Abstract
Virus infection induces the production of type I and type II interferons (IFN-I and IFN-II), cytokines that mediate the antiviral response. IFN-I (IFN-α and IFN-β) induces the assembly of IFN-stimulated gene factor 3 (ISGF3), a multimeric transcriptional activation complex composed of STAT1, STAT2, and IFN regulatory factor 9. IFN-II (IFN-γ) induces the homodimerization of STAT1 to form the gamma-activated factor (GAF) complex. ISGF3 and GAF bind specifically to unique regulatory DNA sequences located upstream of IFN-I– and IFN-II–inducible genes, respectively, and activate the expression of distinct sets of antiviral genes. The balance between type I and type II IFN pathways plays a critical role in orchestrating the innate and adaptive immune systems. Here, we show that the phosphorylation of STAT1 by IκB kinase epsilon (IKKε) inhibits STAT1 homodimerization, and thus assembly of GAF, but does not disrupt ISGF3 formation. Therefore, virus and/or IFN-I activation of IKKε suppresses GAF-dependent transcription and promotes ISGF3-dependent transcription. In the absence of IKKε, GAF-dependent transcription is enhanced at the expense of ISGF3-mediated transcription, rendering cells less resistant to infection. We conclude that IKKε plays a critical role in regulating the balance between the IFN-I and IFN-II signaling pathways.
Keywords: JAK/STAT, STAT dimerization, IFN-stimulated response element, gamma-activated sequences
Host immune defenses counteract virus infection by coordinating an intracellular innate immune response with the adaptive immune response. Systemically, the antiviral defense is generally cell-mediated, involving the recruitment and activation of dendritic cells, macrophages, neutrophils, and natural killer cells to the site of infection (1). This initial response is followed by a second wave of specific antiviral defenses involving cytotoxic T cells and antibodies generated from plasma B cells (2). The success of the adaptive immune response is intricately linked to the intracellular innate defenses initiated at the site of infection (1). The signaling required to coordinate the successful induction of the intracellular immune response to virus infection relies on the activation of interferon (IFN) genes, which encode cytokines with antiviral and immunomodulatory activity (3).
The vertebrate type I IFN (IFN-I) genes are arranged in a large gene cluster consisting of a single IFNβ gene and several tandemly arranged IFNα genes encoding distinct isotypes (4, 5). In contrast to type I IFNs, type II IFN (IFN-II) is encoded by a single IFNγ gene. Although IFN-I mediates cellular resistance to virus infection, IFN-II confers limited cellular protection through the induction of a subset of genes that are shared between the IFN-I and IFN-II transcriptomes (6, 7). Virus infection leads to the activation of IFN-I genes and the secretion of IFNα and -β, which bind to IFN receptors at the cell surface. This, in turn, leads to the transcriptional activation of a group of IFN-stimulated genes (ISGs), and these ISGs collectively establish a nonpermissive environment for virus replication (6).
The transcriptional response to IFNs is mediated by activation of the Janus kinase and signal transducer and activator of transcription (JAK/STAT) pathway (8). In this pathway, type I and type II IFNs bind to their respective receptors, leading to receptor dimerization and tyrosine phosphorylation by receptor-bound JAK kinases. These phosphotyrosines serve as docking sites for the STAT proteins via their Src homology 2 (SH2) domains (9–12). The recruitment of STAT1 and STAT2 to the receptors results in their tyrosine phosphorylation by the associated JAK kinases and their dimerization via phosphotyrosine and SH2 domain interactions in each monomer (8, 13, 14). The STAT protein dimers are then released from the receptors and translocate to the nucleus where they bind to specific regulatory DNA sequences and coordinately activate transcription of ISGs.
In the case of the IFN-I signaling pathway, both STAT1 and STAT2 are activated, leading to the formation of heterodimers that associate with the IFN regulatory factor 9 (IRF9) to form a transcription factor complex termed IFN-stimulated gene factor 3 (ISGF3) (15, 16). This complex binds to IFN-stimulated response elements (ISREs) in the promoters of ISGs, and the expression of these ISGs results in the establishment of an antiviral state (17). In contrast, IFN-II signaling results in the recruitment and tyrosine phosphorylation of two STAT1 proteins, leading to the formation of STAT1:STAT1 homodimers, a complex referred to as the gamma-activated factor (GAF) (18). GAF binds to gamma-activated sequences (GAS) found in the promoters of many genes, including a subset that is also induced by IFN-I (19). STAT1 is therefore a critical player in both IFN-I and IFN-II signaling.
We previously demonstrated that the IκB kinase ε (IKKε) phosphorylates serine 708 of STAT1 and that this phosphorylation is required for an effective antiviral response (20). However, the mechanism by which IKKε-mediated phosphorylation confers an optimal antiviral response is not known. IKKε, also known as IKKi, was originally identified on the basis of sequence similarity to the IκB kinases IKKα and IKKβ and its transcriptional induction in response to lipopolysaccharide (21–23). Following the initial discovery, IKKε and TBK1, a ubiquitously expressed homolog of IKKε, were shown to phosphorylate specific serine residues in the transcription factors IRF3 and IRF7 in response to virus infection (24–27). The phosphorylation of both transcription factors induces a conformational change and subsequent nuclear translocation where the factors bind to the IFNβ gene enhancer along with NF-κB and ATF2/cJUN to form the IFNβ enhanceosome (28, 29).
Despite the clear explanation of why the loss of IKKε renders cells more susceptible to virus infection, studies of TBK1 and IKKε knockout mice confirmed a critical role for TBK1 in the induction of IFN-I, suggesting that IKKε is dispensable for this activity, except perhaps in specialized antigen-presenting cells (20, 25). The inducible nature of IKKε led to speculation that the kinase plays a redundant role to TBK1 and is responsible for activation of IRF7 late during infection; however, IKKε knockout mice failed to demonstrate defects in the induction of IRF7-depedendent IFNα isotypes (20).
