Abstract
Cotton is remarkable among our major crops in that four species were independently domesticated, two allopolyploids and two diploids. In each case thousands of years of human selection transformed sparsely flowering, perennial shrubs into highly productive crops with seeds bearing the vastly elongated and abundant single-celled hairs that comprise modern cotton fiber. The genetic underpinnings of these transformations are largely unknown, but comparative gene expression profiling experiments have demonstrated up-regulation of profilin accompanying domestication in all three species for which wild forms are known. Profilins are actin monomer binding proteins that are important in cytoskeletal dynamics and in cotton fiber elongation. We show that Gossypium diploids contain six profilin genes (GPRF1–GPRF6), located on four different chromosomes (eight chromosomes in the allopolyploid). All but one profilin (GPRF6) are expressed during cotton fiber development, and both homeologs of GPRF1–GPRF5 are expressed in fibers of the allopolyploids. Remarkably, quantitative RT-PCR and RNAseq data demonstrate that GPRF1–GPRF5 are all up-regulated, in parallel, in the three independently domesticated cottons in comparison with their wild counterparts. This result was additionally supported by iTRAQ proteomic data. In the allopolyploids, there This usage of novel should be fine, since it refers to a novel evolutionary process, not a novel discovery has been novel recruitment of the sixth profilin gene (GPRF6) as a result of domestication. This parallel up-regulation of an entire gene family in multiple species in response to strong directional selection is without precedent and suggests unwitting selection on one or more upstream transcription factors or other proteins that coordinately exercise control over profilin expression.
Keywords: polyploidy, crop evolution, fiber, directional selection
With its highly exaggerated, unicellular seed hairs (trichomes), cotton provides the world's most important source of renewable natural fiber. The origin of cultivated cotton is notable among our major crop plants in that four Gossypium species were independently domesticated, in different geographic regions, by separate prehistoric cultures (1). This parallel domestication process involved two allopolyploid (AD genome) species from the New World, Gossypium hirsutum (the source of “Upland” cotton) and Gossypium barbadense (the source of “Pima” or “Egyptian” cotton), and two diploid (A genome) species from the Old World, Gossypium arboreum and Gossypium herbaceum. As a consequence of thousands of years of human-mediated selection and agronomic improvement, each of the cultivated species underwent a series of phenotypic modifications, including transformation from perennial shrubs and small trees to more compact annual plants, loss of photoperiod sensitivity, reduction in seed dormancy, and most importantly, morphological transitions in fibers that dramatically enhanced fiber yield and quality. At present, the genetic bases of these possibly contemporaneous transformations are unknown.
In contrast to the wild progenitors, which have relatively short and coarse, tan-colored fibers, modern cultivars possess much longer, finer, stronger, and whiter fiber (Fig. 1). This similarity among the end-products of strong human directional selection raises a general question about the level and details of the processes governing the evolutionary transformations from wild to domesticated forms. It is of interest to understand the nature of this evolutionary parallelism at various levels of biological organization, from morphology to genotype. With respect to the former, analyses of fiber growth curves reveal that cotton domes-tication at both the diploid and allopolyploid levels is associated with a prolonged period of fiber elongation (2). As to the genomic underpinnings of fiber evolution, a number of recent studies are generating insights and a rich data set based on comparative transcriptome profiling of fiber development in wild vs. domesticated cotton (3–6), and from ongoing comparative proteomic analyses (7). Rapp et al. (2010), for example, reported thousands of genes that are differentially expressed in developing fiber between wild and domesticated G. hirsutum, many associated with key developmental processes presumed to play important roles in primary and secondary wall synthesis and in modulation of reactive oxygen species (3, 5, 6). Thus, the evolutionary process associated with strong directional selection practiced by aboriginal domesticators appears to be extraordinarily complex, at least insofar as perturbation of the transcriptional network, especially in the early stages of fiber development (4).
Fig. 1.
Morphological differences between cultivated cottons (from Left to Right: G. hirsutum, G. barbadense, and G. herbaceum) and their wild counterparts (from Left to Right: G. hirsutum, G. darwinii, and G. herbaceum).
Because fiber length per se is an agronomically important property, much attention has been focused on the molecular mechanisms of fiber elongation (8–10). In plants, a highly dynamic actin cytoskeleton has emerged as a key determinant of cell morphology. Numerous studies have demonstrated that dynamic actin is essential for cell expansion in tip-growing cells, such as root hairs, pollen tubes, and the protonemata of mosses (11–13). Furthermore, actin is also required for expansion of trichomes (14, 15), a category that includes cotton “fiber.”
The dynamic rearrangement of actin filaments involves maintaining a proper balance between filamentous and monomeric actin, which is controlled by a number of actin-binding proteins. Profilin is one of the most abundant actin monomer binding proteins and has been extensively characterized in vitro and in vivo (11, 16, 17). The role of profilin in regulating actin dynamics may be to help maintain a pool of actin monomers competent to polymerize at the fast-growing end of the actin filament (18–21). This is achieved, in part, by the interaction of profilin with the formin class of actin nucleators to promote actin polymerization and elongation (22–24). However, profilin also binds to phosphoinositides and other proteins (25, 26), suggesting that it plays a broad role in modulating the actin cytoskeleton.
Profilin has been shown to affect tip growth in moss, root hairs, pollen tubes, and cotton fiber (15, 27–29). Profilin is expressed during early cotton fiber development (14, 15), and on the basis of the observation that overexpression in transgenic tobacco cells produced elongated cells with thicker and longer microfilament cables, Wang et al. (15) suggested that profilin plays a role in the rapid elongation of cotton fibers by promoting actin polymerization.
