Abstract
During cell entry, enveloped viruses fuse their viral membrane with a cellular membrane in a process driven by energetically favorable, large-scale conformational rearrangements of their fusion proteins. Structures of the pre- and postfusion states of the fusion proteins including paramyxovirus PIV5 F and influenza virus hemagglutinin suggest that this occurs via two intermediates. Following formation of an initial complex, the proteins structurally elongate, driving a hydrophobic N-terminal “fusion peptide” away from the protein surface into the target membrane. Paradoxically, this first conformation change moves the viral and cellular bilayers further apart. Next, the fusion proteins form a hairpin that drives the two membranes into close opposition. While the pre- and postfusion hairpin forms have been characterized crystallographically, the transiently extended prehairpin intermediate has not been visualized. To provide evidence for this extended intermediate we measured the interbilayer spacing of a paramyxovirus trapped in the process of fusing with solid-supported bilayers. A gold-labeled peptide that binds the prehairpin intermediate was used to stabilize and specifically image F-proteins in the prehairpin intermediate. The interbilayer spacing is precisely that predicted from a computational model of the prehairpin, providing strong evidence for its structure and functional role. Moreover, the F-proteins in the prehairpin conformation preferentially localize to a patch between the target and viral membranes, consistent with the fact that the formation of the prehairpin is triggered by local contacts between F- and neighboring viral receptor-binding proteins (HN) only when HN binds lipids in its target membrane.
Keywords: fusion protein refolding, membrane fusion, electron microscopy
Enveloped viruses such as influenza virus, human immunodeficiency virus (HIV), and parainfluenza virus 5 (PIV5) encode class I fusion proteins that mediate coalescence of the viral and target membranes (1–6). Homotrimeric class I fusion proteins are synthesized as biologically inactive precursors that are activated by proteolytic cleavage, which generates a new N-terminal hydrophobic sequence known as the fusion peptide. The crystal structures of the paramyxovirus F-protein in its metastable prefusion form and the human parainfluenza virus 3 (hPIV3) F-protein in its very stable postfusion form, and also the influenza virus HA in pre- and postfusion conformations, reveal a unique protein architecture that undergoes large-scale, irreversible refolding during membrane fusion (7–10).
These structures provided snapshots of the locations of the critical fusion peptide in the initial and final states: In the prefusion form, the fusion peptide is located along the protein surface, near the virus membrane; while in the postfusion state it is positioned to penetrate into the viral membrane through the formation of a hairpin conformation. Nevertheless, photochemical labeling (11) indicates that the fusion peptide engages with the target membrane in an intermediate state known as the “prehairpin” state. This prehairpin state has been hypothesized to be a highly extended parallel coiled coil, which drives the fusion peptide beyond the surface of the virus to allow it to insert deeply into the target membrane. Interestingly, this conformational change would be expected to drive the membranes further apart than in the initial complex, due to the proposed elongated nature of the prehairpin. Next, the formation of the final hairpin form is hypothesized to drive the close apposition of the bilayers required for fusion to proceed. The hypothetical extended prehairpin form has served as a remarkably robust model for the design of peptide drugs that inhibit HIV and PIV5 fusion such as T-20 and C1 (12–14). However, to date, the existence of this prehairpin intermediate has been shown only indirectly (15–17). Direct observation of this state would be a valuable test of current models of viral fusion and the mode of action of entry inhibitors.
Here we characterize the prehairpin intermediate of a paramyxovirus, which fuses with the cytoplasmic membrane and does not require endocytosis or low pH-triggering (2). The initial binding occurs via transient interactions between its receptor-binding protein known as hemagglutinin-neuraminidase (HN) and sialic acid moieties on the cell surface. The engagement of HN with its substrate leads to conformational changes in neighboring F-proteins on the viral envelope, which then convert to the prehairpin form. We built a computational model for F in this metastable intermediate, which predicted a relatively long bilayer-to-bilayer spacing of 21 nm. We used electron microscopy (EM) to characterize the fusion of virions with nano-bead supported bilayers (18), which were expected to be less deformable than liposomes or cells and hence slower to fuse with virus. A previously characterized prehairpin-binding peptide was used to further trap this intermediate and enable immuno-gold labeling. Direct measurement of the interbilayer distance shows that the separation closely matches that predicted from the computational model for the prehairpin intermediate. Additionally, the F-proteins in the prehairpin state on the surface of the virus specifically localize to the contact area between the virus and the target membrane. These experiments provide direct evidence for this prehairpin intermediate in an intact cell, and suggest that the fusion protein does travel through this large, extended intermediate state on the path to fusion.
Results and Discussion
A Computational Modeling of the Prehairpin Intermediate State.