We previously reported that IKKε is not required for the activation of IFN-I in response to virus infection (20). Rather, IKKε plays an important role in the IFN-I–signaling pathway. Specifically, we found that IKKε knockout mice express normal levels of IFN-I in response to influenza A virus (IAV) infection, presumably due to the presence of TBK1. However, these mice display an abnormal transcriptional response to IFN-I stimulation, leading to the loss of a critical subset of IKKε-dependent antiviral proteins and the inability to clear virus infection. Here we describe a role for IKKε in the assembly of STAT1-containing transcriptional activation complexes. Specifically, the phosophorylation of STAT1 by IKKε inhibits STAT1 homodimerization, thereby increasing the pool of STAT1 available to associate with STAT2 and IRF9 to optimally assemble the critical IFN-I transcription factor complex ISGF3.
Results
Regulation of IFN-I Signaling by IKKε Is Required for an Effective Response to IAV Infection.
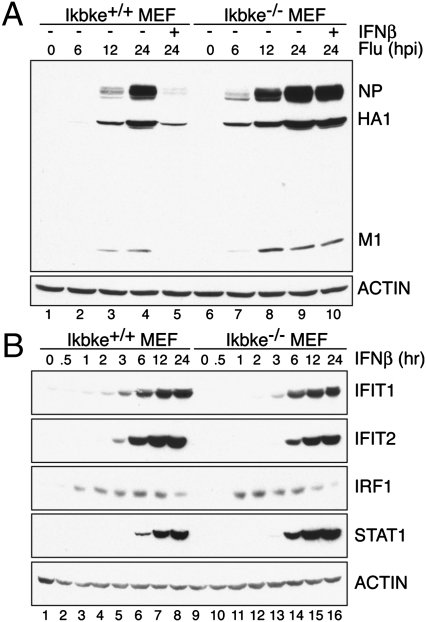
We infected Ikbke+/+ and Ikbke−/− murine embryonic fibroblasts (MEFs) with influenza A virus (A/Puerto Rico/8/34) in the presence or absence of recombinant IFNβ (Fig. 1A). IAV replication in Ikbke+/+ MEFs was detected 12 h post infection (hpi), as measured by nucleocapsid, hemagglutinin, and matrix protein levels, and replication was inhibited by IFNβ pretreatment. In contrast, virus replication in Ikbke−/− MEFs was detected as early as 6 hpi, revealing a significant increase in the kinetics of viral infection in the absence of IKKε. In addition, when Ikbke−/− MEFs were treated with IFNβ, there was no significant decrease in viral load, suggesting that increased IAV replication in the knockout cells is due to a defect in IFN-I signaling. These results corroborate in vivo data and support the notion that IKKε plays an essential role in coordinating the IFN-I–mediated antiviral response to IAV infection.
Fig. 1.
IKKε knockout MEFs are susceptible to IAV infection due to an IFN-I–signaling defect. (A) Ikbke+/+ and Ikbke−/− MEFs were infected with A/Puerto Rico/8/34 at a multiplicity of infection of 1. Cells were harvested 0, 6, 12, and 24 h post infection (hpi). Recombinant type I IFNβ was added to the media at the beginning of the 24-h infection time point. Cell extracts were analyzed by Western blot for nucleocapsid (NP), hemaggluttin (HA1), and matrix (M1) protein expression. Actin was used as a loading control. (B) Western blot analysis of Ikbke+/+ and Ikbke−/− MEFs stimulated with IFNβ at the time points indicated. Blots depict protein levels of IFIT1, IFIT2, IRF1, and STAT1. Actin was used as a loading control. Experiments were independently repeated at least three times.
To examine IFN-I signaling in Ikbke−/− MEFs, we analyzed the expression of ISGs by Western blot analysis. We observed a decrease in the production of the IFN-I–dependent antiviral proteins IFIT1 (ISG56) and IFIT2 (ISG54) in Ikbke−/− MEFs compared with Ikbke+/+ MEFs (Fig. 1B). Analysis of the kinetics of IFIT1 and IFIT2 expression reveal that these two proteins are expressed later and at reduced levels in Ikbke−/− MEFs compared with Ikbke+/+ MEFs. In contrast, there was no decrease in the expression of IRF1, a protein that is primarily regulated by IFN-II and whose expression requires the GAF regulatory complex (Fig. 1B). However, we note that the expression of IRF1 was elevated earlier in Ikbke−/− MEFs. STAT1, another protein whose expression is predominantly GAF-regulated, was also increased in Ikbke−/− MEFs relative to wild type, an observation previously reported (20). Taken together, these data confirm that Ikbke−/− MEFs have reduced ISGF3-dependent gene expression and suggest an increase in GAF-mediated transcription.
Loss of IKKε Results in Distinct IFN-I– and IFN-II–Dependent Signaling Profiles.
The transcriptomes induced by IFN-I and IFN-II display considerable overlap, but are distinct (6, 7). For example, expression of Ifit1 and Ifit2 are primarily induced by IFN-I (30–32). By contrast, Irf1, Icam1, and Irf8 are primarily IFN-II–inducible genes, but lower levels can be induced by IFN-I. The overlap in the type I and II IFN transcriptomes is due in part to the activation of common factors within the IFN-I and IFN-II pathways, as well as the presence of multiple ISRE or GAS elements in many antiviral gene promoters (3, 33, 34).