In previous investigations of cotton domestication based on microarray data, profilin genes ranked among the most highly differentially expressed genes (4, 14, 30), suggesting that they, or one or more upstream regulators, were targets of human selection. Moreover, this observation was repeated for three different, independently domesticated cotton species. Here, we study this evolutionary parallelism in more detail, describing the cotton profilin gene family and its expression in fiber collected from all three of the domesticated species for which wild forms are known. A total of six profilin gene members (12 homeologs) were identified in Gossypium, and all but one of these are expressed during cotton fiber development. Remarkably, all five of the genes expressed in fiber are up-regulated in all three independently domesticated cottons in comparison with their wild counterparts, a result congruent with proteomic data. This parallel up-regulation of a gene family, instead of a single gene, is without precedent and suggests commonalities in the upstream targets of human-mediated directional selection among three species of domesticated cotton.
Results
Characterization of the Cotton Profilin Gene Family.
To understand the gene family structure of profilin in Gossypium, we first analyzed profilin genes in other plants. Profilin is a protein of 130–134 aa encoded by a small gene family. Plant profilins fall into two classes, one thought to be constitutively expressed and the other predominantly expressed in anthers or pollen (31–34). The size of the profilin gene family varies among plants. Only one profilin gene is annotated in algae (Chlamydomonas reinhardtii and Volvox carteri), whereas 2–14 gene members have been detected in land plants. No length variation among genes is revealed in Physcomitrella (132 aa for four genes), Selaginella (132 aa for two genes), or monocots (131 aa for three genes in Oryza and five genes in Zea), but most eudicot genomes encode profilin proteins of varying lengths. In Arabidopsis, for example, the five profilin genes encode protein products with either 131 or 134 aa, the length difference reflecting a three-codon indel in the first exon. At the same position, a two-codon indel is apparent in other eudicots (such as Ambrosia, Carica, Glycine, Gossypium, Lotus, Manihot, Medicago, Ricinus, and Vitis).
PCR amplification followed by cloning and sequencing in Gossypium led to the identification of six profilin gene family members (GPRF1–GPRF6) in all diploid cotton species surveyed. For each of these six genes in the allopolyploids, both copies from the two resident genomes were recovered, as expected; thus, a pair of homeologs exists for each of GPRF1–GPRF6, denoted by subscripts representing their genome of origin (AT deriving from the A-genome progenitor and DT deriving from the D-genome progenitor).
All six profilin genes share a similar structure, including three exons and two introns (Fig. S1). Gene lengths vary widely; although from 569 to 1623 bp, this variability is due mostly to intron size variation. Both introns of GPRF1 and GPRF6 are much shorter (65–96 bp) than introns of the other four profilins (382–633 bp); the longest intron (633 bp), in GPRF3, is almost 10-fold longer than the shortest one (65 bp in GPRF6). Interestingly, this level of variation in profilin intron sizes has not been detected in other plants. Alignment of protein sequences (Fig. S1) revealed that a 6-bp indel in the first exon is responsible for the longer protein products of GPRF1 and GPRF6 (134 aa) than of the other four genes (132 aa). As described above, this indel was also observed in other eudicots, suggesting that the co-occurrence of long and short profilin types originated early in eudicot evolution. Amino acids required for interaction with actin (35), poly-l-prolines (36) and PI(4,5)P2 (37) are conserved in the six Gossypium profilin genes.
GPRF1 and GPRF6 are more similar to each other than they are to the remaining four genes, structurally and with respect to composition, although they differ significantly at the nucleotide level (e.g., Ka = 0.075; Ks = 0.401 for the A-genome paralogs) (Table S1). Intergenic comparisons within the A- and D-genome diploids show that all six genes are divergent from one another at the nucleotide level, although they retain 75–95% amino acid identity (Table S1). Nonsynonymous differences (Ka) among the six profilin genes vary 8- (A genome) to 10-fold (D genome) within a species. GPRF2 and GPRF5 have higher amino acid identity to one another than either does to GPRF3 and GPRF4, with the latter gene being the most divergent of this group of four profilin genes containing long introns. Paralleling this distinction is the observation that the GPRF4 introns are substantially shorter than those of GPRF2, GPRF3, and GPRF5. Synonymous substitution rates (Ks) between A and D orthologs fall within the range typically observed in cotton (38), ranging from rather slowly evolving genes (GPRF2 and GPRF5; Ks = 0.015) to faster-evolving genes (GPRF3, Ks = 0.099). Sequence divergence is similar between the parental diploid species and their respective genomes in the allopolyploids; that is, orthologs from diploids (A vs. D) have Ks values similar to those of homeologs (AT vs. DT) in the allopolyploids, consistent with previous observations in Gossypium (38).
To investigate the history of gene duplication events that led to the observed profilin gene family structure, we constructed a maximum likelihood tree of profilins from sequenced plant genomes and data from other plants (Fig. S2). Rooted with the green alga sequences (Chlamydomonas and Volvox), sequences partitioned into two major clades (I and II, Fig. S2), a pattern suggestive of a duplication event early in angiosperm evolution. Six Gossypium genes, four long intron, 132 aa (i.e., GPRF2–GPRF5), and two short intron, 134 aa (GPRF1 and GPRF6), fell into these same two clades, as did sequences from other eudicots and monocots. These data indicate that the two major clades of profilins originated from a duplication event before the divergence of monocots and eudicots. For Gossypium, phylogenetic analysis yielded the topology expected, with no evidence of gene conversion (39).