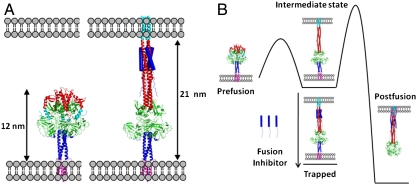
An atomic-scale model was constructed to determine the overall length of the aqueous domain, and hence the expected interbilayer distance of the intermediate. Type I fusion proteins, including PIV5 F, influenza virus HA, and HIV Env, have two helical repeat (HRA and HRB) regions that form coiled coils during different stages of fusion. In the prefusion state of PIV5, HRA lies along the protein surface, while HRB forms a trimeric coiled coil stalk adjacent to the viral membrane (7). In the prefusion state, HRA is believed to detach from the protein surface and refold into a long, extended three-stranded coiled coil projecting towards the target bilayer, while HRB remains essentially intact and anchored near the virus surface. During formation of the hairpin intermediate, the HRA trimer remains constant, while the original HRB bundle dissociates and its individual chains bind along the exterior of the HRA trimer, pulling the viral membrane toward the target bilayer until the postfusion trimer of helical hairpins is fully formed (8, 19). Thus, the prehairpin intermediate shows hybrid character, having HRA and HRB in the starting and postfusion conformations, respectively. One domain of F (residues 290–357) is largely invariant in the pre- and postfusion structures; it appears to form a hinged hub about which HRA and HRB pivot during these conformational rearrangements. Thus, by superposing this domain in the pre- and postfusion states, it was possible to define the positions of HRA and HRB in the prehairpin intermediate (see Methods and Dataset S1). The end-to-end distance of the resulting model (Fig. 1 A and B) is 21 nm with an associated error of approximately 1 nm, primarily reflecting the error in predicting where the transmembrane (TM) and fusion peptides enter the membranes.
Fig. 1.
Structures of the fusion protein in fusion states and their thermodynamics. (A) Models of the prefusion (7, 27) (left) and the prehairpin intermediate bound by the fusion inhibitor (right) show the distance expected to be observed in EM. The fusion peptide is shown in cyan, heptad repeat A in red, the globular head domain in green, heptad repeat B in dark blue (cartoon), and the TM domain in magenta. The fusion-inhibitory peptide, C1 (12), is shown in dark blue (filled cylinders). (B) Schematic diagram illustrating the free energy changes in the system. The canonical pathway moves from prefusion, to the intermediate state, through a high barrier to the postfusion state (8). Introduction of the fusion-inhibitory peptide, C1, traps the protein in the intermediate state conformation (12).
Bilayer-Supported Silica and Polystyrene Nanobeads.
For microscopic studies, lipid bilayers were supported on nanosilica or polystyrene (PS) nanobeads. The silica-supported bilayers maintain, by tethering the bilayer to the nanobead with poly(ethylene glycol) (PEG), an internal space that allows for incorporation of membrane proteins containing cytoplasmic domains. For experiments requiring thin-sectioning we used the softer polystyrene (PS) as the support. An electron micrograph (Fig. S1) shows the structure of silica and PS nanobeads stained using a mixture of 2% uranyl acetate and 2% osmium tetroxide. The membrane surface of the silica-supported nanobeads is approximately 50 Å from the nanobead core because of the flexible PEG linkages, as compared to the 40 Å that would be observed if the membrane was directly on the surface of the solid support. To control the size homogeneity, curvature of membranes, and structural properties of the nanobeads, 100–150 nm silica and ≈120 nm polystyrene beads (average diameter) were selected.
To induce fusion of PIV5 particles (virons) with nanobead-supported bilayers, the bilayer was modified with sialogangliosides to serve as a receptor for binding by the PIV5 attachment protein, HN. PIV5 HN binding to sialic acid is believed to lead to activation of F-protein, priming the conformational transition from the prefusion state to intermediates along the fusion reaction pathway (20, 21).
Fusion Between Viruses and Nanobead-Supported Bilayers.
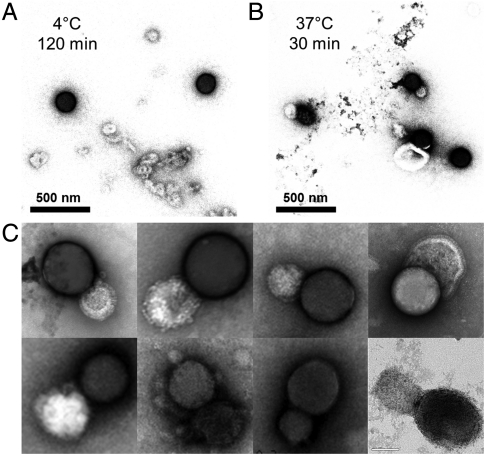
For paramyxoviruses, binding of virus to surface sialic acids at 37 °C to 42 °C triggers the conformational changes leading to fusion (22, 23). As expected, fusion was observed at 37° and 42 °C by EM after 30 min incubation (Fig. 2B), but not at 4 °C and 25 °C (Fig. 2A). Darker, nanobead-supported bilayers are readily distinguished from the virus particles.
Fig. 2.