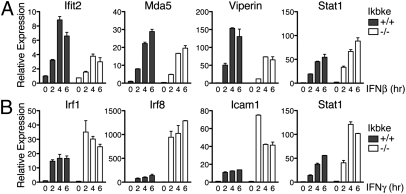
To examine IFN-I– and IFN-II–induced gene transcription, we performed quantitative PCR (qPCR) analysis of Ikbke+/+ and Ikbke−/− MEFs stimulated with IFNβ or IFNγ (Fig. 2). Consistent with the Western blot analysis of Fig. 1B, Ifit2 mRNA expression is reduced in Ikbke−/− MEFs (Fig. 2A). We also observed a similar decrease in levels of expression of other IFN-I–regulated genes such as Mda5 and Viperin in Ikbke−/− MEFs. In contrast, the predominantly IFN-II–stimulated genes Irf1, Irf8, and Icam1 are expressed at higher levels in Ikbke−/− MEFs compared with Ikbke+/+ MEFs (Fig. 2B). Furthermore, qPCR analysis demonstrated that Stat1 mRNA levels are higher in Ikbke−/− MEFs when treated with either IFNβ or IFNγ, although this increase is more pronounced with IFNγ stimulation (Fig. 2). Stat1 gene expression is inducible by both IFN-I and IFN-II, but is primarily regulated by the GAF complex (6, 7). These data clearly show that the increase in transcription of GAF-regulated genes in Ikbke−/− MEFs is inversely proportional to a decrease in expression of ISGF3-regulated genes, consistent with the hypothesis that IKKε regulates the balance in assembly and DNA binding of ISGF3 and GAF complexes.
Fig. 2.
Loss of IKKε impacts IFN-I– and IFN-II–mediated transcription. qPCR from RNA derived from Ikbke+/+ and Ikbke−/− MEFs stimulated with IFNβ (A) or IFNγ (B) at the time points indicated. Genes analyzed include Ifit2, Mda5, Viperin, Stat1, Irf1, Irf8, and Icam1. All samples were normalized to Actin and standardized to unstimulated wild-type cells. Error bars represent SDs from triplicate measurements. Experiments were independently repeated at least three times.
ISGF3 Formation and Binding Decrease as GAF Formation and Binding Increase.
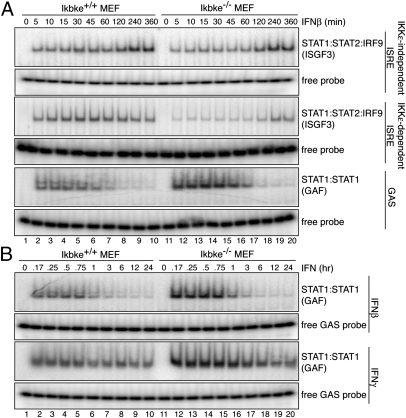
Electrophoretic mobility shift assays were performed to examine ISGF3 and GAF complex binding in extracts from Ikbke+/+ and Ikbke−/− MEFs. Cells were stimulated with IFNβ or IFNγ and analyzed for binding to either ISRE or GAS-containing DNA sequences (Fig. 3). There are at least two classes of ISREs: IKKε-dependent and IKKε-independent elements (20). As representatives of IKKε-dependent and -independent ISREs, we used elements found in the Adar1 and Irf7 gene promoters, respectively. The GAS sequence derives from an element in the promoter of the Irf1 gene.
Fig. 3.
ISGF3 binding decreases, whereas GAF binding increases in IKKε knockout MEFs. (A) EMSA with extracts derived from Ikbke+/+ and Ikbke−/− MEFs stimulated with IFNβ for the time points indicated. EMSA probes include IKKε-independent ISRE, IKKε-dependent ISRE, or IRF GAS element. (B) EMSA with extracts derived from Ikbke+/+ and Ikbke−/− MEFs stimulated with IFNβ or IFNγ for time points indicated and analyzed as in A. Unbound probe was used as loading control. Experiments were independently repeated at least three times.
IFNβ stimulation of wild-type IKKε MEFs results in the preferential assembly and binding of ISGF3 to the ISRE probes, binding as early as 5 min post induction and maintained up to 6 h post induction (Fig. 3A). The kinetics of ISGF3 binding to the IKKε-independent ISRE were indistinguishable in extracts from Ikbke+/+ and Ikbke−/− MEFs. In addition, competition binding studies using increasing amounts of unlabeled DNA probe showed no differences in affinity of ISGF3 to the IKKε-independent ISRE in Ikbke+/+ and Ikbke−/− extracts (Fig. S1). However, consistent with the observed decrease in ISG expression, the level of ISGF3 binding to the IKKε-dependent ISRE probe was significantly reduced in Ikbke−/− MEFs compared with the Ikbke+/+ MEFs (Fig. 3A). These observations suggest that the level of ISGF3 is lower in the Ikbke−/− extract and that the IKKε-independent ISREs have a higher affinity for ISGF3 than IKKε-dependent ISREs.
IFN-I stimulation can also activate GAF formation and binding to a GAS probe; however, GAF binding decreases 1–2 h post stimulation (Fig. 3). In contrast to ISGF3 binding, GAF assembly and binding to the GAS element increased in Ikbke−/− MEFs relative to the wild-type control. This increase in GAF binding is observed in Ikbke−/− MEFs upon either IFNβ or IFNγ stimulation (Fig. 3B). Thus, an increase in GAF binding is observed in cells lacking IKKε, whereas ISGF3 binding is reduced. These differences would be expected to significantly alter the IFN-inducible transcriptome in Ikbke−/− MEFs.
The observed decrease in ISGF3 binding and concomitant increase in GAF binding may be a consequence of competition for STAT1 during formation of the ISGF3 and GAF complexes. To investigate this possibility, size-exclusion chromatography and Western blot analysis were performed to determine the relative ratio of ISGF3 and GAF in extracts from IFNβ-stimulated Ikbke+/+ and Ikbke−/− MEFs (Fig. S2). Gel filtration column fractions C12 and C8 showed enriched levels of STAT1α/β, and their elution correlated with the anticipated molecular weights of ISGF3 and GAF, respectively. Higher levels of ISGF3 formation compared with GAF were observed in extracts from Ikbke+/+ MEFs, whereas the ratio shifted in Ikbke−/− MEFs. We conclude that the formation and binding of ISGF3 and GAF are inversely proportional and are significantly influenced by the presence of IKKε, leading to the observed misregulation of IFN signaling.