We mapped all of the profilin genes on the Gossypium raimondii genome sequence and linked these to the tetraploid map (Table S2). The mapping results showed that we identified all genomic copies, and that the six profilin genes (GPRF1–GPRF6) are located on four different chromosomes (eight in the allopolyploid, i.e., both homeologs) (Table S2). Notably, tandem pairs of Gossypium profilin genes, each containing one gene from the two basal clades (I and II) are located on two different chromosomes (9 and 6), indicating a second round of gene duplication.
Profilin Expression in Cotton Fiber.
Reverse transcriptase PCR (RT-PCR) analysis indicated that GPRF6 was not transcribed in fiber from any Gossypium species studied. Additionally, GPRF1 was not detected in fibers from the D-genome diploid, but was expressed both in the A genome and the allopolyploid species (Fig. S3). In all allopolyploids, both homeologs of GPRF1 to GPRF5 (10 sequences total) were identified by amplification, cloning, and Sanger sequencing and through Illumina “RNASeq” transcriptome sequencing.
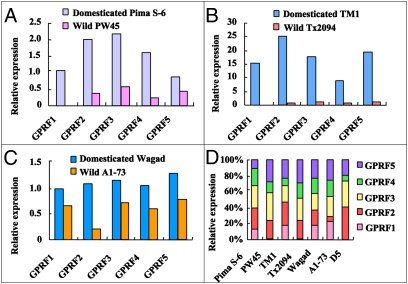
Gene-specific (although not homeolog-specific) primers for GPRF1–GPRF5 were used to assess relative expression levels of profilins between wild and domesticated cottons using real-time, quantitative RT-PCR. Cultivated accessions of both diploid and allopolyploid species exhibited up-regulation relative to their wild counterparts of not just one profilin gene but all five genes (GPRF1–GPRF5) in each of the three species studied (Fig. 2). The magnitude of up-regulation varied among genes and species, ranging from 2- to 106-fold, 11- to 390-fold, and 1.5- to 5.2-fold in G. barbadense (Pima S-6 vs. PW45), G. hirsutum (TM1 vs. Tx2094), and G. herbaceum (Wagad vs. A1-73), respectively (Fig. 2 A–C). Thus, there was far greater profilin up-regulation in the allopolyploids than in the diploid. We estimated the proportional expression of each of the five profilin genes in each sample (Fig. 2D). In G. barbadense and its wild relative Gossypium darwinii, the highest expression was found for GPRF3 (28–35% of total), whereas GPRF2 and GPRF5 were the most highly expressed profilins in cultivated and wild G. hirsutum (29% of total). In G. herbaceum, GPRF5 is the most highly expressed gene (∼25% of total).
Fig. 2.
Quantitative RT-PCR estimates of expression for five profilin genes in cotton fiber. (A) Comparison between domesticated G. barbadense (Pima S-6) and its wild counterpart, G. darwinii (PW45). (B) Comparison between domesticated (TM1) and wild (Tx2094) G. hirsutum. (C) Comparison between domesticated (Wagad) and wild (A1-73) G. herbaceum. (D) Relative expression of each gene in each accession.
Fiber transcriptome RNAseq datasets yield data (Table 1) congruent with the results from quantitative RT-PCR: (i) Expression of GPRF6 was not detected in fibers; (ii) GPRF1 was expressed at low levels, especially in the wild forms; and (iii) convergent up-regulation of profilin genes was observed in domesticated cottons, with the sole exception that GPRF5 was more highly expressed in G. barbadense accession K101 than in Pima S-6. GPRF1 was expressed at a low level compared with the other profilins, but still was up-regulated by domestication (3.9-fold and 420-fold) in G. barbadense and G. hirsutum, respectively (Fig. 3 and Table 1).
Table 1.
Count reads and ratios for profilin genes of domesticated and wild cottons at 10-dpa fiber cDNA libraries
| Profilin gene | Domesticated count | Wild count | Total domesticated | Total wild | Ratio of domesticated to wild | |
| Maxxa vs. Tx2094 | GPRF1 | 448 | 1 | 8,343,842 | 7,823,250 | 420.0482* |
| GPRF2 | 4,391 | 2,564 | 8,343,842 | 7,823,250 | 1.6057* | |
| GPRF3 | 4,630 | 2,340 | 8,343,842 | 7,823,250 | 1.8552* | |
| GPRF4 | 3,405 | 1,793 | 8,343,842 | 7,823,250 | 1.7806* | |
| GPRF5 | 1,446 | 1,065 | 8,343,842 | 7,823,250 | 1.2730* | |
| Pima-6 vs. K101 | GPRF1 | 439 | 129 | 4,863,645 | 5,515,575 | 3.8593* |
| GPRF2 | 3,728 | 3,487 | 4,863,645 | 5,515,575 | 1.2124* | |
| GPRF3 | 2,952 | 3,032 | 4,863,645 | 5,515,575 | 1.1041* | |
| GPRF4 | 3,417 | 3,528 | 4,863,645 | 5,515,575 | 1.0984* | |
| GPRF5 | 700 | 1,505 | 4,863,645 | 5,515,575 | 0.5275* |
Ratio of domesticated to wild = (domesticated count/total domesticated count)/(wild count/total wild count).
