP, Lear JD, Cooperman BS. A comprehensive model for the allosteric regulation of mammalian ribonucleotide reductase. Functional consequences of ATP- and dATP-induced oligomerization of the large subunit. Biochemistry. 2002;41:462–474. doi: 10.1021/bi011653a. [DOI] [PubMed] [Google Scholar]
- 22.Rofougaran R, Vodnala M, Hofer A. Enzymatically active mammalian ribonucleotide reductase exists primarily as an alpha6beta2 octamer. J Biol Chem. 2006;281:27705–27711. doi: 10.1074/jbc.M605573200. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Lohman GJ, Stubbe J. Enhanced subunit interactions with gemcitabine-5′-diphosphate inhibit ribonucleotide reductases. Proc Natl Acad Sci USA. 2007;104:14324–14329. doi: 10.1073/pnas.0706803104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bochner BR, Ames BN. Complete analysis of cellular nucleotides by two-dimensional thin layer chromatography. J Biol Chem. 1982;257:9759–9769. [PubMed] [Google Scholar]
- 25.Buckstein MH, He J, Rubin H. Characterization of nucleotide pools as a function of physiological state in Escherichia coli. J Bacteriol. 2008;190:718–726. doi: 10.1128/JB.01020-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eriksson S, Sjöberg B-M, Hahne S. Ribonucleoside diphosphate reductase from Escherichia coli. An immunological assay and a novel purification from an overproducing strain lysogenic for phage lambdadnrd. J Biol Chem. 1977;252:6132–6138. [PubMed] [Google Scholar]
- 27.Hristova D, Wu CH, Jiang W, Krebs C, Stubbe J. Importance of the maintenance pathway in the regulation of the activity of Escherichia coli ribonucleotide reductase. Biochemistry. 2008;47:3989–3999. doi: 10.1021/bi702408k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nordlund P, Eklund H. Structure and function of the Escherichia coli ribonucleotide reductase protein R2. J Mol Biol. 1993;232:123–164. doi: 10.1006/jmbi.1993.1374. [DOI] [PubMed] [Google Scholar]
- 29.Stafford WF. Protein-protein and ligand-protein interations studied by analytical ultracentrifugation. Methods Mol Biol. 2009;490:83–113. doi: 10.1007/978-1-59745-367-7_4. [DOI] [PubMed] [Google Scholar]
- 30.Philo JS. Improved methods for fitting sedimentation coefficient distributions derived by time-derivative techniques. Anal Biochem. 2006;354:238–246. doi: 10.1016/j.ab.2006.04.053. [DOI] [PubMed] [Google Scholar]
- 31.Erickson HP. Size and shape of protein molecules at the nanometer level determined by sedimentation, gel filtration, and electron microscopy. Biol Proced Online. 2009;11:32–51. doi: 10.1007/s12575-009-9008-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Climent I, Sjöberg B-M, Huang CY. Site-directed mutagenesis and deletion of the carboxyl terminus of Escherichia coli ribonucleotide reductase protein R2. Effects on catalytic activity and subunit interaction. Biochemistry. 1992;31:4801–4807. doi: 10.1021/bi00135a009. [DOI] [PubMed] [Google Scholar]
- 33.Svergun DaK M. Small-angle scattering studies of biological macromolecules in solution. Rep Prog Phys. 2003;66:1735–1782. [Google Scholar]
- 34.Ormo M, Sjöberg B-M. An ultrafiltration assay for nucleotide binding to ribonucleotide reductase. Anal Biochem. 1990;189:138–141. doi: 10.1016/0003-2697(90)90059-i. [DOI] [PubMed] [Google Scholar]
- 35.Segel DJ, Fink AL, Hodgson KO, Doniach S. Protein denaturation: A small-angle X-ray scattering study of the ensemble of unfolded states of cytochrome c. Biochemistry. 1998;37:12443–12451. doi: 10.1021/bi980535t. [DOI] [PubMed] [Google Scholar]
- 36.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40:191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 37.Mylonas E, Svergun DI. Accuracy of molecular mass determination of proteins in solution by small-angle X-ray scattering. J Appl Crystallogr. 2007;40:s245–s249. [Google Scholar]
- 38.Uppsten M, Farnegardh M, Domkin V, Uhlin U. The first holocomplex structure of ribonucleotide reductase gives new insight into its mechanism of action. J Mol Biol. 2006;359:365–377. doi: 10.1016/j.jmb.2006.03.035. [DOI] [PubMed] [Google Scholar]
- 39.Nordlund P, Sjöberg B-M, Eklund H. Three-dimensional structure of the free radical protein of ribonucleotide reductase. Nature. 1990;345:593–598. doi: 10.1038/345593a0. [DOI] [PubMed] [Google Scholar]
- 40.Salowe SP, Stubbe J. Cloning, overproduction, and purification of the B2 subunit of ribonucleoside-diphosphate reductase. J Bacteriol. 1986;165:363–366. doi: 10.1128/jb.165.2.363-366.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salowe SP, Ator MA, Stubbe J. Products of the inactivation of ribonucleoside diphosphate reductase from Escherichia coli with 2′-azido-2′-deoxyuridine 5′-diphosphate. Biochemistry. 1987;26:3408–3416. doi: 10.1021/bi00386a024. [DOI] [PubMed] [Google Scholar]
- 42.Hogbom M, et al. Displacement of the tyrosyl radical cofactor in ribonucleotide reductase obtained by single-crystal high-field EPR and 1.4-angstrom X-ray data. Proc Natl Acad Sci USA. 2003;100:3209–3214. doi: 10.1073/pnas.0536684100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ando N, Chenevier P, Novak M, Tate MW, Gruner SM. High hydrostatic pressure small-angle X-ray scattering cell for protein solution studies featuring diamond windows and disposable sample cells. J Appl Crystallogr. 2008;41:167–175. [Google Scholar]
- 44.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Crystallogr. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78:1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Laue TM, Shah BD, Ridgeway TM, Pelletier SL. Computer-aided interpretation of analytical sedimentation data for proteins. In: Harding SE, et al., editors. Analytical Ultracentrifugation in Biochemistry and Polymer Science. Cambridge, UK: Royal Soc Chemistry; 1992. pp. 90–125. [Google Scholar]
- 47.Garcia de la Torre J. Building hydrodynamic bead-shell models for rigid bioparticles of arbitrary shape. Biophys Chem. 2001;94:265–274. doi: 10.1016/s0301-4622(01)00244-7. [DOI] [PubMed] [Google Scholar]
- 48.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 49.Voss NR, Yoshioka CK, Radermacher M, Potter CS, Carragher B. DoG Picker and TiltPicker: software tools to facilitate particle selection in single particle electron microscopy. J Struct Biol. 2009;166:205–213. doi: 10.1016/j.jsb.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brignole EJ, Smith S, Asturias FJ. Conformational flexibility of metazoan fatty acid synthase enables catalysis. Nat Struct Mol Biol. 2009;16:190–197. doi: 10.1038/nsmb.1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Dobeln U, Reichard P. Binding of substrates to Escherichia coli ribonucleotide reductase. J Biol Chem. 1976;251:3616–3622. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.