IKKε Promotes IFN-I Signaling and Diminishes IFN-II Signaling.
We have previously shown that IKKε phosphorylates the C terminus of STAT1 on serines S708, S744, and S747 (20). We therefore carried out studies to determine the effect of IKKε phosphorylation of STAT1 on the binding of ISGF3 and/or GAF complexes to the IKKε-dependent ISRE and GAS elements, respectively. We used adenovirus vectors to express either luciferase (AdV Luc) or IKKε (AdV IKKε) in E1A-expressing HeLa cells (Fig. S3). We chose these cells because E1A antagonizes IRF3-dependent activation of IFN-I gene expression (35), thereby making it possible to control IFN stimulation by exogenous introduction of IFN. The expression of AdV Luc or AdV IKKε alone did not lead to the binding of ISGF3 or GAF to an ISRE or GAS element, respectively. However, ISGF3 binding was observed upon IFN-I stimulation, and AdV IKKε did not decrease ISGF3 binding, confirming that the phosphorylation of STAT1 by IKKε does not inhibit the binding of ISGF3 to the ISRE (Fig. S3A). By contrast, overexpression of IKKε decreased the binding of GAF to the GAS element (Fig. S3B). In addition, an inverse correlation between the levels of IKKε and GAS binding was observed (Fig. S3C). These data provide additional evidence that IKKε reduces the level of GAF complex assembly in response to IFN stimulation.
To determine the effect of IKKε on gene expression, AdV-treated cell extracts were analyzed for expression of ISGF3-induced IFIT2 or GAS-induced IRF1 (Fig. S4). Cells treated with IFN-I or IFN-II expressed moderate levels of IFIT2 and high levels of IRF1, respectively (Fig. S4A). These levels were not affected by the expression of AdV Luc. In contrast, AdV IKKε treatment resulted in the induction of IFIT2 in response to IFN-II, whereas the level of IRF1 dramatically decreased following IFN-II treatment. These data provide additional evidence that IKKε plays a central role in establishing the cellular antiviral response by decreasing the expression of GAS-dependent genes and increasing the expression of ISRE-driven genes. In addition, there is an inverse correlation between the levels of IKKε and the expression of IRF1 (Fig. S4B), consistent with the observed decrease in GAF binding. Furthermore, as the level of IRF1 decreases in the presence of high levels of IKKε, there is a corresponding increase in the level of IFIT2. We conclude that the phosphorylation of STAT1 by IKKε shifts the cellular response toward an IFN-I–mediated transcriptome even in the presence of IFN-II.
IKKε Suppresses the Formation of Activated STAT1 Homodimers but Not STAT1:STAT2 Heterodimers.
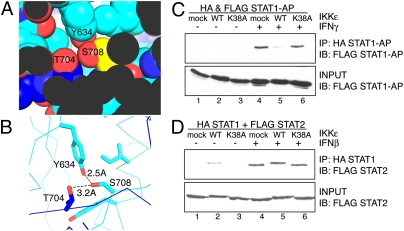
The 3D structure of the GAF complex (a STAT1 homodimer) bound to the GAS element is known (36). This structure reveals that the STAT1 S708 residue is located within a key dimerization interface between the two STAT1 molecules. S708 engages in critical hydrogen-bonding interactions within this tightly packed interface (Fig. 4 A and B). On the basis of this structure and our previous results, we reasoned that the phosphorylation of STAT1 by IKKε may prevent STAT1:STAT1 homodimerization (in GAF formation) and thus inhibit GAS binding. However, this phosphorylation must not interfere with the interaction between STAT1 and STAT2 in the ISGF3 complex. To determine if decreased GAS binding and reduced IRF1 production results from the inability of STAT1 to homodimerize, we carried out coimmunoprecipitation experiments. However, as previously established, we found that STAT1 dimerizes in the absence of IFN, likely through an antiparallel conformation, requiring N-terminal domain interactions (37, 38). Upon IFN stimulation, STAT1 phosphorylation on Y701 results in a spatial reorientation from the antiparallel conformation to a parallel configuration, with the phosphotyrosine binding to the SH2 domain of the partner STAT1 (37, 38).
Fig. 4.
IKKε phosphorylation disrupts the STAT1 homodimer, but not the STAT1:STAT2 heterodimer interface. (A) Space-filling diagram of the S708 (yellow) region and (B) hydrogen-bonding interactions of S708 in the STAT1:STAT1 homodimerization interface (light and dark blue). The diagram is adapted from the 3D structure published by Chen et al. (36) using PyMol. (C) 293T cells were transfected with HA STAT1 and FLAG STAT1 antiparallel (AP) mutants and GFP (mock transfected), IKKε WT, or IKKε K38A. Cells were unstimulated or stimulated with IFNγ. (D) 293T cells were transfected with HA STAT1 and FLAG STAT2 and GFP, IKKε WT, or IKKε K38A. Cells were unstimulated or stimulated with IFNβ. Coimmunoprecipitation experiments were performed using anti-HA matrix beads for the immunoprecipitation (IP) and immunoblotted (IB) for anti-FLAG tagged protein. Input samples were loaded as a control. Experiments were independently repeated at least three times.