*P value <0.01 (Fisher's exact test).
Fig. 3.
Relative expression counts for five profilin genes based on ∼16 million Illumina reads per accession. Maxxa and Tx2094 are cultivated and wild forms of G. hirsutum, whereas Pima S-6 and K101 are cultivated and primitive accessions of G. barbadense, respectively. The y axis is expressed in gene counts per million reads, and (*) represents significant expression divergence between domesticated and wild accessions (Fisher's exact test; P < 0.01).
In addition to estimating expression using transcriptomic estimates, we also estimated profilin expression during fiber development using iTRAQ proteomic analysis, which allows simultaneous protein identification and comparative quantification of multiple samples. From the G. hirsutum fiber proteomic data, peptides corresponding to profilin gene family members were detected. Among those, gene member-specific peptides were identified for GPRF1 and GPRF4, respectively, which allowed the expression analysis for these two profilin genes. For GPRF2, GPRF3, and GPRF5, only peptides corresponding to their conserved region were detected, which were used to measure the combined expression of the three proteins as GPRFX. As shown in Table 2, all numbers are greater than 1, indicating greater protein expression in fibers from domesticated than in wild G. hirsutum. Ratios varied among stages and genes, from near parity to a 4.6-fold increase. Accordingly, and also because of high variance in some cases among different peptides of the same protein, statistical significance was not obtained in every comparison. Notably, and with the exception of GPRF1, for which only a single read was recorded in the RNAseq data in wild cotton (leading to a spuriously high domesticated-to-wild transcript ratio), these protein expression ratios closely mirror the transcript expression data (proteomic and transcript data are positively correlated; r = 0.999, P < 0.01).
Table 2.
Protein ratios of profilins in fiber extracts from domesticated and wild Gossypium hirsutum
| 5 dpa | 10 dpa | 20 dpa | 25 dpa | |
| GPRF1 | 1.1323 | 2.4878* | 4.6008* | 2.0378 |
| GPRFX | 1.1559 | 1.0140 | 1.7368 | 1.4000 |
| GPRF4 | 1.2253 | 1.1013 | 1.3937** | 1.1334 |
Dpa, days postanthesis.
*P value <0.05;
**P value <0.01 (Student's t test).
Transcript Levels of Other Proteins Involved in Actin Cytoskeleton Remodeling.
We examined the RNAseq data for expression of other proteins implicated in regulating actin or actin-mediated processes. Contigs detected are listed in Table S3, and total expression for several genes is shown for G. hirsutum in Fig. S4. The expression of actin was the highest in both 10-d postanthesis (dpa) and 20-dpa fibers of G. hirsutum, followed by profilin, and then actin-depolymerizing factor (ADF), whereas cyclase-associated protein (CAP) was expressed at relatively lower levels.
Discussion
Profilin Gene Family in Gossypium.
Here we describe six profilin genes (GPRF1–GPRF6) from five species of Gossypium; four of these are newly recognized, but GPRF1 and GPRF3 were reported earlier from G. hirsutum (14, 15, 30, 40). All six cotton profilin genes in all species share high amino acid sequence identity (75–95%), gene structures (two introns and three exons), and conserved motifs, but fall into two distinct and ancient clades. In one clade (containing GPRF2–GPRF5), the first exon is missing two amino acids and has longer introns, whereas in the other clade (containing GPRF1 and GPRF6) the first exon is longer but the genes have shorter introns. Phylogenetic analysis further shows that these two classes of profilins are descendants of an ancient duplication that occurred early in angiosperm evolution, perhaps before the separation between eudicots and monocots (Fig. S2).
Plant profilins generally are classified into two groups by their expression patterns. Kandasamy et al. (41) characterized the regulation of five Arabidopsis profilins in different organs and during microspore development, reporting that PRF1, PRF2, and PRF3 are expressed constitutively in all organs, whereas PRF4 and PRF5 are expressed specifically in pollen. As shown here, these two classes of Arabidopsis profilin genes fall into two clades, grouping with different cotton profilins. PRF1–PRF3 are grouped with GPRF2–GPRF5, whereas PRF4 and PRF5 are grouped with GPRF1 and GPRF6. This topological structure implies a possible functional correlation between these classes in Arabidopsis and Gossypium. Recently, Wang et al. (30) identified a profilin homolog, GhPFN2 (same as GPRF3 in the present study), from a cultivated variety of G. hirsutum. They found that GhPFN2 was expressed constitutively in multiple organs and preferentially in fiber cells. Expression was significantly induced during the period of rapid fiber elongation and secondary wall synthesis. Our expression analysis reveals that this class of four profilin genes, including GPRF2–GPRF5, are all expressed in fibers from three different cotton species.
The second class of cotton profilin genes (GPRF1 and GPRF6) differs from the other cotton profilins in expression. RT-PCR and RNAseq results show that GPRF6 is not transcribed in 10-dpa fibers. Similarly, with the exception of G. herbaceum, GPRF1 is either not expressed or is weakly expressed in developing fibers from wild cottons, although it is up-regulated in the domesticated allopolyploids (Figs. 2 and 3). Combined with the phylogenetic analyses, which indicate a homology between these two cotton genes and the pollen-specific genes of Arabidopsis, we speculate that little to no expression in fibers is ancestral for this group of profilins, and this is largely reflected in wild cottons. However, Ji et al. (14) recovered a profilin gene that was significantly up-regulated early in cultivated cotton (G. hirsutum) fiber development that corresponds to our GPRF1. This gene was later shown (15) to be predominantly expressed in rapidly elongating cotton fibers and that transcript abundance declined sharply with the onset of secondary wall synthesis. Our data indicate that GPRF1 has become associated with fiber cell elongation in cultivated but not wild cotton, suggesting novel gene recruitment associated with domestication.