Temperature triggers viral fusion with nanobead-supported bilayers. EM images of the samples. (A) At 4 °C, nanobead-supported bilayers made up of silica nanobeads were combined with purified PIV5 particles that express the fusion (F) protein spike on their surface. At this temperature there is little association between the virus and the nanobead-supported bilayers. Dark spheres represent silica nanobead and light vesicles are viruses. Silica-supported bilayers appear darker under these conditions. (B) After warming to 37 °C for 30 min viral particles fuse to nanobead-supported bilayers. Samples in (A and B) were stained with 4% uranyl acetate. (C) EM gallery of fused virus/nanobead-supported bilayers pairs in higher magnification. PIV5 virions with a concentration of 1.0 × 1010 plaque forming units (PFU)/mL were mixed with silica nanobeads with an approximate concentration of 5 × 109 particles/mL into 100 μL total volumes and incubated at the desired temperature. The concentration of silica nanobeads was calculated from weight, assuming a 120 nm mean diameter of silica and its density of 2.2 g/cm3.
Measurement of the Spacing Between Viral Membranes and Nanobead-Supported Bilayers.
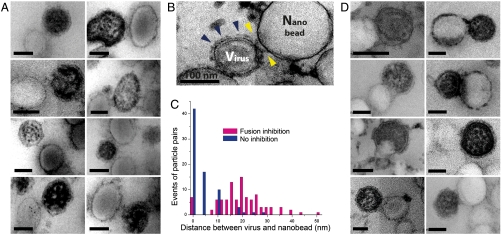
The computational model predicts a bilayer separation of 21 nm in the prehairpin intermediate (Fig. 1A). Using thin-sectioning EM with staining allowed the surface spike glycoproteins of PIV5 to be observed. To trap the F-protein in the prehairpin conformation, we included an inhibitory peptide, C-1, derived from the HRB region of F (12). The C-1 peptide binds specifically to the prehairpin form of F, and is believed to inhibit PIV5 fusion by trapping the protein in this state and preventing it from proceeding to the final hairpin form (Fig. 3A). More than 150 images of virus/nano-particle mimic pairs were analyzed. In both the presence and absence of the fusion inhibitor, the distance between the viral and nanobead-supported bilayer was measured (Fig. 3 A and D). Unlike unsectioned silica nanobead-supported samples (Fig. 2), the contrast of sectioned samples (Fig. 3) resulted in the viruses being more darkly stained due to the internal ribonucleoprotein core. In the absence of the fusion-inhibitory peptide, fusion was observed (Fig. 3D) and most virus/nanobead-supported bilayer pairs are separated by a short distance (Fig. 3C). In the presence of the fusion-inhibitory peptide, the distribution of interparticle distances peaked at approximately 20 nm (Fig. 3C), very closely matching the value predicted by the computational model (Fig. 1A).
Fig. 3.
Observation of the prehairpin intermediate by thin-sectioning and EM. EM gallery of viral particles and PS nanobead-supported bilayers made up of polystyrene beads from thin sectioned samples (A) at 42 °C in the presence of the fusion-inhibitory peptide C-1 and (B) in the absence of the fusion-inhibitory peptide, C-1, under the same condition as in (A). Under these staining conditions viral particles are darker than the PS nanobeads. (B) Blue arrows show the putative prefusion state in the absence of the fusion-inhibitory peptide C-1, while yellow arrows show putative prehairpin intermediates. (C) Distance distribution between the edge of the viral particle and nanobead with and without fusion inhibitor at 42 °C. Note the peak near 20 nm for the inhibited state. (D) EM gallery of viral particles fusing with nanobead-supported bilayers at 42 °C in the absence of fusion-inhibitory peptides C-1. PIV5 virions with a concentration of 1.0 × 1010 plaque forming units/mL were mixed with amino polystyrene nanobeads with a concentration of 5 × 1010 particles/mL. The faint appearance of sections in EM images is an artifact arising from four independent quadrants in the camera used for image capture.
The conformation of the trapped prehairpin intermediate is suggested by significant changes of proteins adjacent to the nanobead-supported bilayer surface (Fig. 3 A and B). While regions of the virus far from the interface show densely packed protein (presumably a mixture of HN and F in its prefusion conformation, blue arrows, Fig. 3B), regions close to the interface appear to be less densely packed, (yellow arrows, Fig. 3B) consistent with a transition between the prefusion conformation and the prehairpin intermediate (Fig. 3B).
Immuno-Gold Labeling Shows Localization of Inhibited PIV5 F-Protein to the Interfacial Region.
We next localized the proteins that had reached the prehairpin intermediate by using a fusion-inhibitory peptide coupled to colloidal gold and visualized by EM. The nanobead-supported bilayers and PIV5 virions were first incubated in the presence of the C-1 peptide that binds to the prehairpin intermediate state (12). To allow gold-labeling a biotin was incorporated attached to the N terminus of the peptide via a short PEG linker. Next, streptavidin-labeled gold particles were added to label the location of the C-1 peptides and the F-protein prehairpin intermediate. The nanobead-supported bilayers, virus particles, and gold particles were then displayed by thin-sectioning and EM (Fig. 4A).