To distinguish IFN-activated parallel dimers from the unstimulated and antiparallel dimers, mutants were generated that either lacked the N-terminal 135 amino acids of STAT1 or were disrupted specifically at residues critical for preassociation (36, 37). Using these antiparallel STAT1 mutants, coimmunoprecipitation experiments were performed with cell extracts from fibroblasts exogenously expressing differentially tagged STAT1 mutants (Fig. 4C). Antiparallel mutant STAT1 did not associate before IFN stimulation, and dimerization was induced upon IFNγ stimulation. This dimerization was disrupted by the expression of IKKε, but not by the expression of the GFP control or the catalytically dead mutant IKKε K38A.
In contrast to STAT1 homodimerization, IKKε does not disrupt STAT1:STAT2 heterodimerization (Fig. 4D). It is interesting to note that IKKε expression results in a phosphorylation-dependent shift in STAT2 mobility, suggesting that IKKε may phosphorylate STAT2 in addition to STAT1. In the absence of IFN stimulation, STAT1 and STAT2 can weakly preassociate (39), and we noted that IKKε expression stabilizes this asso-ciation before IFN stimulation (Fig. 4D, lane 2). These data suggest that phosphorylation of S708 disrupts the parallel STAT1 homodimer and facilitates a STAT1:STAT2 dimerization interface, which must differ from that of the STAT1 homodimer. The structural differences, if any, between the components in the ISGF3 complex assembled in the presence or absence of IKKε are currently unknown.
Next, we performed electrophoretic mobility shift assays using purified recombinant proteins that are modified by phosphorylation and assayed for their ability to bind either the GAS or the ISRE DNA element (Fig. S5). Recombinant transcription factors were phosphorylated by the recombinant kinases JAK or IKKε. The phosphorylation of STAT1 by JAK resulted in GAF formation and binding to DNA; however, when STAT1 was both JAK- and IKKε-phosphorylated, it failed to bind to DNA. By contrast, IKKε phosphorylation of STAT1 did not disrupt ISGF3 complex formation. These data provide further evidence that IKKε phosphorylation directly regulates the STAT-mediated antiviral cellular response induced by IFN-I and IFN-II, acting as a molecular switch and integrating two independent signaling cues into a single cellular transcriptional output.
Loss of IKKε Results in Aberrant ISG Induction in Bone Marrow-Derived Macrophages.
We analyzed ISG expression in bone marrow-derived macrophages (BMMs) from Ikbke+/+ and Ikbke−/− mice by RNA deep sequencing (RNA-seq). Analysis of the RNA-seq data from Ikbke+/+ BMMs identified a set of 538 ISGs whose levels increased at least threefold upon IFNβ or IFNγ induction. We ordered these ISGs by their beta–gamma mixture (BGM), which provides a number that reflects the fold induction in response to IFNβ and IFNγ, ranging from −1 for a purely beta response, 0 for equal response to beta and gamma, and +1 for a purely gamma response (Fig. S6A). The expression of individual ISGs, such as IFIT2 and IRF1, is shown, as well as the relative order of the ISGs in the BGM scale. In Ikbke−/− BMMs, IFNβ stimulation resulted in a relative decrease of previously characterized IFN-I–mediated genes and a corresponding increase in genes responsive to IFN-II.
We then examined the relationship between BGM- and IKKε-dependent gene expression (Fig. S6B). Although an increase in the basal level of transcription was observed for most ISGs in Ikbke−/− BMMs, genes primarily induced by IFNγ showed a significant increase in baseline expression. We also observed a positive correlation between BGM- and IKKε-dependent gene expression changes in IFNβ-stimulated cells. ISGs with negative BGM (indicating stronger IFNβ responses) were expressed at lower levels in Ikbke−/− BMMs, consistent with our observations of MEFs (20). By contrast, those ISGs with a positive BGM (indicating stronger IFNγ responses) were expressed at a higher level in Ikbke−/− BMMs. We did not observe a correlation between BGM- and IKKε-dependent changes in IFNγ-stimulated cells, but it is interesting to note that there is a general increase in IFNγ-stimulated expression of ISGs in Ikbke−/− BMMs compared with Ikbke+/+ BMMs. We conclude that IKKε biases the transcriptome away from the IFN-II–signaling pathway and toward the IFN-I pathway.
Altered STAT1α Binding in the Promoters of ISGs in IKKε Knockout BMMs.
To correlate transcriptional activation with STAT1-complex binding in vivo, we performed STAT1α ChIP-seq analyses on BMMs from Ikbke+/+ and Ikbke−/− mice. Bone marrow from Ikbke+/+ and Ikbke−/− mice were isolated, differentiated into macrophages, and either not treated or treated with IFNβ or IFNγ for 6 h before cross-linking for ChIP-seq analysis. ChIP reactions were performed with an anti-STAT1α antibody, and libraries of STAT1-bound DNA were generated for high-throughput sequencing. The ChIP-seq data confirm that IKKε-deficient BMMs have a different STAT1α-binding profile compared with wild-type BMMs, as seen in representative type I and II IFN-induced ISGs (Fig. S7). For example, the density of reads in STAT1α peaks associated with Tlr9 and Ifit2 is higher in Ikbke+/+ BMMs compared with Ikbke−/− BMMs, indicating that STAT1α binding, in the form of ISGF3, is diminished in IKKε-deficient cells. In contrast, reads in STAT1α peaks associated with Nos2, Gbp2, and Irf1, ISGs that are predominantly activated by IFN-II, do not have reduced read density in Ikbke−/− BMMs. These data confirm that STAT1α binding as the GAF complex is not inhibited in IKKε-deficient cells. In addition, Nos2 has a higher read density in Ikbke−/− BMMs compared with Ikbke+/+ BMMs. However, we did not observe a significant increase in GAF binding in Gbp2 and Irf1, suggesting that STAT1α binding as the GAF complex is robust in macrophages. Thus, the primary level of regulation by IKKε appears to be in the enhancement of ISGF3 formation, thus increasing the ratio of ISGF3/GAF complex formation.