Expression Alteration of Other Cytoskeletal Proteins.
Actin binding proteins regulate the dynamics of the actin cytoskeleton by controlling the balance between monomeric, filamentous, and bundled actin. On the basis of RNAseq data derived from ongoing studies of the fiber transcriptome in wild and domesticated cotton, several proteins involved in actin cytoskeleton dynamics demonstrated up-regulation in parallel with the profilins, including actin, ADF, and CAP (Fig. S4). Interestingly, profilin, ADF, and CAP are important for regulating the concentration of actin monomers. These data suggest that upon up-regulation of actin, control of the actin monomer pool is critical for the dramatic expansion in cotton fibers in domesticated cotton.
Parallel Up-Regulation of a Gene Family Under Domestication.
As a specialized unicellular trichome with a greatly exaggerated length, cotton fiber represents a masterpiece of human domestication, made all the more remarkable by its parallel, independent origin in four cultivated species (three studied here). Little is known about the genetic, genomic, or metabolomic transformations that mediate these independent transformations, although insights are emerging from comparative expression profiling experiments (3–6). The present study sheds light on one aspect of this general question, implicating the up-regulation of the profilin gene family concomitant with strong directional selection under human domestication. Not only was the same protein family up-regulated by aboriginal domesticators in multiple species, but the effects have been widespread across the profilin gene family members, as opposed to affecting a single gene in each domesticate. Gene expression data revealed that expression of five genes was signif-icantly enhanced in all cultivated cottons in comparison with their wild counterparts, including novel gene recruitment associated with domestication.
To our knowledge this observation of up-regulation of a gene family is without precedent in evolutionary biology, let alone parallel up-regulation in multiple species. Insights into the genetic basis of morphological change in nature are often facilitated using crop models, as Darwin famously noted in the introduction to On the Origin of Species when he wrote “At the commencement of my observations it seemed to me probable that a careful study of domesticated animals and of cultivated plants would offer the best chance of making out this obscure problem.” Since Darwin's time, numerous mutations have been identified that control traits selected by humans during the domestication process, including loss-of-function alleles, changes in coding sequence, or altered levels or domains of expression (42–46). Notwithstanding these striking discoveries, relatively little is understood about the downstream effects on domestication mutations or transcriptional and physiological networks, nor how these are propagated into the phenotypes being subjected to directional selection. Here we have illustrated one likely dimension of this process involved the altered regulation of a suite of related proteins, which simultaneously and in parallel in multiple species become up-regulated.
An exciting prospect for future work will be to isolate the causative lesions responsible for profilin gene family up-regulation, which likely comprise the hidden targets of human selection. In principle, these unknown targets of selection likely comprise one or more upstream transcription factors or other proteins that coordinately exercise control over profilin expression. It will be of considerable interest to reveal the degree of parallelism experienced in each species (47) and the effects of each species-specific mutation (or mutations) on the underlying transcriptional, proteomic, and metabolomic architecture of cotton fiber development. Insights into these and related questions will likely derive from a combination of forward genetic (e.g., introgression lines), population genetic (e.g., testing for selective sweeps), and genomic (e.g., expression profiling) approaches.
A final aspect of our results that merits highlighting is the extent to which the different profilin genes were up-regulated in each domesticated species. For example, in cultivated G. hirsutum, each of the five genes GPRF1–GPRF5 contributes substantially and relatively equitably to the total profilin transcriptome (18, 29, 21, 10, and 22%, respectively), whereas in the wild form of the same species, there is more variation among genes (1, 23, 28, 19, and 29%, respectively; Fig. 2D). The relative proportions and responses to selection in G. barbadense and G. herbaceum each pres-ent rather different patterns from this. When more is understood about the causative lesions in each species, it will be of considerable interest to study the complex rewiring of the transcriptional network that has responded similarly, but differently among these three species.
Materials and Methods
More detailed descriptions of all methods are provided in SI Materials and Methods. We used one domesticated and one wild accession for each of the three cotton species. The modern domesticated lines chosen were Pima S-6 (elite cultivar of G. barbadense), Texas Marker Stock 1 (TM1, the genetic and cytogenetic standard of G. hirsutum), and Wagad (an Indian cultivar of G. herbaceum). Choice of wild accessions was based on previous morphological and molecular evidence (48): for G. barbadense, because truly wild rather than feral forms are difficult to verify, its sister taxon from the Galapagos Islands G. darwinii (accession PW45) was chosen, as it previously was treated as conspecific with G. barbadense (49); for G. hirsutum, an unambiguously wild, sprawling shrub from the north coast of the Yucatan Peninsula, var. yucatanense accession Tx2094 (US Department of Agriculture GRIN accession PI 501501), was used; and for G. herbaceum, we chose a wild form from Botswana, G. herbaceum subsp. africanum (accession A1-73). We also included the best living model of the D-genome diploid progenitor, G. raimondii.