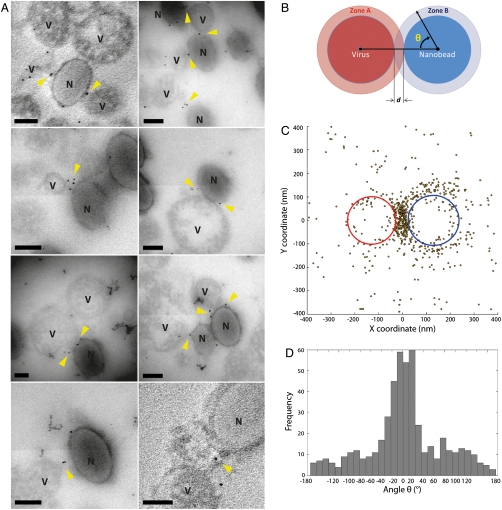
Fig. 4.
Immuno-gold labeling of the fusion inhibitor shows it is concentrated between the viral particles and nanobead-supported bilayers in the trapped state. (A) Gallery of thin sectioned EM images of viral particles and nanobead-supported bilayers labeled using immuno-gold. The small black spheres are 5 nm gold particles. “V” labels viral particles, and “N” labels nanobeads. The thin sections containing immune-gold are approximately 70 nm thick, and produced a silver interference color. (B) Illustration of how the angle θ is calculated for a virus and nanobead pair. (C) Superimposition of the positions of all gold particles around interacting viral particles and nanobead-supported bilayers. The average virus ellipse is in red and the average nanobead-supported bilayer ellipse is in blue. Gold particles are shown as yellow dark circles. (D) The distribution of the angle θ. Note the large peak at 0° the angle at which a gold particle is in the space between the virus and the nanobead-supported bilayer. All scale bars in (A) are 100 nm. The faint appearance of sections in EM images is an artifact arising from four independent quadrants in the camera used for image capture.
Approximately 300 virus/nanobead-supported bilayer images were then overlaid to show the position of the gold particles relative to the virions and nanobead-supported bilayers (Fig. 4C). To allow quantitative comparison of the distributions, all EM images were superimposed along a line between the center of the viral particle ellipse and the center of the PS nanobead-supported bilayers; the midpoint of the virus-nanobead vector defines the origin of the superimposed structures. The location of the gold particles relative to the virus particles and nanobead-supported bilayers are shown in red and blue circles, respectively (Fig. 4C). The position of the gold particles clearly localize to the contact area between the virus and the nanobead-supported bilayer (Fig. 4A), indicating the accumulation of F-proteins in the prehairpin intermediate state at the interface. The angle distribution of gold particles around the particles (Fig. 4D) supports this conclusion, and shows the particles are localized in a narrow range within 35° of the line between the centers of the two particles. The counts within this angular peak are 5 to 20-fold greater than at other angles, which represent locations outside of the contact zone. Thus, the prehairpin conformation segregates within the contact zone, consistent with previous findings that formation of this intermediate is triggered by contact between the virus and the target at permissive temperatures.
Conclusions
Here we confirm the existence of the viral fusion prehairpin intermediate and provide direct images of the bilayer spacing and distribution of the population of F-protein in this intermediate state. Previous evidence for the existence of the prehairpin intermediate in this and other viruses has been indirect. For example, peptides with the sequence of HRB strongly inhibit fusion despite the occlusion of its postfusion binding partner, HRA, in the prefusion state (12–14). While this result is consistent with an extended prehairpin intermediate that exposes HRA during the fusion pathway, the direct measurements made here provide strong new support for this hypothesis. Combined with computational modeling, these images also bring us much closer to an atomic level structure that could be used in the design of future entry inhibitors.
Materials and Methods
Computational Modeling of the Prehairpin Intermediate.
Computational modeling of the prehairpin intermediate involved primarily four steps. First, the crystal structures of the F prefusion state of PIV5 and F postfusion states of hPIV3 were structurally aligned to create an extended model that contains both HRA and HRB as trimeric coiled coils. Second, the hPIV3 sequence was replaced by that of PIV5 and repacked. Third, the TM and fusion peptide domains were added to the model. Finally, the combined model was minimized to remove clashes.
To estimate the interbilayer spacing of the prehairpin intermediate we built a detailed atomic model of this structure (24, 25), in which the N-terminal region (residues 103–289) has extended into a trimeric coiled coil projecting towards the target bilayer, and the C-terminal residue (residues 358–511) undergoes its conformational change primarily at the late stages of fusion (going from the prehairpin intermediate to the postfusion states). This hypothesis is consistent with the fusion peptide from the N-terminal region inserting during the early stages of fusion, prior to the zippering of HRB that helps bring the bilayers together. The crystal structures of the prefusion (7) and postfusion (8) states were aligned using a region of the globular head domain, residues 290–357 (PIV5 numbering). This region is largely constant within each chain between the two structures (Cα rmsd = 1.7 Å when comparing individual chains) and presumably is also constrained in the prehairpin intermediate. While the rmsd is larger when all chains are considered because of relative rotation of the chains (Cα rmsd = 4.4 Å), a structural alignment of Cα positions places residues 357 of the postfusion structure and 358 of the prefusion structure within close proximity, allowing connection of the two regions. Therefore, the N-terminal region was taken from the postfusion structure, and the C-terminal region taken from the prefusion structure.