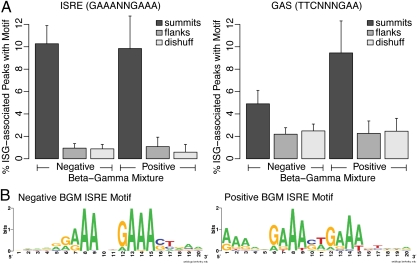
To examine the DNA sequence motifs recognized by STAT1α, we determined whether the ISRE consensus motif GAAANNGAAA or the GAS motif TTCNNNGAA was enriched in 100-bp regions surrounding the summits of STAT1α-bound peaks (Fig. 5A). As controls, we used random dinucleotide shuffles of the same sequences, as well as 100-bp regions flanking the called peaks. Although both motifs were enriched in summits relative to the two controls, significant differences were not observed between the percentage of ISG-associated peaks with the consensus ISRE in negative and positive BGM genes. In contrast, the GAS motif was found in a higher percentage of peaks associated with positive BGM genes than with negative BGM genes. Because the use of predefined consensus motifs may provide some limitations to the analysis, we also used an unbiased motif-finding approach to detect the motifs enriched in STAT1α peaks associated with negative and positive BGM genes (Fig. 5B). This method identified ISRE motifs, with the motif from the positive BGM genes containing additional purine-rich contacts. This analysis confirms the motif preference previously predicted for IKKε-independent ISREs, in which additional DNA contact points may compensate for the lack of an IKKε-modified ISGF3 complex (20, 40). Taken together, the ChIP-seq data suggest that IKKε regulates ISG transcription by promoting the assembly of STAT1 into the ISGF3 complex and suppressing STAT1 homodimerization to form GAF. We conclude that IKKε deficiency increases STAT1 incorporation into GAF, leading to the loss of crucial ISGF3-regulated antiviral gene transcription and increased susceptibility to virus infection.
Fig. 5.
Stat1α peaks near type-I and type-II ISGs are associated with different sequence motifs. (A) Motif frequencies in negative BGM (type-I) and positive BGM (type-II) ISG-associated peaks and control regions. For each peak associated with an ISG, a 100-bp region centered on the peak summit was extracted. As controls, each of these regions was dinucleotide shuffled (42), and also the 100 bp flanking each called peak was taken. The fraction of dinucleotide shuffled (dishuff) or flanking region (flank) controls, or real summits, containing at least one ISRE motif or GAS motif is shown. (B) Motifs discovered by unbiased search using BioProspector in summit regions (100 bp) associated with negative and positive BGM ISGs.
Discussion
Modeling of ISGF3 based on the IRF DNA-binding domain and STAT dimers in the parallel configuration suggests that both major and minor grooves of the ISRE sequence are completely occupied by IRF9 and STAT1 on opposing strands (20). Furthermore, the ISGF3-binding specificity appears to depend on the ISRE core as well as on the surrounding sequences. We propose that the structure of the STAT1/STAT2 heterodimer within the ISGF3 complex is distinct from that of the STAT1/STAT1 homodimer. Specifically, we propose that phosphorylation of serine 708 of STAT1, located at the homodimer interface, blocks the formation of a STAT1/STAT1 homodimer but does not block the formation of the STAT1/STAT2 heterodimer within the ISGF3 complex. On the basis of the 3D structure of the STAT1 homodimer, we propose that any modification of serine, including phosphorylation, would disrupt its hydrogen-bonding interactions within this homodimerization interface. However, it appears that ISGF3 can accommodate S708 phosphorylation in the context of the trimeric complex, suggesting that, in combination with IRF9, the STAT1/STAT2 interface differs from that of the heterodimer.
Here, we demonstrate that phosphorylation of STAT1 by IKKε suppresses the formation of a parallel STAT1 dimer, thus preventing the formation of the GAF complex. This, in turn, results in the loss of GAF binding to GAS elements and a decrease in expression of GAS-dependent genes. Thus, in the context of virus infection where both IFN-I and IFN-II are produced, IKKε shifts the STAT1 pool toward the formation of ISGF3 and away from GAF. Presumably, the relative levels of phosphorylated and unphosphorylated STAT1 in cells treated with IFN-I depend on the ratio of activated IKKε to STAT1. If activated IKKε is limiting, the loss of IKKε would not affect the switch from the type I to the type II pathway. Rather, the presence of IKKε ensures that the cellular response to IFN-I and IFN-II includes the optimal production of ISGF3-dependent genes. In addition, as STAT1 forms an antiparallel configuration before JAK/STAT activation (37, 38), IKKε-mediated phosphorylation of STAT1 may be one mechanism to ensure that sufficient STAT1 is available to dimerize with other partners such as STAT2.
In addition, we show that STAT1 phosphorylation can affect the binding of ISGF3 to different types of ISREs. Specifically, we propose that the binding of ISGF3 to an IKKε-dependent minimal ISRE requires S708 phosphorylation-mediated conformational changes in ISGF3 (20). In contrast, a longer ISRE that provides additional purine-rich contact points for ISGF3 binding is IKKε independent. In the absence of IKKε, those ISGs that are regulated by a minimal ISRE are the first to be affected. Thus, IKKε can modify IFN signaling by the regulation of STAT1 incorporation into either ISGF3 or GAF.
During the course of virus infection, IKKε expression is inducible, and IFN-I stimulation is thought to activate IKKε via p38 kinase signaling (3, 20). It is tempting to speculate that the up-regulation of IKKε serves to ensure that virus-infected cells respond to both IFN-I and IFN-II with the formation of ISGF3 to generate a cellular antiviral environment. On the basis of the evidence presented here, we propose that IKKε regulates IFN signaling by mediating STAT1 complex assembly and DNA-binding specificity. This regulation would fine-tune the ISGF3-induced gene expression profile and ensure the establishment of an effective cellular antiviral response.