Using profilin genes from GenBank and Gossypium as query sequences, we searched our EST assemblies to design degenerate PCR primers (Table S4) to amplify profilin genes from Gossypium. PCR reactions were performed as described (50), and amplicons were cloned, sequenced (deposited in GenBank under accession numbers HM484221–HM484292), and mapped as described (SI Materials and Methods). Protein annotation and identification of conserved domains were facilitated using the Conserved Domains Database (51). We conducted phylogenetic analysis using species with available genome sequence, including 10 eudicot species, two monocots, Selaginella moellendorffii and Physcomitrella patens, and using two algae as the outgroup. Additionally, 66 plant profilins with annotations of sequence and functional information were also included in the analysis. Sequences were aligned using Jalview 2.5.1 (52). Maximum likelihood analysis was conducted using the default option as implemented in PhyML_3.0 (53). Confidence of the tree topology was assessed by a bootstrap set of 1,000 replicates. Estimation of nonsynonymous (Ka) and synonymous (Ks) substitution rates was performed within and between Gossypium species using DnaSP version 5 (54).
Fibers were harvested at 10 dpa because microarray data from G. hirsutum (4) showed that the expression level of profilin transcripts peaks at this stage. Total RNA was extracted as described (55). Profilin cDNA sequences were submitted to GenBank (HM543080–HM543138). To estimate transcript accumulation levels, we used quantitative real time RT-PCR analyses and Illumina RNAseq data. For the latter, we used fiber transcriptome data from 10- and 20-dpa fibers from wild and domesticated representatives of both domesticated polyploids (National Center for Biotechnology Information Sequence Read Archive Study SRP001603). We estimated expression levels for other actin binding proteins involved in actin cytoskeleton dynamics, including actin, ADF, and CAP using published cotton and Arabidopsis genes as queries and RNAseq data.
Total proteins were extracted from developing cotton fibers (56) using a liquid nitrogen/glass beads shearing method (57). Isolated proteins from wild and domesticated G. hirsutum were subjected to a comparative proteomic analysis using isobaric tags for relative and absolute quantification (iTRAQ) followed by strong-cation exchange fractionation and tandem mass spectrometry (10). The resulting mass spectrometry data from three replicates were processed and statistically analyzed using the ProteinPilot 4.0 software suite (AB SCIEX).
Supplementary Material
Acknowledgments
We thank Lei Gong, Kara Grupp, Chunming Xu, and Corrinne Grover for technical assistance. Financial support was provided by the National Science Foundation Plant Genome Program (to J.F.W.) and Program for New Century Excellent Talents in University Grant NCET-06-0609 (to Y.B.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Profilin cDNA sequences reported in this paper have been deposited in the GenBank database (accession nos. HM484221–HM484292 and HM543080–HM543138).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1115926109/-/DCSupplemental.
References
- 1.Wendel J, Cronn R. Polyploidy and the evolutionary history of cotton. Adv Agron. 2003;78:139–186. [Google Scholar]
- 2.Applequist WL, Cronn R, Wendel JF. Comparative development of fiber in wild and cultivated cotton. Evol Dev. 2001;3:3–17. doi: 10.1046/j.1525-142x.2001.00079.x. [DOI] [PubMed] [Google Scholar]
- 3.Chaudhary B, Hovav R, Flagel L, Mittler R, Wendel JF. Parallel expression evolution of oxidative stress-related genes in fiber from wild and domesticated diploid and polyploid cotton (Gossypium) BMC Genomics. 2009;10:378. doi: 10.1186/1471-2164-10-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rapp RA, et al. Gene expression in developing fibres of Upland cotton (Gossypium hirsutum L.) was massively altered by domestication. BMC Biol. 2010;8:139. doi: 10.1186/1741-7007-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chaudhary B, et al. Global analysis of gene expression in cotton fibers from wild and domesticated Gossypium barbadense. Evol Dev. 2008;10:567–582. doi: 10.1111/j.1525-142X.2008.00272.x. [DOI] [PubMed] [Google Scholar]
- 6.Hovav R, et al. The evolution of spinnable cotton fiber entailed prolonged development and a novel metabolism. PLoS Genet. 2008;4:e25. doi: 10.1371/journal.pgen.0040025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu G, et al. Genomically biased accumulation of seed storage proteins in allopolyploid cotton. Genetics. 2011;189:1103–1115. doi: 10.1534/genetics.111.132407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shi YH, et al. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li XB, Fan XP, Wang XL, Cai L, Yang WC. The cotton ACTIN1 gene is functionally expressed in fibers and participates in fiber elongation. Plant Cell. 2005;17:859–875. doi: 10.1105/tpc.104.029629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mei W, Qin Y, Song W, Li J, Zhu Y. Cotton GhPOX1 encoding plant class III peroxidase may be responsible for the high level of reactive oxygen species production that is related to cotton fiber elongation. J Genet Genomics. 2009;36:141–150. doi: 10.1016/S1673-8527(08)60101-0. [DOI] [PubMed] [Google Scholar]