Second, the sequence of PIV5 was threaded onto the postfusion structure using the Rosetta Design software package to repack the new side chains (26). Repacking occurred using the default parameters. Third, for the TM domain adjacent to HRB, the model was taken from that obtained from disulfide cross linking (27). For the fusion peptide adjacent to HRA, sequence conservation data (28) supports a coiled coil structure. The coiled coil from HRA was extended into the fusion peptide to model this region, placing the most conserved residues at the core of the coiled coil. The structure was then briefly minimized (200 steps of steepest decent) in Gromacs (29) using the all-atom OPLS force field (30, 31) to relax any steric clashes in the new structure. Minimization using other force fields, such as CHARMM (32), gave very similar results.
Preparation of Nanobead-Supported Bilayers.
Small unilamellar vesicles (SUVs) containing a 4∶4∶2 molar ratio of 1-palmitoyl-2-oleyl-sn-glycero-3-phosphocholine (POPC), 1,2-dioleoyl-sn-glycero-3-[phosphor-L-serine] (DOPC) and cholesterol (Avanti Polar Lipids) doped with 1 M% bovine brain disialoganglioside GD1a (Sigma Aldrich) at a lipid concentration of 5 mM were prepared by tip sonication of vacuum dried lipid films in PBS buffer (pH 7.5, 100 mM NaCl) for 30 min. PEG-tethered silica nanobeads were prepared by the reaction of spherical bare silicas with (C2H5O)3Si(CH2)3O[OCH2CH2]8–12-OH (Gelest) in water (33). The silica particles were synthesized by Stöber’s method (34). The mean particle diameter ∼120 nm was assessed by EM and its surface area was determined by BET method (35). Overnight incubation of SUVs with silica and polystyrene nanobeads formed lipid-bilayers on the hydrophilic surface of silica and polystyrene nanobeads. Silica nanobeads were treated with lipid concentrations sufficient to provide coverage of ∼2.5 lipids/nm2 of the silica surfaces, which corresponds to a bilayer. Coverage was confirmed by adsorption isotherms (33), and verified by phosphorus analysis and fluorescence of NBD-doped lipids. Polystyrene nanobeads (Polyscience, amino PSs used with 5.68 × 1012 particles/mL in concentration) combined with 1 mL of 5 mM SUV stock solution formed lipid-bilayers on the hydrophilic surface of nanobeads. After encapsulation by bilayers, the silica nanobeads were rinsed at least three times with PBS buffer followed by vortexing and centrifugation at 10,000 rpm for 2 min and the supernatant was discarded. The resulting lipid-coated nanobeads were reconstituted in PBS buffer (pH 7.5). Additional sonication of the lipid-coated nanobeads was avoided so as to not disrupt the nanobead lipid bilayer. Silica nanobeads with a mean diameter of 120 nm were used. Each tube of lipid-coated silica nanobeads contains 2.8 mg of 120 nm silica nanobeads in PBS buffer. Nanobeads were vortexed or sonicated until a homogeneous suspension formed. PS nanobeads were used for the advanced EM measurements of the interparticle distance. Organic beads such as PS provide excellent contrast levels and better brightness to highlight proteins in EM. In addition, the softness of PS provides allows sectioning. For the EM measurements, aqueous suspensions were stained using 2% uranyl acetate (20) and observed using a 80 kV FEI-Tecnai T12 to display the lipid membrane on the nanobeads.
Production and Purification of PIV5 Virions.
PIV5 was grown in Madin-Darby bovine kidney cells as described previously (36, 37). Virus was purified essentially as described (38) on 15–60% sucrose/NTE (100 mM NaCl, 10 mM Tris pH 7.4 and 1 mM EDTA) gradients by ultracentrifugation (24,000 rpm for 2 h at 4 °C) in a Beckman SW 32 rotor. The virus band was collected, diluted in NTE buffer, and virus pelleted at 100,000 × g for 1 h in a Beckman Ti60 rotor. The viral pellet was resuspended in NTE buffer and Dounce homogenized. Purified PIV5 virions were aliquoted in with a concentration of 1.0 × 1010 plaque forming units (PFU) per mL and stored at -80 °C.
Electron Microscopy in Combination with Sectioning and Staining Techniques.
Electron microscopy.
EM images were taken on a JEOL 2010 microscope operating at 120 kV or 200 kV for sectioned samples and a FEI-Tecnai G12 operating at 80 kV for silica nanobead-coated bilayers. The imaging device was a Gatan image filter, and the Gatan Digital Micrograph software was used to record the images. In the case of lipid-coated silica nanobeads (Fig. 2), droplets of an aqueous suspension of a mixture of silica nanobead-supported bilayers and PIV5 viruses were dropcast onto a carbon grid (Electron Microscopy Sciences, 400-mesh) and visualized on an FEI-Tecnai G12 Transmission electron microscope at 80 kV.