Materials and Methods
Cell Culture, Viral Infections, and Reagents.
Cells were cultured as described in SI Materials and Methods. Viral infections and reagents used are also described in SI Materials and Methods.
Western Blot Analysis, Coimmunoprecipitation, and Size-Exclusion Chromatog-raphy.
Western blot analysis, coimmunoprecipiation, and size exclusion chro-matography are described in SI Materials and Methods.
EMSA and Recombinant Kinase Assay.
EMSA and recombinant kinase assay are described in SI Materials and Methods.
RNA Isolation and qPCR.
RNA was isolated and qPCR was performed as described in SI Materials and Methods.
High-Throughput Sequencing Data Analysis.
RNA-seq and ChIP-seq were performed and analyzed as described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank J. Ye and M. Zhao for manuscript review; D. Panne and A. García-Sastre for reagents; and E. Procko, D. Pakotiprapha, M. Lorini, and the R.M.M. laboratory for technical support. This work was sup-ported by the National Science Foundation and Soros Fellowships (to S.-L.N.); National Institutes of Health Grant AI20642 (to T.M.); National Institute of Allergy and Infectious Diseases Grant R01AI080624 (to B.R.T.); and HudsonAlpha Institute funds (to R.M.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the European Nucleotide Archive database (accession nos. ERR033735, ERR033736, ERR033738, ERR033739, ERR033740, and ERR033741) and the Gene Expression Omnibus database, www.ncbi.nlm.nih.gov/geo (accession no. GSE33913).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1119137109/-/DCSupplemental.
References
- 1.Thompson AJ, Locarnini SA. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunol Cell Biol. 2007;85:435–445. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- 2.Baumgarth N, Choi YS, Rothaeusler K, Yang Y, Herzenberg LA. B cell lineage contributions to antiviral host responses. Curr Top Microbiol Immunol. 2008;319:41–61. doi: 10.1007/978-3-540-73900-5_3. [DOI] [PubMed] [Google Scholar]
- 3.Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 4.Hardy MP, Owczarek CM, Jermiin LS, Ejdebäck M, Hertzog PJ. Characterization of the type I interferon locus and identification of novel genes. Genomics. 2004;84:331–345. doi: 10.1016/j.ygeno.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 5.van Pesch V, Lanaya H, Renauld JC, Michiels T. Characterization of the murine alpha interferon gene family. J Virol. 2004;78:8219–8228. doi: 10.1128/JVI.78.15.8219-8228.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Veer MJ, et al. Functional classification of interferon-stimulated genes identified using microarrays. J Leukoc Biol. 2001;69:912–920. [PubMed] [Google Scholar]
- 7.Der SD, Zhou A, Williams BR, Silverman RH. Identification of genes differentially regulated by interferon alpha, beta, or gamma using oligonucleotide arrays. Proc Natl Acad Sci USA. 1998;95:15623–15628. doi: 10.1073/pnas.95.26.15623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darnell JE, Jr, Kerr IM, Stark GR. Jak-STAT pathways and transcriptional activation in response to IFNs and other extracellular signaling proteins. Science. 1994;264:1415–1421. doi: 10.1126/science.8197455. [DOI] [PubMed] [Google Scholar]
- 9.Greenlund AC, Farrar MA, Viviano BL, Schreiber RD. Ligand-induced IFN gamma receptor tyrosine phosphorylation couples the receptor to its signal transduction system (p91) EMBO J. 1994;13:1591–1600. doi: 10.1002/j.1460-2075.1994.tb06422.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krishnan K, Singh B, Krolewski JJ. Identification of amino acid residues critical for the Src-homology 2 domain-dependent docking of Stat2 to the interferon alpha receptor. J Biol Chem. 1998;273:19495–19501. doi: 10.1074/jbc.273.31.19495. [DOI] [PubMed] [Google Scholar]
- 11.Nguyen VP, et al. Stat2 binding to the interferon-alpha receptor 2 subunit is not required for interferon-alpha signaling. J Biol Chem. 2002;277:9713–9721. doi: 10.1074/jbc.M111161200. [DOI] [PubMed] [Google Scholar]
- 12.Yan H, et al. Phosphorylated interferon-alpha receptor 1 subunit (IFNaR1) acts as a docking site for the latent form of the 113 kDa STAT2 protein. EMBO J. 1996;15:1064–1074. [PMC free article] [PubMed] [Google Scholar]
- 13.Schindler C, Shuai K, Prezioso VR, Darnell JE., Jr Interferon-dependent tyrosine phosphorylation of a latent cytoplasmic transcription factor. Science. 1992;257:809–813. doi: 10.1126/science.1496401. [DOI] [PubMed] [Google Scholar]
- 14.Shuai K, et al. Interferon activation of the transcription factor Stat91 involves dimerization through SH2-phosphotyrosyl peptide interactions. Cell. 1994;76:821–828. doi: 10.1016/0092-8674(94)90357-3. [DOI] [PubMed] [Google Scholar]
- 15.Fu XY, Kessler DS, Veals SA, Levy DE, Darnell JE., Jr ISGF3, the transcriptional activator induced by interferon alpha, consists of multiple interacting polypeptide chains. Proc Natl Acad Sci USA. 1990;87:8555–8559. doi: 10.1073/pnas.87.21.8555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Levy DE, Kessler DS, Pine R, Darnell JE., Jr Cytoplasmic activation of ISGF3, the positive regulator of interferon-alpha-stimulated transcription, reconstituted in vitro. Genes Dev. 1989;3:1362–1371. doi: 10.1101/gad.3.9.1362. [DOI] [PubMed] [Google Scholar]