- 11.Baluska F, et al. Root hair formation: F-actin-dependent tip growth is initiated by local assembly of profilin-supported F-actin meshworks accumulated within expansin-enriched bulges. Dev Biol. 2000;227:618–632. doi: 10.1006/dbio.2000.9908. [DOI] [PubMed] [Google Scholar]
- 12.Augustine RC, Vidali L, Kleinman KP, Bezanilla M. Actin depolymerizing factor is essential for viability in plants, and its phosphoregulation is important for tip growth. Plant J. 2008;54:863–875. doi: 10.1111/j.1365-313X.2008.03451.x. [DOI] [PubMed] [Google Scholar]
- 13.Vidali L, et al. Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc Natl Acad Sci USA. 2009;106:13341–13346. doi: 10.1073/pnas.0901170106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ji SJ, et al. Isolation and analyses of genes preferentially expressed during early cotton fiber development by subtractive PCR and cDNA array. Nucleic Acids Res. 2003;31:2534–2543. doi: 10.1093/nar/gkg358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang HY, Yu Y, Chen ZL, Xia GX. Functional characterization of Gossypium hirsutum profilin 1 gene (GhPFN1) in tobacco suspension cells. Characterization of in vivo functions of a cotton profilin gene. Planta. 2005;222:594–603. doi: 10.1007/s00425-005-0005-2. [DOI] [PubMed] [Google Scholar]
- 16.Böttcher RT, et al. Profilin 1 is required for abscission during late cytokinesis of chondrocytes. EMBO J. 2009;28:1157–1169. doi: 10.1038/emboj.2009.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tanaka M, Shibata H. Poly(L-proline)-binding proteins from chick embryos are a profilin and a profilactin. Eur J Biochem. 1985;151:291–297. doi: 10.1111/j.1432-1033.1985.tb09099.x. [DOI] [PubMed] [Google Scholar]
- 18.Carlsson L, Nyström LE, Sundkvist I, Markey F, Lindberg U. Actin polymerizability is influenced by profilin, a low molecular weight protein in non-muscle cells. J Mol Biol. 1977;115:465–483. doi: 10.1016/0022-2836(77)90166-8. [DOI] [PubMed] [Google Scholar]
- 19.Pantaloni D, Carlier MF. How profilin promotes actin filament assembly in the presence of thymosin beta 4. Cell. 1993;75:1007–1014. doi: 10.1016/0092-8674(93)90544-z. [DOI] [PubMed] [Google Scholar]
- 20.Pollard TD, Borisy GG. Cellular motility driven by assembly and disassembly of actin filaments. Cell. 2003;112:453–465. doi: 10.1016/s0092-8674(03)00120-x. [DOI] [PubMed] [Google Scholar]
- 21.Yarmola EG, Dranishnikov DA, Bubb MR. Effect of profilin on actin critical concentration: a theoretical analysis. Biophys J. 2008;95:5544–5573. doi: 10.1529/biophysj.108.134569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sagot I, Rodal AA, Moseley J, Goode BL, Pellman D. An actin nucleation mechanism mediated by Bni1 and profilin. Nat Cell Biol. 2002;4:626–631. doi: 10.1038/ncb834. [DOI] [PubMed] [Google Scholar]
- 23.Romero S, et al. Formin is a processive motor that requires profilin to accelerate actin assembly and associated ATP hydrolysis. Cell. 2004;119:419–429. doi: 10.1016/j.cell.2004.09.039. [DOI] [PubMed] [Google Scholar]
- 24.Pruyne D, et al. Role of formins in actin assembly: Nucleation and barbed-end association. Science. 2002;297:612–615. doi: 10.1126/science.1072309. [DOI] [PubMed] [Google Scholar]
- 25.Goldschmidt-Clermont PJ, Machesky LM, Baldassare JJ, Pollard TD. The actin-binding protein profilin binds to PIP2 and inhibits its hydrolysis by phospholipase C. Science. 1990;247:1575–1578. doi: 10.1126/science.2157283. [DOI] [PubMed] [Google Scholar]
- 26.Bae YH, et al. Profilin1 regulates PI(3,4)P2 and lamellipodin accumulation at the leading edge thus influencing motility of MDA-MB-231 cells. Proc Natl Acad Sci USA. 2010;107:21547–21552. doi: 10.1073/pnas.1002309107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vidali L, Augustine RC, Kleinman KP, Bezanilla M. Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell. 2007;19:3705–3722. doi: 10.1105/tpc.107.053413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pang CY, et al. Comparative proteomics indicates that biosynthesis of pectic precursors is important for cotton fiber and Arabidopsis root hair elongation. Mol Cell Proteomics. 2010;9:2019–2033. doi: 10.1074/mcp.M110.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhao PM, et al. Proteomic identification of differentially expressed proteins in the Ligon lintless mutant of upland cotton (Gossypium hirsutum L.) J Proteome Res. 2010;9:1076–1087. doi: 10.1021/pr900975t. [DOI] [PubMed] [Google Scholar]
- 30.Wang J, et al. Overexpression of a profilin (GhPFN2) promotes the progression of developmental phases in cotton fibers. Plant Cell Physiol. 2010;51:1276–1290. doi: 10.1093/pcp/pcq086. [DOI] [PubMed] [Google Scholar]
- 31.Huang S, McDowell JM, Weise MJ, Meagher RB. The Arabidopsis profilin gene family. Evidence for an ancient split between constitutive and pollen-specific profilin genes. Plant Physiol. 1996;111:115–126. doi: 10.1104/pp.111.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Morales S, Jiménez-López JC, Castro AJ, Rodríguez-García MI, Alché JD. Olive pollen profilin (Ole e 2 allergen) co-localizes with highly active areas of the actin cytoskeleton and is released to the culture medium during in vitro pollen germination. J Microsc. 2008;231:332–341. doi: 10.1111/j.1365-2818.2008.02044.x. [DOI] [PubMed] [Google Scholar]