Negative/positive staining and sectioning protocols.
The standard process of sample fixation with paraformaldehyde or glutaraldehyde caused artifacts in the EM examination after sectioning and was not used. Samples were fixed in 2% osmium tetroxide (OsO4) with 1.5% potassium ferricyanide in 0.1 M sodium cacodylate buffer for 1 h at room temperature. OsO4 is known to stabilize many proteins by transforming them into gels without destroying structural features (39). Proteins that are stabilized by OsO4 are not aggregated by alcohols during dehydration. After washes in dH2O, samples were stained en bloc with 2% aqueous uranyl acetate (20) prior to dehydration in a graded ethanol series. Samples were embedded in a PolyBed 812 bed (Polysciences Inc.). The embedded samples were sectioned on an ultramicrotome. Thin sections were transferred to coated 300 mesh grids and were additionally stained with uranyl acetate, OsO4, tannic acid, phosphotungstic acid (PTA), and bismuth subnitrite (40). Staining times varied from 5 min to 30 min.
Optimizing contrast of proteins and membranes against other compartments in PIV5 virus.
The sectioning and staining protocols were optimized to increase the visibility of proteins on the viral surface (Fig. S2). OsO4 embeds into membranes, creating a high secondary electron emission without the need for coating the membrane with a layer of metal which could obscure details of the cell membrane (41). In staining the viral surface spike proteins (F and HN), tannic acid-UA is commonly used as both a negative and positive stain to improve resolution (42). The positively stained EM presents protein compartments and nucleocapsids when combined with tannic acid and UA, thus creating contrast. Depending on the presence of immuno-gold labels, the staining conditions were found to be optimal at condition A (no immuno-gold): 1% tannic acid (30 min )/2% UA(30 min) (Fig. 3 A and D) or condition B (with immuno-gold): 2% OsO4 (30 min )/1% tannic acid(30 min )/2% UA(10 min) (Fig. 3B).
Synthesis and Purification of Inhibitory Fusion Peptide.
Previous studies have demonstrated that the inhibitory peptide C-1, derived from the heptad repeat region B (HRB) of PIV5 F-protein, displays antiviral activity (43). The sequence is: KLESSQILSIDPLDISQNLAAVNKSLSDALQHLAQSDTYLSAI. The C-1 was synthesized by solid-phase synthesis using Fmoc chemistry with HBTU (O-benzotriazole-N,N,N′,N′-tetramethyl-uronium-hexafluoro-phosphate) as a coupling agent; biotin was attached via a flexible linker by coupling first Fmoc-8-amino-3,6-dioxaoctanoic acid (PEPTIDES Internationals) then a single cysteine residue to biotin (5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno [3,4-d] imidazol-4-yl] pentanoic acid, Sigma-Aldrich). The products were cleaved from the rink amide MBHA resin (Novabiochem, substitution level of 0.56 mmole/g) in a mixture of trifluoroacetic acid (TFA)/triisopropyl silane/H2O (95∶2.5∶2.5 vol/vol) at room temperature under N2 flow for 2 h. The crude peptides, precipitated with cold diethyl ether (Aldrich), were dried in vacuo. Purification proceeded by preparative reverse phase high performance liquid chromatography (HPLC, Varian ProStar 210) using a preparative C4 column (Vydac) and a linear gradient of buffer A (99.9% H2O and 0.1% trifluoroacetic acid) and buffer B (90% acetonitrile, 9.9% H2O and 0.1% trifluoroacetic acid). Purity was assessed using an HP1100 analytical HPLC system (Hewlett Packard) with a C4 column (Vydac). Molecular mass of all peptides was confirmed by matrix-assisted laser desorption/ionization-time of flight (MALDI-TOF) mass spectrometry using a Bruker Microflex 3.1.
Immuno-Gold Assay for Tracking the Prehairpin Intermediate State.
Immuno-gold labeling EM was performed using a JEOL 2010 microscope operated at an acceleration voltage of 120 kV. The PIV5 virus particles were incubated with lipid-coated PS nanobeads at 42 °C in the presence of biotinylated C-1 peptide using varied incubation time from 2 min to 1 h, followed by incubation with streptavidin-coated 5 nm gold particles (Ted Pella) for 10 min. The posttreatment with 5 nm gold particles labels the inhibitory peptide to show the location of the inhibited fusion proteins. The same thin-sectioning protocols were followed as in the other fusion assay. In the case of the gold labels, the staining protocol allows for higher contrast against the light membrane of the viruses and nanobeads.
Angular Distribution of Gold Particle Binding to Fusion Proteins in Immuno-Gold Assay.