- 17.Levy DE, Kessler DS, Pine R, Reich N, Darnell JE., Jr Interferon-induced nuclear factors that bind a shared promoter element correlate with positive and negative transcriptional control. Genes Dev. 1988;2:383–393. doi: 10.1101/gad.2.4.383. [DOI] [PubMed] [Google Scholar]
- 18.Shuai K, Schindler C, Prezioso VR, Darnell JE., Jr Activation of transcription by IFN-gamma: Tyrosine phosphorylation of a 91-kD DNA binding protein. Science. 1992;258:1808–1812. doi: 10.1126/science.1281555. [DOI] [PubMed] [Google Scholar]
- 19.Decker T, Kovarik P, Meinke A. GAS elements: A few nucleotides with a major impact on cytokine-induced gene expression. J Interferon Cytokine Res. 1997;17:121–134. doi: 10.1089/jir.1997.17.121. [DOI] [PubMed] [Google Scholar]
- 20.Tenoever BR, et al. Multiple functions of the IKK-related kinase IKKepsilon in interferon-mediated antiviral immunity. Science. 2007;315:1274–1278. doi: 10.1126/science.1136567. [DOI] [PubMed] [Google Scholar]
- 21.Peters RT, Liao SM, Maniatis T. IKKepsilon is part of a novel PMA-inducible IkappaB kinase complex. Mol Cell. 2000;5:513–522. doi: 10.1016/s1097-2765(00)80445-1. [DOI] [PubMed] [Google Scholar]
- 22.Peters RT, Maniatis T. A new family of IKK-related kinases may function as I kappa B kinase kinases. Biochim Biophys Acta. 2001;1471:M57–M62. doi: 10.1016/s0304-419x(00)00024-x. [DOI] [PubMed] [Google Scholar]
- 23.Shimada T, et al. IKK-i, a novel lipopolysaccharide-inducible kinase that is related to IkappaB kinases. Int Immunol. 1999;11:1357–1362. doi: 10.1093/intimm/11.8.1357. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald KA, et al. IKKepsilon and TBK1 are essential components of the IRF3 signaling pathway. Nat Immunol. 2003;4:491–496. doi: 10.1038/ni921. [DOI] [PubMed] [Google Scholar]
- 25.Hemmi H, et al. The roles of two IkappaB kinase-related kinases in lipopolysaccharide and double stranded RNA signaling and viral infection. J Exp Med. 2004;199:1641–1650. doi: 10.1084/jem.20040520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry AK, Chow EK, Goodnough JB, Yeh WC, Cheng G. Differential requirement for TANK-binding kinase-1 in type I interferon responses to toll-like receptor activation and viral infection. J Exp Med. 2004;199:1651–1658. doi: 10.1084/jem.20040528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sharma S, et al. Triggering the interferon antiviral response through an IKK-related pathway. Science. 2003;300:1148–1151. doi: 10.1126/science.1081315. [DOI] [PubMed] [Google Scholar]
- 28.Hiscott J. Convergence of the NF-kappaB and IRF pathways in the regulation of the innate antiviral response. Cytokine Growth Factor Rev. 2007;18:483–490. doi: 10.1016/j.cytogfr.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Maniatis T, et al. Structure and function of the interferon-beta enhanceosome. Cold Spring Harb Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 30.Berchtold S, et al. Forced IFIT-2 expression represses LPS induced TNF-alpha expression at posttranscriptional levels. BMC Immunol. 2008;9:75. doi: 10.1186/1471-2172-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matsumoto M, et al. Activation of the transcription factor ISGF3 by interferon-gamma. Biol Chem. 1999;380:699–703. doi: 10.1515/BC.1999.087. [DOI] [PubMed] [Google Scholar]
- 32.Zimmermann A, et al. A cytomegaloviral protein reveals a dual role for STAT2 in IFN-gamma signaling and antiviral responses. J Exp Med. 2005;201:1543–1553. doi: 10.1084/jem.20041401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Levy DE, Darnell JE., Jr Stats: Transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 34.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 35.Juang YT, et al. Primary activation of interferon A and interferon B gene transcription by interferon regulatory factor 3. Proc Natl Acad Sci USA. 1998;95:9837–9842. doi: 10.1073/pnas.95.17.9837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen X, et al. Crystal structure of a tyrosine phosphorylated STAT-1 dimer bound to DNA. Cell. 1998;93:827–839. doi: 10.1016/s0092-8674(00)81443-9. [DOI] [PubMed] [Google Scholar]
- 37.Mao X, et al. Structural bases of unphosphorylated STAT1 association and receptor binding. Mol Cell. 2005;17:761–771. doi: 10.1016/j.molcel.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 38.Mertens C, et al. Dephosphorylation of phosphotyrosine on STAT1 dimers requires extensive spatial reorientation of the monomers facilitated by the N-terminal domain. Genes Dev. 2006;20:3372–3381. doi: 10.1101/gad.1485406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stancato LF, David M, Carter-Su C, Larner AC, Pratt WB. Preassociation of STAT1 with STAT2 and STAT3 in separate signalling complexes prior to cytokine stimulation. J Biol Chem. 1996;271:4134–4137. doi: 10.1074/jbc.271.8.4134. [DOI] [PubMed] [Google Scholar]
- 40.Schmid S, Mordstein M, Kochs G, García-Sastre A, Tenoever BR. Transcription factor redundancy ensures induction of the antiviral state. J Biol Chem. 2010;285:42013–42022. doi: 10.1074/jbc.M110.165936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Becker S, Groner B, Müller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394:145–151. doi: 10.1038/28101. [DOI] [PubMed] [Google Scholar]
- 42.Altschul SF, Erickson BW. Significance of nucleotide sequence alignments: A method for random sequence permutation that preserves dinucleotide and codon usage. Mol Biol Evol. 1985;2:526–538. doi: 10.1093/oxfordjournals.molbev.a040370. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.