- 33.Liu Q, Guo Z. Molecular cloning and characterization of a profilin gene BnPFN from Brassica nigra that expressing in a pollen-specific manner. Mol Biol Rep. 2009;36:135–139. doi: 10.1007/s11033-007-9161-8. [DOI] [PubMed] [Google Scholar]
- 34.Staiger CJ, et al. The profilin multigene family of maize: Differential expression of three isoforms. Plant J. 1993;4:631–641. doi: 10.1046/j.1365-313x.1993.04040631.x. [DOI] [PubMed] [Google Scholar]
- 35.Schutt CE, Myslik JC, Rozycki MD, Goonesekere NC, Lindberg U. The structure of crystalline profilin-beta-actin. Nature. 1993;365:810–816. doi: 10.1038/365810a0. [DOI] [PubMed] [Google Scholar]
- 36.Mahoney NM, Janmey PA, Almo SC. Structure of the profilin-poly-L-proline complex involved in morphogenesis and cytoskeletal regulation. Nat Struct Biol. 1997;4:953–960. doi: 10.1038/nsb1197-953. [DOI] [PubMed] [Google Scholar]
- 37.Lassing I, Lindberg U. Specific interaction between phosphatidylinositol 4,5-bisphosphate and profilactin. Nature. 1985;314:472–474. doi: 10.1038/314472a0. [DOI] [PubMed] [Google Scholar]
- 38.Senchina DS, et al. Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol Biol Evol. 2003;20:633–643. doi: 10.1093/molbev/msg065. [DOI] [PubMed] [Google Scholar]
- 39.Salmon A, Flagel L, Ying B, Udall JA, Wendel JF. Homoeologous nonreciprocal recombination in polyploid cotton. New Phytol. 2010;186:123–134. doi: 10.1111/j.1469-8137.2009.03093.x. [DOI] [PubMed] [Google Scholar]
- 40.Argiriou A, Kalivas A, Michailidis G, Tsaftaris A. Characterization of PROFILIN genes from allotetraploid (Gossypium hirsutum) cotton and its diploid progenitors and expression analysis in cotton genotypes differing in fiber characteristics. Mol Biol Rep. 2011 doi: 10.1007/s11033-011-1125-3. [DOI] [PubMed] [Google Scholar]
- 41.Kandasamy MK, McKinney EC, Meagher RB. Plant profilin isovariants are distinctly regulated in vegetative and reproductive tissues. Cell Motil Cytoskeleton. 2002;52:22–32. doi: 10.1002/cm.10029. [DOI] [PubMed] [Google Scholar]
- 42.Burger JC, Chapman MA, Burke JM. Molecular insights into the evolution of crop plants. Am J Bot. 2008;95:113–122. doi: 10.3732/ajb.95.2.113. [DOI] [PubMed] [Google Scholar]
- 43.Burke JM, Burger JC, Chapman MA. Crop evolution: From genetics to genomics. Curr Opin Genet Dev. 2007;17:525–532. doi: 10.1016/j.gde.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 44.Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell. 2006;127:1309–1321. doi: 10.1016/j.cell.2006.12.006. [DOI] [PubMed] [Google Scholar]
- 45.Tan L, et al. Control of a key transition from prostrate to erect growth in rice domestication. Nat Genet. 2008;40:1360–1364. doi: 10.1038/ng.197. [DOI] [PubMed] [Google Scholar]
- 46.Vollbrecht E, Springer PS, Goh L, Buckler ES, 4th, Martienssen R. Architecture of floral branch systems in maize and related grasses. Nature. 2005;436:1119–1126. doi: 10.1038/nature03892. [DOI] [PubMed] [Google Scholar]
- 47.McGrath PT, et al. Parallel evolution of domesticated Caenorhabditis species targets pheromone receptor genes. Nature. 2011;477:321–325. doi: 10.1038/nature10378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wendel J, Brubaker C, Seelanan T. In: The Origin and Evolution of Gossypium. Physiology of Cotton. Stewart J, Oosterhuis D, Heitholt J, Mauney J, editors. The Netherlands: Springer; 2010. pp. 1–18. [Google Scholar]
- 49.Wendel J, Percy R. Allozyme diversity and introgression in the Galapagos endemic Gossypium darwinii and its relationship to continental G. barbadense. Biochem Syst Ecol. 1990;18:517–528. [Google Scholar]
- 50.Bao Y, Ge S. Origin and phylogeny of Oryza species with the CD genome based on multiple gene sequence data. Plant Syst Evol. 2004;249:55–66. [Google Scholar]
- 51.Marchler-Bauer A, et al. CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 2005;33(Database issue):D192–D196. doi: 10.1093/nar/gki069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview version 2—a multiple sequence alignment. analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guindon S, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 54.Librado P, Rozas J. DnaSP v5: A software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- 55.Wan CY, Wilkins TA. A modified hot borate method significantly enhances the yield of high-quality RNA from cotton (Gossypium hirsutum L.) Anal Biochem. 1994;223:7–12. doi: 10.1006/abio.1994.1538. [DOI] [PubMed] [Google Scholar]
- 56.Yao Y, Yang YW, Liu JY. An efficient protein preparation for proteomic analysis of developing cotton fibers by 2-DE. Electrophoresis. 2006;27:4559–4569. doi: 10.1002/elps.200600111. [DOI] [PubMed] [Google Scholar]
- 57.Hovav R, et al. A majority of cotton genes are expressed in single-celled fiber. Planta. 2008;227:319–329. doi: 10.1007/s00425-007-0619-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.