For the localization of the prehairpin intermediate, EM images of viral particle/PS nanobead pairs including immuno-gold were analyzed using a JEOL 2010 microscope with over 200 images (about 300 pairs) in five sectioned samples. In order to present a full distribution of gold particles binding to fusion protein in PIV5 virus, all EM images were superimposed such that the midpoint of the line between the center of the viral particle ellipse and the center of the PS nanobead-supported bilayers ellipse are at the origin, the viral particle is on the negative x axis, and the nanobead-supported bilayer is on the positive x axis (Fig. 4C). The alignments were made using the Matlab computing language. The angles of gold particles within 200 nm of the nanobead-supported bilayer center were then quantified (Fig. 4D).
Supplementary Material
Acknowledgments.
We thank Dong Kuyn Ko and Taejong Baik for EM measurements. We also thank Raymond Meade and Biao Zuo in BioMedical Imaging Core at University of Pennsylvania for preparation of all sectioned and stained samples for EM analysis. This work was supported by National Institutes of Health (NIH) grants (GM54616, AI-23173) and the Materials Research Science and Engineering Centers (MRSEC) program of the National Science Foundation (NSF). G.P.L. is a Research Specialist and R.A.L. is an Investigator of the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1116034108/-/DCSupplemental.
References
- 1.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15:690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lamb RA, Jardetzky TS. Structural basis of viral invasion: lessons from paramyxovirus F. Curr Opin Struct Biol. 2007;17:427–436. doi: 10.1016/j.sbi.2007.08.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Colman PM, Lawrence MC. The structural biology of type I viral membrane fusion. Nat Rev Mol Cell Biol. 2003;4:309–319. doi: 10.1038/nrm1076. [DOI] [PubMed] [Google Scholar]
- 4.Weissenhorn W, et al. Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol. 1999;16:3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 5.Cross KJ, Burleigh LM, Steinhauer DA. Mechanisms of cell entry by influenza virus. Expert Reviews in Molecular Medicine. 2001;3:1–18. doi: 10.1017/S1462399401003453. [DOI] [PubMed] [Google Scholar]
- 6.White JM, Delos SE, Brecher M, Schornberg K. Structures and mechanisms of viral membrane fusion proteins: multiple variations on a common theme. Crit Rev Biochem Mol Biol. 2008;43:189–219. doi: 10.1080/10409230802058320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yin HS, Wen X, Paterson RG, Lamb RA, Jardetzky TS. Structure of the parainfluenza virus 5 F protein in its metastable, prefusion conformation. Nature. 2006;439:38–44. doi: 10.1038/nature04322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin HS, Paterson RG, Wen X, Lamb RA, Jardetzky TS. Structure of the uncleaved ectodomain of the paramyxovirus (hPIV3) fusion protein. Proc Natl Acad Sci USA. 2005;102:9288–9293. doi: 10.1073/pnas.0503989102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature. 1981;289:366–373. doi: 10.1038/289366a0. [DOI] [PubMed] [Google Scholar]
- 10.Bullough PA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature. 1994;371:37–43. doi: 10.1038/371037a0. [DOI] [PubMed] [Google Scholar]
- 11.Brunner J, Zugliani C, Mischler R. Fusion activity of influenza virus PR8/34 correlates with a temperature-induced conformational change within the hemagglutinin ectodomain detected by photochemical labeling. Biochemistry. 1991;30:2432–2438. doi: 10.1021/bi00223a019. [DOI] [PubMed] [Google Scholar]
- 12.Russell CJ, Jardetzky TS, Lamb RA. Membrane fusion machines of paramyxoviruses: capture of intermediates of fusion. EMBO J. 2001;20:4024–4034. doi: 10.1093/emboj/20.15.4024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chan DC, Kim PS. HIV entry and its inhibition. Cell. 1998;93:681–684. doi: 10.1016/s0092-8674(00)81430-0. [DOI] [PubMed] [Google Scholar]
- 14.Jiang S, Lin K, Strick N, Neurath AR. HIV-1 inhibition by a peptide. Nature. 1993;365:113. doi: 10.1038/365113a0. [DOI] [PubMed] [Google Scholar]
- 15.Lee KK. Architecture of a nascent viral fusion pore. EMBO J. 2010;29:1299–1311. doi: 10.1038/emboj.2010.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Damico RL, Crane J, Bates P. Receptor-triggered membrane association of a model retroviral glycoprotein. Proc Natl Acad Sci USA. 1998;95:2580–2585. doi: 10.1073/pnas.95.5.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furuta RA, Wild CT, Weng Y, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka M, Sackmann E. Polymer-supported membranes as models of the cell surface. Nature. 2005;437:656–663. doi: 10.1038/nature04164. [DOI] [PubMed] [Google Scholar]
- 19.Baker KA, Dutch RE, Lamb RA, Jardetzky TS. Structural basis for paramyxovirus-mediated membrane fusion. Mol Cell. 1999;3:309–319. doi: 10.1016/s1097-2765(00)80458-x. [DOI] [PubMed] [Google Scholar]
- 20.Yuan P, et al. Structural studies of the parainfluenza virus 5 hemagglutinin-neuraminidase tetramer in complex with its receptor, sialyllactose. Structure. 2005;13:803–815. doi: 10.1016/j.str.2005.02.019. [DOI] [PubMed] [Google Scholar]
- 21.Crennell S, Takimoto T, Portner A, Taylor G. Crystal structure of the multifunctional paramyxovirus hemagglutinin-neuraminidase. Nat Struct Biol. 2000;7:1068–1074. doi: 10.1038/81002. [DOI] [PubMed] [Google Scholar]
- 22.Lamb RA, Parks GD. In: Fields Virology. 5th edition. Knipe DM, Howley PM, editors. 2007. pp. 1449–1496. [Google Scholar]
- 23.Paterson RG, Russell CJ, Lamb RA. Fusion protein of the paramyxovirus SV5: destabilizing and stabilizing mutants of fusion activation. Virology. 2000;270:17–30. doi: 10.1006/viro.2000.0267. [DOI] [PubMed] [Google Scholar]
- 24.Lamb RA. Paramyxovirus fusion: a hypothesis for changes. Virology. 1993;197:1–11. doi: 10.1006/viro.1993.1561. [DOI] [PubMed] [Google Scholar]
- 25.Carr CM, Kim PS. A spring-loaded mechanism for the conformational change of influenza hemagglutinin. Cell. 1993;73:823–832. doi: 10.1016/0092-8674(93)90260-w. [DOI] [PubMed] [Google Scholar]
- 26.Rohl CA, Strauss CE, Misura KM, Baker D. Protein structure prediction using Rosetta. Methods Enzymol. 2004;383:66–93. doi: 10.1016/S0076-6879(04)83004-0. [DOI] [PubMed] [Google Scholar]
- 27.Bissonnette ML, Donald JE, DeGrado WF, Jardetzky TS, Lamb RA. Functional analysis of the transmembrane domain in paramyxovirus F protein-mediated membrane fusion. J Mol Biol. 2009;386:14–36. doi: 10.1016/j.jmb.2008.12.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Donald JE, et al. From the cover: transmembrane orientation and possible role of the fusogenic peptide from parainfluenza virus 5 (PIV5) in promoting fusion. Proc Natl Acad Sci USA. 108:3958–3963. doi: 10.1073/pnas.1019668108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Der Spoel D, et al. GROMACS: fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 30.Jorgensen WL, Maxwell DS, Tirado-Rives J. Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J Am Chem Soc. 1996;118:11225–11236. [Google Scholar]
- 31.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and reparametrization of the OPLS-AA force field for proteins via comparison with accurate quantum chemical calculations on peptides. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 32.MacKerell AD, Jr, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 33.Fadeev AY, DeGrado WF. Lipid membranes supported on optically transparent nanosilicas: synthesis and application in characterization of protein-membrane interactions. J Colloid Interface Sci. 2011;355:265–268. doi: 10.1016/j.jcis.2010.11.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stöber W, Fink A, Bohn E. Controlled growth of monodisperse silica spheres in the micron size range. J Colloid Interf Sci. 1968;26:62–69. [Google Scholar]
- 35.Gregg SJ, Sing KSW. Adsorption, Surface Area, and Porosity. London: Academic Press; 1982. [Google Scholar]
- 36.Peluso RW, Lamb RA, Choppin PW. Polypeptide synthesis in simian virus 5-infected cells. J Virol. 1977;23:177–187. doi: 10.1128/jvi.23.1.177-187.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Paterson RG, Harris TJ, Lamb RA. Analysis and gene assignment of mRNAs of a paramyxovirus, simian virus 5. Virology. 1984;138:310–323. doi: 10.1016/0042-6822(84)90354-4. [DOI] [PubMed] [Google Scholar]
- 38.Lamb RA, Mahy BW, Choppin PW. The synthesis of sendai virus polypeptides in infected cells. Virology. 1976;69:116–131. doi: 10.1016/0042-6822(76)90199-9. [DOI] [PubMed] [Google Scholar]
- 39.Hayat MA. Principles and Techniques of Electron Microscopy: Biological Applications. Cambridge United Kingdom; New York: Cambridge University Press; 2000. [Google Scholar]
- 40.Yamaguchi K, Suzuki K, Tanaka K. Examination of electron stains as a substitute for uranyl acetate for the ultrathin sections of bacterial cells. J Electron Microsc (Tokyo) 2010;59:113–118. doi: 10.1093/jmicro/dfp045. [DOI] [PubMed] [Google Scholar]
- 41.Bozzola JJ, Russell LD. Electron Microscopy: Principles and Techniques for Biologists. Sudbury, Mass: Jones and Bartlett; 1999. [Google Scholar]
- 42.Haidar A, Ryder TA, Mobberley MA, Wigglesworth JS. Two techniques for electron opaque staining of elastic fibres using tannic acid in fresh and formalin fixed tissue. J Clin Pathol. 1992;45:633–635. doi: 10.1136/jcp.45.7.633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Joshi SB, Dutch RE, Lamb RA. A core trimer of the paramyxovirus fusion protein: parallels to influenza virus hemagglutinin and HIV-1 gp41. Virology. 1998;248:20–34. doi: 10.1006/viro.1998.9242. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.